Abstract
Head and neck squamous cell carcinomas (HNSCC) promote inflammation in the tumor microenvironment through aberrant NF-κB activation, but the genomic alterations and pathway networks that modulate NF-κB signaling have not been fully dissected. Here, we analyzed genome and transcriptome alterations of 279 HNSCC specimens from The Cancer Genome Atlas (TCGA) cohort and identified 61 genes involved in NF-κB and inflammatory pathways. The top 30 altered genes were distributed across 96% of HNSCC samples, and their expression was often correlated with genomic copy number alterations (CNA). Ten of the amplified genes were associated with human papilloma virus (HPV) status. We sequenced 15 HPV(−) and 11 HPV(+) human HNSCC cell lines, and three oral mucosa keratinocyte lines, and supervised clustering revealed that 28/61 genes exhibit altered expression patterns concordant with HNSCC tissues, and distinct signatures related to their HPV status. RNAi screening using an NF-κB reporter line identified 16 genes that are induced by TNF-α or Lymphotoxin β (LTβ) and implicated in the classical and/or alternative NF-κB pathways. Knockdown of TNFR, LTΒR, or selected downstream signaling components established cross-talk between the classical and alternative NF-κB pathways. TNF-α and LTβ induced differential gene expression involving the NF-κB, IFN-γ and STAT pathways, inflammatory cytokines, and metastasis related genes. Improved survival was observed in HNSCC patients with elevated gene expression in T cell activation, immune checkpoints, and IFN-γ and STAT pathways. These gene signatures of NF-κB activation, which modulate inflammation and responses to the immune therapy, could serve as potential biomarkers in future clinical trials.
Keywords: Inflammation, NF-κB, HPV, checkpoint, function genomics, The Cancer Genome Atlas (TCGA), HNSCC
Introduction
Genomic instability and inflammation are two critical hallmarks of cancer development (1). Genomic instability can manifest as chromosomal rearrangements, gene copy number alterations (CNAs) and mutations (1). Proteins expressed as a result of these genomic alterations may mediate inflammatory and stromal cell infiltration, and could be recognized as neoantigens by immune cells (2). These inflammatory responses in the tumor microenvironment (TME) may block or promote tumor progression at different developmental stages, and subsequently affect therapeutic responses, especially to immune checkpoint inhibitors and immunotherapies (2, 3). The interaction of infiltrating and tumor cells in the TME is communicated through production of cytokines, chemokines, and growth factors, which can be regulated through NF-κB signaling pathways in a variety of cancers (4).
The NF-κB pathway in solid tumors and in head and neck squamous cell carcinoma (HNSCC)is often aberrantly activated (5–7). Our early studies of HNSCC focused on molecular mechanisms involved in the classical NF-κB pathway, including the activation of RELA, NF-κB1(p105/p50) and cREL, which are mainly modulated by TNF-α through Inhibitor-kappaB kinase (IKK)α/β/γ activation (8–10). The classical pathway plays an important role in cancer cell proliferation and survival, and in therapeutic resistance (4–7). In HNSCC, we also showed that components of the alternative NF-κB pathway, RELB and NF-κB2 (p100/p52), are modulated by lymphotoxin-β (LTβ) and other ligands through the activation of NF-κB inducing kinase (NIK) and IKKα (CHUK)(11, 12). The alternative pathway promotes tumor cell migration, an important feature underlying malignant spread and metastasis.
Among the major genomic and transcriptomic alterations identified in The Cancer Genome Atlas (TCGA) project, ~50% of HNSCC tumors exhibit alterations in immune and cell death pathways, including the NF-κB signaling pathways (13, 14). These genetic and expression alterations include amplification of FADD (Fas-associated via death domain, 11q13), BIRC2/3 (Baculoviral IAP repeat-containing protein 2/3, also called IAP1/2, inhibitor of apoptosis protein 1/2, 11q22), mutation of caspase-8 (CASP8), altered RIPKs (Receptor-interacting serine/threonine kinases) in HPV(–), and deletion of TRAF3 (TNF receptor associated factor 3) in HPV(+) HNSCC tissues. However, the wider repertoire of molecules that functionally mediate activation of the classical and the alternative NF-κB pathways individually or together in HPV(+) and (–) HNSCC models has not been investigated.
To augment and explore potential links between the alterations found in the NF-κB pathways and inflammatory signal network uncovered by TCGA, we utilized a powerful protein docking algorithm, PRISM (PRotein Interactions by Structural Matching) (15, 16). PRISM enables modeling the 3D interactome of potential protein partners, that can be integrated with the experimentally defined protein network linked to classical and alternative NF-κB pathways from the literature. We focused on the interactions of FADD, BIRC2/3, TRAF3, CASP8, and RIPK1 proteins, which display frequent genetic and expression alterations in HNSCC TCGA datasets (13). We identified 61 proteins that interact with these genetically altered molecules, or are known to be involved in TNFR, NF-κB, inflammation, and death pathways. To further validate the effects of genetic alterations on the expression of these genes, we performed genome-wide exome DNA sequencing (exome DNA-seq) and whole transcriptome sequencing (RNA-seq) in 15 HPV(–) and 11 HPV(+) HNSCC cell lines. We observed consistent gene amplifications and expression patterns in cell lines as those detected in the HNSCC TCGA project. Using the NF-κB reporter cell lines developed in our laboratory, we performed large scale RNAi screening to assess the regulatory function of signaling molecules involved in the NF-κB and death pathways. We linked the NF-κB gene signatures to checkpoint molecules, which are co-regulated by the STAT and IFN pathways. The function and mechanistic validation of these molecules provide candidate therapeutic and prognostic targets for further preclinical and clinical investigation.
Materials and Methods
Analysis of Genomic Alterations and Immune Gene Signatures Using HNSCC TCGA Datasets
The Cancer Genome Atlas (TCGA) project of head and neck squamous cell carcinoma (HNSCC) has undertaken a comprehensive characterization of first 279 tumors with complete data analyses (13). The tumors were harvested predominantly from surgical patients, including mostly oral cavity (n=172/279, 61%) and laryngeal 34 tumors (n=72/279, 26%). The majority of patients were male (n=203/279, 73%) and heavy smokers (mean pack years=51). Among them, 36 patients are identified as HPV(+), and 244 patients are HPV(–), by genomic sequencing of HPV. Details about IRB approval, informed consent, sample collection, tissue-specific sample selection criteria, clinical annotations, and the genomic data pipelines can be found in the HNSCC TCGA publication (13). Data for genomic copy number, mutations, and RNA expression alterations were extracted from c-bioportal for oncoprint (https://www.cbioportal.org).
To analysis of immune gene signatures, data for RNA expression and CNV from 279 HNSCC patients were extracted from the TCGA datasets (13), (dbGaP Study Accession: phs000178.v5.p5) and downloaded from the Broad Institute, FireBrowse website (http://firebrowse.org/). This included level 3 RNA-Seq data (presented as log2 transformed RNA-Seq by Expectation Maximization [RSEM]) and clinical data (HPV status, tumor stage, and tumor source site). RNA-Seq data was subjected to unsupervised hierarchical clustering. IFN-gamma pathway genes were selected based on a previous publication (17).
Immune cell subset and checkpoint-associated genes were selected based on earlier studies (18–21). Data filtering was run using R package (version 3.4.1) as below: The gene lists were filtered using a custom R-script for the following criteria: genes with 75% samples with 0 or missing expression values were removed; 0 was replaced by minimum expression values; log2 transformation, median centered, genes with standard deviation > 50% quantile in all samples were included. In total, 44 immune pathway genes expression levels of 279 TCGA_HNSCC cohort entered into analysis. Unsupervised clustering by Manhattan distance columns, Euclidean distance rows, and complete linkage were performed using the Pheatmap (version 1.0.8) R software package. Samples contained in clustering were divided into three subgroups based on their clustering pattern, which includes 70 cases in subgroup 1, 75 cases in subgroup 2, and 134 cases in subgroup 3. These three subgroups were further analyzed for their survival preferences and clinic features using GraphPad Prism (version 7.0) The Kaplan-Meier plot for survival analysis was performed by the Log-Rank (Mantel-Cox) test. Distributions of clinical parameters were analyzed for significance by χ-square analysis. Plots were also produced by using GraphPad Prism (version 7.0)
Structural Modeling of Protein-Protein Interactions
The TCGA project of the HNSCC uncovered significant genomic and expression alterations of key molecules involved NF-κB and death signaling, including FADD, BIRC2/3, CASP8, RIPK, and TRAF3 (13). We modeled the 3D structures of these binary interactions via PRISM (15, 16, 22), which is a powerful docking tool. We found their experimentally known interaction partners from BioGRID (23) and STRING (24) databases. We modeled the complex (bound) structures of 19 TRAF3 interactions; 8 CASP8 interactions; and 9 RIPK1 interactions, as well as multiple interactions with FADD and IAPs. Furthermore, we included additional molecules involved in TNFR and NF-κB pathways based on our previous knowledge and generated a list of 61 proteins (Supplemental Table S2, Supplemental Figure S1B).
Cell Culture
HPV(–) HNSCC cell lines UM-SCC 1, 6, 9 11A, 11B, 22A, 22B, 38, 46 and 74A, and 74B and HPV(+) cell lines UM-SCC 103–110, SCC-90, 93VU147T, UM-SCC 47 were kindly provided by Drs. Thomas Carey or Mark Prince of University of Michigan in 2010 (Table S1). Authentication of these lines was done at the University of Michigan by DNA genotyping of alleles for 9 loci, D3S1358, D5S818, D7S820, D8S1179, D13S317, D18S51, D21S11, FGA, vWA, and the amelogenin locus as reported previously (25). HPV(+) lines VUMCSCC 4149, 4755, 4975, and 4697 cells were kindly provided by Dr. Wendell Yarbrough of Yale University in 2010 (Table S1). All cell lines were established as frozen stocks that were used for experiments within 3 months of re-passage. All cell line stocks were independently re-authenticated as unique lines in our laboratory by whole exome sequencing as reported in 2018 (26), and confirmed to be mycoplasma free. The HPV(–) cells were grown in MEM (Life Tech. #11095080), and HPV(+) cells except VUMC lines were grown in DMEM (Life Tech. #11965–092), supplemented with 10% FBS (Life Tech. #16000–044, heat inactivated at 56 °C for 30min.), penicillin-streptomycin (Life Tech. #15140148), and glutamine (Life Tech. #25030149). VUMC cell lines were cultured in keratinocyte growth medium (KGM) with included supplements (Life Tech. 17005–042), penicillin-streptomycin and glutamine as described above. Primary human oral keratinocytes (HOK) were cultured in KGM and used within 5 passages in accordance with the manufacturer’s protocol (Science Cell).
RNA Extraction
Total RNA of all the tumor cell lines and HOK cells was isolated from 1–10 × 106 cells using Trizol (Invitrogen, Cat#15596018) and RNeasy Mini Kit (QIAgen, Cat# 74104) combined method as per the manufacturer’s protocol. RNAs were quantified using the Nanodrop 2000 spectrophotometer (Thermo Scientific) and/or Agilent 2100 Bioanalyzer and stored at −80°C. All samples had a RIN of 9.0 or higher.
Whole Transcriptome RNA sequencing
For sequencing of the RNA from the cell lines, Ribo-Zero™ rRNA Removal Kits (Human/Mouse/Rat) (Epicentre by Illumina; Cat#: MRZH11124) were used to remove rRNA from total RNA. Multiplexed libraries for Whole Transcriptome RNA sequencing were constructed with rRNA-depleted total RNA utilizing SOLiD™ Total RNA-Seq Kit for Whole Transcriptome Libraries (Ambion by Life Technologies, Cat#:4445374) by following the manufacturer’s protocol. The data was deposited and available under accession number as reported (SRA:PRJNA453457; dbGaP:phs001581; (26)).
Gene expression analysis of RNA-seq data and heatmap generation for cell lines
The reads of RNA-seq data for the cell lines were filtered using mapping quality (threshold 10). On average, we obtained about 168 million mapped reads per sample. The read count of the genes were normalized using DESeq (estimate Size Factors) (27) R package (version 3.2.0) and upper quartile normalization method (28). The heatmap was generated using hierarchically clustered fold changes (FCs) in expression between HNSCC cell line and the mean of three oral keratinocyte lines as normal control. The column orders were predefined based on the unsupervised hierarchical clustering results and HPV status. The colors of the heatmaps were nonlinearly mapped between colors and FC values to avoid the dominance of extreme values.
Parallel analyses of HNSCC TCGA datasets and molecular features of cell lines
Datasets of the 279 HNSCC tumor plus 16 normal mucosa tissues in the TCGA cohorts including level 3 RNA sequencing data (log2 transformed RPKM value, as reads per kilobase of transcript per million mapped reads), HPV status, and clinic data were downloaded from the Broad Institute Firebrowse website (http://firebrowse.org/). The clinic data for each HNSCC tumor tissue in the TCGA cohorts were extracted based on their unique TCGA barcode. For individual genes, the association between copy number variation (CNVs) (the CNV number associated with each gene was defined as the segmented GISTIC (29) value at the corresponding genomic location) and RNA expression levels was assessed by Pearson correlation test and presented as scattered box plot using the R package (version 3.4.1). The distribution of various CNVs categories between HPV(–) and HPV(+) groups was analyzed by Fisher exact test and presented as grouped bar graphs using GraphPad Prism (version 7.0).
Molecular features of the 26 cell lines include CNVs, RNA expression levels and mutational spectrum were obtained through transcriptome and whole exome sequencing followed by bioinformatic analysis on our in-house sequencing platform (26). The RNA expression profiles were first normalized to normal controls and the comparisons between various pathologic groups were made using Student’s t-test with a two-tailed distribution and calculated at 95% confidence. P values of less than 0.05 were considered statistically significant. The stacked bar graphs were produced using GraphPad Prism. The RNA-seq data has been deposited in the Short Read Archive (SRA:http://www.ncbi.nlm.nih.gov/sra/) of NCBI under BioProject (SRA:PRJNA453457; dbGaP:phs001581).
cDNA Synthesis and qRT-PCR Analysis
For experiments including quantitative RT-PCR determination of mRNA expression, cDNA synthesis was achieved using the High Capacity cDNA Reverse Transcription Kit (ABI, Cat#: 4368814) and qRT-PCR was performed on a ViiA7 Real-Time PCR system (Applied Biosystems). Predesigned Taqman primer/probe set were purchased from Life Technologies (Cat#:4331182). Relative gene expression was normalized to 18s rRNA as an internal control, and fold changes were adjusted to the control samples. Each experiment was done in duplicates, and each sample was assayed in triplicates. 2−ΔΔCt was calculated and used as an indication of the relative expression levels. Data were presented as mean ± standard deviation (SD), and statistical analyses were performed using the unpaired Student t-test and two tailed comparisons.
TNF-α or LTβ regulated gene expression measured by microarray
UM-SCC 46 cells were treated with TNF-α (20ng/ml) or LTβ (100ng/ml, both from R&D Systems) for different time points. Total RNAs were isolated with Trizol and QIAgen RNeasy Kit combined method. Biotin labeled cRNA were synthesized using Illumina TotalPrep RNA Amplification Kit (Part No. AMIL1791). The Biotin labeled cRNA were hybridized onto the Bead Chips using Illumina HumanWG-6 V3 Bead ChIP Kit (Part No. BD-101–0203) base on the manufacture procedure, and the chips were scanned with Illumina Bead Array Reader. Raw data were imported to the GenomeStudio software (GenomeStudio, Gene Expression Module, Illumina), and normalized data were imported to Partek (http://www.partek.com/partek-flow/) to detect differentially expressed genes by using an ANOVA test. mRNA expression of LTβ upregulated genes was verified by qRT-PCR. The three time points selected for verification were based on the greatest increase of gene expression from the bead array. The data were calculated as mean + SD. An asterisk indicates a P-value < 0.05 compared to control using Student’s t-test. The complete microarray datasets were submitted to NCBI Gene Expression Omnibus (GEO, GSE126905).
Ingenuity Pathway Analysis (IPA)
Ingenuity Pathway Analysis was used to detect the significantly enriched signaling pathways within the top upregulated genes (https://www.qiagenbioinformatics.com/ products/ingenuity-pathway-analysis/). Two network connections were established using the upregulated genes involved in depicted pathways in responding to TNF-α or LTβ treatment.
NF-κB Reporter Assay
A stable NF-κB reporter cell line was established using UM-SCC 1 cells transfected with p-Lenti based vector containing repeated DNA binding sites for NF-κB upstream of a β-lactamase reporter gene and a blasticidin resistance gene (30). The β-lactamase reporter enzyme cleaves a fluorescent FRET substrate (Life Technologies, Cat#: K1085), which disrupts FRET and results in blue fluorescence. The reporter cell lines were selected and sorted by flow cytometry based on the green to blue fluorescence shift in responding to TNF-α treatment (10ng/ml). The reporter activity was measured 72 hours after gene knockdown by siRNA transfection (Silencer® Select siRNA, Life Technologies, Cat#: 4390771), with stimulation by TNF-α (10ng/ml) for 16 hours or LTβ for 24 hours before harvest. Thirty-five genes from the sixty-one gene list generated in this study (Supplemental Table S2) were selected from the initial genome-wide RNAi screening based on their knockdown effects of three independent siRNAs (Supplemental Table S3). The three siRNAs for gene knockdown were designed and validated by National Center for Advancing Translational Sciences (NCAT/NIH) RNAi screening core facility (30). To investigate the effects of 35 genes on NF-κB reporter activity in responding to TNF-α and LTβ, two of the three siRNAs were selected based on their best effects of knockdown activities (Supplemental Table S3). The two siRNAs were independently transfected into the reporter cell line using the same culture and transfection conditions as conducted in the NCAT RNAi screening experiments (30).
XTT Proliferation Assay
Cells were seeded in 2×103/well in 96-well plates and reverse transfected with siRNA oligonucleotides for 48 hours with Lipofectamine RNAiMAX (Life Technologies, Cat#:13778150) following the manufacturer’s protocol. Cell proliferation was assayed on the indicated days with sodium 3’-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulfonic acid hydrate (XTT) Cell Proliferation Kit (Roche Diagnostics, Cat#: 11465 015 001), following the manufacturer’s instructions. XTT assay reagent was added for 4 hours prior to assay. At each time point, absorbance was read with BioTek Synergy Microplate Reader at 450 nm and 655 nm. The data represent the mean of 6 replicates in each experimental condition.
Results
Top genomic alterations involved in TNFR and NF-κB pathways in HNSCC
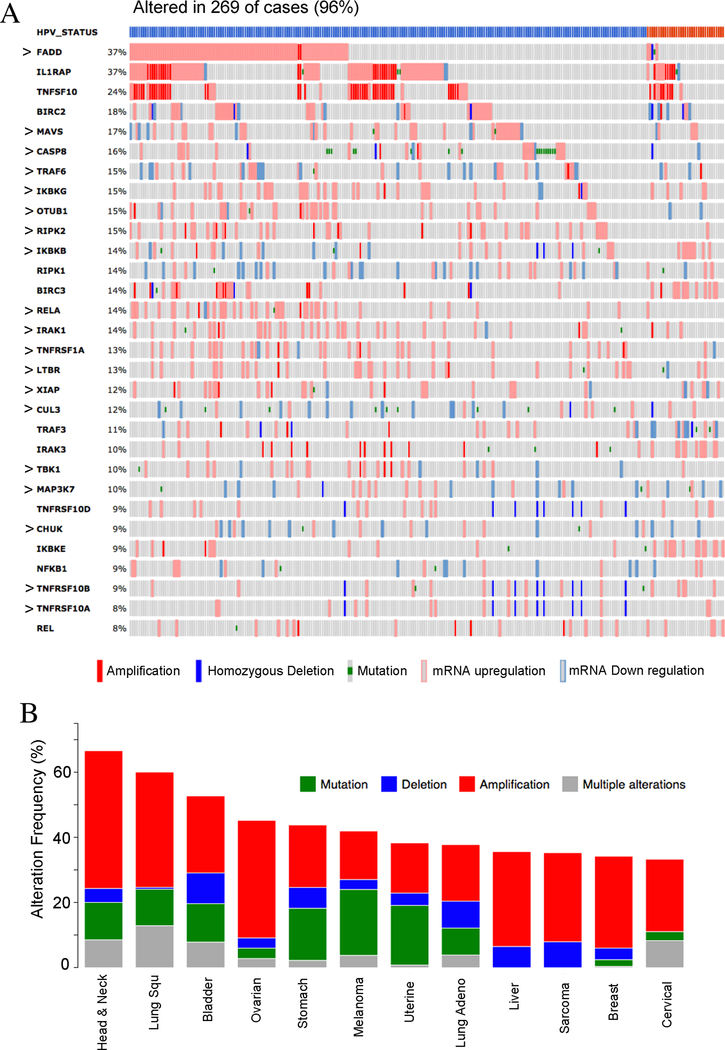
The TCGA project of the HNSCC uncovered significant genomic alterations of key molecules involved in NF-κB and death signaling, including alterations of FADD, BIRC2/3, CASP8, RIPKs, and TRAF3 (13). To explore the detailed mechanisms of NF-κB and death signaling, we identified proteins with experimental interaction of these genetically altered molecules, and modeled 3D structures of protein-protein interaction complexes by PRISM ((15, 16, 22), Supplemental Fig. S1A). We also included molecules involved in the TNFR and NF-κB pathways (Supplemental Fig. S1B) and generated a list of 61 proteins for further investigation (Supplemental Fig. S1C and Table S1). We ranked the altered top 30 genes obtained from the HNSCC TCGA dataset in cBioportal (Supplemental Fig. S1D) and present these as an oncoprint, ranging from 37% to 8% patients for the individual genes (Fig. 1A). In total, 96% of HNSCC patients contain one or multiple alterations of these genes. The top 3 genes altered in HNSCC are FADD, IL1RAP, and TRAFSF10, mainly showing gene amplification and overexpression (Fig. 1A). Altered expression of 57% of these genes was also observed in HNSCC cell lines examined by RNA-sequencing in our laboratory (Figure 1A, “>” annotation). Next, we surveyed altered genes across different cancer types from TCGA datasets and identified the top 12 major cancer types (Fig. 1B and Supplemental Fig. S1E). HNSCC ranked among the top cancers, followed by lung SCC, bladder cancer, and ovarian cancer. Gene amplification was the dominant alteration in the top four cancer types, including HNSCC (Fig. 1B). However, the genetic and expression alterations varied among cancer types, with the top alterations of TNFSF10, FADD, TNFRSF1A, RIPK1/2, TNFRSF10B (TRAILR2/DR5), and BIRC2 genes observed in lung and bladder SCCs, and ovarian cancer. (Supplemental Figure S2A–C). Together, the majority of the genes involved in TNFR and NF-κB pathways were amplified and overexpressed in HNSCC, lung SCC, and bladder cancers.
Figure 1. Genetic and expression alterations of 30 genes related to NF-κB, inflammation and death pathways in HNSCC and other cancer types.
(A) Genetic and expression alterations of 30 genes involved in NF-κB, inflammation, and death pathways were identified in the TCGA HNSCC database from the c-Bioportal website and presented as an OncoPrint. Alterations were observed in 96% of cancer patients (269/279), including amplifications, deletions, mutations, and mRNA up or down regulation. The top bars represents HPV status for tumors, blue: HPV negative; red: HPV positive. Percentage of each gene’s alteration in total patients was represented and each bar represents as individual patient. The symbol “>“ indicates that the gene expression alteration was also observed in the HNSCC lines. (B) The copy number variation and mutations of the 30 genes were ranked across all cancer types from TCGA data, and the top 12 cancers with the highest frequency of genetic alterations were presented in x axis. Percentages of the genetic alterations are presented in y axis. Red: gene amplification, blue: gene deletion, green: mutation, gray: multiple genetic variations. HNSCC is the top cancer type with the highest alteration ratio of 30 genes.
DNA copy number and gene expression alterations differ by HPV status in HNSCC
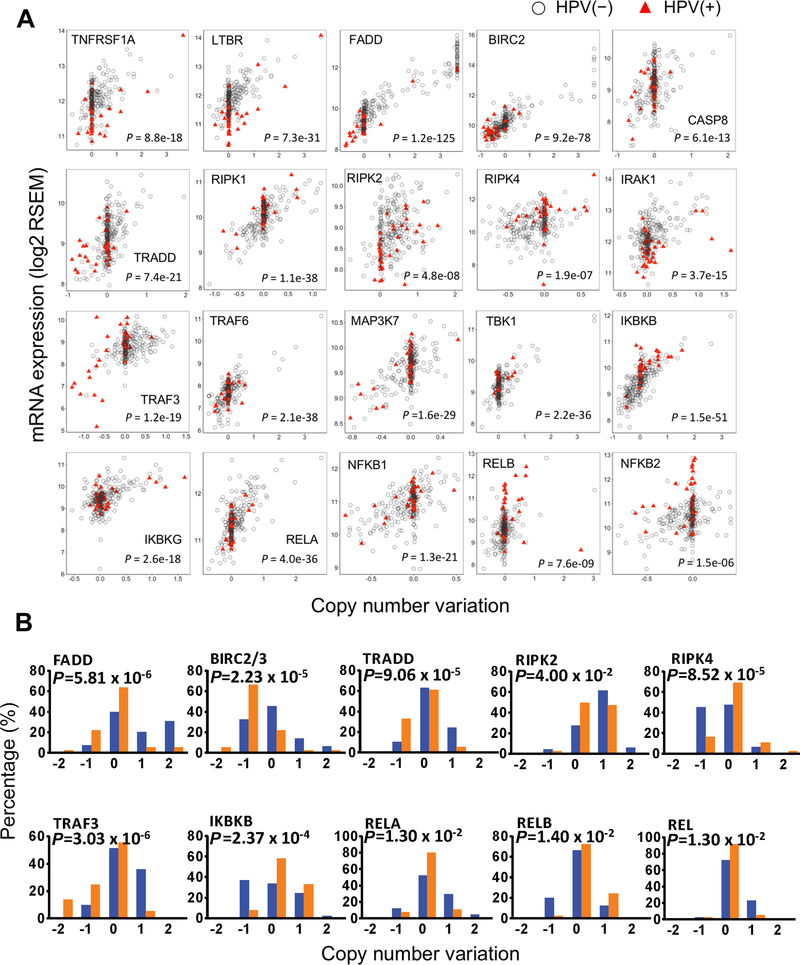
Of the 61 genes involved in the NF-κB and cell death signaling pathways, about 1/3 exhibited significant correlation between RNA expression levels and their corresponding copy number variation (CNVs) (Fig. 2A). The patient data were separately depicted by HPV status, and the statistical significance of comparisons between CNV and expression was presented by Pearson correlation coefficients (r) and P values (Table S3). These genes included several functional categories, such as receptors, signal adaptors, kinases, ubiquitin ligases and caspase proteins, and NF-κB/REL family members. In addition, CNVs of ten genes varied in tumors with different HPV statuses (Fig. 2B). Notably, frequent amplifications of FADD, TRADD, RIPK2, TRAF3, RELA and REL were observed in HPV(–) tumors, whereas frequent deletions of BIRC2/3, TRADD and TRAF3, and gains in RELB were observed in HPV(+) samples (Fig. 2B). These findings revealed different gene CNVs as well as distinct patterns of aberrant gene expression in inflammatory and death pathways between the two HNSCC subgroups differing in HPV status.
Figure 2. Alterations of copy number variation (CNV) and gene expression in HNSCC with different HPV status.
(A) Correlation between (CNV) and mRNA expression in HNSCC with different HPV status. Scatterplots for 20 selected genes showing significant CNV and gene expression correlation in HNSCC TCGA datasets (n = 279), including 243 HPV(–) (gray circles), and 36 HPV(+) specimens (red triangles). Pearson correlation coefficient significance test, with P values presented. X axis, log2 CN ratio; 0 is diploid, −1 is one-copy loss, 0.585 is one-copy gain, and values larger than 1 are amplifications. Y axis, log2 transformed RNA-Seq RSEM values. (B). The genes with statistical significance in distribution of various CNV between HPV(–) samples (blue bar) and HPV(+) samples (orange bar). The percentage of each CNV types in their respective HPV status groups were calculated based on the HNSCC sample counts. Statistical analysis was conducted by Fisher exact test.
Altered gene expression in HNSCC specimens and cell lines differing by HPV status
To model the genomic alterations in HNSCC tissues, we conducted whole exome and transcriptome sequencing of 15 HPV(–) and 11 HPV(+) tumor cell lines, plus three normal human oral keratinocytes cell lines (Supplemental Fig. S1F, Supplemental Table S1). To explore the expression patterns of the 61 genes involved in inflammatory and NF-κB pathways in HNSCC cell lines, we conducted supervised hierarchical clustering of gene expression (Supplemental Fig. S3). The clusters were dominated by up-regulated genes clusters across both HPV(–) and HPV(+) cell lines on the top, and the down-regulated genes at the bottom (Supplemental Fig. S3).
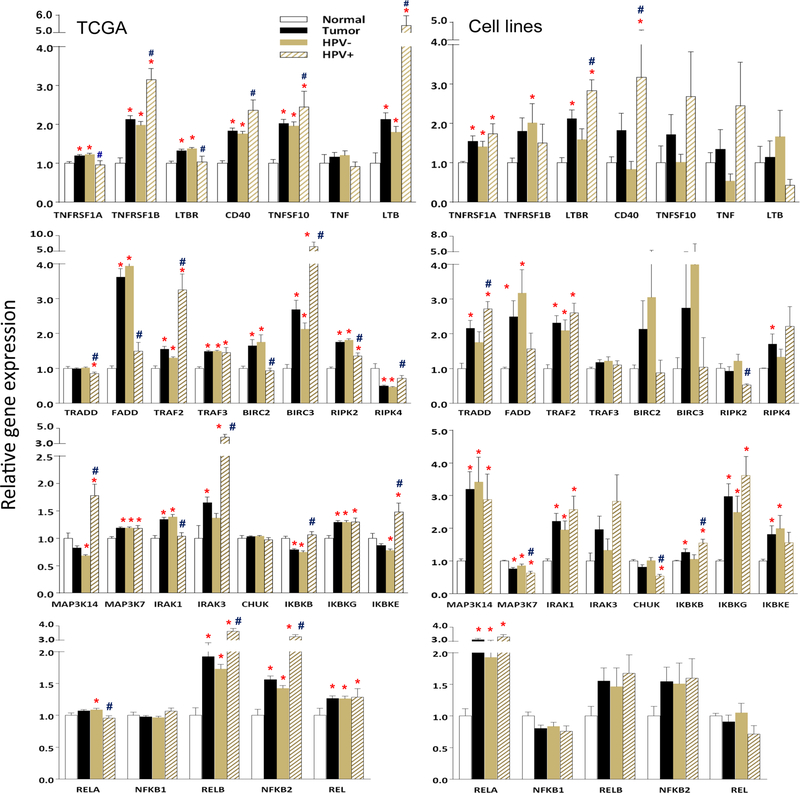
Using HNSCC datasets from TCGA, we compared 28 genes in all 279 tumor samples, including 243 HPV(–) and 36 HPV(+) tumors, with 16 patient mucosa tissues adjacent to tumors (Fig. 3, left panels). We compared tumors with normal mucosa tissues (indicating the significant changes as * by Student T-test, P<0.05), while differences observed between tumors with different HPV status were indicated as # (P<0.05). Genes were selected and grouped as follows: cytokines and receptors (top), signaling adaptor proteins (second row), kinases (third row), and five NF-κB subunits (bottom). In addition, we also determined the significance of differences in the gene expression by HNSCC cell lines with different HPV statuses (indicated as #), as well as compared tumors with HOK cells (indicated as *, Fig. 3, right panels). There were 18 genes overlapped with those listed in Fig. 1A, and 19/30 genes presented in Fig. 1A (displayed with a “>“ symbol) also had altered gene expression in HNSCC cell lines (Fig. 3 right panels). When we compared the altered gene expression between HNSCC tissues with cell lines, more mRNAs exhibited significant differences between the subgroups in the TCGA tissue datasets than observed in the HNSCC cell lines, which could have resulted from expression by the non-tumor infiltrating cells, or tumor cells modulated by the TME. Notably, the genes involved in the alternative NF-κB pathway were highly expressed in the HPV(+) group, including TNFRSF1B, CD40, TNFSF10, LTB, TRAF2, BIRC3, MAP3K14(NIK), IRAK3, RELB, and NF-κB2, indicating the importance of different NF-κB pathways in HNSCC with different HPV statuses.
Figure 3. Altered gene expression in HNSCC specimens and cell lines differing by HPV status.
Altered gene expression between 279 HNSCC specimens and 16 normal mucosa specimens from TCGA (left panels), as well as 26 HNSCC cell lines and 3 HOK lines (right panels) were compared from RNA-seq. Mean expression levels of genes were compared between tumor [including both HPV(–) and HPV(+) samples] and normal tissues, as well as between tumors with different HPV status. The 28 most altered genes are presented by the relative gene expression levels as mean± standard errors (SE). Statistical difference was calculated by Student t-test (P<0.05). * indicates statistical significances when compared mean expression of genes between tumor [including both HPV(–) and HPV(+) samples] with normal tissues, and # indicates significance when the comparison between tumor samples of different HPV status. The gene expression of 26 HNSCC cell lines were compared with the mean of the three HOK lines, as well as between tumor lines with different HPV status (right panels). The data analysis was performed in the same way as for HNSCC TCGA data.
NF-κB pathway activity was differentially induced by TNF-α and LTβ
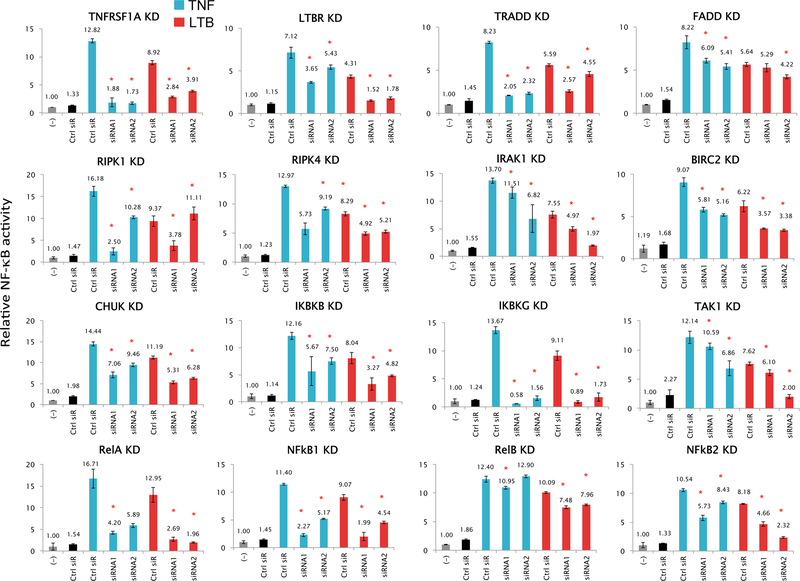
The role of the 35 top altered molecules in mediating functional NF-κB activation was investigated based on the initial results from a genome-wide siRNA screening (Supplemental Table S3). The RNAi screening was conducted using three independent siRNAs provided by NCAT/NIH RNAi core facility in a stable NF-κB reporter cell line of HPV(–) UM-SCC1 cells treated with TNF-α (30). We validated significant effects for 16 of these genes after knockdown with two independent siRNAs on reporter responses to treatment with TNF-α or LTβ, known inducers of the classical and alternative pathways (Fig. 4 and Supplemental Fig. S1F). In general, TNF-α mediated stronger induction of gene expression than LTβ. Knockdown of TNF receptor superfamily member 1A (TNFRSF1A) or lymphotoxin β (LTBR) significantly decreased NF-κB reporter activity in responding to the own ligands, but also significantly affected activity induced by the ligand for the other receptor to a lesser degree (Fig. 4, top two left panels). These data suggested possible cross-activation between the classical and alternative NF-κB pathways, which we confirmed was not due to off-target effects of the siRNAs, by verifying the specificity of knockdown of TNFRSF1 and LTBR on their mRNA targets by qRT-PCR (Supplemental Fig. S4). SiRNAs targeting known classical pathway components, TRADD and FADD, exhibited more consistent inhibitory effects on TNF-α induced activity, whereas knockdown of classical components IKBKG, RELA and NF-κB1, known as inducers of mRNAs for NF-κB2 and RELB with activity to modulate the NF-κB alternative pathway (5), exhibited strong inhibitory effects on both TNF-α and LTβ-induced reporter activity. Thus, our data supported preferential activation and cross-talk involving different components of TNF-α modulated classical NF-κB and LTβ modulated alternative pathway in HNSCC cells.
Figure 4. Effects of gene knockdown on NF-κB reporter activity induced by TNF-α and LTβ.
Gene knockdown were performed in UM-SCC 1 cell line stably transfected with a β-lactamase NF-κB reporter gene (GeneBLAzer). Two independent siRNAs were tested for each gene knockdown along with a negative control siRNA (black). Transfections were done in 96-well plates with 6 replicates of each condition. Assay was performed 72 hours after siRNA transfection, following stimulation for the final 16hr with TNF-α (blue) or 24hr with LTβ (red). Statistical significance was calculated by t-test (P<0.05). Data are from two independent transfections of 2 independent siRNAs as the biological replicates, and 6 replicates for each siRNA knockdown condition. The two siRNAs were selected from the three validated siRNAs provided by NCAT RNAi core facility. The data for TNFα induced NF-κB activity have been replicated in a genome-wide RNAi screening experiment using the three siRNAs for 35 genes (Supplemental Table S3).
TNF-α or LTβ stimulates overlapping and distinct signaling networks
To investigate the regulatory mechanisms and signaling networks of NF-κB and inflammatory pathways, we compared the gene expression induced with TNF-α or LTβ in HPV(–) UM-SCC 46 cells (Supplemental Fig. S1F, S5). TNF-α uniquely induced 172 genes, LTβ specifically induced 202 genes, and 155 genes were overlapping in both experiments (Supplemental Fig. S5A). As TNF-α induced gene expression profiling was partially presented previously (31), we focused on LTβ effects analyzed by Ingenuity Pathway Analysis (IPA, Supplemental Fig. S5B). The top pathways included IL-6 signaling, acute phase responses, macrophage function, and CD40 alternative NF-κB pathway. Conversely, alternative pathway inducer LTβ also induced several genes involved in classical TNFR signaling, such as TNFRSF1A, CHUK(IKKα), IKKβ, IKKγ, RELA, NF-κB1, supporting bi-directional cross-talk between the classical and alterative NF-κB pathways. However, TNF-α mainly regulated mRNAs encoding canonical NF-κB mediated oncogenic and inflammatory signaling components (Supplemental Fig. S5C), whereas LTβ induced mRNAs were often involved in both classical and alternative NF-κB pathways (Supplemental Fig. S5D). Genes involved in inflammatory responses, NF-κB signaling, cell survival, and tumor metastasis were further validated by quantitative RT-PCR (Supplemental Fig. S6).
TNF-α and LTβ induced genes in HNSCC cell lines differing by HPV status
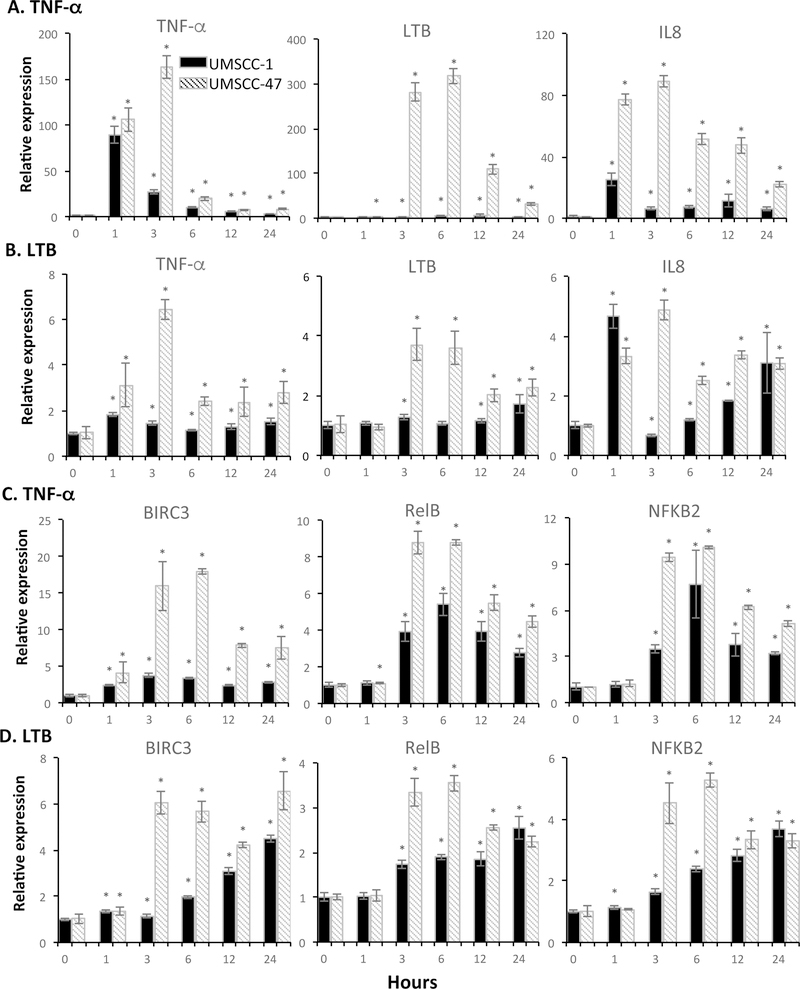
We assessed gene expression induced by TNF-α or LTβ in a HPV(–) UM-SCC1, and a HPV(+) UM-SCC47 line (Fig. 5 and Supplemental Fig. S7). In general, TNF-α induced a stronger gene expression than that induced by LTβ in both lines (Fig. 5 and Supplemental Fig. S7). The peak gene expression induced by TNF-α was around 3–6 hours, whereas LTβ induced a slower pattern. The higher gene induction in UM-SCC 47 cells than UM-SCC 1 cells were observed by both TNF-α or LTβ, which could not simply be explained by lower basal gene expression in UM-SCC 47 cells (Supplemental Fig. S8). We observed TNF-α induced LTΒ, RELB, and NF-κB2 expression, whereas LTβ induced TNF-α and NF-κB1 expression, supporting the functional cross-talk observed between canonical and alternative NF-κB pathways (Fig. 5 and Supplemental Fig. S7). Furthermore, we observed TNF-α or LTB-induced gene expression of metastasis molecules COX2, SERPINE1, PLAU, and IL-6 in both UM-SCC1 and UM-SCC47 cells (Supplemental Fig. S7). Thus, we observed TNF-α and LTβ induced cross-talk between the canonical and alternative NF-κB pathways and downstream inflammatory cytokine and metastasis-related genes.
Figure 5. TNF-α and LTβ modulated differential gene expression in HNSCC cells differing in HPV status.
UM-SCC 1 (Grey) or UM-SCC 47 cells (dash) were treated with TNF-α (20ng/ml, left) or LTβ (100ng/ml, right) for 1, 3, 6, 12, and 24 hours. Shown is the relative mRNA expression of TNF-α or LTβ upregulated genes was verified by qRT-PCR, and compared with time 0 without treatment. A and C showed genes induced by TNF-α, and B and D showed genes induced by LTB. The data were calculated as mean + SD from three replicates of representative of two repeated experiments. An asterisk indicates a P-value <0.05 compared to control using a t-test.
Knockdown of IKKs/NF-κBs inhibited gene expression and cell proliferation
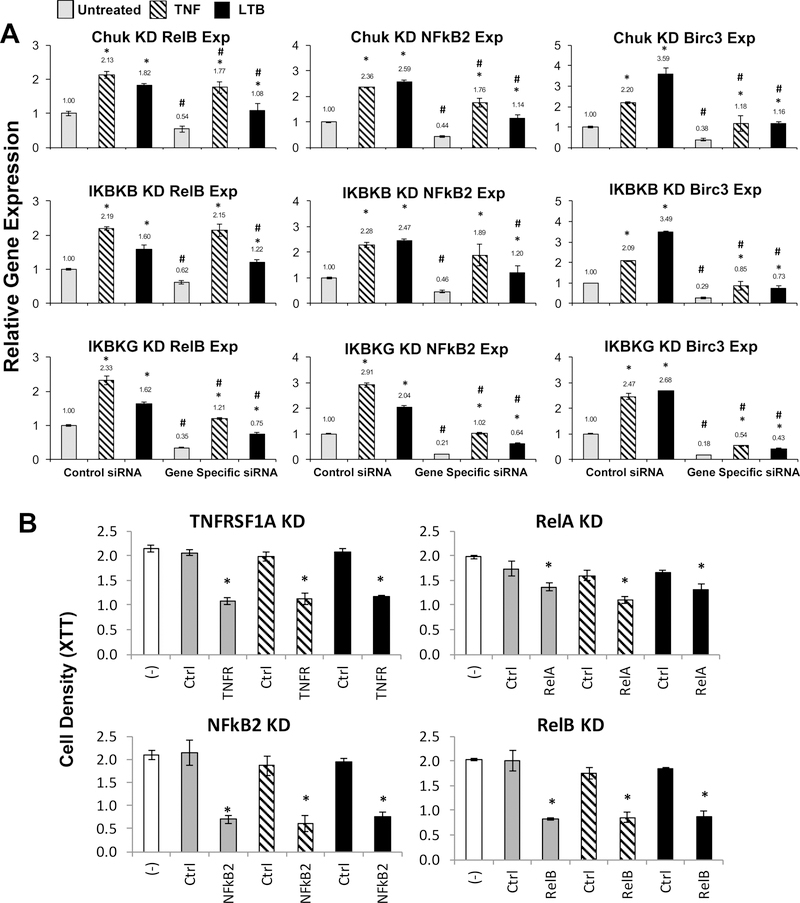
As the classical and alternative pathways share IKKα (CHUK) but no other IKK subunits of the classical trimeric complex, we further examined how IKKα, IKKβ and IKKγ regulate the expression of RELB and NF-κB2 mRNAs, which encode alternative NF-κB transcription factor subunits, and BIRC3, a target of the alternative NF-κB pathway (Fig. 6A, Supplemental Fig. S1F). Compare to the control siRNAs, knockdown of all three kinases resulted in significant reduction in expression of these mRNAs, with the effect of IKKγ knockdown being the strongest among the three, whereas that of IKKβ was stronger than that of CHUK (IKKα) (Fig. 6A, indicated as #). TNF-α induced RELB expression more potently than LTβ, whereas TNF-α and LTβ induced NF-κB2 expression at similar levels (indicated as *). However, LTβ consistently induced higher BIRC3 expression when compared to TNF-α (indicated as *). Next, we knocked down the mRNAs using three independent siRNAs, and observed that knockdown of TNFRSF1A, RELB, or NF-κB2 significantly decreased cell density, whereas knockdown of RELA showed a less profound effect (Fig. 6B).
Figure 6. Effects of knocking down IKKs, TNFRSF1A, and NF-κB family members altered gene expression and cell proliferation in UM-SCC 1 cells.
(A) CHUK (IKKα), IKKβ, IKKγ were knocked down with two siRNA each in UM-SCC1 cells for 48 hours, then treated with TNF-α (10ng/ml) or LTβ (100ng/ml) 6 hours prior to harvesting. Relative gene expression of RELB, NF-κB2 and BIRC3 were verified by qRT-PCR in triplicates compared to cells transfected with control siRNA without treatment. The data were calculated as mean ± SD. * indicates statistical significance compared to no treatment, and # indicates statistical significance compared to control siRNA with same treatment (student t-test, P-value≤0.05). The two siRNAs were transfected independently and served as the biological replicates. (B) Cell density was measured after gene knockdown in UM-SCC 1 cell line in 6 replicates. XTT assay was performed 72 hours after siRNA transfection, without treatment (grey), with stimulation by TNF-α for final 16 hour (dash), or by LTβ for final 24 hour (black) before adding XTT reagents. White bar indicates the control condition without siRNA transfection. The data are from one representative siRNA knockdown of two repeated experiments, and statistical significance was calculated from six replicates (t test, P<0.05).
TNF-α modulated checkpoint molecules and IFN-γ/STAT pathways
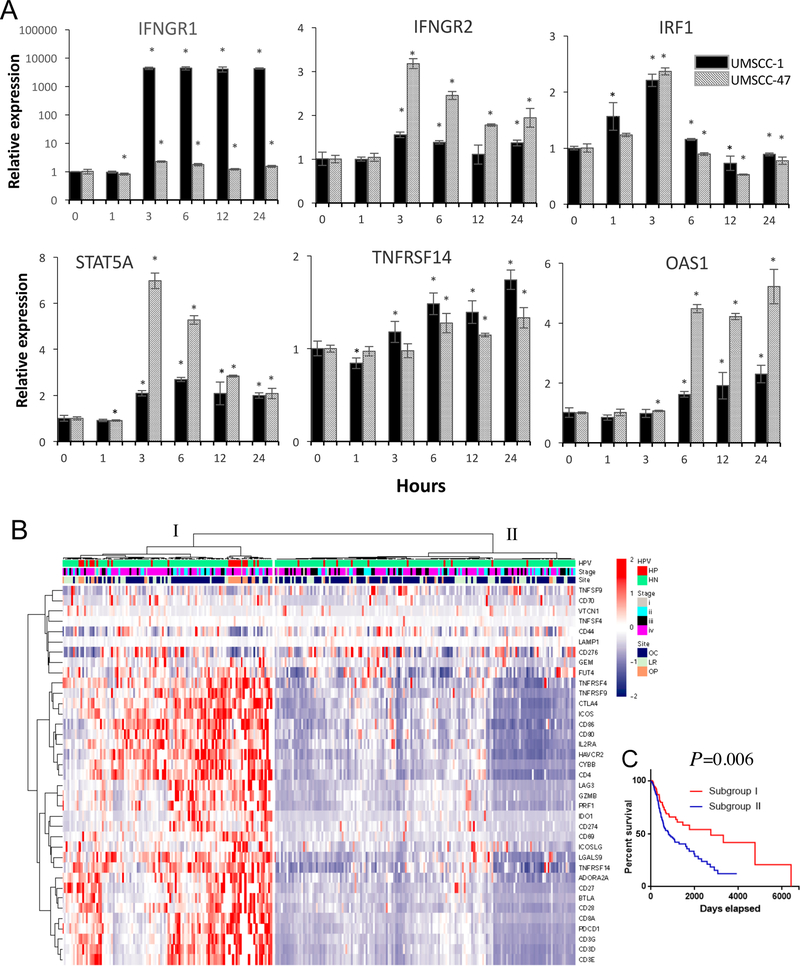
Checkpoint inhibitors have become an effective treatment strategy for HNSCC and other cancer types (32, 33). We hypothesized that TNF-α and LTβ and NF-κB signaling could modulate the expression of immune checkpoint proteins by regulating modulators of the IFN-γ and STAT pathways (17, 18). After screening TCGA HNSCC dataset, we identified 44 genes from the IFN-γ and STAT pathways (17) with significant expression alterations among three patient subgroups (Supplemental Fig. S9A). Patients in subgroup I (n=70) and II (n=75) exhibited higher gene expression levels of IFN-γ and STAT pathway-related mRNAs. The case number distributions of the three patient subgroups was examined based on patient HPV status and primary tumor origins, where subgroup II had more HPV(+) cases from oropharyngeal sites (Supplemental Fig. S9B and C), but no statistical significance was observed in the case numbers from different tumor stages (Supplemental Fig. S9D). Subgroups I and II exhibited more favorable prognosis than group III (Supplemental Fig. S9E). We next tested if the above 44 genes could be induced by TNF-α or LTβ in HNSCC cell lines. TNF-α potently induced IFNGR1, IFNGR2, IRF1, STAT5A, TNFRSF14, and OAS1 gene expression in both cell lines, which are involved in IFN-γ and STAT pathway signaling (Fig. 7A). However, after screening the microarray data, we did not observe significantly induced gene expression after LTβ treatment in UM-SCC 46 cells.
Figure 7. TNF-α modulated gene expression involved in IFN-γ and STAT mediated checkpoint modulation.
(A) UM-SCC 1 (black) or UM-SCC 47 cells (gray) were treated TNF-α (20ng/ml, left) or LTβ (100ng/ml, right) for 1, 3, 6, 12, and 24 hours. Relative mRNA expression of molecules involved in IFN-γ and STAT mediated checkpoint modulation were verified by qRT-PCR, and compared with time 0 without treatment. The data were calculated as mean + SD of 6 replicates from a representative experiment. The same experiment was independently repeated in UM-SCC46 cells using microarray. An asterisk indicates a P-value <0.05 compared to control using a student t-test. (B) Unsupervised hierarchical clustering analysis of immune checkpoint gene expression was performed using RNASeq data of 279 HNSCC patient samples from the TCGA cohort. Heatmap of the expression levels with a list of filtered immune and checkpoint response-associated genes on the y-axis, red indicates relative gene overexpression and blue indicates relative gene underexpression compared to means for each gene). Subgroup of the patients I (n=115) and II (n=164) with clinical characterization indicated at the top of the heatmap, such as HPV status (HP, HPV positive, red; HN, HPV negative, green), staging (I-IV), and primary tumor site (OC, oral cavity; LR, larynx; OP, oropharynx). (C) Survival analysis of 279 HNSCC patient samples from the TCGA cohort, with statistical significance between two subgroups was indicated.
Since IFN-γ and STAT pathways directly modulate checkpoint molecule expression and thereby regulate the therapeutic responses to checkpoint inhibitors, we next examined T cell activation and immune checkpoint-associated gene expression in HNSCC patients from the TCGA dataset. Unsupervised cluster analysis of expression patterns of 37 genes involved in T cell activation and checkpoint molecules was performed using the TCGA HNSCC datasets (20, 21). These expression patterns fell into Subgroup I (n=115), dominated by up-regulated expression of T cell activation and immune checkpoint-associated genes, whereas subgroup II (n=164) displayed a relatively lower expression of these proinflammatory genes in tumors, reflecting with a “cold” immune TME, with potential lower anti-tumor activity (Fig. 7B). The two subgroups of patients exhibited statistical differences with HPV status and staging, but not the primary tumor origins. We noticed that most HPV(+) cancer samples were classified into subgroup I, which was associated with favorable prognosis in the survival analysis (Figure 7C, P=0.006). These data support the hypothesis that TNF-α mediated signaling can affect gene expression involved in the regulation of immune signaling and checkpoint molecule signatures.
Discussion
Here, we analyzed the landscape of genomic and transcriptome alterations from the TCGA HNSCC dataset and identified 61 candidates exhibiting alterations or predicted interactions with inflammatory and NF-κB signaling molecules. Among these, the 30 most frequently altered genes were observed in 96% of tumors, and 60% of HNSCC exhibited genetic alterations in these pathways. The dataset exhibited a significant association of gene expression with CNV, and a subset of these varied with HPV status. Among genes concordantly overexpressed in HNSCC tissues and cell lines, siRNA knockdown of several that encode components of the classical or alternative pathway significantly altered NF-κB activity and gene expression in response to TNF-α or LTβ, supporting cross-talk between these pathways in HNSCC. Knockdown of genes involved in these NF-κB pathways affected downstream gene expression and decreased cell proliferation. TNF-α predominantly induced genes involved in IFN-γ and STAT pathways, which mediate immune checkpoint molecule expression. Thus, our data provide genetic, expression, and functional evidence of aberrant activation and cross-talk between NF-κB classical and alternative pathways in HNSCC with different HPV status (34, 35).
Most studies of the inflammatory and NF-κB pathways focused on transcriptional regulation and post-translational modifications (7, 35). However, genetic alterations driving the activation of NF-κB signaling have not been well studied in solid tumors. Here we showed that ~2/3 of the 30 top altered genes presented in Fig. 1A are associated with gene CNV. Two-copy gain of the FADD gene was observed in 80 cases of 279 HNSCC (28.7%), whereas one copy gain or loss was observed in most other genes. The strong statistical association of gene expression with genetic alterations indicated genetic influence on gene expression. An association of 11q copy gains encoding FADD and BIRC genes correlates with worse survival (36) suggesting possible use as prognostic biomarkers or as therapeutic targets. This point is well supported by our preclinical study of the anti-tumor activity of birinapant, which targets Inhibitor of Apoptosis 1 encoded by BIRC2 (37). Birinapant exhibited strong inhibitory activity in combination with radiotherapy, eradicating tumor xenografts with copy gain and overexpression of FADD and BIRC2. The 30 most altered genes in HNSCC also exhibited high frequencies in lung SCC, bladder and ovarian cancers. The top amplification and overexpression of TNFSF10 (TRAIL), FADD, TNFRSF1A (TNFR1), and LTBR in lung SCC, and the top ranked genes FADD and RIPK1/2 in bladder cancers, suggested targeted therapy incorporating IAP antagonists also warrants investigation in these cancers.
In this study, we observed differences in genomic CNV and expression between HPV(–) and HPV(+) HNSCC, which often differs in patients that smoke or have HPV viral infections (38). Among ten genes that exhibited different CNV patterns, TRAF3 a negative regulator of the alternative NF-κB pathway, exhibited more deletions in HPV(+) tissues, consistent with our previous demonstration of aberrant activation of the NF-κB alternative pathway in oropharyngeal and HPV(+) HNSCC (8, 39). Several genes involved in the NF-kB alternative pathway, such as LTB, CD40, BIRC3, MAP3K14, RELB, and NF-κB2, exhibited significantly higher expression in HPV(+) tissues compared with HPV(–) tissues. In addition, genes involved in the alternative NF-κB pathway such as BIRC3 were highly inducible by LTβ in a HPV(+) cell line. The latter findings might guide future targeted and immune therapies in HPV(+) HNSCC, where inhibition of the alternative NF-κB pathway or prosurvival targets such as BIRC3 (IAP2) combined with activation of the death pathway by death receptor agonists could be more effective (12, 40–42).
Here, we observed a consistent pattern of cross-activation of the canonical and alternative NF-κB pathways in response to TNF-α and LTβ, the typical ligands for either pathways (43–46). Although NFKB2 and RELB proteins are the transcription factors subunits of the alternative NF-κB pathway, their gene expression is regulated in part by the classical NF-κB pathway, as they are RELA inducible genes (47). Co-processing of the precursors of NF-κB1 and NF-κB2 proteins has also been linked to bi-directional cross-talk between these pathways (48). In addition, LTβ binding to LTβR results in TRAF2 and TRAF3 recruitment to the LTβ receptor, which destabilizes the TRAF-cIAP complex, protecting NIK from constitutive degradation, and allowing NIK accumulation and activation (49, 50). NIK has been implicated in cross-activation of the classical pathway (51). LTβR in the presence of limiting amounts of TRAF3 has been shown to enable TRAF2-IKK1 cross-activation of the classical pathway (52). Together, our and others’ data showed the cross-activation of the canonical and alternative NF-κB pathways through multiple molecular components and mechanisms.
Immune checkpoint blockade has become a new paradigm in cancer therapy, especially for those cancers with high mutation rates and neoantigens, including HNSCC (2, 53, 54). Although expression of PD1, PD-L1, CTLA4, and other checkpoint proteins on the immune or tumor cells appear to be required for response to immune checkpoint therapies (19), it is puzzling that these drugs induce poor efficacy in many patients. Mandal et al. have investigated limited immune signatures from TCGA (55), and identified HNSCC as the highly immune-infiltrated cancer type, where CD8+ T cell infiltration and CD56dim NK cell infiltration each correlated with superior survival. IFN-γ and STAT pathways are regulatory mechanisms involved in T cell responses and expression of PD1/PDL1 and other checkpoint molecules. Melanoma patients who harbor a higher rate of genomic defects in the IFN-γ pathway genes exhibit poor clinical responses to ipilimumab (17). We interrogated the gene set of the aforementioned melanoma study focusing on the differential expression of molecules involved in the IFN-γ and STAT regulatory pathways in HNSCC. Several molecules involved in these pathways can be regulated by TNF-α in HNSCC cell lines, and patients with active expression of genes involved in the IFN-γ and STAT regulatory pathways exhibited a better survival in the clinical trials with the conventional therapies, as shown in this study (13). In contrast, in the melanoma tumor microenvironment, the prolonged interferon signaling exhausts the T cells and promotes resistance to checkpoint inhibitors and combinations with radiation plus anti-CTLA4 (25). The effects of TNF-α modulated IFN-γ and STAT regulatory pathways involved in the responses to immunotherapies and checkpoint inhibitors and identifying potential targets for molecular therapy that that could synergize with these therapies merit future investigation.
In summary, these findings have implications for the novel biomarkers or targets for treatment of HNSCC in combination with immune checkpoint inhibitor, radiation and chemotherapy. Notably, the top amplification and overexpression genes FADD and BIRC2 were identified in the HNSCC tumors and cell lines. These are sensitive to SMAC (second mitochondria-derived activator of caspases) mimetic and IAP inhibitors when combined with radiation or immune checkpoint inhibitors that induce TNF-α in vivo, inhibiting or eradicating tumors in xenograft or syngeneic models (37, 40, 41). TNFSF10 is another top amplified and overexpressed gene coding for the death agonists TRAIL, which exhibited synergistic anti-tumor activity of IAP inhibitor birinapant when tested in a panel of HNSCC cell lines in vitro (26, 42). In addition, in HPV(+) HNSCC cells, which are lacking FADD/BIRC2 amplification and overexpression, antibody agonist to TRAILR2 plus TRAIL and birinapant exhibited strong induction of cell death in vitro (42). Furthermore, the gene panels involved in immune and STAT/IFN pathways predicted the HNSCC patients’ survival to conventional therapies, which supported the clinical significance of these genes serving as the potential biomarkers. The genetic and expression alternations involved in the NF-κB and inflammatory pathways are also prevalent and may be of broader relevance to the mechanisms underpinning pathogenesis and therapeutic sensitivity of squamous cell carcinomas and cancers from other sites (56).
Supplementary Material
Acknowledgement
This study was supported by the Intramural Research Program of the National Institute on Deafness and Other Communication Disorders, projects ZIA-DC-000016, 73 and 74 (XPY, HC, JC, AS, HS, SL, CVW, ZC). This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E (EGM, RN). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This work used the computational resources of the NIH High-Performance Computing (HPC) Biowulf cluster (http://hpc.nih.gov), and the authors thank the NIH Helix systems staff for the management and supports. The authors thank Dr. Clint Allen (Head and Neck Surgery Branch, NIDCD, NIH), Dr. Vassiliki Saloura (Thoracic and Gastrointestinal Malignancies Branch, NCI, NIH), and Dr. Cheng-Ming Chiang (UT Southwestern Medical Center) for their helpful comments and suggestions of this manuscript and project.
Footnotes
Conflict of Interest Statement: The authors have declared that no conflict of interest exists.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 2.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. [DOI] [PubMed] [Google Scholar]
- 3.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Waes C Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clin Cancer Res. 2007;13(4):1076–82. [DOI] [PubMed] [Google Scholar]
- 5.Brown M, Cohen J, Arun P, Chen Z, Van Waes C. NF-kappaB in carcinoma therapy and prevention. Expert Opin Ther Targets. 2008;12(9):1109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Yan B, Van Waes C. The Role of the NF-kappaB Transcriptome and Proteome as Biomarkers in Human Head and Neck Squamous Cell Carcinomas. Biomark Med. 2008;2(4):409–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2(4):301–10. [DOI] [PubMed] [Google Scholar]
- 8.Allen C, Saigal K, Nottingham L, Arun P, Chen Z, Van Waes C. Bortezomib-induced apoptosis with limited clinical response is accompanied by inhibition of canonical but not alternative nuclear factor-{kappa}B subunits in head and neck cancer. Clin Cancer Res. 2008;14(13):4175–85. [DOI] [PubMed] [Google Scholar]
- 9.Duffey DC, Chen Z, Dong G, Ondrey FG, Wolf JS, Brown K, et al. Expression of a dominant-negative mutant inhibitor-kappaBalpha of nuclear factor-kappaB in human head and neck squamous cell carcinoma inhibits survival, proinflammatory cytokine expression, and tumor growth in vivo. Cancer Res. 1999;59(14):3468–74. [PubMed] [Google Scholar]
- 10.Yang X, Lu H, Yan B, Romano RA, Bian Y, Friedman J, et al. DeltaNp63 versatilely regulates a Broad NF-kappaB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011;71(10):3688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nottingham LK, Yan CH, Yang X, Si H, Coupar J, Bian Y, et al. Aberrant IKKalpha and IKKbeta cooperatively activate NF-kappaB and induce EGFR/AP1 signaling to promote survival and migration of head and neck cancer. Oncogene. 2014;33(9):1135–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das R, Coupar J, Clavijo PE, Saleh A, Cheng TF, Yang X, et al. Lymphotoxin-beta receptor-NIK signaling induces alternative RELB/NF-kappaB2 activation to promote metastatic gene expression and cell migration in head and neck cancer. Mol Carcinog. 2019;58(3):411–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas N Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc. 2011;6(9):1341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baspinar A, Cukuroglu E, Nussinov R, Keskin O, Gursoy A. PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014;42(Web Server issue):W285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167(2):397–404 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell.167(6):1540–54.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18(1):248–62. [DOI] [PubMed] [Google Scholar]
- 21.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33(Web Server issue):W331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45(D1):D369–D79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, et al. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck. 2010;32(4):417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H, Yang X, Si H, Saleh AD, Xiao W, Coupar J, et al. Genomic and Transcriptomic Characterization Links Cell Lines with Aggressive Head and Neck Cancers. Cell Rep. 2018;25(5):1332–45 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleh AD, Cornelius S, Cheng H, Martin S, Ormanoglu P, Yang X, et al. Abstract 3400: Integrated functional RNAi screening and structural genomics identifies inverse co-modulators of TP53 family and NF-kB transitional activation as potential therapeutic targets in head and neck squamous cell carcinoma. Cancer Research. 2014;74(19 Supplement):3400-. [Google Scholar]
- 31.Lu H, Yang X, Duggal P, Allen CT, Yan B, Cohen J, et al. TNF-alpha promotes c-REL/DeltaNp63alpha interaction and TAp73 dissociation from key genes that mediate growth arrest and apoptosis in head and neck cancer. Cancer Res. 2011;71(21):6867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen CT, Clavijo PE, Van Waes C, Chen Z. Anti-Tumor Immunity in Head and Neck Cancer: Understanding the Evidence, How Tumors Escape and Immunotherapeutic Approaches. Cancers. 2015;7(4):2397–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis RJ, Ferris RL, Schmitt NC. Costimulatory and coinhibitory immune checkpoint receptors in head and neck cancer: unleashing immune responses through therapeutic combinations. Cancers of the Head & Neck. 2016;1(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen C, Duffy S, Teknos T, Islam M, Chen Z, Albert PS, et al. Nuclear factor-kappaB-related serum factors as longitudinal biomarkers of response and survival in advanced oropharyngeal carcinoma. Clin Cancer Res. 2007;13(11):3182–90. [DOI] [PubMed] [Google Scholar]
- 35.Van Waes C, Yu M, Nottingham L, Karin M. Inhibitor-kappaB kinase in tumor promotion and suppression during progression of squamous cell carcinoma. Clin Cancer Res. 2007;13(17):4956–9. [DOI] [PubMed] [Google Scholar]
- 36.Noorlag R, van Kempen PM, Stegeman I, Koole R, van Es RJ, Willems SM. The diagnostic value of 11q13 amplification and protein expression in the detection of nodal metastasis from oral squamous cell carcinoma: a systematic review and meta-analysis. Virchows Arch. 2015;466(4):363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eytan DF, Snow GE, Carlson S, Derakhshan A, Saleh A, Schiltz S, et al. SMAC mimetic birinapant plus radiation eradicates human head and neck cancers with genomic amplifications of cell death genes FADD and BIRC2. Cancer Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11(8):781–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Chen T, Yang X, Cheng H, Spath SS, Clavijo PE, et al. Attenuated TRAF3 Fosters Activation of Alternative NF-kappaB and Reduced Expression of Antiviral Interferon, TP53, and RB to Promote HPV-Positive Head and Neck Cancers. Cancer Res. 2018;78(16):4613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao R, An Y, Ye W, Derakhshan A, Cheng H, Yang X, et al. Dual Antagonist of cIAP/XIAP ASTX660 Sensitizes HPV(−) and HPV(+) Head and Neck Cancers To TNFalpha, TRAIL, and Radiation Therapy. Clin Cancer Res. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao R, Allen CT, Tran L, Patel P, Park SJ, Chen Z, et al. Antagonist of cIAP1/2 and XIAP enhances anti-tumor immunity when combined with radiation and PD-1 blockade in a syngeneic model of head and neck cancer. Oncoimmunology. 2018;7(9):e1471440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An Y, Sun L, Derakhshan A, Carlson S, Das R, Chen Z, et al. Abstract 1061: Modulation of death pathways by TRAILR2 agonist antibody and NF-κB pathway by NIK inhibitor in HPV-positive head and neck squamous cell carcinomas. Cancer Research. 2017;77(13 Supplement):1061-. [Google Scholar]
- 43.Brenner D, Blaser H, Mak TW. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol. 2015;15(6):362–74. [DOI] [PubMed] [Google Scholar]
- 44.Sedger LM, McDermott MF. TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants – past, present and future. Cytokine & Growth Factor Reviews. 2014;25(4):453–72. [DOI] [PubMed] [Google Scholar]
- 45.Bjordahl RL, Steidl C, Gascoyne RD, Ware CF. Lymphotoxin network pathways shape the tumor microenvironment. Curr Opin Immunol. 2013;25(2):222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun SC. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21(1):71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao M, Joy J, Zhou W, De S, Wood WH 3rd, Becker KG, et al. Transcriptional outcomes and kinetic patterning of gene expression in response to NF-kappaB activation. PLoS Biol. 2018;16(9):e2006347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yilmaz ZB, Kofahl B, Beaudette P, Baum K, Ipenberg I, Weih F, et al. Quantitative dissection and modeling of the NF-kappaB p100-p105 module reveals interdependent precursor proteolysis. Cell Rep. 2014;9(5):1756–69. [DOI] [PubMed] [Google Scholar]
- 49.Razani B, Reichardt AD, Cheng G. Non-canonical NF-kappaB signaling activation and regulation: principles and perspectives. Immunol Rev. 2011;244(1):44–54. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi N, Oyama M, Kozuka-Hata H, Inoue J. Involvement of A20 in the molecular switch that activates the non-canonical NF-small ka, CyrillicB pathway. Sci Rep. 2013;3:2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. Classical and/or alternative NF-kappaB pathway activation in multiple myeloma. Blood. 2010;115(17):3541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bista P, Zeng W, Ryan S, Bailly V, Browning JL, Lukashev ME. TRAF3 controls activation of the canonical and alternative NFkappaB by the lymphotoxin beta receptor. J Biol Chem. 2010;285(17):12971–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandal R, Senbabaoglu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1(17):e89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, et al. Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas. Cell Rep. 2018;23(1):194–212 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.