Figure 1.
Transcriptomic Analysis Reveals Major Differences between Human Lymphocytes Activated in HPLMdFBS Relative to RPMIdFBS
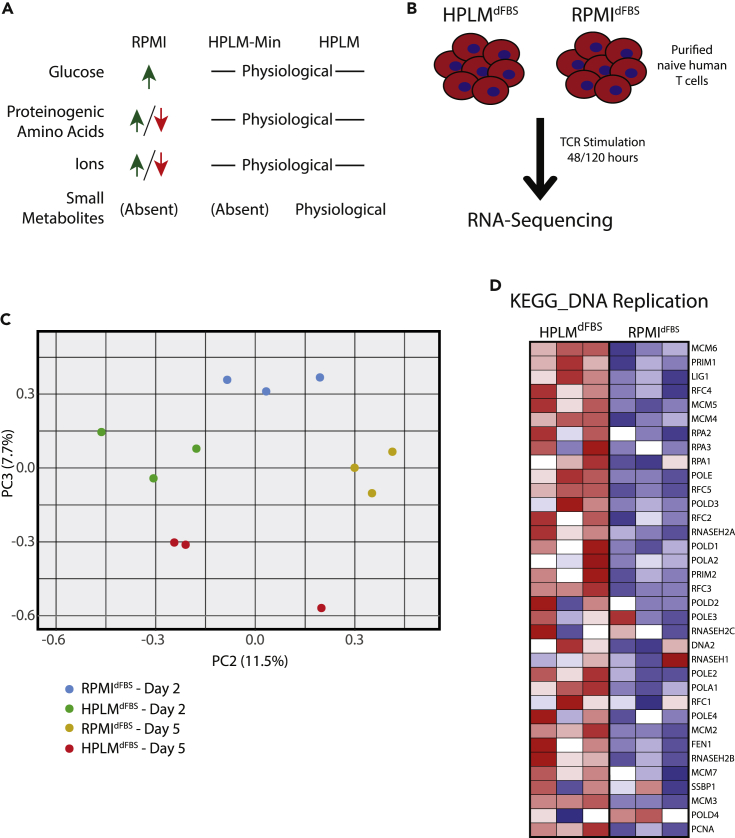
(A) Schematic of the comparative compositions of RPMIdFBS, HPLMdFBS, and HPLMdFBS-Min. Detailed composition can be found in Table S1.
(B) Experimental outline for T cell activation in either HPLMdFBS or RPMIdFBS and downstream transcriptome analysis.
(C) Transcriptional differences between primary naive human mixed CD4+ and CD8+ T cells from three different donors activated in HPLMdFBS or RPMIdFBS as depicted in (B) were measured via RNA-sequencing. These data were used to generate PCA plots showing principal components 2 and 3 at the indicated timepoints.
(D) Heatmap showing the Log2(fold change) in transcript abundance for genes involved in the KEGG DNA replication pathway using the RNA-sequencing data generated as described in (B).