Abstract
The bacterial effector protein RavZ is secreted by the intracellular pathogen Legionella pneumophila and inhibits host autophagy through an irreversible deconjugation of mammalian ATG8 (mATG8) proteins from autophagosome membranes. However, the roles of the LC3 interacting region (LIR) motifs in RavZ function remain unclear. In this study, we show that a membrane-targeting (MT) domain or the LIR motifs of RavZ play major or minor roles in RavZ function. A RavZ mutant that does not bind to mATG8 delipidated all forms of mATG8-phosphatidylethanolamine (PE) as efficiently as did wild-type RavZ. However, a RavZ mutant with a deletion of the MT domain selectively delipidated mATG8-PE less efficiently than did wild-type RavZ. Taken together, our results suggest that the effects of LIR motifs and the MT domain on RavZ activity are complementary and work through independent pathways.
Keywords: Autophagosome, LIR motif, MT domain, RavZ, Xenophagy
INTRODUCTION
Autophagy, or macroautophagy, is a cellular lysosomal degradation mechanism for breaking down cytosolic components, such as proteins, lipids, DNAs, RNAs, and organelles, to maintain cellular homeostasis under physiological conditions or different types of stress, such as starvation (1). The autophagy process is tightly regulated by many autophagy-related (ATG) proteins, including ATG8. In yeast, ATG8 is a small ubiquitin-like (Ubl) protein that is covalently conjugated to phosphatidylethanolamine (PE) in autophagosomes after proteolytic cleavage of its C-terminus by ATG4 cysteine protease, which is also involved in delipidation of ATG8-PE to release ATG8 from autophagosomes (2, 3). Although there is only one ATG8 protein in yeast, mammalian ATG8 (mATG8) proteins belong to one of two subgroups: a family of microtubule-associated protein light-chain 3 (LC3) proteins, LC3A, LC3B, and LC3C, and a family of γ-aminobutyric acid receptor-associated proteins (GABARAPs), GABARAP, GABARAP-L1, and GABARAP-L2 (2, 3). These proteins are also lipidated by PE conjugation to their C-terminal regions.
Since its discovery, autophagy has been considered to be a non-selective degradation pathway in the lysosome. However, recent evidence suggests that autophagy could be a selective process guided by specific autophagy receptors and adaptor proteins for substance selection, recruitment, and degradation. Selective autophagy has been classified into different categories, including mitophagy, pexophagy, aggrephagy, reticulophagy, and xenophagy (4, 5). Xenophagy is a host-cell immune response responsible for the removal of intracellular pathogens, such as cytoplasmic bacteria, viruses, and fungi (6). Intracellular pathogens are ubiquitinated and targeted for autophagic degradation via autophagy adaptors or autophagy receptors, such as p62, NDP52, or CALCOCO2, which commonly have LC3-interacting region (LIR) motifs and Ub-interacting domains, including UBA, UBZ, and ZnF (6, 7). However, many bacteria have evolved mechanisms to inhibit host autophagy. Legionella pneumophila can impair host autophagy via the bacterial effector protein, RavZ, which induces irreversible delipidation of mATG8-PE proteins on autophagosome membranes (8, 9). RavZ, therefore, functions as a cysteine protease similarly to the endogenous ATG4B in host cells. However, the two proteins differ in several ways (9–11). Mammalian ATG4B hydrolyzes the amide bond linking glycine and PE, whereas RavZ hydrolyzes the amide bond between the C-terminal glycine residue and an adjacent aromatic residue. Therefore, RavZ renders its target resistant to being re-conjugated to PE by the host machinery. Additionally, ATG4B cleaves both soluble and membrane-anchored mATG8 proteins, whereas RavZ cleaves only membrane-anchored mATG8 proteins. Therefore, unlike ATG4B, RavZ proteins delipidate mATG8-PE on autophagic membranes irreversibly. Moreover, ATG4B depletes mATG8 proteins more slowly than does RavZ.
Both ATG4 and RavZ contain LIR motifs, which are well-known consensus sequences in mATG8 proteins. Mammalian ATG4B contains a functional C-terminal LIR motif, which binds efficiently to mATG8 proteins and cleaves them (12). The C-terminal LIR motif of ATG4B is also involved in the depletion of mATG8 from the membrane (13). In yeast, the LIR motifs of ATG4 are involved in ATG8-PE binding and the depletion of ATG8-PE from the autophagosome membrane (14). ATG4 has two LIR motifs; one is the N-terminal LIR motif, APEAR (ATG8-PE association region), and the other is the C-terminal LIR, CLIR. The APEAR of ATG4 is involved in the binding and deletion of ATG8-PE on the autophagic membrane, whereas the CLIR participates in constitutive binding to ATG8 (14). The RavZ protein contains three LIR motifs: an N-terminal LIR1 with two motifs (LIR1/2), and a C-terminal LIR3 motif. RavZ binds to two LC3B proteins through its N-terminal and C-terminal LIR motifs, leading to autophagic membrane localization (15). Therefore, mutations in any of the LIR motifs prevent the delipidation of mATG8-PE proteins (15). Other studies report that the phosphatidylinositol 3-phosphate (PI3P) binding MT domain of RavZ plays an essential role in autophagic membrane targeting, and on autophagic membranes, the LIR2 motif of RavZ is involved in the initial recognition of LC3B-PE (10, 11). Therefore, the contribution of LIR motifs to autophagic membrane targeting and substrate recognition of RavZ is controversial.
In this study, we found that RavZ mutants with mutations in all LIR motifs retained the ability to delipidate of all forms of mATG8-PE proteins on autophagic membranes as efficiently as did wild-type RavZ. This process was mediated by the MT domain in an mATG8 binding-independent manner. We also discovered that a RavZ mutant with an MT domain deletion was still able to selectively delipidate mATG8-PE proteins on autophagic membranes. This activity was mediated by the LIR motifs in an mATG8 binding-dependent manner, but with less efficiency than that of wild-type RavZ. Together, the LIR motifs or the MT domain played minor or major roles in RavZ function in mATG8 binding-dependent and -independent manners, respectively.
RESULTS AND DISCUSSION
Contribution of the LIR motifs to the RavZ wild type function
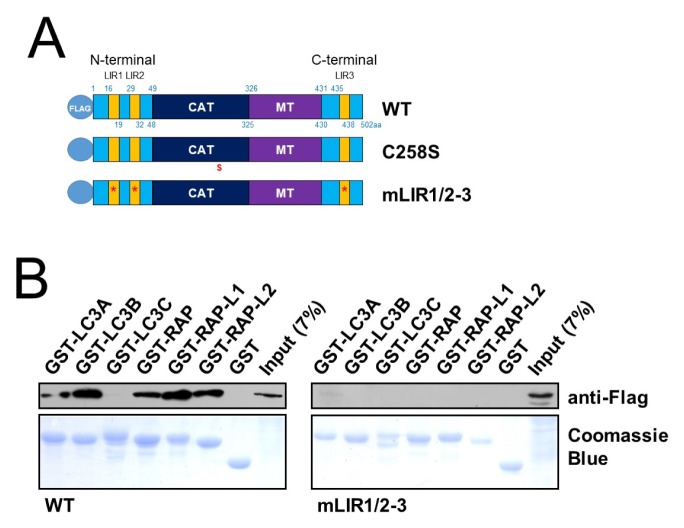
RavZ has previously been found to delipidate mATG8-PE on autophagic membranes in an LIR motif-dependent manner (15). The LIR2 motif is also involved in the initial recognition of LC3B-PE on autophagic membranes in vitro (11). In light of these reports, we expected that a RavZ mutant that could not bind to mATG8 would not delipidate mATG8-PE. First, we examined the contribution of each LIR motif to RavZ functionality. To do this, we did GST-pulldown assays to find out the mATG8 binding properties of the wild-type RavZ protein and RavZ proteins with LIR motif mutations (Fig. 1A). We fused a 3xFLAG motif to the full-length RavZ protein (3xFLAG-RavZ) and found that this protein binds to GST-LC3A, GST-LC3B, GST-GABARAP, GST-GABARAP-L1, and GSTGABARAP-L2, but not to GST-LC3C (Fig. 1B). However, 3xFLAG-RavZmLIR1/2–3, which has a point mutation in the consensus sequences (W/F/Y-X-X-I/L/V → A-X-X-A) in all three LIR motifs, does not bind to any of the mATG8 proteins (Fig. 1B). This result indicates that the intact RavZ protein can bind to LC3A/B and GABARAP/-L1/-L2, but mutations in the three LIR motifs of the RavZ protein abrogated its binding to mATG8 proteins.
Fig. 1.
Binding efficiency of the RavZ protein and RavZ mutants to mATG8 proteins in vitro. (A) Schematic diagram of 3xFLAG-RavZ protein and its mutants. LIR, LC3 interacting region; CAT, Catalytic domain; MT, Membrane targeting domain. WT, 3xFLAG-RavZ wild-type; C258S, 3xFLAG-RavZC258S, a catalytically inactive mutant; mLIR1/2–3, 3xFLAG-RavZmLIR1/2–3, a point mutation in the consensus sequences (W/F/Y-X-X-I/L/V → A-X-X-A) within all LIR motifs from 3xFLAG-RavZ. (B) mATG8 protein-binding property of 3xFLAG-RavZ using GST pulldown assays. 3xFLAG-RavZ binds to GST-LC3A/B and GST-GABARAP/-L1/L2 proteins, whereas 3xFLAG-RavZmLIR1/2–3 did not bind to any of the mATG8 proteins. Upper panel: anti-FLAG, Lower panel: Coomassie blue staining.
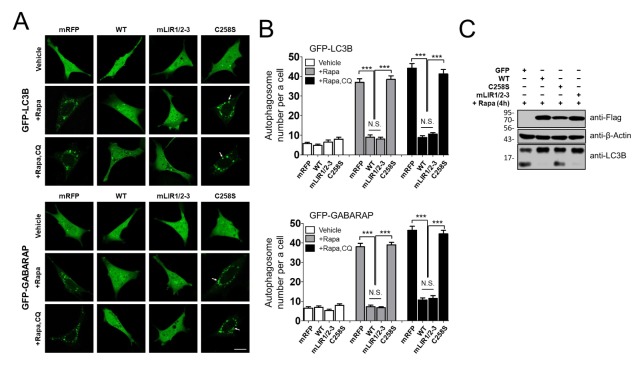
In mammals, mATG8 proteins belong to one of two subgroups, either the LC3 or GABARAP subfamily (2). Therefore, we investigated LC3B− and GABARAP-positive autophagic membranes as representatives of each subgroup by introducing either 3xFLAG-RavZ or RavZ LIR mutant proteins using the GFP-LC3B or GFP-GABARAP proteins. To exclude the possibility that the decrease of mATG8-II results from the rapid degradation of the autophagic membranes, we treated cells with rapamycin and CQ, a lysosome inhibitor. The expression of 3xFLAG-RavZ shows that there are few GFP-LC3B or GFP-GABARAP-positive autophagic membranes in rapamycin or rapamycin + CQ-treated cells compared to either monomeric red fluorescent protein (mRFP) or the 3xFLAG-RavZC258S catalytic mutant (Fig. 2A and 2B). However, overexpression of 3xFLAG-RavZmLIR1/2–3, the mutant that cannot bind mATG8, reduces the number of GFP-LC3B or GFP-GABARAP-positive autophagic membranes in rapamycin or rapamycin + CQ-treated cell, which is a response similar to that seen with 3xFLAG-RavZ expression (Fig. 2A and 2B). These results suggest that 3xFLAG-RavZmLIR1/2–3 may still delipidate GFP-LCB-PE and GFP-GABARAP-PE on autophagosome membranes independently of mATG8 binding. Overall, these results indicate that the binding of mATG8 proteins to RavZ LIR motifs might not be essential for the targeting of RavZ to autophagic membranes or the ability of RavZ to delipidate mATG8-PE on autophagic membranes.
Fig. 2.
Role of the RavZ LIR motifs on the delipidation of LC3B or GARARAP protein in rapamycin-induced autophagy. (A, B) Confocal images (A) showing cellular localization of GFP-LC3B or GFP-GABARAP, co-expressed with 3xFLAG-RavZ protein or the indicated RavZ mutants in MEFs upon autophagy induction (100 nM rapamycin (Rapa) + 50 μM CQ, 4 h). Arrows are representative of autophagic membranes. Scale bar: 10 μm. The bar graphs (B) illustrate the autophagosome spot numbers per cell (n = 20 for each group). 3xFLAG-RavZ and 3xFLAG-RavZmLIR1/2–3 reduce the number of GFP-LC3B or GFP-GABARAP-positive autophagic membranes in rapamycin or rapamycin + CQ-treated cells compared to either mRFP or the 3xFLAG-RavZC258S catalytic mutant. Data are presented as mean ± SEM. N.S., not significant. ***P < 0.001 (one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test). (C) Depletion of endogenous LC3B-II by RavZ protein and the indicated mutants using Western blot analysis in HEK293T cells. Representative Western blots of three independent experiments of endogenous LC3B in cells expressing 3xFLAG-RavZ protein or the indicated mutants in MEFs upon autophagy induction (100 nM rapamycin). 3xFLAG-RavZ or 3xFLAG-RavZmLIR1/2–3 reduces the LC3B-II levels more than does either GFP or 3xFLAG-RavZC258S.
To further investigate the contribution of LIR motifs to RavZ function, we measured the depletion of LC3B-PE in RavZ-expressing cells using Western blot analysis in rapamycin-treated cells HEK293T cells. Overexpression of 3xFLAG-RavZ or 3xFLAG-RavZmLIR1/2–3 reduces LC3B-II levels more than does either GFP or the 3xFLAG-RavZC258S catalytic mutant (Fig. 2C). Our results suggest that RavZ proteins can delipidate all forms of mATG8-PE on autophagic membranes during rapamycin-induced autophagy independently of mATG8 binding.
Contribution of the LIR motifs to the MT domain-deletion RavZ mutant
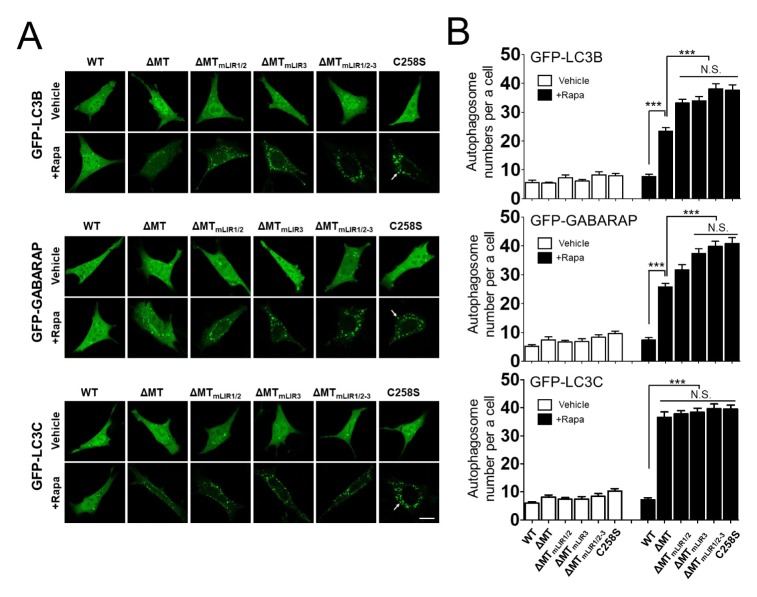
To find out how LIR motifs contribute to the functionality of RavZ, we generated an MT domain-deletion RavZ mutant, 3xFLAG-RavZ(ΔMT), to remove any compensatory effects of the MT domain. We then investigated LC3B− and GABARAP-positive autophagic membranes as well as LC3C-positive autophagic membranes, because the LIR motifs of RavZ do not bind to LC3C. 3xFLAG-RavZ(ΔMT) reduces the number of LC3B− and GABARAP-positive autophagosome numbers more than does 3xFLAG-RavZC258S, but is less effective than 3xFLAG-RavZ, whereas 3xFLAG-RavZ(ΔMT) does not reduce LC3C-positive autophagosome numbers more than does 3xFLAG-RavZC258S (Fig. 3A and 3B). In addition, the expression of 3xFLAG-RavZ(ΔMT)mLIR1/2–3, which both lacks an MT domain and has a point mutation in the consensus sequences (W/F/Y-X-X-I/L/V → A-X-X-A) within all three LIR motifs, does not reduce the number of LC3B-, GABARAP-, or LC3C-positive autophagic membranes, a response similar to that seen with mRFP or 3xFLAG-RavZC258S (Fig. 3A and 3B). These findings reveal that a RavZ protein without an MT domain can selectively delipidate mATG8-PE on autophagic membranes in an LIR motif-dependent manner, but less efficiently than wild-type RavZ does.
Fig. 3.
Role of the LIR motifs of RavZ(ΔMT), an MT domain-deletion mutant, on delipidation of LC3B or GARARAP protein in rapamycin-induced autophagy. (A) Confocal images showing cellular localization of GFP-LC3B, GFP-GABARAP, or GFP-LC3C co-expressed with 3xFLAG-RavZ(ΔMT) protein or the indicated mutants in MEFs upon autophagy induction (100 nM rapamycin). (B) The bar graphs illustrate the GFP-LC3B-, GFP-GABARAP-, or GFP-LC3C-positive autophagosome spot numbers per cell (n = 20 for each group). The expression of 3xFLAG-RavZ(ΔMT), but not 3xFLAG-RavZ(ΔMT)mLIR1/2–3, significantly reduces the number of GFP-LC3B− or GFP-GABARAP-positive autophagic membranes, whereas only the expression of 3xFLAG-RavZ, but not 3xFLAG-RavZ(ΔMT) or the indicated mutants, affects the number of GFP-LC3C-positive autophagic membranes in rapamycin or rapamycin + CQ-treated cells compared to either mRFP or the 3xFLAG-RavZC258S catalytic mutant. Data are presented as mean ± SEM. N.S. (not significant). *P < 0.05, ***P < 0.001 (one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test). WT, 3xFLAG-RavZ wild-type; C258S, 3xFLAG-RavZC258S, a catalytically inactive mutant; ΔMT, 3xFLAG-RavZ(ΔMT), a MT-domain deletion mutant; ΔMTmLIR1/2, 3xFLAG-RavZ(ΔMT)mLIR1/2, a point mutation in the LIR1 and 2 motifs in 3xFLAG-RavZ(ΔMT); ΔMTmLIR3, 3xFLAG-RavZ(ΔMT)mLIR3, a point mutation in the LIR3 motif in 3xFLAG-RavZ(ΔMT); ΔMTmLIR1/2–3, 3xFLAG-RavZ(ΔMT)LIRm1/2–3, a point mutation in all LIR motifs.
The LIR2 motif plays a key role in the initial recognition of LC3 on autophagic membranes in vitro (11). To elucidate which LIR motifs are involved in the RavZ(ΔMT) function in cells, we generated LIR1/2 motifs or an LIR3 motif mutant from 3xFLAG-RavZ(ΔMT). 3xFLAG-3xFLAG-RavZ(ΔMT)mLIR1/2, which has a point mutation in the LIR1/2 motifs, and 3xFLAG-3xFLAG-RavZ(ΔMT)mLIR3, which has a point mutation in the LIR3 motif, increase the number of GFP-LC3B and GFPGABARAP-positive autophagic membranes more than does 3xFLAG-RavZ(ΔMT), indicating that both the LIR1/2 and LIR3 motifs are involved in RavZ(ΔMT) function (Fig. 3A and 3B). Overall, our results suggest that the MT domain plays a major role in RavZ function and is involved in the delipidation of all forms of mATG8-PE on autophagosome membranes, whereas LIR motifs play minor roles in RavZ function and are involved in the selective delipidation mATG8-PE on autophagic membranes.
The results of this investigation provide clear evidence that in RavZ, LIR motifs have activities that are mediated by direct mATG8 binding. In addition, RavZ has two complementary pathways for autophagic membrane targeting: via PI3P or mATG8 binding. PI3P and mATG8 proteins are key players in autophagosome functions, including autophagosome biogenesis and fusion (16–18). PI3P is enriched in early autophagic membranes, whereas mATG8 proteins are stable and remain localized to the autophagic membrane until the membrane is degraded by the lysosome. It appears that RavZ has strategies for early and late autophagic membrane targeting to maximize efficiency for impairing autophagic membrane formation in host cells.
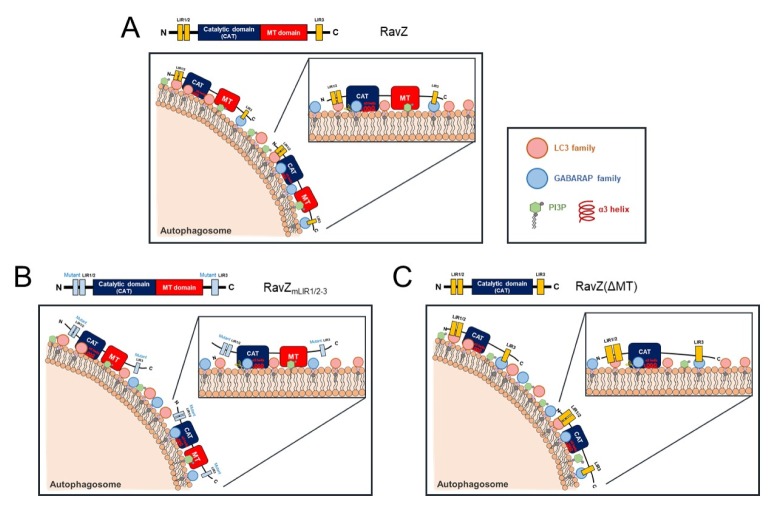
Two working hypotheses have been proposed for RavZ protein localization and activity within host cells. One is known as the “Tethering and Cut” model (7), in which the N-terminal LIR1/2 motifs and the C-terminal LIR3 motif tether mATG8-PE molecules on the autophagosome membrane, and the catalytic domain of RavZ cuts one or the other of the tethered mATG8-PE proteins. In this model, RavZ binds to LC3 proteins via the N- and C-terminal LIR motifs of RavZ. The other model is the “Lift and Cut” model (19), in which RavZ localizes to the autophagic membrane via the MT domain and the α3 helix in the catalytic domain (10). The LIR2 motif near the catalytic domain recognizes and recruits mATG8-PE proteins on the autophagic membrane. The catalytic domain of RavZ then recognizes and cuts the mATG8-PE proteins (11). Here, we propose an updated model of RavZ functionality (Fig. 4). The RavZ protein can target autophagosome membranes using two complementary pathways. One pathway is mediated by the PI3P-positive membrane association via an MT domain for PI3P binding. The other pathway is mediated by direct mATG8 binding by the LIR motifs. On autophagic membranes, a catalytic domain depletes all forms of mATG8 proteins (Fig. 4A). Therefore, in the absence of functional LIR motifs, a RavZ mutant (RavZmLIR1/2–3) can localize to the autophagic membranes by its PI3P-positive membrane association mediated by the MT domain, and delipidate all forms of mATG8-PE on the autophagic membranes (Fig. 4B). In the absence of an MT domain, RavZ(ΔMT) can target the autophagic membranes by direct selective mATG8 binding mediated by the LIR motifs, and delipidate LC3B-PE or GABARAP-PE on autophagosome membranes (Fig. 4C). Thus, it appears that the effects of LIR motifs and the MT domain are complementary and work through independent pathways in RavZ function.
Fig. 4.
Schematic model of the molecular mechanism of autophagosome targeting of RavZ proteins and RavZ mutants. (A) RavZ proteins can target autophagic membranes via two complementary pathways. One major pathway is mediated by the α3 helix in the catalytic domain and the MT domain via weak membrane association and PI3P binding. Another pathway is mediated by the α3 helix in the catalytic domain and the LIR motifs, via weak membrane association and mATG8 binding. The catalytic domain of RavZ hydrolyzes the amide bond between the C-terminal glycine residue and an adjacent aromatic residue of all forms of mATG8 proteins on autophagic membranes. Therefore, RavZ proteins delipidate mATG8-PE irreversibly. However, it is not clear whether the LIR motifs ahead of the catalytic domain may be involved in substrate recognition on autophagic membranes. (B) The mATG8 binding-deficient RavZ mutant (RavZmLIR1/2–3) can target autophagic membranes by the α3 helix in the catalytic domain and the MT domain via weak membrane association and PI3P binding. The catalytic domain of RavZmLIR1/2–3 can delipidate all forms of mATG8-PE on autophagic membranes. (C) RavZ(ΔMT), the MT domain deletion mutant of RavZ, can target autophagic membranes by the α3 helix in the catalytic domain and the LIR motifs, via weak membrane association and mATG8 binding. The catalytic domain of RavZ(ΔMT) can delipidate LC3B-PE or GABARAP-PE, but not LC3C-PE, on autophagic membranes in an LIR motif-dependent manner.
MATERIALS AND METHODS
DNA constructs
All primers used in these experiments are listed in Supplemental Table 1. We generated the region encoding RavZ or RavZC258S by PCR amplification of pcDNA3.1(−)-FLAG-RavZ or RavZC258S vectors and inserted it into the C1–3xFlag vector to generate C1–3xFlag-RavZ or RavZC258S using the restriction enzyme, BglII-Apa1. The pcDNA3.1(−)-Flag-RavZ and RavZC258S vectors were kindly provided by Dr. Hyun Kyu Song (Department of Life Sciences, Korea University, Seongbuk-gu, Seoul, Korea) (15). We amplified mutants of the RavZ LIR motifs by PCR, using RavZ LIR1/2 or three mutant primers (Supplemental Table 1) and inserted into the C1–3xFlag-RavZ vectors using the restriction enzymes BglII-HindIII and SalI-ApaI. C1–3xFlag-RavZ(ΔMT) constructs were changed from the membrane-targeting domain-containing catalytic domain to the catalytic domain in C1–3xFlag-RavZ or RavZC258S vectors using the restriction enzyme HindIII-SalI. Additionally, each domain of RavZ—the LIR1/2 motifs, the LIR3 motif, the membrane-targeting domain, and the catalytic domain—was amplified by PCR using appropriate primers (Supplemental Table 1) and inserted into pGFP-N3 or pGFP-C1 vectors with the restriction enzyme sets NheI-KpnI, XhoI-NotI, SalI-NotI, or BglII-SalI. GST-LC3A, GST-LC3B, GST-LC3C, GST-GABARAP, GST-GABARAP-L1, GST-GABARAP-L2, and pMXs-puro GFP-DFCP1 were obtained from Addgene (Cambridge, MA, USA). In this study, we employed DNA constructs that have previously been described: GFP-LC3A, GFP-LC3B, GFP-LC3C, GFP-GABARAP, GFP-GABARAP-L1, and GFP-GABARAP-L2 (20).
GST pulldown assay
For the GST pulldown assays with HEK293T cell lysates, cells were transfected with the DNA encoding the indicated 3xFLAG or GFP constructs using calcium phosphate (Takara Bio) transfection. After transfection, cells were harvested, washed with PBS, lysed in immunoprecipitation lysis buffer solution (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% TX-100, and protease and phosphatase inhibitors) and removed by centrifugation. Cell lysates were incubated with purified GST-mATG8 proteins with glutathione-agarose beads overnight at 4°C. The following day they were washed with an immunoprecipitation lysis buffer solution at 4°C; this step was repeated three to five times. Proteins were then resolved using SDS–PAGE and analyzed using Western blot analysis.
Cell culture, transfection, and confocal microscopy
This method has been previously described (21). We cultured HEK293T cells and mouse embryonic fibroblast (MEF) cells in Dulbecco’s modified Eagle’s medium (DMEM) and supplemented with 10% (v/v) fetal bovine serum (FBS) and penicillin/streptomycin in a humidified atmosphere of 5% (v/v) CO2 at 37°C. Cells were seeded in a sticky-slide eight-well system (Catalog #: 80828; Ibidi, Martinsried, Germany) to obtain 40%–60% confluent cells on the day of imaging. Cells were transfected with the indicated DNA constructs using calcium phosphate or Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) 24–26 h before imaging. The relative amount of each construct was empirically calculated based on the relative expression of each construct combination. We observed cells using an inverted Zeiss LSM-700 scanning laser confocal microscope with ZEN software (Carl Zeiss, Oberkochen, Germany). The laser lines for excitation and the spectral detection windows for fluorochromes were 488 nm (508–543 nm for GFP) and 561 nm (578–649 nm for mRFP). We used appropriate GFP (500–550 nm) and mRFP (575–625 nm) emission filters for sequential imaging of each fluorescent protein. Most images taken were of live cells.
Drug treatment
Rapamycin and chloroquine (CQ) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Autophagy was induced by incubating cells with 100 nM rapamycin in the presence or absence of 50 μM CQ in DMEM + 10% FBS for 4 h. We did all treatments and assays at 37°C unless otherwise indicated.
Western blots
We prepared transfected HEK293T cell lysates by adding cells to lysis buffer solution (50 mM Tris pH 7.5, 150 mM NaCl, 1% TX100, 5 mM EDTA, and protease inhibitors). Equal amounts of protein were resolved by SDS-PAGE, transferred to PVDF membranes, and incubated with primary antibodies overnight at 4°C. After three washes, we incubated membranes with secondary antibodies and conjugated with horseradish peroxidase for 1 h. Signals were visualized with ECL with an Advansta WesternBright ECL kit (K-12045-D50). We used FLAG (Sigma, F1804) antibody at 1:20,000, the LC3 antibody (Signaling Technology, #2775) at 1:1000, the β-actin antibody (Santa Cruz Biotechnology, sc-47778) at 1:1000, and the goat anti-mouse HRP antibody (Santa Cruz Biotechnology, sc-2005) or goat anti-rabbit HRP antibody (Santa Cruz Biotechnology, sc-2313) at 1:10,000.
Spot number analysis
To find the reduction in mATG8-positive autophagosomes by enzyme activation of RavZ in autophagy-induced cells, we counted the number of spots over a certain size in a single cell using the Image-J software. The cell image was changed to an 8-bit image and inverted. The background was then removed so that only the spots remained visible. Finally, we counted the number of spots using the “Analyze particles” function of Image-J. A minimum of 20 cells was quantified using this approach. All statistical data were calculated and graphed using GraphPad Prism5.
Supplementary Information
ACKNOWLEDGEMENTS
This work was supported by National Research Foundation (NRF-2017R1D1A3B03030972), National Scientist Honor Program, and Korea Health Technology R&D Project (HI18C0158), the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (2017M3A9G7073521) to J.-A L; and by the National Research Foundation under grant (NRF-2017R1D1A3B03028657) to D.-J.J.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slobodkin MR, Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 2013;55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 3.Lee YK, Lee JA. Role of the Mammalian ATG8/LC3 Family in Autophagy: Differential and Compensatory Roles in the Spatiotemporal Regulation of Autophagy. BMB Rep. 2016;49:424–430. doi: 10.5483/BMBRep.2016.49.8.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun J, Um JH. Emerging role of mitophagy in human diseases and physiology. BMB Rep. 2017;50:299–307. doi: 10.5483/BMBRep.2017.50.6.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma V, Verma S, Seranova E, Sarkar S, Kumar D. Selective Autophagy and Xenophagy in Infection and Disease. Front Cell Dev Biol. 2018;6:147. doi: 10.3389/fcell.2018.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon DH, Song HK. A Structural View of Xenophagy, a Battle between Host and Microbes. Mol Cells. 2018;41:27–34. doi: 10.14348/molcells.2018.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherwood RK, Roy CR. Autophagy Evasion and Endoplasmic Reticulum Subversion: The Yin and Yang of Legionella Intracellular Infection. Annu Rev Microbiol. 2016;70:413–433. doi: 10.1146/annurev-micro-102215-095557. [DOI] [PubMed] [Google Scholar]
- 9.Choy A, Dancourt J, Mugo B, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horenkamp FA, Kauffman KJ, Kohler LJ, et al. The Legionella Anti-autophagy Effector RavZ Targets the Autophagosome via PI3P− and Curvature-Sensing Motifs. Dev Cell. 2015;34:569–576. doi: 10.1016/j.devcel.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang A, Pantoom S. Elucidation of the anti-autophagy mechanism of the Legionella effector RavZ using semisynthetic LC3 proteins. Elife. 2017;6:e23905. doi: 10.7554/eLife.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skytte Rasmussen M, Mouilleron S, Kumar Shrestha B, et al. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy. 2017;13:834–853. doi: 10.1080/15548627.2017.1287651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kauffman KJ, Yu S, Jin J, et al. Delipidation of mammalian Atg8-family proteins by each of the four ATG4 proteases. Autophagy. 2018;14:992–1010. doi: 10.1080/15548627.2018.1437341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abreu S, Kriegenburg F, Gomez-Sanchez R. Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation. EMBO Rep. 2017;18:765–780. doi: 10.15252/embr.201643146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon DH, Kim S, Jung YO, et al. The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane. Autophagy. 2017;13:70–81. doi: 10.1080/15548627.2016.1243199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nascimbeni AC, Codogno P, Morel E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 2017;284:1267–1278. doi: 10.1111/febs.13987. [DOI] [PubMed] [Google Scholar]
- 17.Jang DJ, Lee JA. The roles of phosphoinositides in mammalian autophagy. Arch Pharm Res. 2016;39:1129–1136. doi: 10.1007/s12272-016-0777-x. [DOI] [PubMed] [Google Scholar]
- 18.Carlsson SR, Simonsen A. Membrane dynamics in autophagosome biogenesis. J Cell Sci. 2015;128:193–205. doi: 10.1242/jcs.141036. [DOI] [PubMed] [Google Scholar]
- 19.Pantoom S, Yang A, Wu YW. Lift and cut: Anti-host autophagy mechanism of Legionella pneumophila. Autophagy. 2017;13:1467–1469. doi: 10.1080/15548627.2017.1327943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YK, Jun YW, Choi HE, et al. Development of LC3/GABARAP sensors containing a LIR and a hydrophobic domain to monitor autophagy. EMBO J. 2017;36:1100–1116. doi: 10.15252/embj.201696315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SW, Jun YW, Choi HE, Lee JA, Jang DJ. Deciphering the molecular mechanisms underlying the plasma membrane targeting of PRMT8. BMB Rep. 2019;52:601–606. doi: 10.5483/BMBRep.2019.52.10.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.