SUMMARY
Whether mouse visual cortex contains orderly feature maps is debated. The overlapping pattern of geniculocortical (dLGN) inputs with M2 muscarinic acetylcholine receptor-rich patches in layer 1 (L1) suggests a non-random architecture. Here, we found that L1 inputs from the lateral posterior thalamus (LP) avoid patches and target interpatches. Channelrhodopsin-2-assisted mapping of EPSCs in L2/3 shows that the relative excitation of parvalbumin-expressing interneurons (PVs) and pyramidal neurons (PNs) by dLGN, LP and cortical feedback are distinct and depend on whether the neurons reside in clusters aligned with patches or interpatches. Paired recordings from PVs and PNs shows that unitary uIPSCs are larger in interpatches than patches. The spatial clustering of inhibition is matched by dense clustering of PV-terminals in interpatches. The results show that the excitation/inhibition balance across V1 is organized into patch and interpatch subnetworks which receive distinct long-range inputs and are specialized for the processing of distinct spatiotemporal features.
Keywords: visual cortex, inhibition, parvalbumin interneurons, thalamocortical connections, intracortical feedback
In Brief
D’Souza, Bista et al., show that parvalbumin interneuron-mediated inhibition in mouse primary visual cortex is spatially clustered and that these modules receive differential inputs to layer 1 from the first and second order thalamus and extrastriate visual cortex.
INTRODUCTION
The modernist’s maxim that form follows function manifests itself in neuroscience as functional architecture. The mesoscale description of the spatial relationship between neuronal responses, layers, columns, and areas has driven much of what is known about the matrix of the cortical network (Hubel and Wiesel, 1962, 1974). The concepts that emerged from these studies include: the representation of space in topographic maps, the complexification of receptive fields, functionally distinct columns, hierarchical processing along specialized streams, the exponential distance rule, and the counterstream connectivity between areas in which feedforward inputs convey prediction errors to improve internal models of the world (Felleman and Van Essen, 1991; Horvát et al., 2016; Hubel and Wiesel, 1962; Rao and Ballard, 1999). Although the functional architecture implies that the spatiotemporal tuning of neurons is determined by connectivity and physical layout, the networks of modular processing have been difficult to define (DaCosta and Martin, 2013). The challenge has been greatest in rodents because of the view that cortex lacks columnar organization (Ohki et al., 2005; 2007). However, recent studies in mouse primary visual cortex (V1) have found that subcortically and intracortically projecting PNs are clustered and are aligned to microcolumns with distinct tuning preferences (Kondo et al, 2016; Maruoka et al., 2017).
Previously, we found an interdigitating pattern of M2 muscarinic acetylcholine receptor-expressing (M2+) patches and M2− interpatches in L1 of mouse V1, which aligns with distinct cell clusters in L2/3 tuned for high spatial frequency (SF) and high temporal frequency (TF), respectively (Ji et al., 2015). This finding provides structural evidence for functionally discrete modules and raises the question whether the excitation (E) / inhibition (I) balance in patches and interpatches is spatially organized. That inhibition is not a uniform blanket across V1 (Karnani et al., 2014), but is deployed in clusters of PV and somatostatin- (SOM) expressing GABAergic neurons (Ebina et al., 2014; Maruoka et al., 2017), and that activation of these cells including those which express vasoactive intestinal peptide can shape stimulus selectivities of PNs is gaining acceptance (Ayzenshtat et al., 2016; Lee, et al., 2012; Lee et al., 2014; Zhu et al., 2015). Whether the inhibitory network is tied to the clustered patch/interpatch system in V1 and provides distinct subnetworks for processing visual information remains unknown.
To determine whether inhibition across V1 is modular we measured the strength of synaptic long-range input to L2/3 PNs and PVs in patches and interpatches by using subcellular Channelrhodopsin-2 (ChR2) assisted circuit mapping (sCRACM) (Mao et al, 2011; Petreanu et al., 2009). We recorded from pairs of PVs and PNs and analyzed unitary excitatory (uEPSCs) and inhibitory (uIPSCs) postsynaptic currents in patches and interpatches. The results show that V1 contains two different circuit motifs in which patches and interpatches have distinct thalamocortical and interareal inputs to PNs and PVs, and that PVs in interpatches provide stronger inhibition to PNs than in patches. The modular organization of inhibition is consistent with the notion that neurons in interpatches are more sensitive to rapidly changing visual inputs (Ji et al., 2015).
RESULTS
Clustering of thalamocortical inputs to L1 of V1
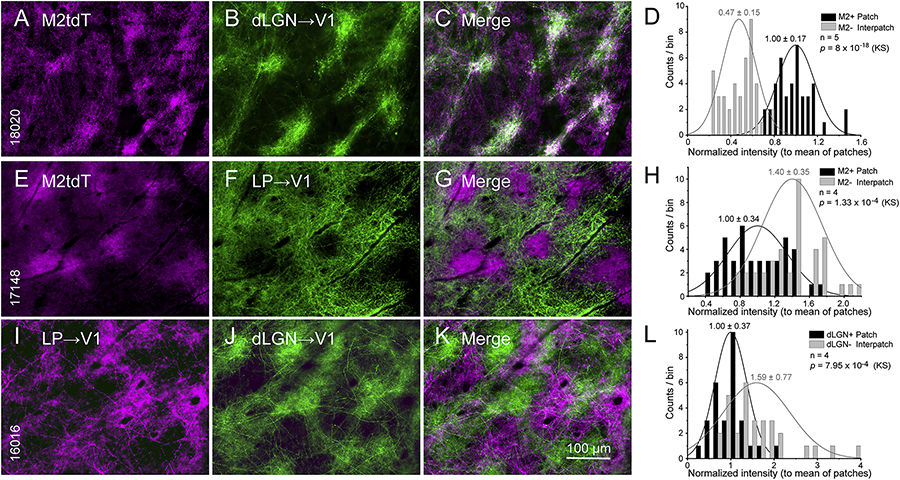
We have shown that inputs from the dLGN to L1 of V1 are clustered and overlap with the patchy pattern of M2 immunostaining (Ji et al., 2015). Here, we show in flatmounted cortex of Chrm2tdT mice that dLGN→V1 axons labeled by anterograde tracing with AAV2/1.hSyn.EGFP preferentially terminate in M2+ patches of L1 and avoid M2− interpatches (Figures 1A–C). For quantitative analysis we made contour maps (Sincich and Horton, 2005) of the distribution of fluoresence which demarcates patches (top 33% quantile) and interpatches (bottom 33% quantile), and compared M2 expression with dLGN→V1 intensity in regions of interest (ROIs) at the center of the projection (Figures S1A–B). Random shuffling of pixel values in each ROI showed a greater than chance overlap of M2+ and dLGN projections (Figure S1C). We normalized the values in interpatches to the mean intensity in patches across 5 mice and plotted the counts in different bins. We found that the intensity in patches was 2.1 ± 0.024-fold (p = 8 × 10−18, Kolmogorov-Smirnov test [KS]) higher than in interpatches (Figure 1D). Patches and interpatches in L1 were 60–80 μm wide and centroids were 120–140 μm apart. dLGN projections to L3–4 (Figure 4A) and 5/6 appeared uniform. Inputs to L1 from the lateral posterior thalamic nucleus (LP) exhibited a different pattern, showing on average (N = 4 mice) 1.4 ± 0.14-fold (p = 1.33 × 10−4, KS) stronger projections to M2− interpatches (Figures 1E–H). Simultaneous tracing of dLGN→V1 and LP→V1 inputs with AAV2/1hSyn.tdT and AAV2/1.hSyn.EGFP, respectively, confirmed the interdigitating pattern of projections (patch/interpatch ratio = 0.5, Figures S1D–F) showing denser LP input to interpatches (N = 4 mice, p = 7.95 × 10−4, KS) (Figures 1I–L).
Figure 1. Patchy dLGN→V1 and LP→V1 projections to L1.
Tangential sections through L1 of V1 of Chrm2tdT (A-C, E-G) and C57BL/6J mice (I-K). (A-C) Spatially clustered dLGN→V1 projections anterogradely traced with AAV2/1.hSyn.EGFP (green) overlap with M2tdT+ patches (purple). (D) Normalized fluorescence intensity of dLGN→V1 input to M2tdT+ patches (for contour maps, Figure S1) is higher than to M2tdT-interpatches. (E-G) Patchy LP→V1 projections traced with AAV2/1.hSyn.EGFP (green) overlap with M2tdT- interpatches. (H) Normalized fluorescence intensity of LP inputs to M2tdT-interpatches is higher than to M2tdT+ patches. (I-K) Interdigitating patchy LP→V1 and dLGN→V1 projections traced in the same mouse with AAV2/1hSyn.tdTomato (purple) and AAV2/1.hSyn.EGFP (green), respectively. (L) LP→V1 input is weaker in dLGN+ patches (proxies of M2tdT+ patches; A-C) than in dLGN- interpatches. Mean ± SD, KS (Kolmogorov-Smirnov test), N = number of mice.
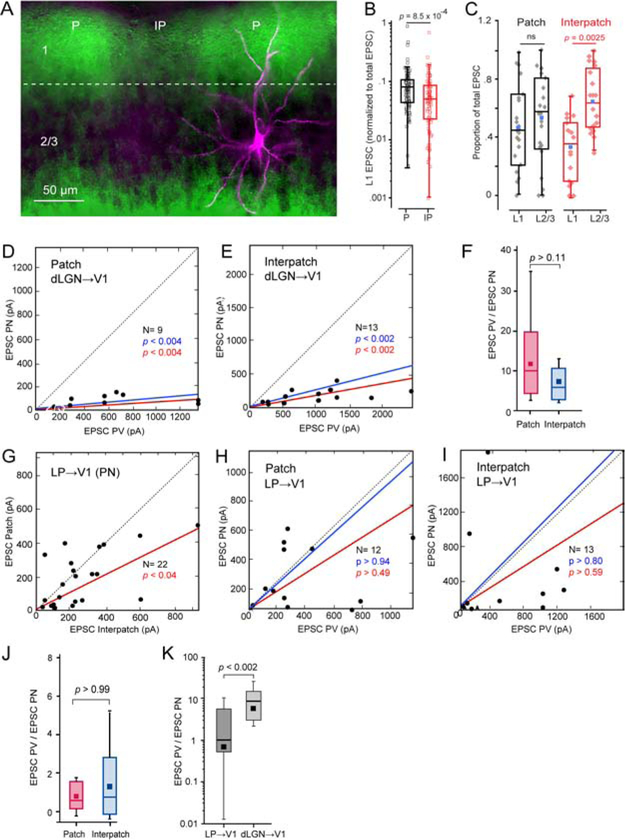
Figure 4. Coronal slices: sCRACM of dLGN→V1 and LP→V1 input to L2/3 PNs and PVs in patches and interpatches.
(A) dLGN→V1 projections (green) traced with AAV2/1.hSyn.ChR2(H134R).EYFP, showing dense terminations in patches (P) and sparse input to interpatches (IP) in L1. Biocytin-filled L2/3 PN (magenta) recorded in patch, showing apical dendrites projecting to patch and basal dendrites descending to uniformly labeled dLGN-recipient deep L2/3. (B) Box plots of recordings from L2/3 PNs showing that EPSCs in L1 (normalized to total EPSCs) in IP (N = 20) are significantly smaller than in P (N = 23). (C) Box plots of recordings from L2/3 PNs showing that the proportion of EPSCs (normalized to total EPSC) in L1 of interpatches is smaller (N = 20, p = 0.0025, paired t-test) than in L2/3. No significant (ns) laminar differences in patches (N = 23). Blue dot denotes mean L1 or L2/3 EPSC. (D, E) Relative strengths of EPSCs from neighboring pairs of L2/3 PNs and PVs evoked by dLGN→V1 input. Recordings from patches (D) and interpatches (E) obtained in different slices. Mean slope of currents from zero (red), slope after normalization to conductance (blue). (F) Similar EPSCPV/EPSCPN ratio in patches and interpatches. (G) EPSCs evoked by LP→V1 input to pairs of neighboring PNs (dots) in patches and interpatches recorded in the same slice, showing stronger inputs to interpatches. (H, I) EPSCs evoked by LP→V1 inputs to pairs of PNs and PVs in patches and interpatches recorded in different slices, showing similar inputs to PNs and PVs. (J) EPSCPV/EPSCPN ratio in patches and interpatches. (K) EPSCPV/EPSCPN ratio evoked by stimulation of dLGN→V1 and LP→V1 axons. Pooled responses from patches and interpatches.
Clustering of intracortical inputs to L1 of V1
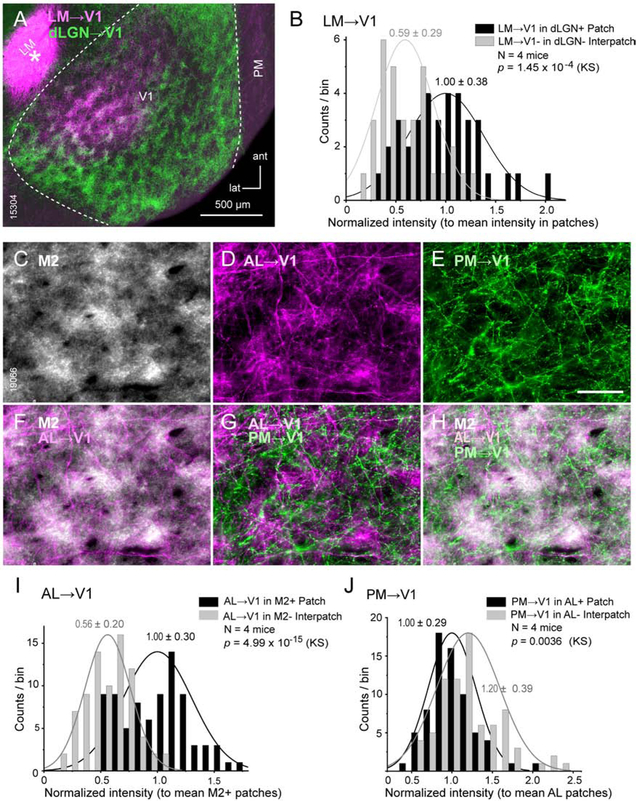
We next compared feedback projections to L1 of V1 from the higher ventral stream lateromedial area, LM, with inputs from the dLGN. Double viral tracings from the dLGN and LM showed that inputs from both sources overlapped in presumptive M2+ patches of L1 (Figure 2A). On average (N = 4 mice) LM inputs to patches were 1.7 ± 0.05-fold denser than to interpatches (p = 1.45 × 10−4, KS) (Figure 2B). We have shown previously that V1 inputs from the dorsal stream anterolateral area, AL, terminate in M2+ patches of L1 (Ji et al., 2015), raising the question whether M2+ patches are the preferred targets of cortical feedback. To address this, we traced the connections from the posteromedial area (PM), another member of the dorsal stream (Wang et al. 2012), to V1. PM→V1 projections showed a preference for M2− interpatches (N = 3 mice, Figures S1G–I), and differed from simultaneously labeled AL→V1 inputs, which on average (N = 4 mice) preferred M2+ patches (Figures 2C–I). Direct comparison of patterns in L1 showed that PM→V1 inputs to interpatches are largely excluded from patches targeted by AL→V1 inputs (Figure 2J, N = 4 mice), demonstrating that feedback projections by the two dorsal stream areas terminate at distinct sites.
Figure 2. Patchy LM→V1, AL→V1 and PM→V1 projections to L1.
Tangential sections through L1 of V1 of C57BL/6J mice (A, C-H). (A) Overlapping clusters of dLGN→V1 (green, AAV2/1.hSyn.EGFP) and LM→V1 (purple, AAV2/1hSyn.tdTomato injection [*]) projections in patches (dLGN+ patches are proxies of M2+ patches) of L1. (B) Frequency distribution of fluorescence intensity of LM→V1 projections (normalized to mean in patches) shows stronger inputs to dLGN+ patches. (C-H) Interdigitating patterns of AL→V1 (D, purple, tracing with tdTomato) and PM→V1 (E, green, tracing with EGFP) projections. Immunostaining for M2 (C) showing that AL→V1 overlap with M2+ patches (F). PM→V1 input to M2-interpatches alternates with AL→V1 to M2+ patches (G, H). (I) Frequency distribution of fluorescence intensity of AL→V1 projections indicates stronger input to M2+ patches. (J) Distribution of PM→V1 intensity indicates stronger input to interpatches where AL→V1 is weak. Same conventions as in Figure 1.
Clustering of cholinergic inputs to L1 of V1
Classic studies suggest that the cholinergic innervation of cortex is diffuse and that spatial selectivity arises from clustering of receptors and axons (Muñoz and Rudy, 2014). But whether cholinergic fibers are spatially mapped is unknown. To address the question, we stained tangential sections through V1 with antibodies against M2 and choline acetyl transferase ChAT. We found that ChAT labeled axons in L1 were clustered and on average (N = 2 mice) significantly (p = 0.009) denser in M2+ patches (Figures S2A–D).
Development of M2 clusters
Motivated by the report of Maruoka et al., (2017) that L5 neurons in V1 of 6-day-old (P6) mice form 20 μm-wide microcolumns, we have looked for clustered M2 expression in postnatal development. Our results in tangential sections through V1 of P4 Chrm2tdT mice show that M2 expression in L1 is patchy (10 – 30 μm wide, 52–79 μm center-to-center, Figure S3A). In L2/3 patches were smaller and contained 0.8 – 1.5 μm-wide rings, presumably representing membranes of cross-sectioned dendrites (Figure S3B). At P10 the L1 patches were larger (30 – 60 μm wide) and more widely spaced (80–100 μm center-to-center) (Figure S3C), a difference which may be accounted for by the 1.7 fold increase in brain size (Agrawal et al., 1968).
Module- and pathway-specific strengths of inputs to PNs and PVs
We have shown previously that L2/3 neurons in M2− interpatches of V1 are more often tuned to the direction of visual motion and respond to higher speeds and TFs than neurons in M2+ patches (Ji et al., 2015). This non-uniform distribution of selectivities suggested that the two cell clusters differentially process spatiotemporal information. Feedforward inhibition (FFI) mediated by excitation of PVs (D’Souza et al., 2016) and their inhibitory output is known to shape the temporal sensitivity of PNs in auditory cortex (Li et al., 2014). We therefore characterized the precipitating events of FFI by comparing the strength of excitatory inputs to patches and interpatches from thalamocortical and intracortical pathways to L2/3 neurons in V1. We used sCRACM in slices of PVtdT mice in which inputs to V1 were anterogradely labeled with ChR2(H134R).EYFP.
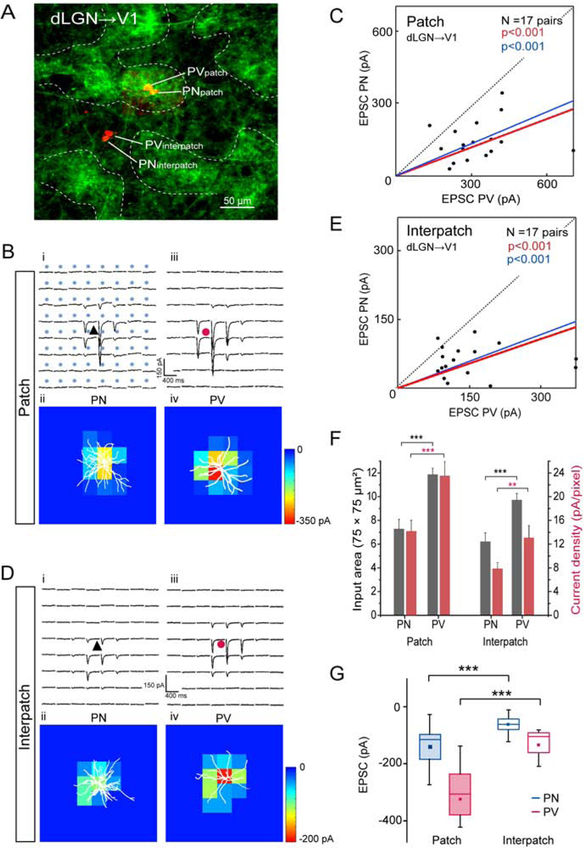
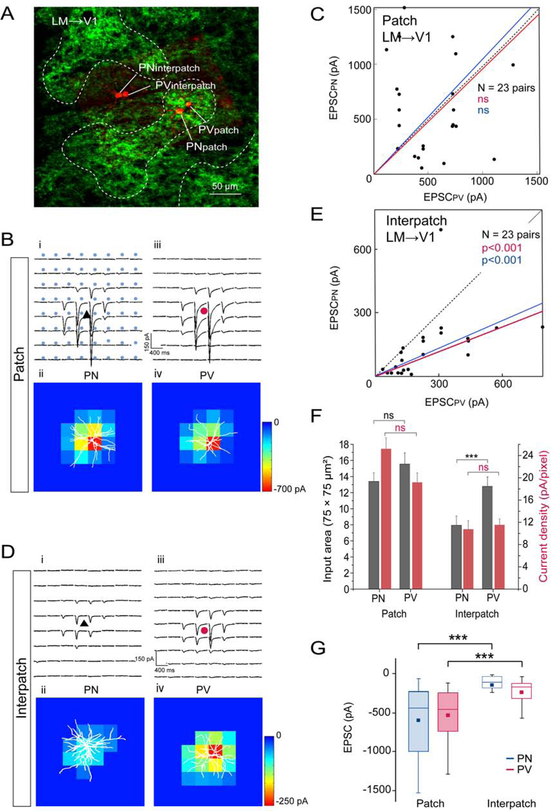
dLGN input in tangential slices.
To select for inputs to distal dendrites of L2/3 neurons in L1–2 and preserve their 3D organization, we obtained tangential slices of V1. Whole cell patch clamp recordings were performed from pairs of PNs and PVs (≤ 40 μm apart) aligned with densely ChR2-EYFP-expressing patches or sparsely labeled interpatches, visualized by viral tracing from the dLGN. Locations of the recorded neurons were determined by intracellular filling with Alexa-594 hydrazide (Figure 3A). Immunostaining of re-sectioned slices for M2 revealed that dLGN→V1 inputs overlapped with M2+ patches (Figures S4A–C). Input strength was measured by EPSCs elicited by laser stimulation of ChR2-expressing terminals. Laser stimulation was delivered in an 8 × 8 grid with 75 μm spacing centered on the cell body. sCRACM in the same slice of dLGN inputs to neighboring PVs and PNs showed stronger synaptic activation in patches than interpatches. Direct comparison of cell pairs in patches and interpatches showed that EPSCs from PVs were larger than from PNs (Figures 3B–E). The average EPSC recorded from PVs (323.6 ± 130 pA) in patches was 2.3-fold larger (p < 0.001, N = 17 pairs, Wilcoxson signed-rank test (Wt)) than from PNs (140.6 ± 81.6 pA) (Figure 3G). A similar 2.3-fold difference (p < 0.001, N = 17 pairs) was found in interpatches between the overall more weakly responsive PVs (147.7 ± 91.2 pA) and PNs (62.9 ± 31.1 pA). Heatmaps of EPSCs in patches and interpatches show that responses were maximal near the cell body and decreased distally (Figures 3B, D). The responsive area was smaller for PNs than PVs (p < 0.001, Wt), but similar in patches and interpatches (Figure 3F). dLGN inputs to PVs evoked higher current densities (p < 0.01, Wt, Figure 3F) and exhibited faster rise times of EPSCs than inputs to PNs, in both patches (PV: 11.6 ± 0.1 ms; PN: 14 ± 0.3 ms) and interpatches (PV: 11.7 ± 0.1 ms; PN: 14.8 ± 0.3 ms) (p < 0.05, Wt). These results indicate that the strength of activation by dLGN inputs to L1–2 is cell type and module specific. To examine whether the shape of the apical dendritic tree of PNs and PVs contributes to this specificity we measured branch lengths outside a 100 μm-diameter circle centered on the soma. For PNs in patches with a total dendritic length of 234 ± 21 μm (N = 6), 82.5% of branches were in patches and 17.5% in interpatches (p < 0.01, t-test). PVs in patches with a total arbor of 157 ± 34 μm (N = 6) showed 91.8% of branches in patches and 8.2% in interpatches (p < 0.001). PNs in interpatches with a total arbor of 235 ± 19 μm (N = 5) showed 81% of branches in interpatches and 10% in patches (p < 0.05). PVs in interpatches with a total arbor of 164.7 ± 23 μm (N = 7) showed 83.3% of branches in interpatches and 16.7 % in patches (p < 0.05).
Figure 3. Tangential slices: sCRACM of dLGN→V1 input to L1 onto L2/3 PNs and PVs in patches and interpatches.
(A) Confocal Z-stack showing ChR2-Venus labeled dLGN→V1 projections in L1 and Alexa 594 hydrazide-filled pairs of L2/3 PNs and PVs in patch and interpatch. (B, D) Whole cell patch clamp recordings from PN (black triangle) and PV (red circle) in patches and interpatches in the same slice. Each trace represents average of EPSCs (3 to 5 per neuron) of PNs and PVs in patches (Bi, Biii) and interpatches (Di, Diii) upon laser stimulation (blue dots, 75 × 75 μm grid) of ChR2-expressing dLGN→V1 terminals. Heatmaps of responses evoked at different locations of the dendritic arbor (white profiles) of PN and PV in patches (Bii, Biv) and interpatches (Dii, Div). (C, E) Each dot represents relative strength of dLGN→V1 input (summed pixels of significant EPSCs) of a pair of L2/3 PNs and PVs in patch (C) and interpatch (E). Red line denotes mean slope from zero, blue line shows mean slope after normalizing currents to mean conductance. (F) Distribution of dLGN→V1 input strength across dendritic tree. Grey bars represent input area as number of pixels with non-zero EPSCs. Red bars EPSC density. (G) Box plots of dLGN→V1 EPSCs from PVs and PNs in patches and interpatches. (C, E, F) Wilcoxon signed-rank test (Wt) (***p < 0.001, **p < 0.01).
dLGN input in coronal slices.
An issue with tangential slices is that they may not always remove inputs to basal dendrites in L2/3 and therefore confound measurements of the strength of L1 input to patches and interpatches. To sort out inputs to apical and basal dendrites we obtained coronal slices in which dLGN→V1 inputs were labeled with ChR2.EYFP. Projections to L1 were denser in patches than interpatches and inputs to the bottom of L2/3 were uniform (Figure 4A). sCRACM was performed by laser stimulation in an 8 × 10 (mediolateral × dorsoventral) 75 μm grid aligned to the pial surface. Recordings from L2/3 PNs showed that EPSCs in L1 normalized to the total EPSCs were larger (p = 8.5 × 10−4, t-test) in patches than in interpatches (Figures 4A, B). Comparison of the proportion of L1 and L2/3 EPSCs of total EPSCs showed that in patches, inputs to L1 and L2/3 were similar (Figure 4C). In contrast, in interpatches the proportion of L1 inputs was lower (p = 0.0025, paired t-test) than in L2/3 (Figure 4C), demonstrating, in support of structural evidence, that inputs to apical dendrites are clustered whereas those to basal dendrites are uniform.
Although recordings from single cells were readily obtained, measurements from two pairs in the same slice were challenging, because one member of a pair in a patch and neighboring interpatch was often buried in the slice where patchy dLGN→V1 inputs were difficult to resolve. We therefore recorded from L2/3 PN/PV patch and interpatch pairs in separate slices. The results showed that EPSCs from PVs were larger compared to PNs and the PV/PN ratios in patches and interpatches were similar (Figures 4D–F). The range of responses was comparable in patches and interpatches, easing the concern of across-slice comparisons. The results indicate that in coronal slices in which dLGN→V1 inputs to L1–5 are preserved, module-specificity of geniculocortical input is revealed by the proportion of L1 to the total strength of input (Figure 4C). The results suggest that tangential slicing truncates basal dendrites of L2/3 cellsand selects for dLGN→V1 input to L1.
LP input in coronal slices.
We used sCRACM in coronal slices to examine responses of LP→V1 inputs to L2/3 cells. Patches and interpatches were identified by tdT-expressing dLGN→V1 projections. Recordings in the same slice showed that EPSCs from PNs were larger (p < 0.04, Wt) in interpatches than in patches (Figure 4G). This pattern is consistent with the increased innervation density of L1 inputs to interpatches and the lack of inputs to L3–4 whose presence in coronal slices obscures the module-specificity of dLGN→V1 input to L2/3 cells (Figures 4D–F). Next, we compared responses to LP→V1 L1 inputs from PNs and PVs in patches and interpatches. Responses from both cell types were matched, with median EPSCPN/EPSCPV ratios in patches and interpatches similar to unity and not significantly different from one another (Figures 4H–J). This differed from the strong preference for PVs found after stimulation of dLGN→V1 inputs in both tangential and coronal slices (Figures 3G, 4D–F), and suggests that dLGN→V1 and LP→V1 inputs to L1 feed distinct subnetworks in patches and interpatches in which PNs and PVs are excited at different strengths (Figure 4K).
Feedback input from LM.
Input from LM→V1 to L1 terminated in periodic clusters which overlapped with dLGN→V1 projections (Figure 2A). Laser stimulation of ChR2-expressing LM→V1 terminals in tangential slices showed that EPSCs from PNs and PVs in patches were larger (p < 0.001, Wt) than in interpatches (Figures 5A, B, D, G), indicating that input strength is correlated with the density of connections to patches in L1 (Figure 2B). Within patches EPSCs from PVs (529.5 ± 305.9 pA, N = 23) and PNs (598.9 ± 433.2 pA, N = 23) were similar (p = 0.38, Wt) (Figures 5C, G). A similar balance was observed in the matching current densities and the spatial extent of activation of PNs and PVs in patches (Figure 5F). In contrast, in interpatches EPSCs from PVs (244 ± 186.4 pA, N = 23) were 1.9-fold larger (p < 0.001, Wt) than from PNs (130.1 ± 145.9 pA, N = 23) (Figures 5E, G). Although in interpatches current densities from PVs and PNs were similar, inputs to PVs covered a larger area, which resulted in the more effective activation of PVs (Figures 5E, F). Together, the comparison of thalamocortical and intracortical inputs to L1 shows that each pathway preferentially targets specific modules and within these modules differentially excites PNs and PVs.
Figure 5. Tangential slices: sCRACM of LM→V1 input to L1 onto L2/3 PNs and PVs in patches and interpatches.
(A) Venus labeled LM→V1 projections in L1 and Alexa 594 hydrazide-filled pairs of L2/3 PNs and PVs in patch and interpatch. (B, D) Whole cell patch clamp recordings in the same slice. Each trace represents average of EPSCs evoked by laser stimulation of ChR2-expressing LM→V1 terminals recorded from PNs and PVs in patches (Bi, Biii) and interpatches (Di, Diii). Heatmaps of responses from PN and PV in patches (Bii, Biv) and interpatches (Dii, Div). (C, E) Dots represent relative strength of LM→V1 input to a pair of L2/3 PNs and PV in patches (C) and interpatches (E). (F) Distribution of LM→V1 input strength across dendritic tree of PNs and PVs in patches and interpatches. Grey bars denote input areas, red bars represent current densities. (G) Box plots of strength of LM→V1-evoked EPSCs from PVs and PNs in patches and interpatches. (C, E, F, G). Wt (***p < 0.001, ns = not significant). Same conventions as in Figure 3.
Distinct inhibitory subnetworks in V1
The clustering in the cell-specific strength of activation by inputs to L1 raised the question whether inhibition is non-uniformly distributed across the sheet of V1.
Clustering of GABAergic neurons.
To determine whether GABAergic neurons are clustered we used VGAT-EYFP mice. As proxy for M2 patches we traced the dLGN→V1 connections with AAV2/1hSyn.tdT and cut tangential sections though V1. We found that VGAT expression in L1–2 was less conspicuously clustered than the dLGN→V1 projections (Figures S5A–E). Normalization of the fluoresence intensity in interpatches to the mean of patches, however, showed that on average (N = 2 mice) labeling in interpatches was brighter (p < 0.035, KS) (Figure S5F).
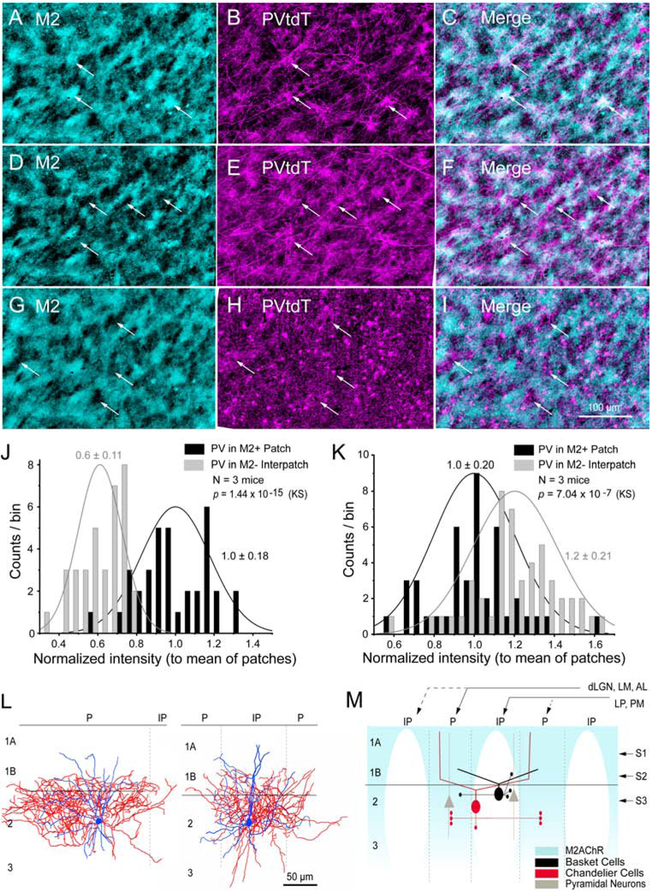
Previous reports have shown that PV- and SOM-expressing GABAergic neurons in L2/3 of mouse V1 are clustered and radially aligned in L5 with subcortically projecting PNs (Ebina et al., 2014; Maruoka et al., 2017). To test spatial clustering we cut tangential sections through V1 of PVtdT mice and found a striking patchiness (120 μm center-to-center spacing) of labeled fibers in L1 (Figure S5G). To determine whether M2 and PVs are spatially registered, we stained tangential sections through V1 with an antibody against M2. We found that in the outer half of L1 (L1A, Figure 6M) clusters of PVtdT+ dendrites overlapped with M2+ patches (Figures 6A–C). In contrast, in L1B and the top of L2/3 the PVtdT clusters switched registration and overlapped with M2− interpatches. PVtdT clusters in L1B contained dendrites and axons to which cell bodies and pericellular boutons of basket cells (BCs) were added in L2 (Figures 6D–I, M). The complementary mapping schemes were confirmed by shifting alignments of PVtdT intensities with M2+ patches in L1A (p =1.44 × 10−15, KS) to M2− interpatches in L1B-2 (p = 7.04 × 10−7, KS) (Figures 6J, K). We further found that the PV cell density in the top 160 μm of L2/3 was 37% higher in interpatches (204/mm2) than in patches (128/mm2). A similar module-selective mapping was found in biocytin-filled PVs in patches (N = 3) and interpatches (N = 5) whose axons were largely confined to 100 μm-wide clusters within the home module (Figure 6L).
Figure 6. Clustering of PV neurons in V1.
(A-C) Tangential 40 μm section through V1 at 40 μm below pial surface (see, S1 in M) showing overlapping pattern of immunolabeled M2+ patches (cyan, arrows) with PVtdT processes (magenta). (D-F) Section ~80 μm below surface (see, S2 in M; section aligned to A) showing overlapping pattern (arrows) of PV processes with M2− interpatches. (G-I) Section ~120 μm below surface (see, S3 in M; aligned to B) showing overlapping pattern (arrows) of PV cell bodies, dendrites and boutons with M2− interpatches. (J) Normalized PVtdT intensity (to mean intensity in patches) in L1A showing higher PV in patches. (K) Normalized PVtdT intensity (to mean intensity in patches) in L2 showing higher PV in interpatches. (J, K) KS, mean ± SD. (L) Biocytin-filled L2/3 PV+ BCs (coronal plane) showing that dendrites (blue) branch in L1B-2. Axons (red) of cells in patches (P, left panel) and interpatches (IP, right panel) branch near the cell body with little spread to neighboring IPs or Ps, respectively. Left panel shows patch-cell with asymmetrical axon arbor, largely contained within P. Elongated shape of axonal arbor is due to oblique section angle across P. PVs are less frequent in P. (M) Diagram of coronal section. Strong (solid lines) dLGN→V1, LM→V1, AL→V1 inputs to M2+ Ps (blue). Strong LP→V1, PM→V1 inputs to M2− IPs. High density of PV+ BC cell bodies, dendrites, axo-somatic and axo-dendritic boutons in L1B-2 of IPs. Putative PV+ ChC dendrites are denser in Ps and extend to L1A. Dense axo-axonic connections in L2/3 of Ps.
Does the shifting alignment of PVtdT and M2 expression across L1 reflect a cell type-specific depth profile similar to prefrontal cortex (Miyamae et al., 2017), where dendrites of chandelier cells (ChCs) project to L1A and BCs to L1B (Figure 6M)? Previous studies have shown that the distribution of L2/3 ChCs is non-random and projections to Ankyrin G immunolabeled axon initial segments (AIS) are spatially clustered (Blázquez-Lorca et al., 2015; Inan et al., 2013; Taniguchi et al., 2013). Thus, we compared the density of PVtdT+ putative contacts with AIS. Counts were made on radially oriented, tapered AISs of presumptive PNs in 75 μm-wide ‘columns’ aligned with M2+ patches and M2− interpatches (Figures S6A–D). We found that the length density of boutons was 52% higher (p < 0.01, paired t-test) in patches than interpatches, which supports that ChCs are clustered and that the connections preferentially innervate PNs in patches (Figures 6M; S6E).
To find out whether clustering of inhibitory neurons in interpatches results in irregular cell densities within L2/3 we determined the density of Ankyrin G expressing, vertical AIS and treated them as proxies of PNs. We found that the AIS density in interpatches was 21% lower (p < 0.01, paired t-test) than in patches (Figure S6E), which suggests that the ‘missing’ PNs in interpatches are offset by interneurons in proportion of their overall abundance (Keller et al., 2018).
Subnetwork-specific inhibition.
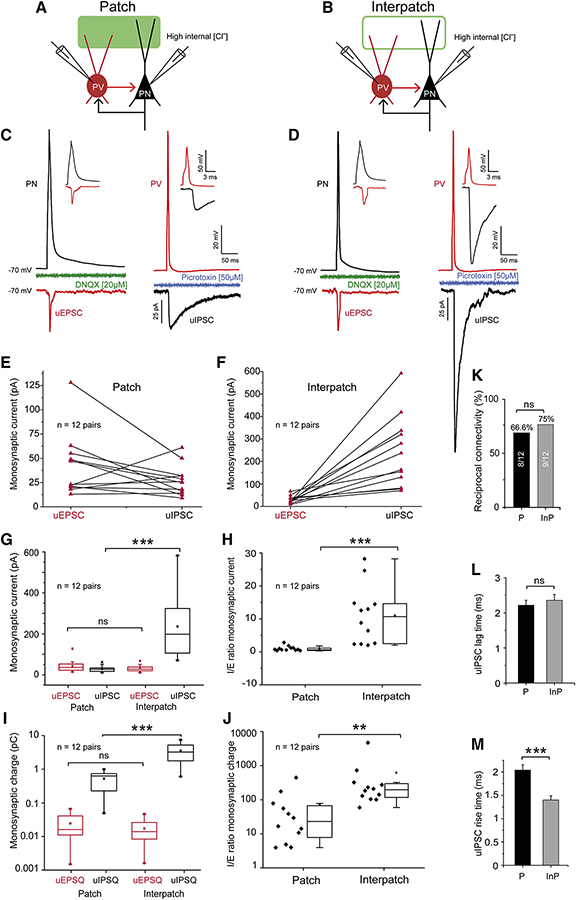
PV-mediated inhibition of PNs depends on the intensity of long-range and local recurrent excitation of PVs as well as the strength of their inhibitory output (Isaacson and Scanziani, 2011). To determine the effectiveness of local patch and interpatch subnetworks we recorded unitary uEPSCs and uIPSCs from synaptically connected pairs of L2/3 PVs and PNs in V1 of PVtdT mice. Patches and interpatches were distinguished by tracing dLGN→V1 input to L1 with AAV2/9.CAG.ChR2.Venus. PVs were identified based on tdT expression and fast spiking properties. Neighboring PNs were selected based on morphology and regular firing (Figures S7A–C). Recordings from PNs were performed at holding potentials of −70 mV with pipettes containing a high [Cl−]. This enhanced the inward-directed (Luo et al., 2013), monosynaptic uIPSCs (latency 2.4 ± 0.3 ms [patches N = 22], 2.3 ± 0.3 ms [interpatches, N = 26]), elicited by spikes (recorded in current clamp) from presynaptic PVs (Figures S7D, E). Recordings from PNs showed that in interpatches uIPSC amplitudes (209 ± 149 pA) and charge transfers (2.79 ± 1.64 pC) were 5–7-fold larger (p < 0.001, two-sample t-test) than in patches (30 ± 15 pA, 0.54 ± 0.29 pC) (Figures S7F, G). Bath-application of the GABAA-receptor antagonist, Picrotoxin (50 μM), abolished uIPSCs in patches and interpatches (Figures S7D–F). Although PVs were less numerous in patches, the percentage of PVs→PNs contacts in patches (73%, 19 of 26) and interpatches (86.4%, 19 of 22 pairs) were similar (Figure S7H), suggesting that subnetwork-specific inhibition is due to the number of contacts per PV and/or the strength of synaptic input.
Subnetwork-specific E/I balance.
PV-mediated inhibition counterbalances mono- and polysynaptic excitation of PNs, which project back onto PVs and in turn provide subnetwork-specific feedback inhibition to PNs. In experiments separate from those shown in Figure S7, we measured uEPSCs and uIPSCs from reciprocally connected PN↔PV-pairs in patches and interpatches. The procedures were similar to those in Figure S7, except that we also recorded uEPSCs from PVs elicited by presynaptic spikes from PNs (Figures 7A–D). We found that uEPSC amplitudes and charge transfers from PVs in patches (42.2 ± 9.2 pA) and interpatches (29.1 ± 4.7 pA) were similar (p = ns, two-sample t-test) (Figures 7C–F, G, I). In contrast, mean uIPSCs and uIPSQs recorded from PNs in interpatches (238.5 ± 45.9 pA, N =12 pairs) were 8.8-fold larger (p < 0.001, two-sample t-test) than in patches (27 ± 4.5 pA (N = 12 pairs) (Figures 7C–F, H, J). Bath application of Picrotoxin (50 μM) blocked uIPSCs in patches (10/10) and interpatches (11/11) (Figures 7C, D). In the returning excitatory connections, bath application of the AMPA receptor antagonist, DNQX (20 μM), blocked uEPSCs in patches (9/9) and interpatches (10/10) (Figures 7C, D). In patches the uIPSCs/uEPSCs ratio was 0.89 ± 0.21 (N = 12 pairs) and differed significantly (p < 0.001, two-sample t-test) from 10.9 ± 2.5 (N = 12 pairs) in interpatches (Figure 7H). Similar I/E ratios were obtained for synaptic charge transfer (Figure 7J).
Figure 7. Stronger inhibition in interpatches than patches.
(A, B) Diagram of recordings in coronal slices of unitary uIPSCs and uEPSCs in synaptically reciprocally connected pairs of L2/3 PNs and PVs (PN↔PV) aligned with ChR2.Venus-expressing dLGN→V1 patches (solid green) and interpatches (green outline) in L1. Recordings of inward uIPSCs from PNs with high [Cl−] internal solution at −70 mV holding potential. Recordings from PVs with K-gluconate internal solution. (C, D) uEPSCs (inward red trace) from PVs and uIPSCs (inward black trace) from PNs, after presynaptic spike from PNs or PVs, respectively, in patch (C) and interpatch (D). uEPSC in patches and interpatches have similar amplitudes and are blocked by DNQX (green C, D). In the reverse connection uIPSCs (black trace) in interpatches (D) were larger than in patches (C), and were blocked by Picrotoxin (blue C, D). The insets in (C) and (D) show that in the PV→PN and PV←PN direction postsynaptic responses exhibit monosynaptic delays. (E, F) uEPSCs and uIPSCs recorded in pairs of PNs and PV in patches and interpatches. In most pairs, uIPSCs in interpatches are larger than in patches. (G) Average uEPSCs from reciprocally connected PV↔PN pairs are similar, whereas uIPSCs in interpatches are larger (***p < 0.001). (H) Average I/E ratios of PV↔PN pairs in patches and interpatches. I/E balance in interpatches is tilted toward inhibition (***p < 0.001, paired t-test). (I) Average charge of uEPSCs and uIPSCs in patches and interpatches, showing that excitatory charge transfer at PN→PV contacts in patches and interpatches is similar. In interpatches the inhibitory charge transfer at PV→PN contacts is larger (*** p < 0.001, one-way ANOVA, Bonferroni correction). (J) I/E ratio showing that in reciprocally connected PV↔PN pairs in interpatches, uEPSCs are more strongly (** p < 0.01, two-sample t-test) opposed by uIPSCs. (K) The percentages of the total number of reciprocally connected PV↔PN pairs in patches (Ps) is similar (ns, Chi square test) in patches and interpatches (IPs). (L) The onset latency of uIPSC in Ps and IPs is similar (ns, Chi square test). (M) The rise time of uIPSCs in IPs is faster than in Ps (***p<0.001, paired t-test).
In both patches and interpatches uIPSCs lagged uEPSCs by 2.21 ± 0.14 ms and 2.35 ± 0.16 ms, respectively (Figure 7L), but the rise times of uIPSCs in interpatches were faster (p < 0.001, two-sample t-test) (Figure 7M). Thus, PV-inhibition of PNs is markedly stronger and faster in interpatches than in patches.
In accordance with the higher density of PVs in interpatches the percentage of reciprocally connected PV↔PNs pairs in interpatches was higher (75%, N = 12) than in patches (N = 12, 66.6%) (Figure 7K). Connections across the patch/interpatch border remain unknown due to the inability to determine the boundary with ≤40 μm accuracy.
DISCUSSION
We have found two interdigitating maps of M2+ patches and M2− interpatches in L1 of mouse V1 and show that PV-mediated inhibition of neighboring L2/3 PNs is stronger in interpatches than in patches. Each module is driven by distinct long-range inputs to dendrites in L1. The weakly-inhibiting patch subnetwork is the preferred target of dLGN, LM and AL inputs, whereas the strongly-inhibiting interpatch subnetwork is favored by inputs from LP and PM. Patch- and interpatch-preferring inputs differentially activate PVs and PNs of each subnetwork. In patch-preferring pathways, inputs from the dLGN to PVs in patches and interpatches are stronger than to PNs. In contrast, inputs from LM to PVs in interpatches, but not in patches, are stronger than to PNs. In the interpatch-preferring LP→V1 pathway, inputs to PVs and PNs are equally strong in patches and interpatches. Taken together, the clustering and strength of long-range inputs to weakly and strongly inhibiting PV↔PN modules suppress the spike output of PNs by distinct amounts of net inhibition which is strongest in the feedforward LP→V1, intermediate in the dLGN→V1 and weakest in the feedback pathway from LM.
Patchy networks in L1 of V1
Finding interdigitating maps of clusters of thalamic and intracortical inputs in L1 was unexpected, given the salt-and-pepper organization of mouse V1 (Ohki and Reid, 2007). However, interdigitating microcolumns of subcortically and intracortically projecting L5 neurons, including clusters in L2/3 and bundled dendrites with similar orientation and SF tuning, were recently found in mouse V1 (Kondo et al., 2016; Maruoka et al., 2017; Ringach et al., 2016). It was puzzling, though, why L5 neurons were mapped in a hexagonal pattern with a periodicity of 30–45 μm, which differed from the quasi-rectangular lattice and the 120 μm periodicity of M2 patches in L1 (Ji et al., 2015). The mismatch may be explained by the 25% larger grid size of supragranular layers (A. Burkhalter, unpublished results) which forces a fan out of ascending L5 dendrites to remain aligned with homotopic points.
Clustered long-range projections to L1 from horizontal and feedback networks have been observed in primate and mouse V1 (Ji et al., 2015; Stettler et al., 2002). Here, we show that L1 projections from dLGN and the LP are organized in interdigitating maps registered to M2+ patches and M2− interpatches. Unlike the canonical core dLGN→V1 pathway to L3–4, projections to L1 originate in the dLGN shell, which receives direction selective retinal and tectal input and renders L2/3 neurons postsynaptic to L1 patches orientation and direction selective (Cruz-Martin et al., 2014; Roth et al., 2016). Recordings from dLGN→V1 terminals have shown that they carry locomotion and saccade related signals and inform dendrites in L1 whether the speed of self-motion is matched to visual flow (Roth et al., 2016). LP input to L1 derives from multiple subnuclei (Bennett et al., 2019), including the V1-recipient anterior portion (Beltramo and Scanziani, 2019) whose projections we have traced here. Direction selectivity is inherited from the superior colliculus (Bennet et al., 2019) and inputs to L1 are tuned to the mismatch of self-motion and visual flow, suggesting a role in detecting unexpected of moving objects (Roth et al., 2016).
Among intracortical feedback connections to L1, patch-targeting inputs from LM and AL interdigitate with interpatch-targeting inputs from PM. Despite AL and PM belonging to the dorsal stream (Wang et al., 2012), they project to distinct compartments in V1. This suggests that the dorsal stream is branched and resembles the occipito-parietal network in primates (Kravitz et al., 2011). The bifurcation suggests that the AL-substream is specialized for visually guided actions whereas the PM-limb is more involved in spatial navigation (Kravitz et al., 2011). The convergence of feedback from LM and AL suggests that patches multiplex inputs from functionally non-matching presynaptic neurons (Glickfeld et al., 2013; Marshel et al., 2011). Alternatively inputs from LM and AL may target different cell types within patches, possibly with tuning-dependent retinotopic specificity (Marques et al., 2018).
Subnetwork-selective targeting of PNs and PVs
By stimulating L1 input from thalamocortical and intracortical pathways and recording EPSCs from L2/3 PNs and PVs in patches and interpatches we have found that the strength of synaptic activation matches the density of spatially clustered projections to L1. Studies in rodent visual and barrel cortex have shown that synaptic inputs to apical dendrites in L1 elicit spikes in L2/3 PNs which enhance orientation selectivity, and improve the detection of whisker deflection and accuracy of tactile perception (Cauller and Connors, 1994; Manita et al., 2015; Smith et al., 2013; Takahashi et al., 2016). Our findings indicate that these functions are compartmentalized, controlled by pathway and module-specific long-range innervation of PNs and PVs, which drive strongly- and weakly inhibitory local subnetworks and generate patch- and interpatch-specific excitatory output.
Consistent with previous demonstrations of patchy patterns of GABAergic neurons in primate V1 (Blümcke et al., 1990; Hendrickson et al., 1981) our results show that PV somata in L2/3 and terminals in L1/2 are clustered (Ichinhoe et al. 2003; Maruoka et al., 2017; Znamenskiy et al., 2018). This architecture is compatible with the clustering of IPSCs (Ebina et al., 2014), which was ~5-fold larger in interpatches than patches. The strong locally generated inhibition may lower the gain of PN spike output and increase the sensitivity and robustness of responses to TF in interpatches (Atallah et al., 2012; Ji et al., 2015; Katzner et al., 2011; Zhu et al., 2015). Together, the clustering of long-range input and local inhibitory subnetworks indicate that inhibition stabilizes the network (Xue et al, 2014) non-uniformly and provides for visual tuning in a pathway- and module-specific fashion. Strong inhibition in the LP→V1 and dLGN→V1 feedforward pathways may select for synchronous inputs and enhance stimulus detection, whereas weaker inhibition in the LM→V1 feedback pathway (D’Souza et al., 2016; Yang et al., 2013) may broaden the integration window of convergent inputs and enhance the discriminability of stimulus features (Gabernet et al., 2005; Wang et al., 2010).
In addition to the PV+ BC dendrites in L1B of interpatches, we found PV+ dendrites in patches of L1A, which may belong to ChCs (Taniguchi et al., 2013; Tasic et al., 2018). Clustering of ChC dendrites is consistent with the patchy innervation of AIS of cortico-cortically projecting L2/3 PNs (Blázquez-Llorca et al., 2015; Fariñas and DeFelipe, 1991) and the high density of PV contacts with AIS in patches. The distinct sublaminar distribution of putative ChCs and BCs in L1, suggests that each cell type is activated by different inputs. Recruitment of ChC-mediated axo-axonic inhibition may cancel spike outputs from L2/3 PNs (Woodruff et al., 2011). In contrast, BC-mediated inhibition, may regulate PN firing in a more nuanced fashion by coordinating the dendritic integration of bottom-up and top-down inputs (Larkum et al., 2007; Larkum, 2013).
What is the functional significance of modules? One possibility is that they provide for differential sensitivity to high visual acuity and temporal precision in a grid that ensures equal representation across the cortical point image (Ji et al., 2015). Direct support for this comes from subcortically projecting L5B PNs which overlap with PV clusters, suggesting that they receive interpatch input onto thick dendrites in L1 (Kim et al., 2015; Maruoka et al., 2017). Notably, these cells are sensitive to high TF and low SF (Kim et al., 2015), precisely the properties we have found in L2/3 of interpatches (Ji et al., 2015). By contrast, intracortically projecting L5A neurons are excluded from PV clusters, are tuned to high SF and resemble the L2/3 cells in patches (Ji et al., 2015; Kim et al., 2015; Maruoka et al., 2017). However, there are many types of L2/3 neurons (Harris et al., 2018) of which most project to multiple targets (Han et al., 2018), making a clean separation into patch and interpatch neurons based on connections alone unlikely.
STAR * METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources, reagents, and data should be directed to the Lead Contact, Andreas Burkhalter (burkhala@wustl.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Experiments were performed on male and female, C57BL/6J, PV-Cre (Bg.129P2-Pvalbtm(cre)Arbr/J) × Ai9 (Gt[ROSA]26Sortm9(CAG-tdTomato)Hze), Chrm2-tdT-D knock-in mice (Bg6.Cg-Chrm2tm1.1Hze/J), Chrm2 M2R−/−(B6N.129S4(Cg)-Chrm2tmJwe/J) and VGAT-ChR2-EYFP (B6.Cg-Tg(Slc32a1-COP4*H134R/EYFP)8Gfng/J) mice. For anatomy we used 4–10 day old and >46 day old animals. Slice recordings were done in 34–46 day-old animals. Thalamocortical and intracortical connections to V1 were visualized by axonal tracing with AAV. sCRACM mapping of long-range input to PNs and PVs and recordings of uEPSCs and uIPSCs between synaptically connected pairs of PNs and PVs were performed in acute slices of V1. All experimental procedures were approved by the Institutional Animal Care and Use Committee at Washington University.
METHOD DETAILS
Tracing of connections
Connections were traced anterogradely by intracerebral injection of AAV2/9.CAG.ChR2.Venus.WPRE.SV40 (Vector Core, University of Pennsylvania), AAV2/1.hSyn.ChR2(H134R).EYFP (Vector Core, University of Pennsylvania), AAV2/1.hSyn.EGFP.WPRE.bGH (Vector Core, University of Pennsylvania) and/or AAV2/1hSyn.tdTomato.WPRE.bGH (Allen Institute) in 18–24 day-old or 8–20 week-old mice. Animals were anesthetized with Ketamine/Xylazine (86/13 mg/kg, IP). Analgesia was performed with Buprenorphine (0.1 mg/kg, SQ). The eyes were protected with ophthalmic ointment. All surgical procedures were performed in a stereotaxic apparatus. Injections (46–92 nl) were made with glass pipettes (tip diameter 15–25 μm) connected to a Nanoject II injector. Stereotaxic injections were made into the (in mm): dLGN (2.35 posterior of bregma, 2.15 lateral of midline and 2.55 below the pial surface), LP (1.85 posterior of bregma, 1.25 lateral of midline and 2.65 below the pial surface), higher visual cortical lateromedial area, LM (1.4 anterior to transverse sinus, 4.1 lateral to midline, 0.3–0.5 below the pial surface) and posteromedial area, PM (1.9 anterior to transverse sinus, 1.6 lateral to midline, 0.3–0.5 below the pial surface). Postsurgical survival was 2–3 weeks.
Immunostaining
Mice were overdosed with Ketamine/Xylazine (500/50 mg/kg, IP) and transcardially perfused with heparinized (10,000 Units) phosphate-buffered saline (PBS), followed by 1% paraformaldehyde (PFA). Brains were extracted from the skull, the left cortical hemisphere was removed, flat mounted, postfixed overnight in 4% PFA and cryoprotected in 30% sucrose. Tangential or parasagittal sections were cut at 40 μm with a freezing microtome. Sections were washed in 0.1 M phosphate buffer (PB), treated in blocking solution containing 10% normal goat serum (NGS), and 0.1% Triton X-100 in PB. Immunolabeling was performed by incubating sections for 48 hours at 4°C with primary antibodies against M2 muscarinic acetylcholine receptor (1:500 rat monoclonal, MAB367 MilliporeSigma) or anti-Ankyrin G (1:1000, mouse monoclonal, clone N106/36, NeuroMab). After washing, sections were treated with Alexa Fluor 647-labeled goat anti-rat IgG secondary antibody (1:500 in 10% NGS; A21247 Invitrogen) or donkey-anti-mouse-Cy5 (1:500, 715-175-151, Jackson Immuno Research). Cholinergic fibers were identified with an antibody against choline acetyl transferase (1:1000, goat anti-ChAT, MilliporeSigma AB144P), detected with a biotinylated donkey-anti goat secondary antibody (1:200, MilliporeSigma AP180B) and visualized with NeutrAvidin Oregon Green 488 (1:400, ThermoFisher A6374). Sections were mounted onto glass slides, coverslipped in PB or Aqua Poly/Mount (Polysciences) and imaged under a fluorescence microscope (Nikon 80i) equipped with a CCD camera (CoolSnap EZ, Roper Scientific or Infinity3S-URM, Lumenera). Confocal imaging was performed with an Olympus Fluoview (FV1200) microscope. The specificity of the M2 primary antibody was validated in C57BL/6J-M2−/− mice (Gomeza et al., 1999) in which we saw no detectable staining.
Slice preparation
Slices of V1 were prepared from 34–46 days-old virus-injected mice. The slices were either cut in the tangential or the coronal plane. Tangential slices were optimally suited for identifying repeating clusters of ChR2.Venus-labeled thalamocortical and intracortical inputs to L1 and preserving the apical dendritic arbors of L2/3 PNs and PVs. Mice were decapitated under isoflurane (2% in oxygen) anesthesia. The brain was rapidly removed from the skull and submerged in ice-cold cutting solution aerated with 95% O2/5% CO2 containing (in mM): 240 sucrose; 2.5 KCl, 1.25 NaH2PO4, 2.1 NaHCO3, 7 MgCl2, 0.5 CaCl2, 10 glucose, adjusted with NaOH to pH 7.35. Next, the cerebellum and anterior third of the brain were removed. With the cut rostral surface towards the base, the lower part of the brain was resected parallel to the surface of V1 and the tissue block was mounted with the cut-side down onto the specimen plate. Single tangential slices (350 μm) were cut in ice-cold cutting solution on a Vibratome (Leica VT 1200). Slices were kept in a holding chamber in which they were submerged in oxygenated artificial cerebrospinal fluid (ACSF; containing [in mM]:125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 1 MgCl2, 2 CaCl2, 25 glucose, pH 7.35) for 1 hour at 32°C before transferring them to the recording chamber maintained at room temperature (22–24°C).
Coronal slices were prepared as described previously (Yang et al., 2013). Coronal slices are optimal for preserving connections and dendrites across layers. Although clustered projections to L1 could be readily identified in coronal and parasagittal slices (Figures 4A, S6) the pattern is more easily resolved in tangential slices.
Subcellular Channelrhodopsin-2 assisted circuit mapping (sCRACM) and tissue processing
For recording, slices were transferred to a submersion chamber mounted on the stage of a modified upright fluorescence microscope (Nikon Eclipse FN1) equipped with a CCD camera (Retiga-2000C; Qimaging). Slices where perfused (1.5 ml/min) with recirculating oxygenated ACSF (22–24°C). Whole cell patch clamp recordings were obtained from pairs of tdT-expressing PVs (identified with fluorescence optics) and nearby (within ≤ 40 μm) unlabeled PNs cells (identified with DIC-IR optics) in L2/3 of V1. Cell pairs were either in-register (i.e. within patches) or out-of-register (i.e. within interpatches) with ChR2.Venus- or ChR2.EGFP-labeled patches of axons projecting from the dLGN, LP or area LM and terminating in L1 of V1. For coronal slice experiments examining LP→V1 synaptic inputs in patches and interpatches, two injections were performed in each mouse: AAV2/1hSyn.tdTomato.WPRE.bGH was injected into the dLGN to label patches, and AAV2/1.hSyn.ChR2(H134R).EYFP injected into the LP. For recording in tangential slices, neurons were approached from the cut surface of the slice, which was mounted with the pial surface facing down. In tangential and coronal slices, recordings were made 30–120 μm below the surface of the slice. Electrical signals were sampled at 10 kHz by Multiclamp 700B amplifiers (Molecular Devices), digitized (NI USB 6363; National instruments) and acquired using Matlab-based (MathWorks, Natick, MA) Ephus software (Suter et al., 2010). Electrodes were pulled from borosilicate glass capillaries (G150F-4, Warner Instruments). Pipettes were filled with (in mM): 128 potassium gluconate, 4 MgCl2, 10 HEPES, 1 EGTA, 4 Na2ATP, 0.4 Na2GTP, 10 sodium phosphocreatine, 3 sodium L-ascorbate, 0.02% Alexa 594 hydrazide (Invitrogen) or 3 mg/ml biocytin, pH 7.25, 290 mOsm. The pipette resistance was 3–5 MΩ. The liquid junction potential was not corrected. The seal resistance was > 2 GΩ. Recordings with access resistance of > 20 MΩ were excluded from the study. Neuron type was assessed by recording spiking patterns (i.e. fast for PVs, regular for PNs) in response to 300 ms pulses of hyperpolarizing and depolarizing current in current clamp mode. For sCRACM mapping (Petreanu et al., 2009), EPSCs were recorded in voltage clamp at a holding potential of −70 mV, with tetrodotoxin (1 μM) and 4-aminopyridine (4-AP; 100 μM; Tocris Bioscience) in the bath to block action potentials and fast repolarizing potassium currents, respectively. After recording, slices were fixed overnight in 4% PFA, cleared in 10% sorbitol, mounted on glass slides in Aqua Poly/Mount and imaged under a confocal microscope (Olympus, Fluoview FV1200) and reconstructed in 3D, using ImageJ.
Photostimulation
Photostimulation was performed with a blue laser (473 nm; CrystaLaser). The light was reflected by a fixed set of mirrors onto galvanometer scanners (Cambridge Scanning) that controlled beam position. The light then passed through an air objective (4 PlanApo, NA 0.2; Nikon), which at 0.25 mW/cm2 laser power formed a beam at half maximal intensity with a diameter of ~20 μm in the specimen plane. The durations and intensities of the light pulses were controlled with a Pockels cell (ConOptics) and a shutter (LS6, Uniblitz). Because the proportion and labeling intensity of ChR2-expressing axons varied across slices and animals, the laser power (0.25–1 mW/cm2) was adjusted in every slice to evoke EPSCs. The laser power was constant for all recordings made that day. Recordings in V1 were performed from pairs of nearby PNs and PVs in patch and interpatch regions of L2/3. Each trial consisted of 100 ms baseline, followed by the photostimulus (1 ms) and 300 ms of response. Photostimulation was performed in an 8 × 8 grid in which individual points were spaced 75 μm apart and the grid was aligned with the recorded soma at the center. For mapping in coronal slices, one side of the grid was aligned with the pial surface. The stimulation sequence was pseudorandom allowing maximal intervals between nearby stimulation sites. sCRACM maps were generated from 3–5 repetitions per neuron.
Recordings from synaptically connected pairs
To examine the presence and strength of synaptic connections between PVs and PNs in patches and interpatches we recorded from synaptically connected pairs in superficial L2/3 of V1. Patchy projections to L1 were identified as clusters of Venus-expressing axon terminals, labeled by tracing dLGN→V1 inputs with AAV2/9.CAG.ChR2.Venus. Recordings were obtained from cell pairs (≤ 40 μm apart) aligned with patches or interpatches in coronal slices. Responses in PVs were recorded with pipettes (4–6 MΩ resistance) filled with (in mM): 128 potassium gluconate, 4 MgCl2, 10 HEPES, 1 EGTA, 4 Na2ATP, 0.4 Na2GTP, 10 sodium phosphocreatine, 3 sodium L-ascorbate, pH 7.25, 290 mOsm. For recording PNs, pipettes were filled with a high [Cl−] solution containing (in mM): 145 KCl, 10 HEPES, 5 NaATP, 0.2 NaGTP and 5 EGTA, pH 7.3, adjusted with KOH, 285 mOsm. Under these conditions the reversal potential of IPSCs is 0 mV and at −70 mV holding potential the currents flow inward (Luo et al., 2013). Monosynaptic uIPSCs were recorded by holding PNs at −70 mV. uIPSCs were elicited by triggering single action potentials from presynaptic PVs with 2 ms depolarizing current pulses. The same stimulation/recording paradigm was used for eliciting and measuring uEPSCs, except that spikes were elicited from PNs and responses were recorded from PVs. Responses were averaged across 50–150 repetitions at 0.5 Hz. The error due to the liquid junction potential was not corrected. Access resistance was monitored throughout the experiment. Cells whose series resistance was >20 MΩ or varied >25% for the duration of the experiment were excluded from the analysis. Series resistance errors were not compensated. To block spontaneous polysynaptic NMDA receptor-mediated excitatory currents, CPP ((RS)-3-(2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid, 50 μM, Tocris) was applied in the bath. uIPSCs were blocked by bath application of the GABAA-receptor antagonist Picrotoxin (50μM, Tocris). uEPSCs were blocked by bath application of the AMPA-receptors antagonist DNQX (6,7-dinitroquinoxaline-2,3-dione, 20 μM, Tocris). After recording, the slices were fixed in 4% PFA, mounted on glass slides, cleared in 10% sorbitol and Alexa 594 hydrazide-filled neurons were imaged under the confocal microscope.
Confocal imaging and Neuron reconstruction
Alexa Fluor 594 hydrazide-filled neurons were reconstructed posthoc and their location in Venus-expressing patches of dLGN or LM inputs determined by imaging under a confocal microscope (Olympus, Fluoview FV 1200), using a 30× silicone oil (UPLSAPO, 1.05 NA, Olympus) objective. Twelve bit 1024 × 1024 pixel images were taken at 1.5× digital zoom. Z-stacks were acquired at 0.80 μM/section (Nyquist volume: 1.6 μM) across the thickness of the slice. Multicolor scanning was done in sequential and frame-by-frame mode. The images were acquired in separate high sensitivity detector channels for each fluorophore. The signals were acquired and averaged by Kalman’s method to increase signal/noise ratio. The neurons were then traced and reconstructed by using the ‘Simple Neurite Tracer’ Plugin of Fiji (ImageJ). PNs were identified by the presence of dendritic spines, whereas PVs have aspinous, beaded dendrites. The same protocol was used for imaging biocytin-filled neurons, except that after overnight fixation in 4% PFA, slices were rinsed in 0.1 PB and incubated overnight in 1:1000 streptavidin Alexa Fluor 647 (Thermo Fisher) or 1:400 Neutralite Avidin-CY5 (SouthernBiotech). For bright field microscopy slices containing biocytin-filled neurons were treated with an ABC reaction and an intensification of the reaction product with AgNO3 and HAuCl2 (Yang et al., 2013). Filled neurons were reconstructed under a 40X oil objective using Neurolucida (MicroBrightField).
The innervation density of PN-AISs by PVtdT expressing boutons was determined by confocal imaging with a 100× oil immersion objective of z-stacks (0.2 μm step size) in ROIs (65×135μm) aligned with M2+ patches and M2− interpatches. To minimize contamination by Ankyrin G-expressing AIS of interneurons we focused the analysis on tapered, vertically (±3° relative to the pial surface) descending profiles. Appositions between boutons and AIS were scored as contacts if there was no detectable gap between pre-and postsynaptic elements and their association remained stable under image rotation.
QUANTIFICATION AND STATISTICAL ANALYSIS Contour plots of patches and interpatches
Automatic patch/interpatch definition followed the general procedures of Sincich and Horton (2005). Fluorescent images of spatially clustered M2 expression or virally traced projection patterns in L1 of V1 were high-pass filtered using an 80 μm filter radius. Images were then blurred using a circular averaging filter of 30 μm radius, with the ‘fspecial’ function in Matlab. All pixels in the resulting images were then divided into six intensity quantiles. The top two quantiles were considered to be patches and the bottom two interpatches. For statistical testing, images in matching fields of view were analyzed. Images were downsampled to have a pixel area of 150 μm2 each. A permutation test was then performed by shuffling fluorescent image pixels within the image and determining the ratio of resulting average patch intensity to average interpatch intensity, maintaining the original patch/interpatch borders derived from M2 or viral tracings. Patch/interpatch ratios in the original image outside the 95% bounds of the randomized distribution from 100,000 shuffling iterations were considered significant deviations from a 1:1 patch/interpatch intensity ratio. These tests were performed using custom scripts in Matlab
Quantification of fluorescence intensity
The intensity of immunofluorescence of M2, fluorescently labeled projections tagged by viral axonal tracing, and fluorescence of transcribed tdT, EGFP and EYFP genes were quantified in images of tangential sections through L1, acquired with a CCD camera (Lumenera Infinity3S-URM) and Metamorph NX2.0 software (Molecular Devices). Gray scale images were opened with Image J, background subtracted to correct for global non-uniformities in brightness and overlaid with contour maps of fluorescence intensity determined by a custom Matlab script. Analyses were performed on the overlapping region between fluorescently labeled axonal projections and M2tdT expression at 8X to 20X magnification, and typically included 2 to 5 ROIs per tangential section. Pixel values in patches and interpatches were measured at multiple sites, normalized to the mean brightness of patches, binned and plotted as counts of normalized fluorescence intensity. Statistical comparisons of intensity distributions were made using the KS test.
EPSCs and IPSCs
The amplitude of significant responses was >4 times the SD of the baseline. Individual pixel values of sCRACM maps were computed from the mean EPSC amplitude in a 75 ms response window after the photostimulus. For each neuron, maps were averaged across 3–5 repetitions. These averages represent synaptic charge transfer. Because the responses were dominated by the current amplitude and small long-lasting currents were negligible, we have adopted the simplification introduced by Petreanu et al., (2009) and represent responses in pA instead of Coulomb. The charge value for each pixel in a 75 ms window was calculated using custom Matlab software. EPSC amplitudes were measured with reference to the soma at the center. To display the scaled magnitudes and spatial distributions of thalamocortical and intracortical inputs from LM to PNs and PVs, maps for each cell class were peak normalized within individual slices and displayed as heatmaps. Comparisons of inputs to PNs and PVs were made by plotting the average responses from pairs of PNs and PVs within layer 2/3 of the same slice and in the same patch or interpatch module. Thalamocortical and intracortical inputs to PNs and PVs recorded in the same layer and same slice were plotted against each other and the relative strengths of excitation was assessed by plotting the mean slope from zero. uIPSC lag time was calculated as the time delay from the onset of the presynaptic PV spike to the onset of the uIPSC recorded from the postsynaptic PNs. uIPSC rise time was measured as the delay between response onset and the peak.
Statistics
Statistical analyses were performed using Origin 9.1 (Origin Laboratory) or customized Matlab software. Normality was assessed using the Shapiro-Wilk normality test to select between parametric and nonparametric tests. Comparisons between two groups were performed with two-tailed Student’s t-test. Neighboring neurons that were recorded sequentially were considered pairs and subjected to a paired t-test or the Wilcoxon signed-rank test (Wt). For comparisons across more than two groups, data were analyzed using one-way ANOVA followed by Bonferroni’s post hoc analysis to correct for multiple comparisons. For data with non-normal distribution, nonparametric KS test or the Mann-Whitney U test was used. Significance was p < 0.05. Data are mean ± SEM, except when otherwise indicated as mean ± SD. Box plots show median, mean, and 25th and 75th percentile values; box plot whiskers represent 5th and 95th percentile values.
DATA AND CODE AVAILABILITY
The codes generated during this study are available at https://github.com/amsmeier/Bista-DSouza-et-al.-2019.
Supplementary Material
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Donkey anti mouse IgG Cy5 | Jackson Immunoresearch | Cat# 715-175-151 |
| Donkey anti goat IgG biotin-SP conjugate | Sigma-Aldrich | Cat# AP180B |
| Goat anti choline acetyl transferase | Sigma-Aldrich | Cat# AB144P |
| Goat anti rat IgG Alexa Fluor 647 | ThermoFisher | Cat# A21247 |
| Mouse monoclonal anti Ankyrin G antibody | NeuroMab | Cat# N106/36 |
| Rat monoclonal anti M2 muscarinic acetylcholine receptor antibody | Sigma-Aldrich | Cat# MAB367 |
| Labeled reagents | ||
| Alexa 594 hydrazide | ThermoFisher | Cat# A10438 |
| NeutrAvidin Oregon Green 488 | ThermoFisher | Cat# A6374 |
| Neutralite Avidin-CY5 | SouthernBiotech | Cat# 7200–15 |
| Streptavidin, Alexa Fluor 647 conjugate | ThermoFisher | Cat #S21374 |
| Biochemical Reagents | ||
| 4-Aminopyridine | Tocris Bioscience | Cat# 0940 |
| DNQX (6,7-dinitroquinoxaline-2,3-dione) | Tocris Biochemical | Cat# 2312 |
| Picrotoxin | Tocris Bioscience | Cat# 1128 |
| (RS) CPP ((RS)-3-(2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid | Tocris Bioscience | Cat# 0173 |
| Tetrodotoxin | Tocris Bioscience | Cat# 1078 |
| Experimental Models: Organisms/Strains | ||
| Ai9 (Gt[ROSA]26Sortm9(CAG-tdTomato)Hze) | Jackson Laboratory | Stock No: 007909 |
| C57BL/6J | Jackson Laboratory | Stock No: 000664 |
| Chrm2-tdT-D knock-in mice (Bg6.Cg-Chrm2tm1.1Hze/J) | Jackson Laboratory | Stock No: 030330 |
| Chrm2 M2R−/− (B6N.129S4(Cg)-Chrm2tmJwe/J) | Jackson Laboratory | Stock No: 030162 |
| PV-Cre (Bg. 129P2-Pvalbtm(cre)Arbr/J) | Jackson Laboratory | Stock No: 017320 |
| VGAT-ChR2-EYFP (B6.Cg-Tg(Slc32a1-COP4*H134R/EYFP) 8Gfng/J) | Jackson Laboratory | Stock No: 014548 |
| Virus strains | ||
| AAV2/9.CAG.ChR2.Venus.WPRE.SV40 | University of Pennsylvania | PennID AV-1–20071P |
| AAV2/1.hSyn.EGFP.WPRE.bGH | University of Pennsylvania | PennID p1696 |
| AAV2/1 phSyn 1 (S)-tdT omato.WPRE | Addgene | Plasmid#51506 |
| Software and Algorithms | ||
| Ephys | Open Ephys | https://www.open-ephys.org |
| Fiji software | GPL v2, Fiji | https://fiji.sc |
| Image J | National Institutes of Health | https://imagej.nih.gov/ij/download.html |
| Matlab 9.1 | Mathworks | https://www.mathworks.com/ |
| Metamorph NX2.0 | Molecular Devices | https://www.moleculardevices.com |
| Neurolucida 11 | MicoBrightField | https://www.mbfbioscience.com/ |
| Origin 9.1 | Origin Laboratory | https://www.originlab.com |
| Photoshop CC 2018 | Adobe Inc. | https://www.adobe.com/products/photoshop |
| Illustrator CC 2018 | Adobe Inc. | https://www.adobe.com/products/illustrator |
Highlights.
Interdigitating organization of dLGN and LP thalamic input to layer 1 of V1
Distinct feedback from ventral and dorsal stream cortical areas to layer 1 of V1
Spatial clustering of PV+ axons, dendrites and cell bodies in layers 1–3 of V1
Spatially clustered differential synaptic inhibition by PV+ interneurons
ACKNOWLEDGEMENTS
We thank Hongkui Zheng of the Allen Institute for Brain Science for the AAV2/1hSyn.tdTomato.WPRE.bGH, James Fitzpatrick and Dennis Oakley (Washington University Center for Cellular Imaging) and Katia Valkova for technical support. Thanks also to Tim Holy for the VGAT-ChR2-EYFP mice. Supported by National Eye Institute grants RO1 EY16184, RO1 EY022090, RO1 EY027383 and the McDonnell Center for Systems Neuroscience.
Footnotes
DECLARTION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agrawal HC, Davis JM, and Himwich WA (1968). Developmental changes in mouse brain: weight, water content and free amino acids. J. Neurochem 15, 917–923. [DOI] [PubMed] [Google Scholar]
- Atallah BV, Burns W, Carandini M, and Scanziani M (2012). Parvalbumin-expressing interneurons linearly transform cortical responses to visual stimuli. Neuron 73, 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayzenshtat I, Karnani MM, Jackson J, and Yuste R (2016). Cortical control of spatial resolution by VIP+ interneurons. J. Neurosci 36, 11498–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramo R, and Scanziani M (2019). A collicular visual cortex: neocortical space for an ancient midbrain visual structure. Science 363, 64–69. [DOI] [PubMed] [Google Scholar]
- Bennett C, Gale SD, Garret ME., Newton ML, Callaway EM., Murphy GJ, and Olsen SR(2019). Higher-order thalamic circuits channel parallel streams of visual information in mice. Neuron 102, 477–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez-Llorca L, Woodruff A, Inan M, Anderson SA, Yuste R, DeFelipe J, and Merchán-Pérez A (2015). Spatial distribution of neurons innervated by chandelier cells. Brain Struct. Funct 220, 2817–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümcke I, Hof PR, Morrison JH, and Celio MR (1990). Distribution of parvalbumin immunoreactivity in the visual cortex of old world monkeys and humans. J. Comp. Neurol 301, 417–432. [DOI] [PubMed] [Google Scholar]
- Cauller LJ, and Connors BW (1994). Synaptic physiology of horizontal afferents to layer 1 in slices of rat S1 neocortex. J. Neurosci 14, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Martín A, El-Danaf RN, Osakada F, Sriram B, Dhande OS, Nguyen PL, Callaway EM, Gosh A, and Huberman AD (2014). A dedicated circuit links direction-selective retinal ganglion cells to the primary visual cortex. Nature 507, 358–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaCosta NM, and Martin KAC (2013). Sparse reconstruction of brain circuits: Or, how to survive without a microscopic connectome. Neuroimage 80, 27–36. [DOI] [PubMed] [Google Scholar]
- D’Souza RD, Meier AM, Bista P, Wang Q, and Burkhalter A (2016). Recruitment of inhibition and excitation across mouse visual cortex depends on the hierarchy of interconnecting areas. eLife 2016; 5:e193332, doi: 10.7554/eLife.19332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina T, Sohya K, Imayoshi I, Yin S-T., Kimura R, Yanagawa Y, Kameda K, Hioki H, Kaneko T, and Tsumoto T (2014). 3D clustering of GABAergic neurons enhances inhibitory actions on excitatory neurons in the mouse visual cortex. Cell Report 9, 1–12. [DOI] [PubMed] [Google Scholar]
- Fariñas I, and DeFelipe J (1991). Patterns of synaptic input on cortico-cortical and corticothalamic cells in cat visual cortex. II. The axon initial segment. J. Comp. Neurol 304, 70–77. [DOI] [PubMed] [Google Scholar]
- Felleman DJ and Van Essen DC (1991). Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex 1, 1–47. [DOI] [PubMed] [Google Scholar]
- Gabernet L, Jadhav SP, Feldman DE, Carandini M, and Scanziani M (2005). Somatosensory integration controlled by dynamic thalamocortical feed-forward inhibition. Neuron 48, 315–327. [DOI] [PubMed] [Google Scholar]
- Glickfeld LL, Andermann M, Bonin V, and Reid RC (2013). Corticocortical projections in mouse visual cortex are functionally target-specific. Nat. Neurosci 16, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomeza J, Shannon H, Kostenis E, Felder C, Zhang L, Brodkin J, Grinberg A, Sheng H, and Wess J (1999). Prononced pharamcologic deficits in M2 muscarinic acetylcholine receptor kockout mice. Proc. Natl. Acad. Sci. USA 96, 1692–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Kebschull JM, Cambell RAA, Cowan D, Imhof F, Zador AM, and Mrsic-Flogel TD (2018). The logic of single-cell projection for visual cortex. Nature 556, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JA, Mihalas S, Hirokawa KE, Whitsell JD, Knox J, Bernard A, Bohn P, Caldejon S, Casal L, Cho A, Feng D, Gaudreault N, Graddis N, Groblewski PA, Henry A, Ho A, Howard R, Kuan L, Lecoq J, Luviano J, McConoghy S, Mortrud MT, Naeemi M, Ng L, Oh SW, Ouellette B, Sorensen SA, Wakeman W, Wang Q, Williford A, Phillips JW, Jones A, Koch C, and Zeng H (2018). The organization of intracortical connections by layer and cell class in the mouse brain. bioRxiv posted Apr. 1, 2018. doi: http://dx.doi.org/10.1.1101/292961. [Google Scholar]
- Hendrickson AE, Hunt SP, and Wu J-Y (1981). Immunocytochemical localization of glutamic acid decarboxylase in monkey striate cortex. Nature 292, 605–607. [DOI] [PubMed] [Google Scholar]
- Horvát S, Gǎmǎnut R, Ercsey-Ravaz M, Magrou L, Gǎmǎnut B, Van Essen DC, Burkhalter A, Knoblauch K, Trorczkai Z, and Kennedy H (2016). Spatial embedding and wiring cost constrain the functional layout of the cortical network of rodents and primates. PLOS Biol doi: 10.1371/journal.pbio.1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubel DH, and Wiesel TN (1962). Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol 160, 106–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubel DH, and Wiesel TN (1974). Uniformity of monkey striate cortex: a parallel relationship between field size, scatter, and magnification factor. J. Comp. Neurol 158, 295–305. [DOI] [PubMed] [Google Scholar]
- Ichinohe N, Fujiyama F, Kaneko T, and Rockland KS (2003). Honeycomb-like mosaic at the border of layers 1 and 2 in cerebral cortex. J. Neurosci 23, 1372–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan M, Blázquez-Llorca L, Merchán-Pérez A, Anderson SA, DeFelipe J, and Yuste R (2013). Dense overlapping innervation of pyramidal neurons by chandelier cells. J. Neurosci 33, 1907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson JS, and Scanziani M (2011). How inhibition shapes cortical activity. Neuron 72, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Gămănut R, Bista P, D’Souza RDD, Wang Q, and Burkhalter A (2015). Modularity in the organization of mouse primary visual cortex. Neuron 87, 632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani MM, Agetsuma M, and Yuste R (2014). A blanket of inhibition: functional inferences from dense inhibitory connectivity. Curr. Opin. Neurobiol 26, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzner S, Busse L, and Carandini M (2011). GABAA inhibition controls response gain in visual cortex. J. Neurosci 31, 5931–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D, Erö C, and Markram H (2018). Cell densities in the mouse brain: A systematic review. Front. Neuroanat 23 Oct 2018; 12:83. doi: 10.3389/fnana.2018.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Juavinett AL, Kyubwa EM, Jacobs MW, and Callaway EM (2015). Three types of cortical layer 5 neurons that differ in brain-wide connectivity and function. Neuron 88, 1253–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Yoshida T, and Ohki K (2016). Mixed functional microarchitectures for orientation selectivity in mouse primary visual cortex. Nat. Commun 7, 13210. doi: 10.1038/ncomms13210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz DJ, and Saleem KS, Baker CI, and Mishkin M(2011). A new neural framework for visuospatial processing. Nat. Rev. Neurosci 12, 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum ME, Waters J, Sakmann B, and Helmchen F (2007). Dendritic spikes in apical dendrites of neocortical layer 2/3 pyramidal neurons. J. Neurosci 27, 8999–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkum ME (2013). A cellular mechanism for cortical associations: an organizing principle for the cerebral cortex. Trends in Neurosci 36, 141–151. [DOI] [PubMed] [Google Scholar]
- Lee S-H, Kwan AC, Zhang S, Phoumthipphavong V, Flannery JG, Masmanidis SC, Taniguchi H, Huang ZJ, Zhang F, Boyden ES, Deisseroth K, and Dan Y (2012). Activation of specific interneurons improves V1 feature selectivity and visual perception. Nature 488, 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-H, Kwan AC, and Dan Y (2014). Interneurons subtypes and orientation tuning. Nature 508, (7494):E1–2. doi: 10.1038/nature13128. [DOI] [PubMed] [Google Scholar]
- Li L-Y, Ji X-Y, Liang, Li Y-T, Xiao Z, Tao HW, and Zhang LI (2014). A feedforward inhibitory circuit mediates lateral refinement of sensory representation in upper layer 2/3 of mouse primary auditory cortex. J. Neurosci 34, 13670–13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Partridge JG, and Vicini S, (2013). Distinct roles and extrasynaptic GABAA receptors in striatal inhibition dynamics. Front Neural Circuits 7, 186 dio: 10.3389/fncir.2013.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manita S, Suzuki T, Homma C, Matsumoto T, Odagawa M, Yamada K, Ota K, Matsubara C, Inutsuka A, Sato M, Ohkura M, Yamanaka A, Yanagawa Y, Nakai J, Hayashi Y, Larkum ME, Murayama M (2015). A top-down cortical circuit for accurate sensory perception. Neuron 86, 1304–1316. [DOI] [PubMed] [Google Scholar]
- Mao T, Kusefoglu D, Hooks BM, Huber D, Petreanu L, and Svoboda K (2011). Long-range neuronal circuits underlying the interaction between sensory and motor cortex. Neuron 72,111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques T, Nguyen J, Fioreze G, and Petreanu L (2018).The functional organization of cortical feedback inputs to primary visual cortex. Nat. Neurosci 21, 757–764. [DOI] [PubMed] [Google Scholar]
- Marshel JH, Garrett ME, Nauhaus J, and Callaway EM (2011). Functional specialization of seven mouse visual cortical areas. Neuron 72, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruoka H, Nagakawa N, Tsuruno S Sakai S, Yoneda T, and Hosoya T (2017). Lattice system of functionally distinct cell types in the neocortex. Science 258, 610–615. [DOI] [PubMed] [Google Scholar]
- Miyamae T, Chen K, Lewis DA, and Gonzalez-Burgos G (2017). Distinct physiological maturation of parvalbumin-positive neuron subtypes in mouse prefrontal cortex. J. Neurosci 37, 4883–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz W, and Rudy B (2014). Spatiotemporal specificity in cholinergic control of neocortical function. Curr. Opin. Neurobiol 26, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki K, Chung S, Ch’ ng YH, Kara P, and Reid RC (2005). Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature 433, 597–603. [DOI] [PubMed] [Google Scholar]
- Ohki K, and Reid RC (2007). Specificity and randomness in the visual cortex. Curr. Opin. Neurobiol 17, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, and Svoboda K (2009). The subcellular organization of neocortical excitatory connections. Nature 457, 1142–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RP, and Ballard DH (1999). Predictive coding in the visual cortex: a functional interpreation of some extra-classical receptive field effects. Nat. Neurosci 2, 79–87. [DOI] [PubMed] [Google Scholar]
- Ringach DL, Mineault PJ, Tring E, Olivas ND, Garcia-Junco-Clemente P, and Trachtenberg JT (2016). Spatial clustering of tuning in mouse primary visual cortex. Nat. Comm. Aug 2; 7, 12270. doi: 10.1038/ncomms1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MM, Dahmen JC, Muir DR, Imhof F, Martini FJ, and Hofer SB (2016). Thalamic nuclei convey diverse contextual information to layer 1 of visual cortex. Nat Neurosci 19, 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SL, Smith IT, Branco T, and Häusser M (2012). Dendritic spikes enhance stimulus selectivity in cortical neurons in vivo. Nature 503, 15–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincich LC, and Horton JC (2005). Input to V2 thin stripes arises from V1 cytochrome oxidase patches. J. Neurosci 25, 10087–10093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler DD, Das A, Bennett J, and Gilbert C (2002). Lateral connectivity and contextual interactions in macaque primary visual cortex. Neuron 36, 739–750. [DOI] [PubMed] [Google Scholar]
- Suter BA, O’Connor T, Iyer V, Petreanu LT, Hooks BM, Kiritani T, Svoboda K, and Shepherd GM (2010). Ephus: multipurpose data acquisition software for neuroscience experiments. Front. Neural Circuits 4, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Oertner TG, Hegemann P, and Larkum ME (2016). Science 354, 1587–1590. [DOI] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, Penn O, Bakken T, Menon V, Miller J, Fong O, Hirokawa KE, Lathia K, Rimorin C, Tieu M, Larsen R, Casper T, Barkan E, Kroll M, Parry S, Shapovalova N,V, Hischstein D, Pendergraft J, Sullivan HA, Lim TK, Szafer A, Dee N, Groblewski P, Wickersham I, Cetin A, Harris JA, Levi BP, Sunkin SM, Madisen L, Daigle T.l., Looger L, Bernard A, Phillips J, Lein E, Hawrylycz M, Svoboda K, Jones AR, Koch C, and Zeng H (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Lu J, and Huang ZJ (2013). The spatial and temporal origin of chandelier cells in mouse neocortex. Science 339, 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sporns O, and Burkhalter A (2012). Network analysis of cortico-cortical connections reveals ventral and dorsal processing streams in mouse visual cortex. J. Neurosci 32, 4386–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Webber RM, and Stanley GB (2010). Thalamic synchrony and the adaptive gating of information flow to cortex. Nat. Neurosci 12, 1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff AR, McGarry LM, Vogels TP, Inan M, Anderson SA and Yuste R (2011). State-dependent function of neocortical chandelier cells. J Neurosci 31, 17872–17886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Atallah BV, and Scanziani M (2014). Equalizing excitation-inhibition ratio across visual cortical neurons. Nature 511, 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Carrasquillo Y, Hooks BM, Nerbonne JM and Burkhalter A (2013). Distinct balance of excitation and inhibition in an interareal feedforward and feedback circuit of mouse visual cortex. J. Neurosci 33, 17373–17384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qiao W, Liu K, Zhong H, and Yao H (2015). Control of response reliability by parvalbumin-expressing interneurons in visual cortex. Nat. Comm 6, 6802 dio: 10.1038/ncomms7802. [DOI] [PubMed] [Google Scholar]
- Znamenskiy P, Kim M, Muir DR, Iacaruso F, Hofer SB, and Mrsic-Flogel TD (2018). Functional selectivity and specific connectivity of inhibitory neurons in primary visual cortex. bioRxiv April 4, 2018; dio: 10.1101/294835. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The codes generated during this study are available at https://github.com/amsmeier/Bista-DSouza-et-al.-2019.