Abstract
Juvenile myelomonocytic leukemia (JMML) is a pediatric myeloproliferative neoplasm that arises from malignant transformation of the stem cell compartment and results in increased production of myeloid cells. Somatic and germline variants in CBL (Casitas B-lineage lymphoma proto-oncogene) have been associated with JMML. We report an incompletely penetrant CBL Y371C mutation discovered by whole-exome sequencing in three individuals with JMML in a large pedigree with 35 years of follow-up. The Y371 residue is highly evolutionarily conserved among CBL orthologs and paralogs. In silico bioinformatics prediction programs suggested that the Y371C mutation is highly deleterious. Protein structural modeling revealed that the Y371C mutation abrogated the ability of the CBL protein to adopt a conformation that is required for ubiquitination. Clinically, the three mutation-positive JMML individuals exhibited variable clinical courses; in two out of three, primary hematologic abnormalities persisted into adulthood with minimal clinical symptoms. The penetrance of the CBL Y371C mutation was 30 % for JMML and 40 % for all leukemia. Of the 8 mutation carriers in the family with available photographs, only one had significant dysmorphic features; we found no evidence of a clinical phenotype consistent with a “CBL syndrome”. Although CBL Y371C has been previously reported in familial JMML, we are the first group to follow a complete pedigree harboring this mutation for an extended period, revealing additional information about this variant’s penetrance, function and natural history.
Introduction
Juvenile myelomonocytic leukemia (JMML) is a pediatric myeloproliferative neoplasm that arises from malignant transformation of the stem cell compartment and results in increased production of myeloid cells. Both germline and somatic mutations that dysregulate the Ras signaling pathway have been linked etiologically to JMML; approximately 60 % of children with JMML carry a mutation in one of several key genes in the Ras pathway (PTPN11, NRAS or KRAS), and an additional 15 % have mutations in the neurofibromatosis type 1 gene (NF1) (Kalra et al. 1994; Loh et al. 2004, 2009; Niemeyer et al. 1997; Shannon et al. 1994; Tartaglia et al. 2003). More recently, somatic (Loh et al. 2009; Sanada et al. 2009) and germline (Niemeyer et al. 2010; Perez et al. 2010) variants in CBL (Casitas B-lineage lymphoma proto-oncogene) have been associated with JMML.
Homozygous somatic mutations in CBL have been reported in 10–15 % of children affected with JMML (Loh et al. 2009; Sanada et al. 2009). CBL is located on chromosome 11q23.3 and encodes the 120-kDa c-CBL protein, a member of the CBL ubiquitin ligase family. In JMML, CBL mutations are often associated with somatic loss of the wild-type allele and duplication of the variant allele, although this is not always the case (Strullu et al. 2013). This acquired isodisomy is associated with a loss of the 11q23 region and homozygosity for the CBL mutation. Heterozygous germline CBL mutations in JMML have been identified by two research groups. In a study by Niemeyer et al. (2010), 17 of 21 children who had homozygous somatic mutations in CBL were found to have germline CBL mutations. A majority of these patients (15 of 21; 71 %) had features consistent with hyperactive Ras/Raf/MEK/ERK signaling, and congenital anomalies that overlap with Noonan syndrome, neurofibromatosis type 1 (NF1) and Legius syndrome (Niemeyer et al. 2010). Perez et al. (2010) identified four individuals with homozygous somatic mutations in CBL; three of these individuals were found to harbor germline heterozygous mutations in the gene. These patients also exhibited features of a novel disorder termed the “CBL syndrome”, which included dysmorphic features.
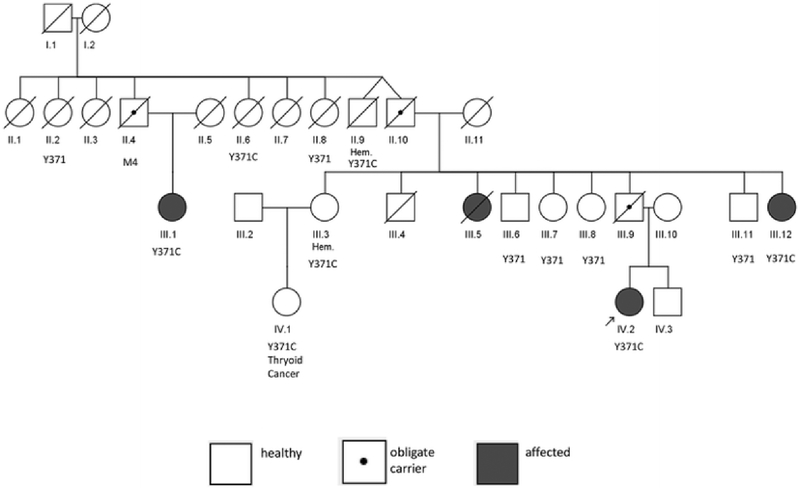
In 1979, an unusual family was evaluated at the NIH Clinical Center (Fig. 1). Four members of this family developed leukemia during the first few years of life; an affected girl (III.5), died at 16 months of age, and the remaining three affected members (III.1, III.12 and IV.2) were evaluated and followed in this study. They appeared to have a rare myeloproliferative disorder characterized by hepatosplenomegaly, anemia and granulocytosis developing in the first year of life, and they were given a working diagnosis of “familial childhood chronic myelogenous leukemia (CML)”. At that time, the term “familial childhood CML” was more a clinical description rather than an evidence-based diagnosis. Patients with this putative disorder were noted to develop long-lasting “CML” or myeloproliferative disease, unless they succumbed to infection at an early age. This disorder was characterized by absence of the Philadelphia chromosome. A recent retrospective re-evaluation of the family led to the diagnosis of JMML; the World Health Organization diagnostic features of JMML are listed in Supplemental Table 1 (Swerdlow et al. 2008). This family did not have clinical features consistent with either NF1 or Noonan syndrome. In the absence of a formal syndrome diagnosis, we conducted whole-exome sequencing of the one unaffected and three affected family members.
Fig. 1.
Pedigree of NCI JMML family. III.4 died in childhood of non-leukemic causes; III.9 died in a motor vehicle accident. CBL Y371 genotyping was performed on all individuals with available DNA. Filled symbols JMML cases; symbol with a dot in the center obligate carrier. Hem hematologic abnormalities without meeting JMML diagnostic criteria, M4 acute myelomonocytic leukemia/M4
Materials and methods
Patient descriptions
Patients from this family were enrolled into NIH protocol “Clinical, Laboratory and Epidemiologic Characterization of Individuals and Families at High Risk of Cancer (#78-C-0039; NCT-00001163)” which was subsequently merged into a specific familial leukemia/lymphoma protocol “Clinical, Laboratory and Epidemiologic Characterization of Individuals and Families at High Risk of Hematologic Cancer (# 02-C-0210; NCT-00039676)”. Clinical histories of the four affected family members are summarized below; additional family members are summarized in “Supplemental Patient Descriptions”. Available patient records from the initial evaluation of the family at the NIH Clinical Center in 1979 and their subsequent intermittent follow-up to the present were reviewed. Death certificates and records of significant hospitalizations and diagnoses were obtained and reviewed whenever possible. The study was approved by the NCI Institutional Review Board and written informed consent was obtained from all participants. Overall, 400 person-years of follow-up were available for this family. Patients or next-of-kin were queried regarding specific clinical features (e.g., optic atrophy, cardiac abnormalities) (Niemeyer et al. 2010) associated with the CBL Y371C mutation (Table 1). Patient III.1 was re-evaluated at NIH in 2012. Given the age of the records and family, we were unable to systematically obtain height, weight, head circumference and café-au-lait macule counts. Supplemental Table 1 summarizes the JMML diagnostic data for selected members of the pedigree. The three leukemia patients in this family for whom records are available (III.12, III.1, IV.2) fulfill two major WHO criteria for a JMML diagnosis (blasts <20 % and lack of a Philadelphia chromosome) and two of three (IV.2 and III.12) had peripheral blood monocytosis >1000/μl (Swerdlow et al. 2008), therefore, meeting full JMML diagnostic criteria. Patient III.1 had a peripheral monocyte count of 924/μl, less than the diagnostic threshold of 1000/μl. However, given her clinical course, need for a splenectomy and limitations of the historical record, we consider her as affected with JMML. In addition, all three affected members of this family met minor criteria of immature granulocytes in the peripheral blood and white blood cell (WBC) count >10,000/μl. One patient also had the minor criterion of having increased fetal hemoglobin. The diagnosis of JMML was made retrospectively based on chart review; the patients met the various diagnostic criteria, but at different time-points. Patient III.5 died in 1951; detailed records were unavailable.
Table 1.
Clinical characteristics of CBL Y371C carriers in NCI JMML family
| Pedigree ID | Current age | Gender | Cell/tissue | NM_005188.2: c.1112A>G(CBL Y371C) | JMML/ leukemia | Age at diagnosis | Clinical information | Hepatosplenomegaly | Dysmorphic features | Cardiac abnormality | Hypertension | Vasculitis | Optic atrophy | Cutaneous xanthomas |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II.6 | Deceased 2000; age 74 years | Female | Fibroblasts | Yes (Het) | No | N/A | No | No | Yes CHF | No | No | No | No | |
| II.9 | Deceased 2004; age 83 years | Male | Fibroblasts | Yes (Het) | No | N/A | Polyclonal gammopathy | No | No | Yes CHF | No | No | No | No |
| III.5 | Deceased 1951; age 16 months | Female | N/A | N/A | Yes (“CML”) | 12 months | Yes | No | No | No | No | No | No | |
| III.12 | 49 years | Female | Fibroblasts | Yes (Het) | Yes (JMML) | 9 months | Yes | No | No | No | No | No | No | |
| III.1 | 67 years | Female | Fibroblasts | Yes (Het) | Yes (JMML) | 20 months | Yes | No | No | No | No | No | No | |
| III.3 | 68 years | Female | Fibroblasts | Yes (Het) | No | N/A | Polyclonal gam- mopathy | No | No | No | No | No | No | No |
| III.9 | Deceased 2004; age 46 years | Male | N/A | N/A | No | N/A | Granulocytic hyperplasia of bone marrow | Yes | No | No | No | No | No | No |
| IV.1 | 48 years | Female | Fibroblasts | Yes (Het) | No | N/A | Thyroid cancer (1992, age 26) | No | No | No | No | No | No | No |
| IV.2 | 36 years | Female | Fibroblasts | Yes (Het) | Yes (JMML) | 9 months | Yes | Yes | No | No | No | No | No | |
| II.4 | Deceased 1991; age 69 years | Male | N/A | Obligate carrier | Acute myelomonocytic leukemia/ M4 | 69 years | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| II.10 | Deceased 2001; 77 years | Male | N/A | Obligate carrier | No | Benign cochlear tumor | No | No | No | No | No | No | No |
N/A not available, JMML juvenile myelomonocytic leukemia, CML chronic myelogenous leukemia
Patient IV.2 (CBL Y371C)
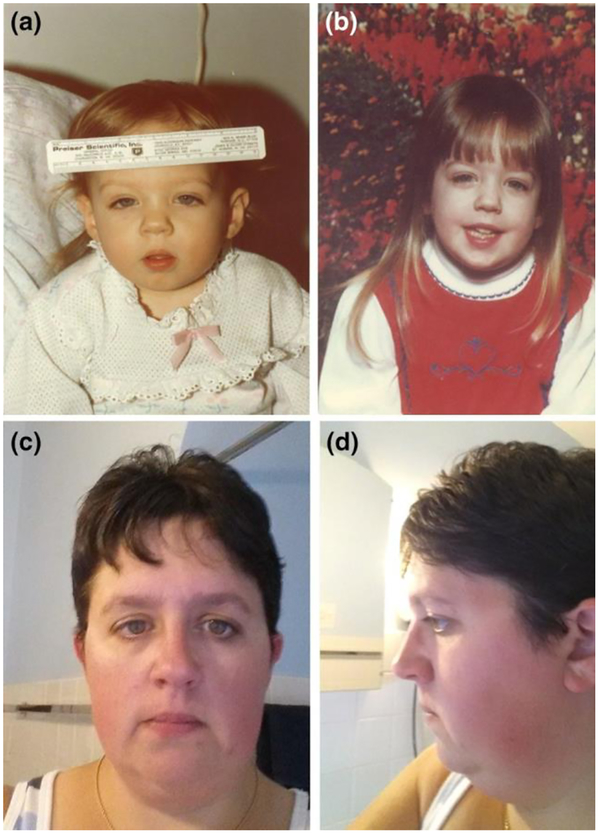
The proband (Fig. 2a), now 36 years old, was first noted to have increasing abdominal girth at 6–7 months of age, while clinically well. At age 9 months, while being treated for an upper respiratory tract infection, hepatosplenomegaly was noted. The patient had modest anemia, an elevated EBV titer, but no lymphadenopathy. At age 15 months, hepatosplenomegaly was unchanged and down-slanting palpebral fissures were apparent (Fig. 2a). A peripheral smear showed white blood cells (WBC) = 31,600 cells/μl with 58 % lymphocytes, 24 % segmented neutrophils, 2 % blasts, 5 % metamyelocytes, 1 % myelocytes and 2 % promyelocytes. Platelets = 120,000/μl. No monocyte count or percentage was reported in the differential. Her hemoglobin was 8.5 g/dl and with 1.2 % reticulocytes. Her spleen and liver were palpable 10 cm and 3 cm below the left and right costal margins, respectively. At a follow-up visit on 7/16/1979, a bone marrow aspirate showed a myeloid:erythroid (M:E) ratio of 20:1 with 8 % blasts. No Philadelphia chromosome was detected. At that time, her working diagnosis was chronic myelogenous leukemia (CML). In August 1979, her bone marrow aspirate showed 17 % blasts and the patient was started on a 4-day course of cytarabine plus 6-mercaptopurine (6-MP). This resulted in rapid clinical response and treatment with daily 6-MP was continued. Cytogenetic tests performed (9/1979), post-induction, on both peripheral blood and bone marrow revealed no clonal abnormality; a sample was collected in 2012 for contemporary cytogenetic analysis, revealing a normal constitutional karyotype. Fetal hemoglobin was increased at 6.7 % (11/1979). The patient also had a splenectomy at 3 years of age, and now has persistent thrombocytosis secondary to asplenia. This patient met two of three major criteria for the World Health Organization (WHO) Clinical Criteria for JMML (Supplemental Table 1) (Swerdlow et al. 2008). In addition, the patient’s recent complete blood counts (CBCs) (2010–2013) indicate a persistently elevated absolute monocyte count, though she is currently clinically asymptomatic off-therapy.
Fig. 2.
Dysmorphic features in the JMML CBL p.Y371C heterozy- gote proband (IV.2). At 18 months of age (a), the proband exhib- its dysmorphic features including down-slanting palpebral fis- sures which are also apparent at 5 years of age (b). In an adult (age 36 years) frontal (c) photograph, the proband continues to have down-slanting palpebral fissures. d Adult profile (side) photograph (Swerdlow et al. 2008). In addition, the patient’s recent complete blood counts (CBCs) (2010–2013) indicate a per- sistently elevated absolute monocyte count, though she is currently clinically asymptomatic off-therapy.
Patient III.12 (CBL Y371C)
Patient III.12 (Supplemental Figure 1a), now 49 years old, was diagnosed with “acute leukemia” at 9 age months; the diagnosis was changed to CML a few months later in 1966. She received 6-MP, cyclophosphamide and prednisone, followed by 6-MP maintenance for 2 years. At age 3, she developed transfusion-dependent anemia and splenomegaly. Bone marrow cytogenetics revealed that she was Philadelphia chromosome negative; a sample was not available for contemporary cytogenetic analysis. The peripheral blood had a few immature cells, but no blasts. The white blood cell count was moderately increased, with significant lymphocytosis. Her treatment was changed to busulfan, followed by splenic irradiation, but neither reduced her splenomegaly. Splenectomy and liver biopsy were performed when she was 4 years old. Her transfusion requirements decreased after splenectomy, but thrombocytosis persisted, with platelets ranging from 200,000 to 900,000/μl. 6-MP was restarted and continued to age 18 years.
The patient was seen at the NIH Clinical Center as part of the familial leukemia study when she was 14 years old. At that time, her bone marrow aspirate showed moderate hypercellularity with an M:E ratio of 6–8:1. Fetal hemoglobin was not detected and the leukocyte acid phosphatase was within normal limits. When she was 28 years old, she developed sternal and rib pain. Work-up showed a normal peripheral blood smear, but her bone scan showed increased uptake in the upper sternum. In addition, the bone marrow aspirate showed hypercellularity with an increased M:E ratio. The patient was also found to have elevated gamma globulins at 1.8 g/100 ml (normal range 0.5–1.5 g/100 ml).
She required chemotherapy until she was 18 years old. A CBC performed in June 1993 at age 28 showed a WBC of 13,600/μl with an absolute monocyte count of 1904/μl and platelets elevated at 776,000/μl. A CBC performed in July 2014 at 49 years of age, showed elevated WBCs at 22,000/μl and elevated monocytes of 3800/μl; this persisted on CBCs drawn 3 and 6 weeks later. She met all three major and two minor criteria for the diagnosis of JMML (Supplemental Table 1) (Swerdlow et al. 2008).
Patient III.1 (CBL Y371C)
This individual is now 67 years old, but was first noticed to have a prominent abdomen and peri-orbital edema at 20 months of age when she was hospitalized for pneumonia in 1949. Exam at that time showed splenomegaly, and leukocytosis and normoblasts were seen in the peripheral blood. Her hemoglobin = 7.6 g/dl, white blood cells = 13,200/μl, with 13 % promyelocytes, 2 % blasts, 26 % lymphocytes, 7 % monocytes, 26 % segmented neutrophils, 14 % bands, 1 % basophils, 2 % eosinophils, and 7 % myelocytes. The total monocyte count was 924/μl, nearly meeting diagnostic criteria for JMML (see Supplemental Table 1). Her working diagnosis at that time was “myeloid leukemia”. Abdominal plain films confirmed an enlarged spleen that occupied the entire left hemi-abdomen. She received splenic irradiation (360 rads in 3 fractions), which resulted in a significant decrease in spleen size.
In 1979, at age 32 years, the patient was evaluated at the NIH Clinical Center (Supplemental Figure 1b). Her medical problems at that time included left sacroiliac joint pain that had begun when she was 16 years old. Physical exam showed the spleen to be 3 cm below the left costal margin. She had no lymphadenopathy. Laboratory tests showed a hemoglobin of 12.6 g/dl with 0.9 % reticulocytes, WBC = 4500/μl, 75 % segmented neutrophils, 14 % lymphocytes, 10 % monocytes and 1 % eosinophils and platelets of 256,000/μl fetal hemoglobin was 0.2 %. Protein electrophoresis also showed a mild polyclonal increase in gamma globulin (IgG 1.8 g/dl). Cytogenetic studies demonstrated a normal 46 XX karyotype and were examined by modern cytogenetic analysis in 2012, also revealing a normal constitutional karyotype. A bone marrow aspirate was also noted to be morphologically normal. A liver–spleen scan revealed an enlarged spleen measuring 20 cm in craniocaudal diameter. A year later, the patient presented with left upper quadrant pain. An exploratory celiotomy and splenectomy were performed, and she was ultimately was found to have toxoplasmosis and was treated with trimethoprim–sulfamethoxazole. At age 37 years, the patient had a hysterectomy due to persistent abdominal pain and uterine enlargement. The patient was also seen again recently at the NIH Clinical Center in 2012 at 65 years of age, and was doing well. Her WBCs = 9230/μl, monocytes were at 820/μl and platelet count = 507,000/μl. At original presentation, the patient nearly met all three major criteria and two minor criteria for the diagnosis of JMML (Supplemental Table 1) (Swerdlow et al. 2008).
Patient III.5 (no DNA available)
This patient developed increasing abdominal girth and hepatosplenomegaly during the first year of life (1950–51). She was treated as “refractory anemia”, and died at 16 months of age without a specific diagnosis being made. Autopsy material was reviewed at the University of Illinois and thought to be consistent with CML. Her death certificate states chronic pneumonitis, myocarditis and malnutrition with severe secondary anemia as causes of death. No biological samples were available for additional study.
Exome sequencing
Whole-exome sequencing for the JMML family was performed at the National Cancer Institute Cancer Genomics Research Laboratory (NCI, CGR), as previously described (Shi et al. 2014). Briefly, 1.1 μg of genomic DNA was extracted by standard methods from low-passage cultured fibroblasts from patients III.1, III.12, III.10 and IV.2. SeqCAP EZ Human Exome Library v3.0 (Roche NimbleGen, Madison, WI, USA) was utilized for exome sequence capture. The captured DNA was then subject to paired-end sequencing utilizing the Illumina HiSeq for 2 × 100-bp sequencing of paired-ends (Illumina, San Diego, CA, USA). Exome sequencing was performed to a sufficient depth to achieve a minimum of 80 % of the coding sequence (UCSC hg19 transcripts) covered by ≥15 reads.
Bioinformatic analysis
Details of the bioinformatics analysis pipeline used in this study have been previously published (Shi et al. 2014; Stewart et al. 2014). Sequencing reads were first trimmed using the Trimmomatic program (Lohse et al. 2012), which marked all low-quality stretches (average <Q15 in a 4-bp sliding window) and reported the longest high-quality stretch of each read. Only read pairs with both ends ≥36 bp were used. Reads were aligned to the hg19 reference genome using the Novoalign software version 2.07.14 (http://www.novocraft.com). Duplicate reads based on paired-ends aligning to the same start locations due to either optical or PCR artifacts were removed from further analysis using the MarkDuplicated module of the Picard software version 1.67 (http://picard.sourceforge.net/). Additionally, our analysis used only properly aligned read pairs, i.e., the two ends of each pair must have mapped to the reference in complementary directions and must reflect a reasonable fragment length (250 ± 30 bp). These high-quality alignments for each individual were refined using a local realignment strategy around known and novel sites of insertion and deletion polymorphisms using the RealignerTargetCreator and IndelRealigner modules from the Genome Analysis Toolkit (GATK) (DePristo et al. 2011). Variant discovery and genotype calling of multi-allelic substitutions, insertions and deletions were performed on all individuals globally using the UnifiedGenotyper module from GATK, with the minimum call quality parameter set to 30, including necessary recalibrations of quality scores and variant quality scores following the “best practice” recommended by the GATK development team. Annotation, fitting genetic models, and filtering of each variant locus was performed using a locally developed custom software pipeline which made use of public data from the UCSC GoldenPath database (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/), the ESP6500 dataset from University of Washington’s Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), the National Center for Biotechnology Information dbSNP database (Sherry et al. 2001) build 137, the 1000 Genomes Project (Abecasis et al. 2010), and dbNSFP (Liu et al. 2013).
Variant filtering
Upon completion of the initial bioinformatics analysis and variant calling, the variants were filtered by the following criteria: (1) present in all 3 affected individuals in the pedigree and not present in the unaffected individual; (2) present in ≤10 individuals in an in-house database (NCI CGR “Out Group”) of 1170 exomes in individuals from families with non-hematologic cancer; (3) present in ≤1.0 % in the NHLBI Exome Sequencing Project (ESP) European American population; and (4) not present in areas of repeats or segmental duplications. In addition to the autosomal dominant filtering described above, the variants were also fit to an autosomal recessive and compound heterozygous model. Further bioinformatics analysis was carried out on the resulting variants to determine which could contribute to leukemia.
In silico analysis
For in silico analysis of the potential functional impact of variants, we utilized SIFT (Kumar et al. 2009), PolyPhen-2 (Version 2.2.2) (Adzhubei et al. 2010), Mutation Taster (Schwarz et al. 2010), Mutation Assessor (Release 2) (Reva et al. 2011), FATHMM (Version 2.3) (Shihab et al. 2013), and the Combined Annotation Dependent Depletion (CADD) scaled C-score (Kircher et al. 2014).
Sanger sequencing
Previously published primers for CBL exon 8 were selected for Sanger sequencing (Loh et al. 2009): Cbl-8F forward primer 5′-ACCCAGACTAGATGCTTTCTG-3′; Cbl-8R reverse primer 5′-AGGCCACCCCTTGTATCAGT-3′. Fibroblast or lymphocyte DNA was sequenced for the family members as outlined in Table 1.
Inactive and active CBL structural model comparison and analysis
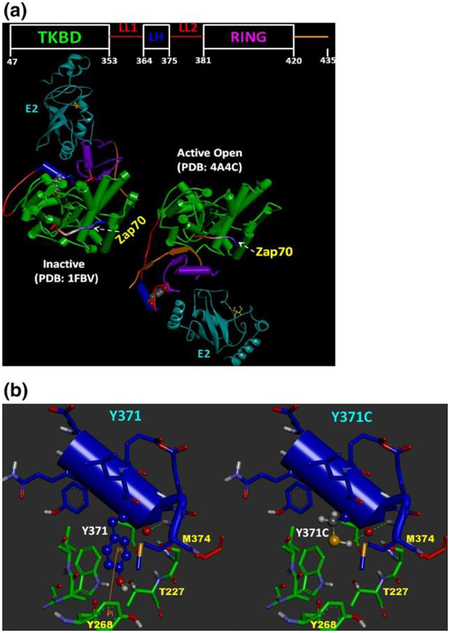
We used experimental 3D structures obtained from the RCSB Protein Data Bank (PDB) (http://www.rcsb.org) to model the impact of the Y371C mutation on phosphorylation-induced CBL activity. Tyrosine 371 belongs to the LH linker region that lies between the N-terminal tyrosine kinase binding domain (TKBD) and the Ring Finger domain (RING) domain. Sequence annotation and domain ranges for CBL were obtained from Uniprot database entry, P22681. The recent work of Dou and colleagues (Dou et al. 2012) hypothesized a two-state mechanism for CBL activation that occurs via Y371 phosphorylation (pY371). Their 3D structure (PDB ID:4A4C) formed the basis of our active phosphorylated Y371 CBL modeling and analysis. For the inactive mode, the Ubiquitin-protein ligase L3 (E2) (Uniprot ID P68036) and Zap-70 (Uniprot ID P43403) substrate-bound CBL structure (PDB ID:1FBV) (Zheng et al. 2000) was used. To compare and analyze the conformational changes between the active and inactive forms of CBL, the Flexible structure AlignmenT by Chaining Aligned fragment pairs allowing Twists (FATCAT) server (Ye and Godzik 2003) was used to align the structures. To visualize the structure around Y371 and the possible impact of the Y371C mutation, we had used the inactive CBL structure (PDB:1FBV). Mutation of Y371C was carried out using Discovery Studio 3.5 Visualizer (Accelrys Software Inc.) to assess the structural impact on the WT structure. Note that we were interested in assessing the effect of the Y → C mutation on CBL, so a restricted energy minimization of just the newly mutated residue was performed. Discovery Studio Visualizer (v. 3.5.0) was used for the analysis and creating the structure-based figures.
Results
Whole-exome sequencing identifies CBL NM_005188.2:c.1112A>G; p.(Tyr371Cys) mutation in affected family members
Exome data were generated for the three affected members with JMML (patients III.1, III.12 and IV.2) and III.10, the unaffected mother of patient IV.2. There were a total of 339,262 variants (both synonymous and non-synonymous) in these four individuals. After restricting analysis to non-synonymous and frameshift variants, 27,162 variants remained. After filtering these data by genetic segregation and population frequency, we detected variants in the following genes: PIK3CD, VWA5B1, TMEM87B, TATDN2, GPR98, TTC37, XKR5, GPT, FREM1, GCNT1, ARRDC1, FAT3, ZW10, CBL, MED4, PCDH20, SLC10A2 and DUSP21. Based on bioinformatics prediction and literature review, the missense variant in CBL (CBL;Ensembl assembly GRCh37, chromosome 11,g.119148892A>G; p.(Try371Cys), NM_005188), was deemed most likely to explain the familial clustering of JMML. This variant has not been previously observed in the Exome Aggregation Consortium, 1000 Genomes or NHLBI ESP databases, and only once in the NCI CGR Out Group of 1170 exomes; but it has been previously reported as a germline variant in a JMML case (Niemeyer et al. 2010).
In silico predictions of CBL Y371C pathogenicity
Eight in silico prediction algorithms classified Y371C as deleterious (Supplemental Table 2).
Sanger validation of CBL (NM_005188.2:c.1112A>G); p.(Tyr371Cys)
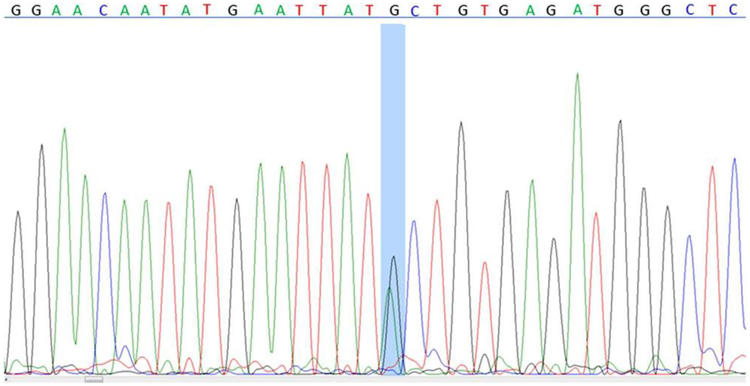
Bi-directional Sanger sequencing (Fig. 3) confirmed the presence of the heterozygous CBL c.1112A>G mutation in the three affected, exome-sequenced family members (III.1, III.12 and IV.2). Four additional family members without JMML were also found to carry the CBL mutation (II.6, II.9, IV.1 and III.3). Six unaffected members of the family carried the wild-type allele at this position. The mutation status of three obligate carriers (III.9, the proband’s father; II.10, proband’s paternal grandfather; II.4 proband’s paternal uncle) could not be directly confirmed due to lack of sample availability. Family member III.5, who died of “CML” in 1951 at 16 months of age, was not genotyped due to lack of a DNA sample. Material was unavailable for III.4 who died in childhood of non-leukemic causes. We did not attempt comprehensive sequencing of CBL in lymphocytes to identify the associated CBL somatic mutation or loss of heterozygosity events.
Fig. 3.
Sequence electrophero- gram demonstrating a germline CBL mutation in the proband (IV.2 of NCI JMML family). There is a chr11:119,148,892, A>G mutation (highlighted in blue) in exon 8 of the CBL gene. Codon 371 is normally TAC (tyrosine 371). The proband is heterozygous for the TGC (cysteine 371) codon
Clinical features of family associated with the CBL c.1112A>G, p.(Tyr371Cys) mutation
We identified seven family members with a CBL Y371C mutation (Fig. 1, Supplemental Figure 1); clinical records were available on all seven individuals. Table 1 summarizes the clinical features of these family members. There were 400 person-years of CBL Y371C natural history follow-up on this family. Photographs of 7 CBL Y371C carriers were available to us; all but one (IV.2) were obtained during adolescence or adulthood.
The photograph of the proband IV.2 at age 18 months (Fig. 2a) shows clear dysmorphic features: down-slanting palpebral fissures, small mouth, long, grooved philtrum, a broad nasal bridge, mild bilateral ptosis, short, upturned nose and facial hypotonia. At age 5 (Fig. 2b), the proband continued to exhibit slanting palpebral fissures, a short upturned nose and long philtrum. The slanting palpebral fissures have persisted into adulthood (Fig. 2c, frontal and 2d profile). Carriers of CBL Y371C in this family had no history of vasculitis, optic atrophy, cutaneous xanthomas or hypertension (Table 1), as noted in a previously reported JMML patient with CBL Y371C (Niemeyer et al. 2010).
Four of the ten (40 %) CBL Y371C carriers developed a leukemia (JMML or acute myelomonocytic leukemia in II.4; obligate carriers included in calculation) but only one (II.4) died from associated complications, although documentation is incomplete. In the other family members with leukemia, there was spontaneous resolution or improvement with minimal treatment. Three of ten carriers developed JMML. It is important to note that patient III.5 died from an unclassified leukemia at age 16 months and may have been a carrier of the CBL Y371C mutation; no DNA was available from this patient. No one in the family has required bone marrow transplantation. The only solid tumor in the family was a thyroid cancer diagnosed at age 26 in carrier IV.1, but no additional details are available. This patient is alive and well and is currently 48 years old. We also observed incomplete penetrance with the CBL Y371C mutation: two of ten (20 %) carriers (III.3, II.9) have not developed leukemia but did have evidence of related hematologic abnormalities including polyclonal gammopathy with monocytosis; the latter patient died of cardiomyopathy at age 83. The proband’s paternal aunt (II.6) was a mutation carrier who died at age 74 years from emphysema, without leukemia, other cancer or dysmorphic features (Supplemental Figure 1D). There were three obligate carriers who could not be genotyped. One, the proband’s father (III.9), had clinically significant splenomegaly requiring splenectomy at age 22 years, monocytosis (13 %) and granulocytic hyperplasia of the bone marrow but no history of leukemia and no evident dysmorphic features (Supplemental Figure 1c). He died at age 46 years in a motor vehicle accident. The second obligate carrier was II.4, father of known carrier III.1. No DNA was available for him. He died from complications of “acute myelomonocytic leukemia”, but pathologic materials were unavailable for NCI review. The third obligate carrier was II.10, grandfather of the proband, who had no history of leukemia or solid tumor and no DNA available for sequencing.
Modeling of CBL protein structure
As observed by (Dou et al. 2012), the phosphorylation of Y371 is the key step in relieving the autoinhibited TKBD-ring finger domain (RING) interaction, leading to the activation of CBL. A similar conclusion was reached using NMR and small-angle X-ray scattering experiments in CBL-B (Kobashigawa et al. 2011). Figure 4a shows the comparison of inactive and active conformations of CBL. The inactive conformation of CBL shows the TKBD and the ring finger domain (RING) in a closed conformation, with Y371 buried inside the protein core. The interaction between TKBD and RING is tighter, not allowing ubiquitin-protein ligase L3 (“E2”) to bind in a favorable conformation. Furthermore, the E2 that is bound to the inactive CBL is not in the optimal position required to transfer ubiquitin to its substrate. There is a significant conformational movement around the linker domain (commonly referred to as the LL1–LL–LL2 segment, see Fig. 4a) upon phosphorylation of the tyrosine 371 and activation of the protein. The right panel of Fig. 4a shows the same protein in the active, phosphorylated Y371 conformation. Figure 4b (left panel) shows the structural arrangement details around Y371, and the right shows the arrangement around Y371C. The mutated residue, cysteine (MW = 103), is smaller in size than the wild-type tyrosine (MW = 163) and, as expected, we did not observe any steric clashes with the surrounding residues. Note in the wild-type (Fig. 4b, left), Y371 is complemented by the surrounding residues specifically using H-bonds (M374, T227) and a Pi–Pi interaction (Y268). Even though the mutated Y371C residue (Fig. 4b, right) has lost the Pi–Pi interaction and possibly an H-bond, structural analysis suggests that it will have no adverse impact on the stability of the closed, inactive structure of CBL. The mutated residue Y371C lacks a hydroxyl group and thus loses the ability to become phosphorylated and generate the active form of the protein.
Fig. 4.
a Structural comparison of inactive CBL-UbcH7(E2)-Zap70 with the active pY371-CBL-UbcH5B(E2)-Zap70 complex to high- light the key role of Y371 phosphorylation (pY371). E2 proteins are colored in cyan and shown in ribbon form. The substrate peptides (Zap70) are displayed in tube format with N-terminal (blue)-to-C- terminal (red) coloring scheme. The Y371 residue in the inactive and the corresponding pY371 in the active forms are shown in CPK mode. Location of catalytic cysteine in E2 is also shown in ball-and- stick and marked in yellow. The inactive conformation of CBL shows the TKBD and the ring finger domain (RING) in a closed conforma- tion and the Y371 is buried inside the protein core. The interaction between TKBD and RING is tighter, not allowing E2 to bind in con- formation required for ubiquitin transfer. In the active conformation (right) there is a significant conformational movement around the linker domain (commonly referred to as the LL1-LL-LL2 segment, see Fig. 4a, right panel). An opening of the TKBD-linker-RING seg- ment creates a favorable CBL conformation that allows E2 to bind favorably to the RING domain while becoming optimally positioned to interact with the substrate. b The possible structural impact of Y371C is shown using the inactive CBL structure (PDB:1FBV). Res- idues surrounding Y371 (left) and Y371C (right) are shown in stick mode. The domain residues are also colored (TKBD: green, LH: blue and LL2: red) to indicate their orientation in the CBL architecture. Residues that can form H-bonds (green dotted lines) or Pi–Pi interac- tion (orange line) are also marked
Discussion
Germline mutations in CBL are unequivocally associated with JMML pathogenesis (Niemeyer et al. 2010; Perez et al. 2010). We detected the Y371C mutation in all three JMML-affected family members, and thus attribute our familial JMML cluster to this specific mutation. Although this mutation has been previously reported in JMML, we are the first group to follow a complete pedigree harboring this mutation over a period of four decades, revealing additional information about this variant’s penetrance and the natural history of disease associated with it. We report that the penetrance of the CBL Y371C mutation was 30 % for JMML and 40 % for all leukemia. We found no evidence of a clinical phenotype consistent with a “CBL syndrome” in the majority of the mutation carriers studied. Protein structural modeling suggested that the Y371C mutation abrogated the ability of the CBL protein to enter a conformation that would permit it to participate in protein ubiquitination.
The CBL protein consists of an N-terminus tyrosine kinase binding domain and a linker-RING domain with E3 ubiquitin ligase activity (Joazeiro et al. 1999). Mutations in CBL are predominantly missense changes affecting the linker-RING domains, coded for in exons 8 and 9 (Hanson et al. 2014). Recruitment of ubiquitin monomers and E2 ubiquitin conjugating enzymes to the linker-RING finger interface are important CBL functions; CBL then multi-ubiquitinylates associated activated receptor tyrosine kinases (RTK) (Ogawa et al. 2010). Upon multi-ubiquitinylation of the RTK, the RTK–CBL complex is then subject to endocytosis (Lee et al. 1999; Levkowitz et al. 1998; Miyake et al. 1998). Its role as a negative regulator of tyrosine kinase signaling suggests that the CBL protein could have an anti-oncogenic or tumor suppressor function (Ogawa et al. 2010). In addition, the CBL knockout mice have splenomegaly, extramedullary hematopoiesis, an expanded hematopoietic progenitor pool, an enlarged thymus, and an increased capacity to repopulate the bone marrow (Murphy et al. 1998; Naramura et al. 1998; Rathinam et al. 2008; Sanada et al. 2009).
Notably, the majority of the disease-associated mutations are located in the linker-RING finger domain, responsible for E3 ubiquitin ligase activity. The tyrosine-371 position in this domain is the most commonly mutated residue in the germline of JMML patients (Kales et al. 2010; Loh et al. 2009). Niemeyer et al. (2010) found 17 germline mutations in JMML cases, with most of the germline mutations located in the linker-RING finger domain. Tyrosine 371 was the most frequently mutated residue, with 7 (41 %) individuals carrying the p.Y371H mutation, 1 (6 %) carrying the p.Y371D mutation and 1 (6 %) carrying the p.Y371C mutation, the mutation detected in this study of familial JMML. Perez et al. (2010) also showed that of three patients with germline CBL mutations, one carried a p.Y371H allele. Furthermore, Loh et al. (2009) showed that 13 of 27 (48 %) somatic mutations in CBL occurred at the Y371 residue in a cohort of 159 JMML samples. While previous studies have shown homozygosity of the Y371 mutation or loss of heterozygosity at the locus, leukemic cells at the time of diagnosis were not available in our study to test for this possibility; this is a limitation of our study.
It has been shown, using mass spectrometry, that the CBL tyrosine 371 position is phosphorylated in vivo following epidermal growth factor (EGF) stimulation (Zhang et al. 2005). In addition, biochemical studies have established that CBL has significantly more E3 ubiquitin ligase activity when it is phosphorylated at tyrosine 371 (Kassenbrock and Anderson 2004; Levkowitz et al. 1999; Ryan et al. 2010). Mutation of the tyrosine 371 residue abrogates CBL’s ability to ubiquitinate receptor tyrosine kinases (Levkowitz et al. 1999; Niemeyer et al. 2010; Sanada et al. 2009; Thien et al. 2001). More specifically, substitution of tyrosine 371 with phenylalanine produces a protein lacking both E3 ligase and oncogenic activity; substitution with a glutamate, which functions as a pseudo-phosphorylated residue, activates the protein’s E3 ligase activity (Kassenbrock and Anderson 2004; Thien et al. 2001).
Normally, phosphorylation of the tyrosine 371 introduces a negatively charged phosphate group which makes the newly modified residue (pY371) incompatible with the hydrophobic environment of the surrounding residues. This promotes the creation of an open conformation in which the ring domain can favorably interact with E2, in turn placing E2 in an optimal orientation to favorably interact with its substrate (Fig. 4a, right panel). Critically, the mutated residue, Y371C, lacks a hydroxyl group and the ability to become phosphorylated. Based on our structural modeling, the CBL Y371C protein lacks the ability to be activated, thereby adversely affecting its function.
The CBL Y371C mutation was first reported in a 6-month-old boy with JMML (Niemeyer et al. 2010). That patient also had café-au-lait spots, cryptorchidism, growth below the third percentile, developmental delay, hearing loss, optic atrophy, hypertension and cardiomyopathy. We specifically queried our family members and reviewed their medical records regarding these features (Table 1). We found minimal evidence of these phenotypes, with the proband (IV.2, Fig. 2) comprising the only exception. She had down-slanting palpebral fissures and other dysmorphic features. This subject had no documented learning difficulties. In general, given the overall paucity of congenital anomalies in our study, the term “CBL syndrome” as previously defined (Perez et al. 2010) is not appropriate for our family.
The CBL Y371C patient from the Niemeyer paper presented at less than 1 year of age and had leukocytosis and hepatosplenomegaly similar to individuals in our family (Niemeyer et al. 2010). This patient had not required hematopoietic stem cell transplantation; he was 7½ years old at the time of the publication. This clinical course is similar to those observed in our family, except that our affected family members exhibited protracted clinical symptoms or laboratory abnormalities. While our proband (IV.2) had self-limiting clinical symptoms during the first year of life, her aunt (III.12) had persistent symptomatic disease, requiring a splenectomy at age 4 and maintenance chemotherapy with 6-MP until she was 18 years old. Both patients still have monocytosis on current CBCs, off chemotherapy. The proband’s first-cousin once removed (III.1) had persistent symptomatic splenomegaly requiring a splenectomy at age 33 years. One individual (III.5) died of an unclassified leukemia at age 16 months and may have been a mutation carrier. This variable expressivity and reduced penetrance, even within the same family, are characteristics of other RASopathies (Rauen 2013). It is important to underscore the relatively good prognosis of the JMML patients in this family, who did not require hematopoietic stem cell transplantation and appeared to have a high rate of spontaneous regression.
Two CBL Y371C mutation-positive family members (II.9; III.3) had polyclonal gammopathy, which has been previously associated with JMML (Castro-Malaspina et al. 1984; Niemeyer et al. 1997). They also exhibited slight peripheral blood monocytosis, suggesting a milder, subclinical phenotype in some mutation carriers. This indicates that part of the CBL Y371C clinical spectrum may include indolent elevation of immunoglobulins and monocytes without clinical features. In addition, the proband’s father, an obligate carrier of the CBL Y371C mutation, exhibited an elevated monocyte percentage on differential. One CBL Y371C mutation carrier (IV.1) developed thyroid cancer at age 26. Her tumor tissue was not available for assessment regarding possible loss of the wild-type CBL allele. It is unclear if this event was related to her mutation or simply coincidental, but her unusually young age-at-diagnosis is suggestive of an underlying cancer susceptibility (Boltz et al. 2013).
In summary, we have detected an incompletely penetrant CBL Y371C mutation in all affected members of a large pedigree with 400 person-years of follow-up. The clinical spectrum included JMML, acute myelomonocytic leukemia/M4, splenomegaly, polyclonal gammopathy and monocytosis. The Y371 residue is highly evolutionarily conserved among CBL orthologs and paralogs. Our bioinformatics analysis demonstrated that the Y371C mutation is predicted to be highly deleterious. Our protein structural modeling suggests that the Y371C mutation abrogates the ability of the protein to adopt a conformation that would permit protein ubiquitination. Clinically, the three JMML patients in the family exhibited a variable clinical course; hematologic abnormalities persisted into adulthood in two out of three cases without overt clinical features and the third individual continued to be symptomatic with splenomegaly into her 30 s. It is interesting to speculate that an unidentified genetic modifier of CBL may be contributing to this divergence of phenotype; in future studies, we plan to investigate the segregation of the 17 other variants identified in this study in the pedigree to assess for modifiers of the phenotype. Although this mutation has been previously reported in JMML, we are the first group to follow a complete, extended pedigree harboring this mutation over a period of four decades, revealing additional information about this variant’s penetrance, functionality and the natural history of disease associated with it.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Supplementary Material
Acknowledgments
This Project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E.
Appendix
NCI DCEG Cancer Genomics Research Laboratory: Sara Bass, Joseph Boland, Laurie Burdette, Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A. Hutchinson, Kristine Jones, Sally Larson, Kerrie Lashley, Hyo Jung Lee, Wen Luo, Michael Malasky, Michelle Manning, Jason Mitchell, David Roberson, Aurelie Vogt, Mingyi Wang, Meredith Yeager, Xijun Zhang.
NCI DCEG Cancer Sequencing Working Group: Bari Ballew, Stephen J. Chanock, Alisa M. Goldstein, Allan Hildesheim, Nan Hu, Maria Teresa Landi, Jennifer Loud, Phuong L. Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Melissa Rotunno, Sharon A. Savage, Philip R. Taylor, Geoffrey S. Tobias, Margaret A. Tucker, Jeannette Wong, Xiaohong R. Yang, Guoqin Yu.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA (2010) A map of human genome var- iation from population-scale sequencing. Nature 467:1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltz MM, Enomoto LM, Ornstein RM, Saunders BD, Hollenbeak CS (2013) Incidence and survival differences of differentiated thyroid cancer among younger women. Clin Oncol Adolesc Young Adults 3:79–88 [Google Scholar]
- Castro-Malaspina H, Schaison G, Passe S, Pasquier A, Berger R, Bayle-Weisgerber C, Miller D, Seligmann M, Bernard J (1984) Subacute and chronic myelomonocytic leukemia in children (juvenile CML). Clinical and hematologic observations, and identification of prognostic factors. Cancer 54:675–686 [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McK- enna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT (2012) Structural basis for autoinhibition and phosphorylation- dependent activation of c-Cbl. Nat Struct Mol Biol 19:184–192. doi: 10.1038/nsmb.2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson HL, Wilson MJ, Short JP, Chioza BA, Crosby AH, Nash RM, Marks KJ, Mansour S (2014) Germline CBL mutation associ- ated with a noonan-like syndrome with primary lymphedema and teratoma associated with acquired uniparental isodisomy of chromosome 11q23. Am J Med Genet Part A 164:1003–1009. doi: 10.1002/ajmg.a.36375 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC (1999) The tyrosine kinase negative regulator c-Cbl as a RING- type, E2-dependent ubiquitin-protein ligase. Science 286:309–312 [DOI] [PubMed] [Google Scholar]
- Kales SC, Ryan PE, Nau MM, Lipkowitz S (2010) Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res 70:4789–4794. doi: 10.1158/0008-5472.CAN-10-0610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra R, Paderanga DC, Olson K, Shannon KM (1994) Genetic anal- ysis is consistent with the hypothesis that NF1 limits myeloid cell growth through p21ras. Blood 84:3435–3439 [PubMed] [Google Scholar]
- Kassenbrock CK, Anderson SM (2004) Regulation of ubiquitin pro- tein ligase activity in c-Cbl by phosphorylation-induced con- formational change and constitutive activation by tyrosine to glutamate point mutations. J Biol Chem 279:28017–28027. doi: 10.1074/jbc.M404114200 [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J (2014) A general framework for estimating the relative path- ogenicity of human genetic variants. Nat Genet 46:310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F (2011) Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc Natl Acad Sci USA 108:20579–20584. doi: 10.1073/pnas.1110712108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, Bow- tell DD, Stanley ER (1999) The Cbl protooncoprotein stimu- lates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J 18:3616–3628. doi: 10.1093/emboj/18.13.3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y (1998) c-Cbl/Sli-1 regu- lates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev 12:3663–3674. doi: 10.1101/gad.12.23.3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y (1999) Ubiquitin ligase activity and tyrosine phos- phorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 4:1029–1040 [DOI] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E (2013) dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 34:E2393–E2402. doi: 10.1002/humu.22376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh ML, Vattikuti S, Schubbert S, Reynolds MG, Carlson E, Lieuw KH, Cheng JW, Lee CM, Stokoe D, Bonifas JM, Curtiss NP,Gotlib J, Meshinchi S, Le Beau MM, Emanuel PD, Shannon KM (2004) Mutations in PTPN11 implicate the SHP-2 phos- phatase in leukemogenesis. Blood 103:2325–2331. doi: 10.1182/blood-2003-09-3287 [DOI] [PubMed] [Google Scholar]
- Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S, Mullighan CG, Chen L, Bergstraesser E, Bueso-Ramos CE, Emanuel PD, Hasle H, Issa JP, van den Heuvel-Eibrink MM, Locatelli F, Stary J, Trebo M, Wlodarski M, Zecca M, Shannon KM, Niemeyer CM (2009) Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood 114:1859–1863. doi: 10.1182/blood-2009-01-198416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B (2012) RobiNA: a user-friendly, integrated software solu- tion for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627. doi: 10.1093/nar/gks540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Lupher ML, Druker B, Band H (1998) The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor α. Proc Natl Acad Sci 95:7927–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CBF, Langdon WY, Bowtell DDL (1998) Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol Cell Biol 18:4872–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramura M, Kole HK, Hu R-J, Gu H (1998) Altered thymic posi- tive selection and intracellular signals in Cbl-deficient mice. Proc Natl Acad Sci 95:15547–15552. doi: 10.1073/pnas.95.26.15547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer CM, Arico M, Basso G, Biondi A, Cantu Rajnoldi A, Creutzig U, Haas O, Harbott J, Hasle H, Kerndrup G, Locatelli F, Mann G, Stollmann-Gibbels B, van’t Veer-Korthof ET, van Wer- ing E, Zimmermann M (1997) Chronic myelomonocytic leuke- mia in childhood: a retrospective analysis of 110 cases. European Working Group on Myelodysplastic Syndromes in Childhood (EWOG-MDS). Blood 89:3534–3543 [PubMed] [Google Scholar]
- Niemeyer CM, Kang MW, Shin DH, Furlan I, Erlacher M, Bunin NJ, Bunda S, Finklestein JZ, Sakamoto KM, Gorr TA, Mehta P, Schmid I, Kropshofer G, Corbacioglu S, Lang PJ, Klein C, Schlegel PG, Heinzmann A, Schneider M, Stary J, van den Heu- vel-Eibrink MM, Hasle H, Locatelli F, Sakai D, Archambeault S, Chen L, Russell RC, Sybingco SS, Ohh M, Braun BS, Flotho C, Loh ML(2010) Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leuke- mia. Nat Genet 42:794–800. doi: 10.1038/ng.641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Shih LY, Suzuki T, Otsu M, Nakauchi H, Koeffler HP, Sanada M (2010) Deregulated intracellular signaling by mutated c-CBL in myeloid neoplasms. Clin Cancer Res 16:3825–3831. doi: 10.1158/1078-0432.CCR-09-2341 [DOI] [PubMed] [Google Scholar]
- Perez B, Mechinaud F, Galambrun C, Ben Romdhane N, Isidor B, Philip N, Derain-Court J, Cassinat B, Lachenaud J, Kaltenbach S, Salmon A, Desiree C, Pereira S, Menot ML, Royer N, Fen- neteau O, Baruchel A, Chomienne C, Verloes A, Cave H (2010) Germline mutations of the CBL gene define a new genetic syn- drome with predisposition to juvenile myelomonocytic leukae- mia. J Med Genet 47:686–691. doi: 10.1136/jmg.2010.076836 [DOI] [PubMed] [Google Scholar]
- Rathinam C, Thien CBF, Langdon WY, Gu H, Flavell RA (2008) The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev 22:992–997. doi: 10.1101/gad.1651408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen KA (2013) The RASopathies. Annu Rev Genomics Hum Genet 14:355–369. doi: 10.1146/annurev-genom-091212-153523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39:e118. doi: 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S (2010) The N terminus of Cbl-c regulates ubiquitin ligase activ- ity by modulating affinity for the ubiquitin-conjugating enzyme. J Biol Chem 285:23687–23698. doi: 10.1074/jbc.M109.091157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, Tamura A, Honda H, Sakata-Yanagimoto M, Kumano K, Oda H, Yama- gata T, Takita J, Gotoh N, Nakazaki K, Kawamata N, Onodera M, Nobuyoshi M, Hayashi Y, Harada H, Kurokawa M, Chiba S, Mori H, Ozawa K, Omine M, Hirai H, Nakauchi H, Koef- fler HP, Ogawa S (2009) Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460:904–908. doi: 10.1038/nature08240 [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D (2010) Muta- tionTaster evaluates disease-causing potential of sequence altera- tions. Nat Methods 7:575–576. doi: 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- Shannon KM, O’Connell P, Martin GA, Paderanga D, Olson K, Dinndorf P, McCormick F (1994) Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med 330:597–601. doi: 10.1056/NEJM199403033300903 [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic varia- tion. Nucleic Acids Res 29:308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, Ghiorzo P, Bressac-de Paillerets B, Nagore E, Avril MF, Caporaso NE, McMaster ML, Cullen M, Wang Z, Zhang X, Bruno W, Pastorino L, Queirolo P, Banuls-Roca J, Garcia- Casado Z, Vaysse A, Mohamdi H, Riazalhosseini Y, Foglio M, Jouenne F, Hua X, Hyland PL, Yin J, Vallabhaneni H, Chai W, Minghetti P, Pellegrini C, Ravichandran S, Eggermont A, Lath- rop M, Peris K, Scarra GB, Landi G, Savage SA, Sampson JN, He J, Yeager M, Goldin LR, Demenais F, Chanock SJ, Tucker MA, Goldstein AM, Liu Y, Landi MT (2014) Rare missense vari- ants in POT1 predispose to familial cutaneous malignant mela- noma. Nat Genet. doi: 10.1038/ng.2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR (2013) Predicting the functional, molecular, and phenotypic consequences of amino acid sub- stitutions using hidden Markov models. Hum Mutat 34:57–65. doi: 10.1002/humu.22225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart DR, Pemov A, Johnston JJ, Sapp JC, Yeager M, He J, Boland JF, Burdett L, Brown C, Gatti RA, Alter BP, Biesecker LG, Sav- age SA (2014) Dubowitz syndrome is a complex comprised of multiple, genetically distinct and phenotypically overlapping dis- orders. PLoS One 9:e98686. doi: 10.1371/journal.pone.0098686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strullu M, Caye A, Cassinat B, Fenneteau O, Touzot F, Blauwblomme T, Rodriguez R, Latour S, Petit A, Barlogis V, Galambrun C, Leblanc T, Baruchel A, Chomienne C, Cave H (2013) In hemat- opoietic cells with a germline mutation of CBL, loss of heterozy- gosity is not a signature of juvenile myelo-monocytic leukemia. Leukemia 27:2404–2407. doi: 10.1038/leu.2013.203 [DOI] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW (2008) World Health Organization classi- fication of tumours of the haematopoietic and lymphoid tissues. 4th edn International Agency for Research on Ancer, Lyon [Google Scholar]
- Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hahlen K, Hasle H, Licht JD, Gelb BD (2003) Somatic muta- tions in PTPN11 in juvenile myelomonocytic leukemia, myelo- dysplastic syndromes and acute myeloid leukemia. Nat Genet 34:148–150. doi: 10.1038/ng1156 [DOI] [PubMed] [Google Scholar]
- Thien CB, Walker F, Langdon WY (2001) RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol Cell 7:355–365 [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A (2003) Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19(Suppl 2):2246–2255 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM (2005) Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics 4:1240–1250. doi: 10.1074/mcp.M500089-MCP200 [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-pro- tein ligases. Cell 102:533–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.