Abstract
Ethical and safety issues have rendered mesenchymal stem cells (MSCs) popular candidates in regenerative medicine, but their therapeutic capacity is lower than that of induced pluripotent stem cells (iPSCs). This study compared original, dental tissue-derived MSCs with re-differentiated MSCs from iPSCs (iPS-MSCs). CD marker expression in iPS-MSCs was similar to original MSCs. iPS-MSCs expressed higher in pluripotent genes, but lower levels in mesodermal genes than MSCs. In addition, iPS-MSCs did not form teratomas. All iPSCs carried mtDNA mutations; some shared with original MSCs and others not previously detected therein. Shared mutations were synonymous, while novel mutations were non-synonymous or located on RNA-encoding genes. iPS-MSCs also harbored mtDNA mutations transmitted from iPSCs. Selected iPS-MSCs displayed lower mitochondrial respiration than original MSCs. In conclusion, screening for mtDNA mutations in iPSC lines for iPS-MSCs can identify mutation-free cell lines for therapeutic applications.
Keywords: hiPSCs, iPSC-derived MSCs, Mitochondrial DNA, Mutations, Regenerative medicine
INTRODUCTION
Mesenchymal stem cells (MSCs) are being explored in a large number of clinical trials due to advantages such as multi-differentiation potency, anti-inflammatory properties, modulation of the host immune system, and safety for allogeneic cell transplants (1, 2). However, populations of MSCs from individuals are limited by a decreasing proliferation potential when propagated for long periods in vitro (3). Because iPSCs are highly expandable in vitro and can be differentiated into almost any cell type, these cells are frequently discussed in regenerative medicine (1, 4). As such, iPSC-derived MSCs (iPS-MSCs) may replace MSCs in stem cell therapies (5). Several investigations have shown that iPS-MSCs were comparable to bone marrow (BM)-MSCs in surface marker expression, differentiation potential, and gene expression profile (6, 7). iPS-MSCs also showed greater regenerative potential, likely due to increased telomerase activity and less senescence than MSCs, leading to superior engraftment and survival after transplantation (7).
Genetic and epigenetic abnormalities in many iPSCs and iPSC-derived MSCs, however, limit their therapeutic use (8). The genomes of iPSCs can contain many anomalies, including aneuploidy, subchromosomal copy number variations, and single nucleotide variations (9–11). Epigenetic variations in iPSCs can be due to incomplete reprogramming or prolonged culture, affecting their ability to differentiate (12). X-chromosome inactivation is reported to vary among iPSCs and can include loss of Xist expression and other repressive chromatin modifications (13).
Mitochondria are the cellular power plants responsible for ATP production (14, 15). The frequency of mitochondrial DNA (mtDNA) mutations is believed to be at least 10- to 20-fold higher than the frequency of nuclear DNA mutations (16). Individual iPSC clones present with mtDNA mutations transmitted from original blood or fibroblasts, resulting in functional abnormalities (17, 18).
This study details the establishment of iPS-MSCs from iPSCs derived from dental tissue MSCs and compares the characteristics and mtDNA instability of MSCs versus iPS-MSCs. MtDNA copy number and mutations were analyzed, and mitochondrial function were compared between iPS-MSCs and original MSCs to evaluate mitochondrial function therein (Fig. 1A). Although many characteristics of iPS-MSCs are reported to be similar to those of MSCs, the nature of these characteristics remains unclear. One study reported differential expression patterns of mesenchymal and pluripotency genes between iPS-MSCs and MSCs and found that iPS-MSCs were less responsive to differentiation in the mesenchymal lineage (19).
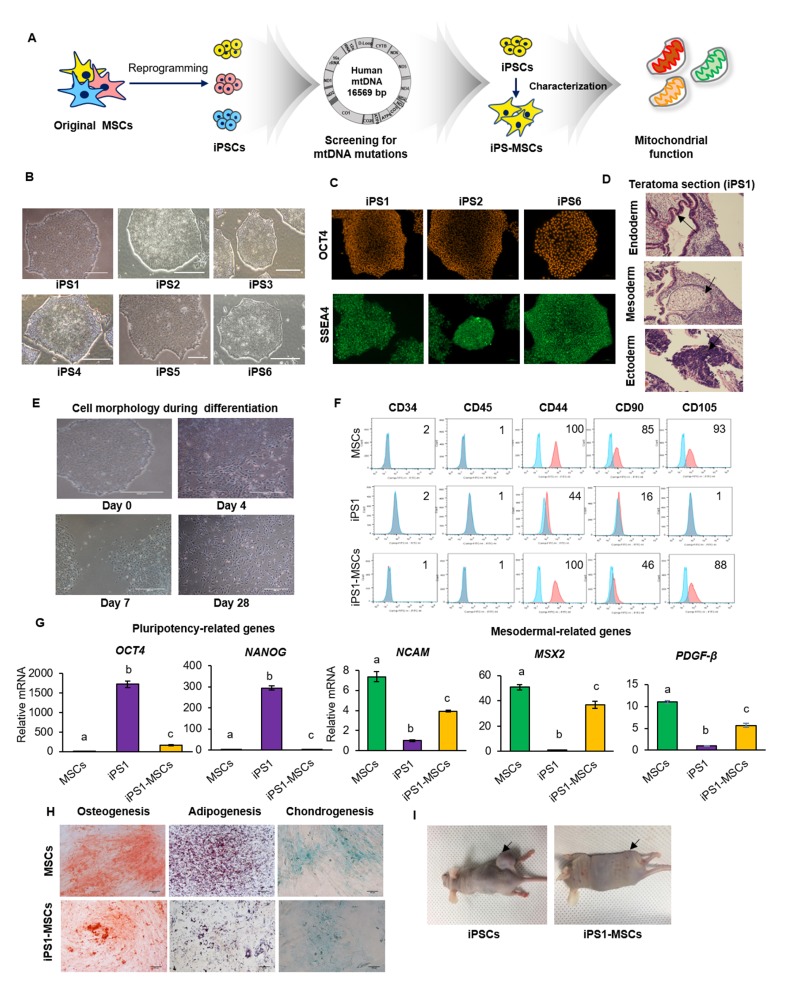
Fig. 1.
Characterization of iPS-MSCs and original MSCs. (A) Experimental design of the study. (B) Morphology of all iPSC lines similar to normal PSC morphology. (C) Characterizations of randomly selected iPSCs. OCT4 and SSEA4 were expressed in iPSC1, 2 and 6. (D) The teratoma formed in the mouse injected with iPSC1. Black arrows indicate three germ layers contained in teratoma. Scale bars = 500 μm. (E) Change in cell morphology to a spindle-like shape during differentiation of iPSC1 to MSCs. Scale bars = 500 μm. (F) Expression of CD markers in MSCs, iPSC1, and iPS1-MSCs. Both of MSCs and iPS1-MSCs were 100% positive in CD44. iPSC1 showed reduced expression of MSC positive markers. Negative MSCs markers, including CD34 and CD45, were expressed at less than 2% in all cell types. (G) Expression of pluripotency and mesodermal related genes in MSCs, iPSC1, and iPS1-MSCs. The level of the pluripotent gene OCT4 was higher in iPS1-MSCs than MSCs, while expression levels of the mesodermal genes NCAM, MSX2, and PDGF-β were lower in iPSC1 than MSCs. (H) Differentiation of MSCs and iPS1-MSCs into adipogenic, osteogenic, and chondrogenic lineages. (I) Teratoma formation by iPSC1 and iPS1-MSCs. No teratomas were observed in the mice injected with iPS1-MSCs. Black arrows indicate teratoma injection sites. The letters a, b, and c indicate significant (P < 0.05) differences among MSCs, iPSCs, and iPS-MSCs. Mean ± SEM. MSCs, mesenchymal stem cells; iPSCs, induced pluripotent stem cells; iPS-MSCs, iPSC-derived MSCs; NCAM, neural cell adhesion molecule; MSX2, Msh homeobox 2; PDGFβ, platelet-derived growth factor subunit B.
RESULTS AND DISCUSSION
Differentiation of dental tissue-derived iPSCs toward MSCs
A total of 6 iPSC lines were generated from dental tissue-derived MSCs, and their pluripotency was characterized (8). All iPSC lines were showed normal PSC morphology (Fig. 1B). Random selected three iPSC lines (iPSC 1, iPSC 2 or iPSC 6) were showed positive expression of OCT4 and SSEA4 (Fig. 1C). Additionally, the iPSC1 line was formed teratoma containing all three germ layers (Fig. 1D).
The selected iPSC1 line was used to test the reported differentiation protocol from iPSCs to MSCs (5–7). The morphology of iPSCs derived from dental-MSCs during differentiation was analyzed to determine whether the shapes formed were similar to those of MSCs. Cells at the edges of iPSC colonies began to change shape on day 7. These cells demonstrated a spindle-like morphology, similar to that of MSCs (Fig. 1E) (20). After 3 more weeks of culture, almost all iPS-MSCs presented with an MSC-like morphology (Fig. 1E). Next, MSC-specific surface markers (CD markers), including CD44, CD90, and CD105 (21), were assayed. Original MSCs were expressed 100%, 85% and 93%, while iPS1-MSCs were 100%, 46%, and 88% for CD44, CD90, and CD105, respectively. It was reported that reduced expression of CD90 in MSCs enhances their ability to differentiate into MSC lineages (22), therefore, reduced CD90 expression in iPS-MSCs in this study did not adversely affect their ability to differentiate. All cell lines were negative for hematopoietic cell markers CD34 and CD45 (Fig. 1F).
Analysis of the expression of pluripotent genes OCT4 and NANOG revealed that their expression was significantly increased in iPSCs over both MSCs and iPS-MSCs (Fig. 1G). The expression of NANOG was similar in MSCs and iPS-MSCs, whereas the expression of OCT4 was significantly increased in iPS-MSCs over MSCs, suggesting greater proliferation and differentiation potential as well as inhibition of spontaneous differentiation (23). The levels of expression of mesodermal genes neural cell adhesion molecule (NCAM), Msh homeobox 2 (MSX2), and platelet-derived growth factor subunit B (PDGF-β) (24, 25), were found to be significantly lower in iPSCs than in MSCs, with those in iPS-MSCs being intermediate. Although expression of mesodermal genes was reduced in iPS-MSCs over MSCs (19, 26), we considered this difference within acceptable parameters for MSC differentiation and focused on the enhanced expression of these genes in iPS-MSCs compared to iPSCs.
Original MSCs and iPS-MSCs were differentiated into adipocytes, osteocytes, or chondrocytes to investigate their ability to differentiate toward mesenchymal lineages (Fig. 1H). Differentiated cells were positive for lineage-specific markers, showing cytoplasmic accumulation of lipid vacuoles, deposition of the calcified extracellular matrix, and formation of proteoglycan. Although we expected iPS-MSCs to show higher differentiation potency due to their increased expression of OCT4 (27), iPS-MSCs did not have greater mesenchymal differentiation ability than MSCs.
To confirm that iPS-MSCs did not have characteristics of pluripotent stem cells (28), iPS-MSCs were injected into SCID mice, and teratoma formation was assessed (Fig. 1I). Injection of iPSCs induced teratoma formation in two of three SCID mice. However, iPS-MSCs did not form teratomas in all injected mice, indicating these cells had lost characteristics of pluripotent stem cells after differentiation.
These results demonstrated that iPS-MSCs showed similar morphology and characteristics to the original MSCs. In particular, CD44 can be used as a specific marker for MSC differentiation (21).
mtDNA mutations and copy number following iPSC reprogramming
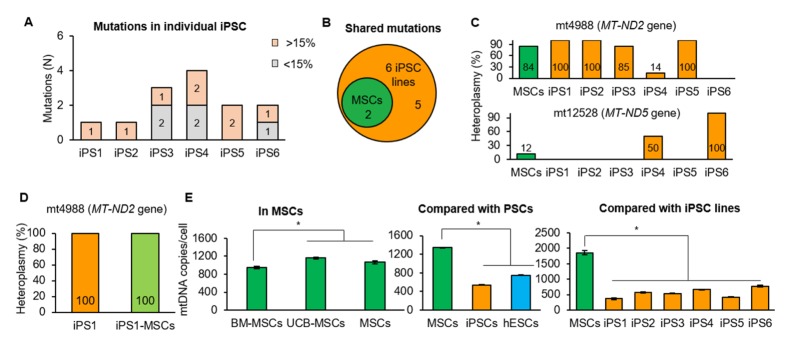
The mtDNA integrity of iPSCs is an important consideration for therapeutic applications (17). mtDNA mutations were therefore screened in MSCs and individual iPSC lines using Illumina MiSeq sequencing. After selecting variants with at least 2% heteroplasmy levels (the relative levels of mutant to wild-type mtDNA), two mutations were identified, mt4988C>T in mitochondrially encoded NADH dehydrogenase 2 (MT-ND2) gene with 84% heteroplasmy and mt12528G>A in mitochondrially encoded NADH dehydrogenase 5 (MT-ND5) gene with 12% heteroplasmy. Six iPSC lines were generated from MSCs, and seven mutations were identified therein (Fig. 2A and B, Supplementary Table 1). All iPSC lines included mtDNA mutations with at least 15% heteroplasmy (Fig. 2A), with each iPSC line harboring one to four mutations. Of the seven mutations detected in iPSC lines, two were transmitted from MSCs and detected in several lines (Fig. 2B). mt4988C>T was detected in five iPSC lines with 14% heteroplasmy to homoplasmy (100% mutant mtDNA), and mt12528G>A was detected in two iPSC lines with 50% heteroplasmy and homoplasmy (Fig. 2C). Among the five mutations in iPSC lines not detected in the original MSCs, one was shared by two iPSC lines but varied in heteroplasmy level. The remaining four mutations were unique and found in only one cell line (Supplementary Table 1). Based on our previous report, the novel mutations, detected only iPSC lines, were transmitted from original MSCs, not generated by reprogramming (17). These results were consistent with previous findings, suggesting the need to screen multiple iPSC lines for mutations, as mutation-free cell lines are required for clinical purposes (17).
Fig. 2.
Mitochondrial genome integrity. (A) All iPSC clones harbored meaningful heteroplasmic (> 15%) mutations. (B) Venn diagram showing two mutations of MSCs shared by iPSC clones. (C) Mutations shared by MSCs and iPSC lines. The mt4988C>T in MSCs was present in five iPSC lines with various heteroplasmy, and the mt12528G>A in MSCs were present in two iPSC lines with 50% heteroplasmy and homoplasmy. (D) Stable mtDNA mutation during differentiation into MSCs. One homoplasmic mutation, detected in iPSC1 line, was transmitted to iPS1-MSCs. (E) mtDNA copy number in iPSCs was similar to hESCs. mtDNA copy number of original MSCs was similar in UCM-MSCs and higher than that in BM-MSCs. mtDNA copy number was similar between iPSCs and hESCs. Asterisks indicate statistically significant differences (*P < 0.05). Mean ± SEM. mtDNA, mitochondrial DNA; UCM-MSCs, umbilical cord matrix MSCs; BM-MSCs, bone marrow MSCs; hESCs, human embryonic stem cells.
mtDNA copy number can change during reprogramming. Theoretically, mtDNA copy number of iPSCs could be reduced compared to the original fibroblasts (29), and we compared mtDNA copy number in various MSCs. BM-MSCs showed a significantly lower mtDNA copy number than umbilical cord matrix (UCM)-MSCs or dental-MSCs, whereas UCM-MSCs and dental-MSCs had similar numbers (Fig. 2E). When we compared MSCs and iPSCs, we found that copy number was significantly lower in iPSCs (P < 0.05, Fig. 2D) and that the average mtDNA copy number of six iPSC lines was comparable to that of human embryonic stem cells (hESCs) (Fig. 2E), suggesting mtDNA copy number may be a physical indicator for reprogramming or differentiation.
Lower mitochondrial respiration in iPS-MSCs
Our results showed that all established iPSC lines harbored mtDNA mutations with over 50% heteroplasmy, which could induce mitochondrial dysfunction (Fig. 2C) (17). We further analyzed iPS-MSCs to determine whether mtDNA mutations were transmitted from iPSCs or arose randomly during differentiation. One homoplasmic mutation (mt4988C>T in MT-ND2 gene) was detected in iPS1-MSCs derived from an iPSC1 line, which carried the same mutation with iPSC1 (Fig. 2C and D, Supplementary Table 1). No novel mutations were detected in iPS1-MSCs, indicating that mtDNA mutations were stable during differentiation (17).
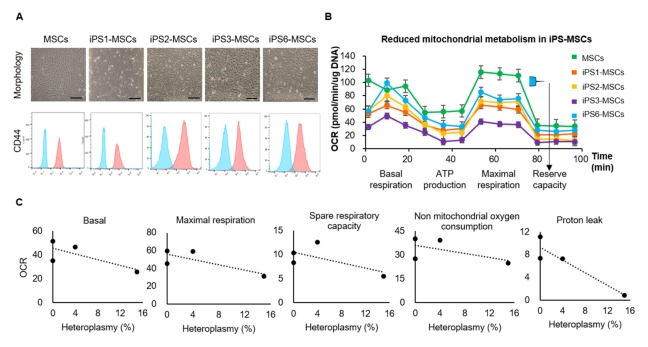
We further evaluated mitochondrial respiration in iPS-MSCs to determine whether the cells displayed adequate metabolic function compared to original MSCs. We performed differentiation of all 6 iPSC lines into MSCs; of these, iPSC5 and iPSC4 went extinct during iPSC maintenance and differentiation, respectively. The highly heteroplasmic non-synonymous mutation in iPSC 5 and rRNA mutation in iPSC4 likely affected cell survival. Differentiated iPS-MSC lines were similar to original MSCs morphologically and showed positive CD44 expression (Fig. 3A). We then measured oxygen consumption rates (OCR) for mitochondrial respiration in iPS1-MSCs, iPS2-MSCs, iPS3-MSCs, and iPS6-MSCs by Seashore platform. The OCR tended to be low in all four iPS-MSC lines compared to original MSCs (Fig. 3B). These results indicate original MSCs could produce more ATP and overcome more oxidative stress than iPS-MSCs (30).
Fig. 3.
Mitochondrial respiration was lower in iPS-MSCs than in MSCs. (A) Cell morphology and expression of CD44 in iPS-MSC lines. iPS-MSC lines were similar to original MSCs morphologically and displayed positive CD44 expression. The results on expression of CD44 in MSCs and iPS1-MSCs was duplicated in Fig. 1F. Scale bars = 250 μm. (B) Lower OCR levels in iPS-MSC lines than in MSCs. Mean ± SM. (C) The correlation of lower OCR to higher heteroplasmy in non-synonymous or RNA gene mutations. Heteroplasmy percentages refer to the sum of heteroplasmy of non-synonymous and RNA gene mutations in each cell line. OCR, oxygen consumption rates.
Although iPS1-MSCs and iPS2-MSCs were harbor fewer mutations, these mutations showed higher heteroplasmy, which could reduce OCR (17). Synonymous mutations can also affect post-transcriptional processing and translation (31), therefore, synonymous mutations in iPS1-MSCs could affect cell metabolism. The iPS3-MSCs, being expected to harbor 2 RNA gene mutations, showed the lowest OCR among all iPS-MSC lines (Fig. 3B). We also analyzed the correlation between OCR and heteroplasmy of non-synonymous or RNA gene mutations and found higher heteroplasmy of non-synonymous or RNA gene mutations resulted in lower OCR in iPS-MSC lines (Fig. 3C).
Although we could not eliminate all nuclear factors, we found that iPS-MSCs derived from single cloned iPSCs with specific mtDNA mutations demonstrated dysfunctional mitochondrial respiration, suggesting limited therapeutic potential and underscoring the need to screen for mutation-free cell lines.
CONCLUSIONS
In general, the characteristics of iPS-MSCs and MSCs were similar, including cell morphology, expression of MSC-specific CD markers and mesodermal genes, and potency of differentiation into mesenchymal lineages. However, expression levels of CD90 and mesodermal genes were lower, and the expression level of OCT4 was higher in iPS-MSCs than MSCs. These findings may be associated with incomplete differentiation due to the inadequate expression of reprogramming factors. Fortunately, iPS-MSCs did not form teratomas, reducing the potential for tumor formation following cell transplantation.
The two mtDNA mutations in MSCs were transmitted to several iPSC lines with various heteroplasmy. Some iPSC lines displayed novel mtDNA mutations not detected in MSCs. mtDNA mutations in iPSCs did not change during differentiation to iPS-MSCs, and iPS-MSCs showed no novel mutations. However, iPS-MSCs demonstrated reduced mitochondrial metabolism and lower OCR associated with higher heteroplasmy of non-synonymous or RNA gene mutations.
Taken together, these results were consistent with a previous study (17), confirming the need to screen multiple iPSC lines to detect mutation-free lines for clinical purposes.
MATERIALS AND METHODS
iPSC reprogramming
iPSCs were generated from MSCs using an Epi5TM Episomal iPSC Reprogramming Kit (Thermo Fisher) according to manufacturer’s instructions. Briefly, plasmid mixtures were electroporated into dental-MSCs with a Neon Transfection System. Transfected cells were seeded onto Geltrex-coated plates (Thermo Fisher). After 3 weeks, colonies were selected and expanded in feeder-free TeSR-E8 medium (Stemcell Technologies).
Immunocytochemistry
Fixed iPSCs were permeabilized with 0.05% Triton X-100 for 30 minutes at room temperature (RT). Cells were incubated with primary antibodies OCT4 and SSEA4 (1:100, Stemgent) during an overnight at 4°C. Cells were washed twice and incubated with secondary goat anti-mouse IgG H&L (1:700, Alexa Fluor 488, Abcam), or goat anti-rabbit IgG H&L (1:700, Alexa Fluor 555, Abcam) antibodies for 1 h at RT.
Differentiation into MSCs from iPSCs
Differentiation of iPSCs toward MSCs was induced as described, with minor modifications (24). Briefly, cells were cultured in ADMEM medium supplemented with 10 μM SB-431542 and 1 ng/μl basic fibroblast growth factor (bFGF) for 7 to 10 days. After 10 days, cells were cultured with ADMEM supplemented with bFGF (5 ng/μl).
Differentiation into mesenchymal lineage
Cells were differentiated into mesenchymal lineage as described previously (21). Adipogenesis was induced by DMEM supplemented with 10% FBS, 100 μM indomethacin, 10 μM insulin, and 1 μM dexamethasone. Osteogenesis was induced by DMEM supplemented with 10% FBS, 200 μM ascorbic acid, 10 mM β-glycerophosphate, and 0.1 μM dexamethasone. For chondrogenesis, cells were induced by STEMPRO® Chondrogenesis Differentiation Kit media with 10% supplement.
RT-qPCR analysis
RNA was extracted using easy-spinTM Total RNA Extraction Kits (iNtRON), and cDNA was synthesized using HisenScriptTM RH(−) RT Premix Kits (iNtRON) according to the manufacturer’s instructions. qRT-PCR was performed using RealMODTM Green AP 5X qPCR mix (iNtRON) according to the qRT-PCR program in the manufacturer’s protocol with minor modifications.
Cell surface marker analysis
Fixed cells were incubated with fluorescein isothiocyanate (FITC) conjugated mouse anti-CD44, CD45, and CD90 (1:100, BD PharmingenTM) at 4°C for 1 hour, or incubated with unconjugated CD34 and CD105 primary antibodies (1:100, BD PharmingenTM) at 4°C for 1 hour followed by incubation with FITC conjugated secondary antibodies at 4°C for 1 hour. Labeled cells were counted by BD FACS Calibur.
Teratoma formation
Approximately 1 × 106 cells were suspended in 30% Matrigel in DMEM/F12. Male SCID mice (6 weeks old) were anesthetized with 30 mg/kg Zoletil (Virbac) and 10 mg/kg Rompun (Bayer), followed by the injection into the right femoral region. Six to eight weeks after injection, mice were euthanized, and teratomas were sectioned and histologically analyzed with hematoxylin and eosin (H&E) staining.
Quantification of mitochondrial copy number
mtDNA copy number was determined by a probe-based, single-tube multiplex qPCR assay on the QuantStudio 6 Flex Real-Time PCR System (Life Technologies), as previously described with minor modifications (32). mtDNA copy number was calculated as the ratio of a number of mitochondria to the number of nuclei.
mtDNA sequencing by MiSeq
The whole mtDNA was amplified using two primer sets as previously described (18): mt3163F-GCCTTCCCCCGTAAAT GATA and mt11599R-TGTTTGTCGTAGGCAGATGG; and mt11506F-TCTCAACCCCCTGACAAAAC and mt3259R-TATG CGATTACCGGGCTCT. The DNA was used for library preparation with the Nextera XT DNA Kit (Illumina). Sequencing was performed on the Illumina MiSeq platform, and the data were analyzed using NextGENe software. The remaining process of mtDNA sequencing was as described previously (17).
Seahorse assay
Mitochondrial respiratory function was measured using an XF Cell MitoStress Test Kit in an XF24 Extracellular Flux Analyzer (Seahorse Biosciences), as described previously (17). Mitochondrial OCR was measured by serial treatment with oligomycin (1.5 μM) for ATP production (oligomycin OCR – basal OCR), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP; 1 μM) for maximal respiration and reserve capacity (maximal OCR – basal OCR), and antimycin A (0.5 μM) and rotenone (0.5 μM) for non-mitochondrial oxygen utilization. Oxygen consumption was normalized to baseline oxygen consumption by measuring cell DNA.
Ethics approval and consent to participate
Human dental tissues were obtained from wisdom teeth extracted from patients at the Department of Oral and Maxillofacial Surgery at Gyeongsang National University Hospital under approved medical guidelines (GNUH IRB-2012-09-004). All subjects were provided written informed consent.
Statistical analysis
Data were presented as mean ± standard deviation (SD) or ± standard error of the mean (SEM). Results were compared by independent-group t-tests for two groups or by ANOVA with Tukey’s test for multiple comparisons. Statistical analyses were performed using GraphPad Prism software.
Supplementary Information
ACKNOWLEDGEMENTS
This study was supported by a grant from the National Research Foundation of Korea, funded by the Ministry of Education, Science, and Technology (2015K1A4A3046807), and by grants from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea (2018-755).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Alan Trounson CM. Stem cell therapies in clinical trials: progress and challenges. Cell Stem Cell. 2015;17:11–22. doi: 10.1016/j.stem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Moussavou G, Kwak DH, Lim MU, et al. Role of gangliosides in the differentiation of human mesenchymal-derived stem cells into osteoblasts and neuronal cells. BMB Rep. 2013;46:527–532. doi: 10.5483/BMBRep.2013.46.11.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Q, Gregory CA, Lee RH, et al. MSCs derived from iPSCs with a modified protocol are tumor-tropic but have much less potential to promote tumors than bone marrow MSCs. Proc Natl Acad Sci U S A. 2015;112:530–535. doi: 10.1073/pnas.1423008112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim C. iPSC technology--Powerful hand for disease modeling and therapeutic screen. BMB Rep. 2015;48:256–265. doi: 10.5483/BMBRep.2015.48.5.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao C, Ikeya M. Generation and applications of induced pluripotent stem cell-derived mesenchymal stem cells. Stem Cells Int. 2018;2018;9601623 doi: 10.1155/2018/9601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuta M, Nakai Y, Kirino K, et al. Derivation of mesenchymal stromal cells from pluripotent stem cells through a neural crest lineage using small molecule compounds with defined media. PLoS One. 2014;9:e112291. doi: 10.1371/journal.pone.0112291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheyn D, Ben-David S, Shapiro G, et al. Human induced pluripotent stem cells differentiate into functional mesenchymal stem cells and repair bone defects. Stem Cells Transl Med. 2016;5:1447–1460. doi: 10.5966/sctm.2015-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamada M, Malureanu LA, Wijshake T, Zhou W, van Deursen JM. Reprogramming to pluripotency can conceal somatic cell chromosomal instability. PLoS Genet. 2012;8:e1002913. doi: 10.1371/journal.pgen.1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussein SM, Batada NN, Vuoristo S, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 11.Gore A, Li Z, Fung HL, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Anguera MC, Sadreyev R, Zhang Z, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Son G, Han J. Roles of mitochondria in neuronal development. BMB Rep. 2018;51:549–556. doi: 10.5483/BMBRep.2018.51.11.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KM, Noh JH, Abdelmohsen K, Gorospe M. Mitochondrial noncoding RNA transport. BMB Rep. 2017;50:164–174. doi: 10.5483/BMBRep.2017.50.4.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace D. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci U S A. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang E, Wang X, Tippner-Hedges R, et al. Age-related accumulation of somatic mitochondrial DNA mutations in adult-derived human iPSCs. Cell Stem Cell. 2016;18:625–636. doi: 10.1016/j.stem.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Ma H, Folmes CD, Wu J, et al. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature. 2015;524:234–238. doi: 10.1038/nature14546. [DOI] [PubMed] [Google Scholar]
- 19.Diederichs S, Tuan RS. Functional comparison of human-induced pluripotent stem cell-derived mesenchymal cells and bone marrow-derived mesenchymal stromal cells from the same donor. Stem Cells Dev. 2014;23:1594–1610. doi: 10.1089/scd.2013.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 21.Park BW, Kang EJ, Byun JH, et al. In vitro and in vivo osteogenesis of human mesenchymal stem cells derived from skin, bone marrow and dental follicle tissues. Differentiation. 2012;83:249–259. doi: 10.1016/j.diff.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Moraes DA, Sibov TT, Pavon LF, et al. A reduction in CD90 (THY-1) expression results in increased differentiation of mesenchymal stromal cells. Stem Cell Res Ther. 2016;7 doi: 10.1186/s13287-016-0359-3. 97-016-0359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai C, Su P, Huang Y, Yew T, Hung S. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol cell. 2012;47:169–182. doi: 10.1016/j.molcel.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Chen YS, Pelekanos RA, Ellis RL, Horne R, Wolvetang EJ, Fisk NM. Small molecule mesengenic induction of human induced pluripotent stem cells to generate mesenchymal stem/stromal cells. Stem Cells Transl Med. 2012;1:83–95. doi: 10.5966/sctm.2011-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eto S, Goto M, Soga M, et al. Mesenchymal stem cells derived from human iPS cells via mesoderm and neuroepithelium have different features and therapeutic potentials. PLoS One. 2018;13:e0200790. doi: 10.1371/journal.pone.0200790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frobel J, Hemeda H, Lenz M, et al. Epigenetic rejuvenation of mesenchymal stromal cells derived from induced pluripotent stem cells. Stem Cell Reports. 2014;3:414–422. doi: 10.1016/j.stemcr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han SM, Han SH, Coh YR, et al. Enhanced proliferation and differentiation of Oct4- and Sox2-overexpressing human adipose tissue mesenchymal stem cells. Exp Mol Med. 2014;46:e101. doi: 10.1038/emm.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 30.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quax TE, Claassens NJ, Soll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell. 2015;59:149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips NR, Sprouse ML, Roby RK. Simultaneous quantification of mitochondrial DNA copy number and deletion ratio: a multiplex real-time PCR assay. Sci Rep. 2014;4:3887. doi: 10.1038/srep03887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.