Abstract
The discovery of hepaciviruses in non-human hosts has accelerated following the advancement of high-throughput sequencing technology. Hepaciviruses have now been described in reptiles, fish, birds, and an extensive array of mammals. Using metagenomic sequencing on pooled samples of field-collected Culex annulirostris mosquitoes, we discovered a divergent hepacivirus-like sequence, named Jogalong virus, from the Kimberley region in northern Western Australia. Using PCR, we screened the same 300 individual mosquitoes and found just a single positive sample (1/300, 0.33%). Phylogenetic analysis of the hepacivirus NS5B protein places Jogalong virus within the genus Hepacivirus but on a distinct and deeply rooted monophyletic branch shared with duck hepacivirus, suggesting a notably different evolutionary history. Vertebrate barcoding PCR targeting two mitochondrial genes, cytochrome c oxidase subunit I and cytochrome b, indicated that the Jogalong virus-positive mosquito had recently fed on the tawny frogmouth (Podargus strigoides), although it is currently unknown whether this bird species contributes to the natural ecology of this virus.
Introduction
Hepaciviruses are positive-sense RNA viruses in the family Flaviviridae. Hepaciviruses are difficult to culture; thus, their diversity was underappreciated until the advent of high throughput sequencing (HTS). The genus Hepacivirus comprises at least fourteen species that infect humans [1], and other mammals including rodents [2–6], cows [7, 8], horses [9], primates [10, 11], and bats [12]. A survey of Australian ticks also identified a hepacivirus from an Ixodes holocyclus tick that fed on a long-nosed bandicoot [13]. Metagenomic analyses of fish and reptiles uncovered the first non-mammalian hepaciviruses [14, 15]. A recent study investigating the etiology of severe disease in ducks identified a highly prevalent and divergent hepaci-like viral sequence in 70% of ducks collected over a wide geographical area [16]. Aside from two turtle hepaciviruses that share a common ancestor with the rodent Hepacivirus J (Myodes gareolus), the remaining non-mammalian viruses represent a diverse and separate clade of hepaciviruses. Despite the remarkable evolutionary distance separating these hosts, hepaciviruses have maintained an affinity for liver infection [14].
Here, we describe the discovery of Jogalong virus (JgV) from a single Culex annulirostris mosquito from the Kimberley region of Western Australia. The unexpected discovery of a hepacivirus sequence in an invertebrate raised suspicion that JgV may represent partially digested material from a blood meal. Subsequent vertebrate barcoding PCRs suggest that the true host may be of avian origin.
Methods
Mosquito collection
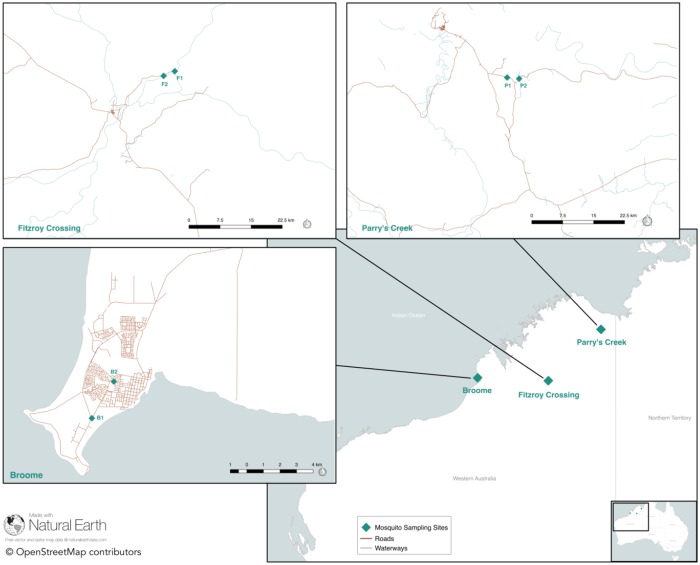
We trapped adult mosquitoes using Encephalitis Virus Surveillance CO2-baited traps [17]. Mosquitoes were collected from three sites located in the Kimberley region of Western Australia during March and April 2018 as part of routine arboviral surveillance [18]. Mosquitoes trapped from the townships of Broome and Fitzroy Crossing were collected from Public land, while mosquitoes collected from the rotunda within Geikie Gorge National Park (Fitzroy Crossing) and from Parry Lagoons Nature Reserve (Parry’s Creek) were collected under Department of Parks and Wildlife, Western Australia, Permit number (08-001839-1) (Fig 1). At each site, two traps were located approximately 2.5 km apart. Mosquitoes were separated by species using morphologic criteria [19], and 50 Cx. annulirostris mosquitoes were set aside from each trap for processing. Mosquitoes with visual evidence of a recent blood meal were excluded from further analysis. Historically, blood fed mosquitoes have been omitted from processing in order to reduce the likelihood of detecting a virus present solely in the blood meal.
Fig 1. Map of northern Western Australia.
Mosquito trap locations for three sites are marked with a green triangle. The inset maps indicate the locations of each of two traps located at each site. Map prepared using QGIS v2.18.15 (http://qgis.osgeo.org), OpenStreetMap, Natural Earth, and Mainroads Western Australia (https://portal-mainroads.opendata.arcgis.com/). The baselayer shapefile was obtained from the Australian Government data portal (https://www.data.gov.au/).
High-throughput sequencing
A total of 300 mosquitoes were individually washed three times using 750 μl refrigerated phosphate buffered saline, prior to homogenization in 750 μl of cold virus transport medium (in-house formulation; [20]) using the TissueLyserLT (Qiagen, Hilden, Germany) set to 50 KHz for 5 min. For unbiased HTS, we enriched pooled supernatants for virus particles. Aliquots of 50 μl supernatant from each of 25 individual mosquitoes were pooled according to trap for a total of 12 pools. An aliquot of 250 μl pooled material was passed through a 0.45 μM filter (EMD Millipore, Bedford, MA, USA); filtrate was treated with 1.5 μl RNase A (Invitrogen, Carlsbad, CA), 1.8 μl benzonase (EMD Millipore, Billerica, MA, USA) and 2.7 μl 1M MgCl2, gently mixed, and left at room temperature for 45 min. Total nucleic acid was extracted from pools using the MagMax Express-96 automated platform (Applied Biosystems, Foster City, CA) with modifications as described by Chidlow et. al. [21]. Nucleic acid concentration and purity was measured on the NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE). Total nucleic acid was reverse transcribed using SuperScript III (Invitrogen) and treated with RNAse H (Invitrogen). Double stranded cDNA was prepared using Klenow fragment (3’– 5’ exo-) (New England Biolabs, Beverly, MA). Fragments approximately 200 nt in length were generated by shearing double stranded cDNA on the Focused-Ultrasonicator E210 (Covaris, Woburn, MA). Each library was uniquely barcoded and prepared for sequencing on one lane of the HiSeq 4000 system (Illumina, San Diego, CA) using the Hyper Prep kit (Kapa Biosystems, Boston, MA). Two negative control libraries were also included; the first introduced during sample extraction, and the second during library preparation. A positive control library consisting of the ERCC spike-in was also included.
Sequencing reads generated from HTS processing were demultiplexed, trimmed, and quality filtered using PRINSEQ v0.20.2 [22]. As the Cx. annulirrostris genome is unavailable, sequences mapping to the Cx. quinquefasciatus complete genome (NCBI Reference Sequence NZ_AAWU00000000.1) were subtracted using Bowtie2 (v2.0.6, http://bowtie-bio.sourceforge.net) [23] prior to assembly (MEGAHIT v1.0) [24]. Resulting contiguous sequences (contigs) and unique singletons were assessed for sequence similarity to viral reference sequences contained within the non-redundant nucleotide or protein sequence databases of Genbank using MegaBLAST and BLASTx. The sequence of JgV was confirmed using overlapping PCR and bidirectional Sanger sequencing.
PCR screening for Jogalong virus
Total nucleic acid was prepared from 250 μl of supernatant from the same 300 individual mosquitoes used for HTS. Supernatants were extracted using the MagMax Express-96 platform (Applied Biosystems) as described above and cDNA was prepared from TNA using SuperScript III (Invitrogen). PCR screening primers were designed in the NS5B region using JgV sequences generated from HTS analyses of mosquito pools for the purposes of individual screening (F: CAGGTCCCTATTCTTACACGG; R: TCTGGTAACCGAGGTGTTGC). The identity of all PCR products was confirmed by Sanger sequencing.
Genome characterization
The hepacivirus polyprotein is co- and post-translationally cleaved using a combination of host proteases (for structural proteins; core, E1, E2, and P7) and viral proteases (for nonstructural proteins; NS2, NS3, NS4A, NS4B, NS5A, and NS5B). We used SignalP 5.0 [25] to identify putative cleavage sites for the structural proteins. To identify the locations of putative cleavage for the nonstructural proteins, we aligned our sequence with other annotated hepacivirus polyproteins and screened for conserved locations.
Phylogenetics
Protein sequences representing all hepaciviruses including recently described reptilian, fish, and bird hepaciviruses [14–16] were obtained from GenBank, as well as representative pegi- and pestiviruses. All hepacivirus names, sequences and associated hosts are detailed in S1 Table. Two members of the genus Flavivirus, represented by Tamana bat virus and yellow fever virus, were included as an outgroup. A conserved region within the NS5 protein [11] was aligned in Geneious 10.2.3 [26] and exported to MEGA6 [27] for phylogenetic analysis. Best-fit model testing was performed within MEGA6 and a maximum likelihood tree was constructed using the Le and Gascuel substitution model [28] with 500 bootstrap repetitions. Newick trees were exported to Figtree (http://tree.bio.ed.ac.uk/software/figtree) for annotation.
Vertebrate barcoding
Reasoning that an hepacivirus was likely to have a vertebrate host, we performed PCR targeting the cytochrome b (cyt b) and cytochrome c oxidase I (COI) genes found in mitochondrial DNA (mtDNA) [29]. We screened all 50 individual mosquitoes from the Parry’s Creek trap that contained the JgV-positive mosquito. All PCR products were sequenced using the Sanger method. We cloned PCR products using the pGem-T easy vector system (Promega, Madison, WI) to resolve mixed bases that were observed in chromatograms obtained from direct sequencing. Ten colonies were screened for each PCR product.
Accession numbers
The Jogalong virus sequence has been deposited in GenBank with the accession number MN133813. Illumina sequence data has been deposited in GenBank under BioProject number PRJNA590265.
Results
Mosquito collection
The majority of all mosquitoes collected from traps placed across the Kimberley region during March–April 2018 were Cx. annulirostris (n = 111,019; 58%) [30]. A total of 20,556 Cx. annulirostris was collected from the six traps located in three sites from across the Kimberley region in the north west of Australia (Table 1, Fig 1); 50 female mosquitoes were randomly selected from each trap for virome analyses.
Table 1. Culex annulirostris trapped from three sites in the Kimberley region.
| Trap | Trap location | Latitude | Longitude | Trap set date | Total trapped |
|---|---|---|---|---|---|
| B1 | Adjacent to caravan park, Broome | -17.97763161 | 122.2125935 | 04/09/18 | 188* |
| B2 | Cemetery, Broome | -17.957757 | 122.224449 | 04/10/18 | 405* |
| F1 | Rotunda, Fitzroy Crossing | -18.10542047 | 125.7016503 | 04/06/18 | 135 |
| F2 | Floodway, Fitzroy Crossing | -18.11554849 | 125.6773817 | 04/06/18 | 170 |
| P1 | Jogalong Billabong, Parry’s Creek | -15.59154698 | 128.261953 | 03/26/18 | 1971* |
| P2 | Mangrove, Parry’s Creek | -15.594099 | 128.288027 | 03/26/18 | 17,687* |
*Total Cx. annulirostris extrapolated based on species identification performed on 600 total mosquitoes per trap
Discovery of Jogalong virus
Sequencing of 12 mosquito pools generated 341 million reads from a single lane of sequencing using the HiSeq 4000 platform (Illumina) (not including controls). A total of 126 million reads were available for assembly following quality filtering and host subtraction. Assembly of reads generated 2.8 million contigs; 32 million unassembled unique singletons remained after assembly. Following BLAST sequence similarity searches, 117,576 (1.7%) sequences (contigs and unique singletons) sourced from mosquito pools shared identity with viral sequences using a minimum MEGABLAST E-value cutoff of 1E-10 or BLASTx cutoff of 1E-3. We identified six contigs (range 201 to 4018 nt; from total of 835 reads) in a single pool from Parry’s Creek that shared low-level identity with hepaciviruses. No hepacivirus-like sequences were observed in negative control samples. We have tentatively named this viral sequence as Jogalong virus (JgV) after the billabong (a seasonal body of water) located near trap P1 at Parry’s Creek.
Incidence of Jogalong virus
We performed direct PCR on all 300 individual samples collected from the 6 traps distributed across three sites to determine the number of JgV-positive mosquitoes. We found a single JgV-positive mosquito (P1-10) from trap P1 at the Parry’s Creek collection site.
Virus characterization
We used PCR on mosquito P1-10 to confirm all contigs that shared identity with hepaciviruses and bridge gaps between sequences obtained from HTS data. PCR primers were designed using assembled HTS data. The complete polyprotein is 8,826 nt (2,941 aa) in length. We identified a hepacivirus-like polyprotein, with putative cellular and viral protease cleavage sites defining 10 co- and post-translationally cleaved proteins (C-E1-E2-p7-NS2-NS3-Ns4a-NS4b-NS5a-NS5b; Table 2, Fig 2). Attempts to identify the complete non-translated genomic regions (NTR) using RACE were unsuccessful. Nonetheless, we were able to confirm 503 nt at the 5’ NTR and 84 nt at the 3’ NTR of the JgV genome using PCR. The presence of a miR-122 binding site could not be located, possibly due to incomplete 5’NTR sequence.
Table 2. Hepacivirus protein length and cleave site sequences.
| Protein | Core | E1 | E2 | P7 | NS2 | NS3 | NS4A | NS4B | NS5A | NS5B | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virus | aa | cleavage | aa | cleavage | aa | cleavage | aa | cleavage | aa | cleavage | aa | cleavage | aa | cleavage | aa | cleavage | aa | cleavage | aa |
| A/Equine hepacivirus/JPN3/Ec | 204 | GEASV | 188 | VSCTD | 335 | AEAYL | 63 | AWAFD | 217 | RLLSP | 631 | TQTNA | 54 | EECFD | 257 | QNCDF | 406 | ESCSL | 588 |
| B/Hepatitis GB virus B | 156 | CSGAR | 193 | TSGNP | 264 | AGLPL | 119* | ASAFD | 208 | AITAP | 620 | VNTSG | 55 | EECAS | 248 | DDCGL | 411 | FSCSM | 590 |
| C/Hepatitis C virus genotype 1 | 191 | ASAYQ | 192 | VDAET | 363 | AEAAL | 63 | AYALD | 217 | RLLAP | 631 | VVTST | 54 | EECSQ | 261 | TPCSG | 448 | VCCSM | 591 |
| D/Guereza hepacivirus/GHV-2/Cg | 181 | GASCV | 211 | VTSTS | 260 | AAAAA | 67 | AVGFD | 208 | SMLNP | 625 | NDCSL | 60 | EECSF | 248 | AQCDG | 882 | AKCAS | 591 |
| E/Rodent hepacivirus/RHV-339/Pm | 168 | ATAVS | 185 | AAAAA | 280 | AYAFT | 54 | AYAAS | 198 | KYTIP | 621 | FFASG | 58 | EECYN | 246 | DLCTP | 355 | HSCSM | 583 |
| F/Hepacivirus F/NLR07/Mg | 149 | AVTNC | 184 | AAAAS | 280 | AFAFT | 54 | TSAYS | 199 | ERTAP | 621 | YFAST | 58 | EECYQ | 246 | EDCSC | 479 | HECSS | 585 |
| G/Norway rat hepacivirus 1/NYC-C12/Rn | 208 | ASAGI | 243 | VAAPV | 271 | VGALE | 55 | EAYEG | 198 | RFTAP | 621 | YFAET | 57 | EECST | 249 | DVCTS | 506 | TDCSW | 583 |
| H/Norway rat hepacivirus 2/NYC-E43/Rn | 165 | AEANL | 216 | SAVAV | 272 | SEAVP | 56 | RAEQF | 197 | QLTKP | 620 | YYCGL | 61 | EECAN | 239 | EICDG | 449 | SSCSK | 580 |
| I/Hepacivirus/SAR-3/Rp | 172 | VEPKP | 197 | SVAAP | 255 | YAQPP | 53 | VEAFS | 204 | QLSSP | 622 | ELASW | 56 | EECAL | 251 | EPCTD | 385 | ETCTY | 586 |
| J/Hepacivirus J/RMU-3382/Mg | 163 | AVSHW | 185 | AEGLP | 287 | ANALV | 44 | AQGGC | 236 | RLTAP | 625 | EEMTD | 58 | EECGF | 258 | AECAG | 555 | TSCNY | 603 |
| K/Hepacivirus K/PDB-829/Hv | 190 | GEASY | 193 | AQANP | 325 | ADAAL | 63 | AVGGP | 217 | RHCSP | 628 | DDTST | 56 | EECLS | 257 | SECAF | 504 | DECSA | 591 |
| L/Hepacivirus L/PDB-112/Hv | 161 | AESVP | 203 | AAAMP | 265 | AWGWP | 59 | AQAAS | 212 | ERNAP | 629 | YSAGG | 57 | EECMQ | 259 | AECDG | 451 | ESCSE | 605 |
| M/Hepacivirus M/PDB-491.1/Om | 189 | VDASF | 193 | SQAAE | 321 | ALAVP | 63 | VDAYT | 217 | RHCSP | 628 | TPTSA | 55 | EECAD | 259 | RNCSC | 456 | SPCSA | 590 |
| N/Bovine hepacivirus/GHC25/Bt | 155 | VSGYR | 190 | VEATT | 267 | ATAAL | 60 | VTALD | 204 | APCAP | 624 | LDVWG | 54 | EECWG | 250 | VPCGF | 390 | KECSY | 579 |
| Hepacivirus P/RHV-GS2015/Cd | 153 | GLAFT | 186 | VAAPV | 270 | AEGAM | 56 | VLGAS | 199 | KRTAP | 622 | YFAST | 58 | EECSL | 245 | DFCSP | 395 | SDCSY | 580 |
| Sifaka hepacivirus | 162 | VGAAF | 194 | AAAAP | 269 | VEAVP | 55 | VEAYT | 198 | RLTAP | 622 | FFTAW | 58 | EECAL | 236 | TLCAS | 386 | QALSQ | 576 |
| Wenling shark virus | 300 | MDSAP | 194 | AVAAP | 214 | AEASV | 71 | ALGDD | 224 | NRCAP | 632 | LVAGL | 56 | DLSGA | 314 | VVMAD | 475 | SPMSH | 606 |
| Duck hepacivirus strain HCL-1/Ap | 290 | ASADH | 195 | GMADR | 274 | AEGML | 74 | VLGAS | 245 | QYTAP | 625 | NCSAA | 55 | EECSA | 250 | YECNS | 1006 | ESCSF | 593 |
| Jogalong virus | 87 | AVAFS | 191 | AQAGT | 306 | IEGAV | 74 | VAGED | 235 | KLAAP | 625 | SAGLT | 52 | EECAS | 247 | TNCTS | 536 | VCCGE | 588 |
*p13 is processed into p6 (57aa) and p7 (62aa) in GBV-B; , location of predicted proteolytic cleavage; further information for each virus can be found in S1 Table.
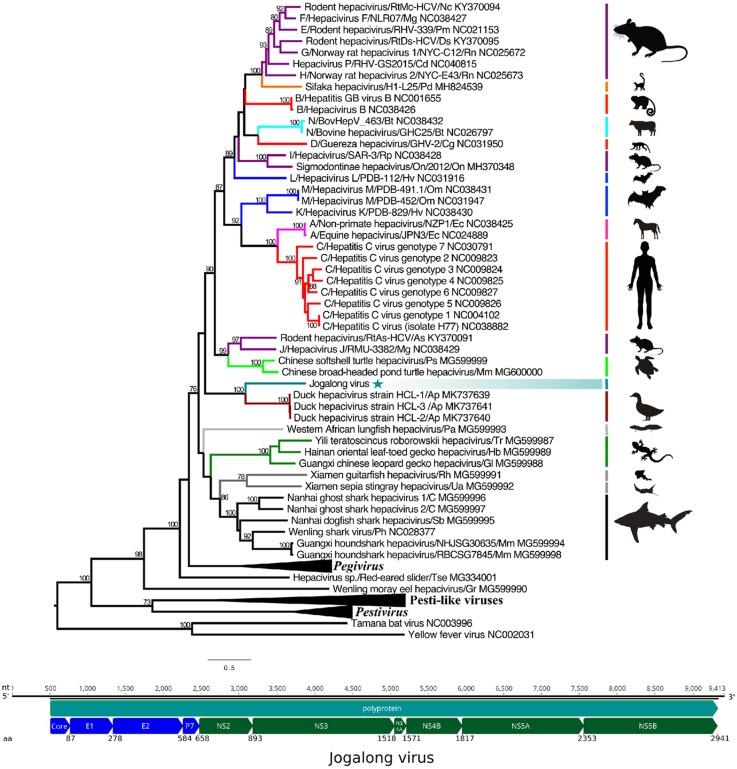
Fig 2. Phylogenetic analysis and putative genome organization of Jogalong virus.
The maximum likelihood tree was constructed using a conserved region within the NS5B protein. The scale bar represents substitutions per site and bootstrap values are displayed when greater than 70%. The genera Pestivirus, Pegivirus, and recently discovered Pesti-like viruses are indicated with a black triangle. Jogalong virus is indicated with a teal star. Hepaciviruses and their associated hosts are indicated by vertical bars. The putative location of the polyprotein (teal arrow) and cleaved structural proteins (blue arrows) and non-structural proteins (dark green arrows) are indicated in the genome map located beneath the tree. Nucleotide (nt) and amino acid (aa) positions can be found above and below the illustration, respectively.
JgV shares greatest identity with duck hepacivirus across all proteins except for the core and NS4A proteins where there was no apparent identity to any viral sequence (Table 3). Amino acid identity across the structural proteins (E1, E2, and P7) was greatest in the E1 protein (39.4%). Within the non-structural proteins, identity was lowest within the NS2 protein (27.3%) and greatest within the NS3 protein (45.0%). The low sequence identity (or lack thereof) to described viral proteins is supported by phylogenetic analysis of a conserved region within the NS5B protein. Analysis of the partial NS5B protein sequence places JgV outside the diversity of all recognized hepaciviruses on a deeply rooted monophyletic branch shared only with duck hepacivirus (Fig 2). JgV shares a closer phylogenetic relationship to the genus Hepacivirus than several recently discovered fish and reptilian hepaciviruses [14, 15]. However, all hepacivirus and hepacivirus-like sequences appear distinct from the clade of viruses belonging to genus Pegivirus.
Table 3. Jogalong virus protein sequence identity.
| Protein | Length (aa) | Closest related viral sequence | Host | Accession | Coverage (%) | E-value | Identity (%) |
|---|---|---|---|---|---|---|---|
| Core | 87 | Nil | |||||
| E1 | 191 | Duck hepacivirus/HCL-2 |
Anas platyrhynchos domesticus Domestic duck |
QDF44087 | 99% | 7.00E-37 | 39.38% |
| E2 | 306 | Duck hepacivirus/HCL-2 |
Anas platyrhynchos domesticus Domestic duck |
QDF44087 | 67% | 9.00E-25 | 31.30% |
| p7 | 74 | Duck hepacivirus/HCL-2 |
Anas platyrhynchos domesticus Domestic duck |
QDF44087 | 79% | 4.00E-05 | 35.59% |
| NS2 | 235 | Duck hepacivirus/HCL-2 |
Anas platyrhynchos domesticus Domestic duck |
QDF44087 | 98% | 8.00E-14 | 27.31% |
| NS3 | 625 | Duck hepacivirus/HCL-2 |
Anas platyrhynchos domesticus Domestic duck |
QDF44087 | 100% | 1.00E-169 | 44.99% |
| NS4A | 52 | Nil | |||||
| NS4B | 247 | Duck hepacivirus/HCL-2 |
Anas platyrhynchos domesticus Domestic duck |
QDF44087 | 90% | 2.00E-31 | 33.19% |
| NS5A | 536 | Duck hepacivirus/HCL-2 |
Anas platyrhynchos domesticus Domestic duck |
QDF44087 | 27% | 2.00E-14 | 36.18% |
| NS5B | 588 | Duck hepacivirus/HCL-2 |
Anas platyrhynchos domesticus Domestic duck |
QDF44087 | 96% | 3.00E-131 | 41.51% |
JgV has a core protein sequence (87aa) that appears to be much shorter than most other hepaciviruses. Strikingly, the NS5A protein from the closest related viral sequence, duck hepacivirus, was nearly twice as long as the same protein of JgV. All remaining proteins were of similar length to other hepaciviruses (Table 2).
Blood meal analysis
During mosquito sorting, we excluded mosquitoes with any amount of abdominal swelling consistent with a recent blood meal. However, blood meals can be difficult to visually detect after approximately 60 hours [31]. To investigate whether the JgV-positive mosquito (P1-10) contained a blood meal, we screened all 50 Cx. annulirostris mosquitoes that were selected from trap P1 at Parry’s Creek. We detected avian mtDNA in a single mosquito, P1-10; the remaining 49 mosquitoes were negative for non-human and non-mosquito mtDNA. The sequence obtained from a single round of COI PCR shared 98.7% nt identity with the tawny frogmouth (Podargus strigoides), a native Australian bird species found throughout the country (Table 4). Sequences obtained from direct sequencing of the cyt b PCR product indicated co-amplification. Cloning and subsequent sequencing of this amplicon identified P. strigoides (98.9% nt identity; 5/10 clones) and Caprimulgus eximius (golden nightjar; 77.5% nt identity; 5/10 clones). We observed a 9-nt deletion within the C. eximius sequence that may indicate the co-amplification of nuclear mtDNA paralogs (numts). As numpts may not be transcribed, we attempted to specifically amplify the COI transcript from P1-10 by DNase-treating total nucleic acid and performing PCR on cDNA [32]. However, we were unable to amplify any product. This may reflect low quality RNA from the digested blood meal. Alternatively, the low identity match to C. eximius may suggest that there is an avian species in the Kimberley region that is yet to be characterized by mtDNA barcoding techniques.
Table 4. Blood meal analysis for mosquito P1-10.
| Gene target | PCR | Length of sequence | Proportion of clones | Closest related host sequence | Accession | Common name | Identity (%) |
|---|---|---|---|---|---|---|---|
| COI | single | 305 | 10/10 (100%) | Podargus strigoides | JQ175917 | Tawny frogmouth | 98.69 |
| Cyt b | single | 435 | 5/10 (50%) | Podargus strigoides | JQ353838 | Tawny frogmouth | 98.85 |
| 426 | 5/10 (50%) | Caprimulgus eximius | LT671509 | Golden nightjar | 77.52 |
COI, cytochrome c oxidase subunit I; cyt b, cytochrome b.
Discussion
We identified nucleic acid sequences of a virus tentatively named Jogalong virus that is related to members of the genus Hepacivirus. The sequence was obtained from a single Cx. annulirostris mosquito collected in the Kimberley region of Western Australia. Large metagenomic surveys of invertebrates are yet to uncover evidence suggesting an invertebrate lineage of hepaciviruses [33]. Thus, all hepaciviruses and hepaci-like viruses identified to date appear to be strictly vertebrate-associated, presumably due to a requirement for the presence of a liver for viral replication. A study of ticks in Australia identified hepacivirus nucleic acid (Collins Beach virus) in an engorged Ixodes hollocyclus tick, but that virus is likely associated with the long-nosed bandicoot from which the tick was removed [13]. To our knowledge, JgV represents the second hepacivirus and the first full hepacivirus polyprotein sequence to be discovered from non-human hosts in Australia.
To investigate whether the detection of JgV was associated with a prior blood meal, we performed vertebrate barcoding PCR on all 50 mosquitoes sampled from the Jogalong trap at Parry’s Creek, the site of JgV detection. We detected avian mitochondrial sequences in one of the mosquitoes from this trap; the positive individual corresponded to the JgV-positive mosquito. While more expansive surveillance is required, these data suggest that JgV may have originated from a non-mosquito host. Sequences from two mitochondrial genes closely match the tawny frogmouth; a native, insectivorous bird of the order Caprimulgiformes that is found throughout Australia [34]. Investigations of the feeding habits of Cx. annulirostris mosquitoes indicate that they are generalist feeders exhibiting high host plasticity that include birds [35]. Our results suggest that the blood meal contained JgV nucleic acid. Given the phylogenetic placement of JgV alongside an avian hepacivirus as well as the lack of invertebrate-associated hepaciviruses described to date, we believe it is unlikely that Cx. annulirostris mosquitoes are the host for this virus.
JgV is a highly divergent hepacivirus and shares only 42% aa identity with its closest relative, duck hepacivirus, across the highly conserved NS5B protein. Duck hepacivirus was recently discovered in China following an investigation of severely diseased ducks. Despite the context of its discovery, the pathogenicity of duck hepacivirus is unclear as the virus was also highly prevalent in healthy ducks [16]. The phylogenetic placement of JgV in a clade shared only with duck hepacivirus offers supporting evidence that JgV is of avian origin. The distant phylogenetic relationship between this potential avian clade (JgV and duck hepacivirus) and all other hepaciviruses is intriguing and suggests a notably different evolutionary history. The vast majority of hepaciviruses identified to date are highly species specific and are thought to have coevolved with their hosts [36]. Thus, a hepacivirus that infects an avian host could be expected to diverge from mammalian, reptilian or fish hepaciviruses. If the blood meal analysis is indicative of the natural host, then JgV may have an avian lineage; however, additional surveillance is required to test this hypothesis.
Supporting information
(DOCX)
Acknowledgments
We are grateful to members of the Surveillance Unit, Pathwest Laboratory Medicine WA, for expert assistance with mosquito processing and shipment, and Ram Lamichhane and Craig Brockway (Environmental Health Directorate, Public and Aboriginal Health Division) for mosquito sorting and advice. The authors also thank members of the Center for Infection and Immunity including Simon Anthony and Rafal Tokarz for manuscript revision, and Stephen Sameroff and Heather Wells for assistance with sequence mapping. The authors are grateful for advice from Cheryl Johansen.
Data Availability
The Jogalong virus sequence has been deposited in GenBank with the accession number MN133813. Illumina sequence data has been deposited in GenBank under BioProject number PRJNA590265.
Funding Statement
This work was supported by a grant from the National Institutes of Health (U19AI109761 Center for Research in Diagnostics and Discovery). The sequencing work was supported by funding from the NHMRC Centre of Research Excellence in Emerging Infectious Diseases (CREID) and the Australian Partnership for Preparedness Research on Infectious Disease Emergencies (APPRISE). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5(9):558–67. 10.1016/S1473-3099(05)70216-4 [DOI] [PubMed] [Google Scholar]
- 2.Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio. 2014;5(5):e01933–14. 10.1128/mBio.01933-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD, et al. Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio. 2013;4(2):e00216–13. 10.1128/mBio.00216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drexler JF, Corman VM, Muller MA, Lukashev AN, Gmyl A, Coutard B, et al. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013;9(6):e1003438 10.1371/journal.ppat.1003438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z, Lu L, Du J, Yang L, Ren X, Liu B, et al. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome. 2018;6(1):178 10.1186/s40168-018-0554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Souza WM, Fumagalli MJ, Sabino-Santos G Jr., Motta Maia FG, Modha S, Teixeira Nunes MR, et al. A Novel Hepacivirus in Wild Rodents from South America. Viruses. 2019;11(3):297 10.3390/v11030297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baechlein C, Fischer N, Grundhoff A, Alawi M, Indenbirken D, Postel A, et al. Identification of a Novel Hepacivirus in Domestic Cattle from Germany. J Virol. 2015;89(14):7007–15. 10.1128/JVI.00534-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corman VM, Grundhoff A, Baechlein C, Fischer N, Gmyl A, Wollny R, et al. Highly divergent hepaciviruses from African cattle. J Virol. 2015;89(11):5876–82. 10.1128/JVI.00393-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, et al. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol. 2012;86(11):6171–8. 10.1128/JVI.00250-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauck M, Sibley SD, Lara J, Purdy MA, Khudyakov Y, Hyeroba D, et al. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old World primate. J Virol. 2013;87(16):8971–81. 10.1128/JVI.00888-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canuti M, Williams CV, Sagan SM, Oude Munnink BB, Gadi S, Verhoeven JTP, et al. Virus discovery reveals frequent infection by diverse novel members of the Flaviviridae in wild lemurs. Arch Virol. 2019;164(2):509–22. 10.1007/s00705-018-4099-9 [DOI] [PubMed] [Google Scholar]
- 12.Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci U S A. 2013;110(20):8194–9. 10.1073/pnas.1303037110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harvey E, Rose K, Eden JS, Lo N, Abeyasuriya T, Shi M, et al. Extensive Diversity of RNA Viruses in Australian Ticks. J Virol. 2019;93(3). 10.1128/JVI.01358-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556(7700):197–202. 10.1038/s41586-018-0012-7 [DOI] [PubMed] [Google Scholar]
- 15.Shi M, Lin XD, Vasilakis N, Tian JH, Li CX, Chen LJ, et al. Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses. J Virol. 2015;90(2):659–69. 10.1128/JVI.02036-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu L, Jin M, Feng C, Wang X, Zhang D. A highly divergent hepacivirus-like flavivirus in domestic ducks. J Gen Virol. 2019; July 8 10.1099/jgv.0.001298 [DOI] [PubMed] [Google Scholar]
- 17.Rohe D, Fall RP. A miniature battery powered CO2 baited light trap for mosquito borne encephalitis surveillance. Bulletin of the Society of Vector Ecology. 1979;4:24–7. [Google Scholar]
- 18.Broom AK, Lindsay MD, Johansen CA, Wright AE, Mackenzie JS. Two possible mechanisms for survival and initiation of Murray Valley encephalitis virus activity in the Kimberley region of Western Australia. Am J Trop Med Hyg. 1995;53(1):95–9. [PubMed] [Google Scholar]
- 19.Liehne PF. An atlas of the mosquitoes of Western Australia. Health Department of Western Australia; Perth, Western Australia: 1991. [Google Scholar]
- 20.Madeley CF, Lennette DA, Halonen P. Specimen collection and transport In: Lennette EH, Halonen P, Murphy FA, editors. Laboratory diagnosis of infectious diseases: principles and practice. 2 New York, NY: Springer-Verlag; 1988. p. 7. [Google Scholar]
- 21.Chidlow G, Harnett G, Williams S, Levy A, Speers D, Smith DW. Duplex real-time reverse transcriptase PCR assays for rapid detection and identification of pandemic (H1N1) 2009 and seasonal influenza A/H1, A/H3, and B viruses. J Clin Microbiol. 2010;48(3):862–6. 10.1128/JCM.01435-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–4. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. 10.1016/j.ymeth.2016.02.020 [DOI] [PubMed] [Google Scholar]
- 25.Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–3. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- 26.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–20. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- 29.Townzen JS, Brower AV, Judd DD. Identification of mosquito bloodmeals using mitochondrial cytochrome oxidase subunit I and cytochrome b gene sequences. Med Vet Entomol. 2008;22(4):386–93. 10.1111/j.1365-2915.2008.00760.x [DOI] [PubMed] [Google Scholar]
- 30.Government of Western Australia. Medical Entomology: 2017/2018 surveillance program annual report Perth, Western Australia: Department of Health; 2018 https://ww2.health.wa.gov.au/~/media/Files/Corporate/generaldocuments/Mosquitoes/PDF/2017-18-WA-ME-Annual-report.pdf.
- 31.Reeves LE, Gillett-Kaufman JL, Kawahara AY, Kaufman PE. Barcoding blood meals: New vertebrate-specific primer sets for assigning taxonomic identities to host DNA from mosquito blood meals. PLoS Negl Trop Dis. 2018;12(8):e0006767 10.1371/journal.pntd.0006767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvignac S, Konecny L, Malard F, Douady CJ. Preventing the pollution of mitochondrial datasets with nuclear mitochondrial paralogs (numts). Mitochondrion. 2011;11(2):246–54. 10.1016/j.mito.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 33.Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–43. 10.1038/nature20167 [DOI] [PubMed] [Google Scholar]
- 34.Serventy DL. Feeding methods of Podargus. Emu. 1936;36(2):74–90. [Google Scholar]
- 35.Stephenson EB, Murphy AK, Jansen CC, Peel AJ, McCallum H. Interpreting mosquito feeding patterns in Australia through an ecological lens: an analysis of blood meal studies. Parasit Vectors. 2019;12(1):156 10.1186/s13071-019-3405-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartlage AS, Cullen JM, Kapoor A. The Strange, Expanding World of Animal Hepaciviruses. Annu Rev Virol. 2016;3(1):53–75. 10.1146/annurev-virology-100114-055104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
The Jogalong virus sequence has been deposited in GenBank with the accession number MN133813. Illumina sequence data has been deposited in GenBank under BioProject number PRJNA590265.