Summary
Despite rapid advances in whole genome sequencing (WGS) technologies, their integration into routine microbiological diagnostics has been hampered by the lack of standardized downstream bioinformatics analysis. We developed a comprehensive and computationally low-resource bioinformatics pipeline (BacPipe) enabling direct analyses of bacterial whole-genome sequences (raw reads or contigs) obtained from second- or third-generation sequencing technologies. A graphical user interface was developed to visualize real-time progression of the analysis. The scalability and speed of BacPipe in handling large datasets was demonstrated using 4,139 Illumina paired-end sequence files of publicly available bacterial genomes (2.9–5.4 Mb) from the European Nucleotide Archive. BacPipe is integrated in EBI-SELECTA, a project-specific portal (H2020-COMPARE), and is available as an independent docker image that can be used across Windows- and Unix-based systems. BacPipe offers a fully automated “one-stop” bacterial WGS analysis pipeline to overcome the major hurdle of WGS data analysis in hospitals and public-health and for infection control monitoring.
Subject Areas: Biological Sciences Research Methodologies, Microbiology, Sequence Analysis
Graphical Abstract

Highlights
-
•
BacPipe is an automated whole genome sequencing pipeline
-
•
Interactive user-friendly GUI
-
•
BacPipe can process raw reads, contigs, or scaffolds
-
•
Time-to-analysis for a 5 Mb genome is ∼30–40 min
Biological Sciences Research Methodologies; Microbiology; Sequence Analysis
Introduction
Next-generation sequencing (NGS) technologies hold the promise to revolutionize the public health sector especially clinical diagnostic microbiology, infection control, outbreak detection, and antibiotic stewardship in hospitals (Arnold, 2015, Kwong et al., 2015, Moran-Gilad, 2017). As costs of sequencing technologies are steadily decreasing and response times getting shorter, their utility as tools for tracking pathogens in real-time for routine hospital epidemiology or as an early warning system for outbreak detection and detecting multi-drug resistant (MDR) pathogens is steadily increasing (Punina et al., 2015). Currently, depending on the pathogen, the identification and characterization process may take one to seven days for culture, an additional one to two days for species identification and susceptibility testing, and one to several weeks for molecular typing. Whole genome sequencing (WGS) of bacterial isolates combines identification, molecular typing, and prediction of antimicrobial susceptibility and virulence, theoretically reducing the time-to-result for these procedures to a few days (Didelot et al., 2012, Joensen et al., 2014, Koser et al., 2012). However, despite rapid advances in WGS workflows and in NGS technologies, their integration into routine microbiological diagnostics and infection control has been hampered by the need for downstream bioinformatics analyses that is challenging and requires considerable expertise (Deurenberg et al., 2017, Muir et al., 2016). WGS analysis comprises different stages, and each stage is crucial for correct data interpretation. Although there are commercial softwares available such as CLC Genomics Workbench (Qiagen), DNA Star (DNASTAR Inc., USA), BioNumerics (Applied Maths), and SeqSphere+ (Ridom GmbH, Münster, Germany), the current licensing costs are very high and cannot be sustained by small to medium laboratories. Furthermore, these tools handle the analysis as a black-box for the user and often lag when it comes to integrating state-of-the-art tools compared with publicly managed software/packages (Lüth et al., 2018). Thus, more extensive use of open-source software for whole-genome sequencing data analysis needs to be advocated (Deurenberg et al., 2017).
Several open access tools are available and are split into two categories, web-based analysis or locally downloadable tools. Few web-based open access pipelines such as Orione (http://orione.crs4.it) (Cuccuru et al., 2014) and the Bacterial analysis pipeline (https://cge.cbs.dtu.dk/services/cge/) (Thomsen et al., 2016) and the microbial genomics virtual laboratory (https://nectar.org.au/) are also available (Afgan et al., 2015). Orione is available in the Galaxy portal (https://usegalaxy.org/), and it offers WGS quality control, assembly and annotation, and variant calling (Cuccuru et al., 2014). The Bacterial analysis pipeline (https://cge.cbs.dtu.dk/services/cge/) offers molecular typing tools as well as resistance and virulence gene predictions and SNP-based phylogeny. However, the performance of web-server based analysis depends on the server load and requires a fast and consistent internet connection to upload large raw data files, which is unreliable when it comes to patient care. Moreover, due to the fact that the analysis is performed remotely, this forms a great barrier for hospital and data protection, which remains a sensitive matter with policies varying between countries (Akgün et al., 2015, Muir et al., 2016).
The second type of the open-source software that those locally installable tools developed specifically for running and managing microbial genomics pipelines includes IRIDA (irida.ca), Innuendo (http://www.innuendoweb.org/project-definition), and nullarbor (https://github.com/tseemann/nullarbor). IRIDA provides a workflow for assembly (SPAdes), annotation (Prokka), SNP phylogeny (SNVPhyl), resistance (CARD), and virulence (Islandviewer) but not for plasmids and MLST typing. INNUENDO, with its INNUca workflow, provides quality control of reads, de novo assembly, and contigs quality assessment. Nullarbor supports Illumina paired-end sequencing data but not single-end reads from either Illumina or Ion Torrent.
To add in this list and overcome the various issues discussed above, we have developed a rapid, “one-stop” bacterial WGS analysis pipeline, BacPipe (Figures 1 and 2). This freely available pipeline offers a graphical user interface, parallel computing for fast execution and a containerized granting it standardization of the results across different hospitals. Its open-source software is capable of performing a plethora of analyses starting from raw data quality check, genome assembly, and annotation, resulting in bacterial typing, resistance, and virulence gene predictions, as well as single nucleotide polymorphisms (SNP)-based phylogeny. BacPipe has been successfully implemented to analyze sample from large-scale projects, such as EBI-SELECTA, a rule-based computational workflow engine developed as part of H2020 COMPARE project (https://www.compare-europe.eu/).
Figure 1.
The Workflow of BacPipe
Complete overview of NGS workflow and analysis performed within BacPipe.
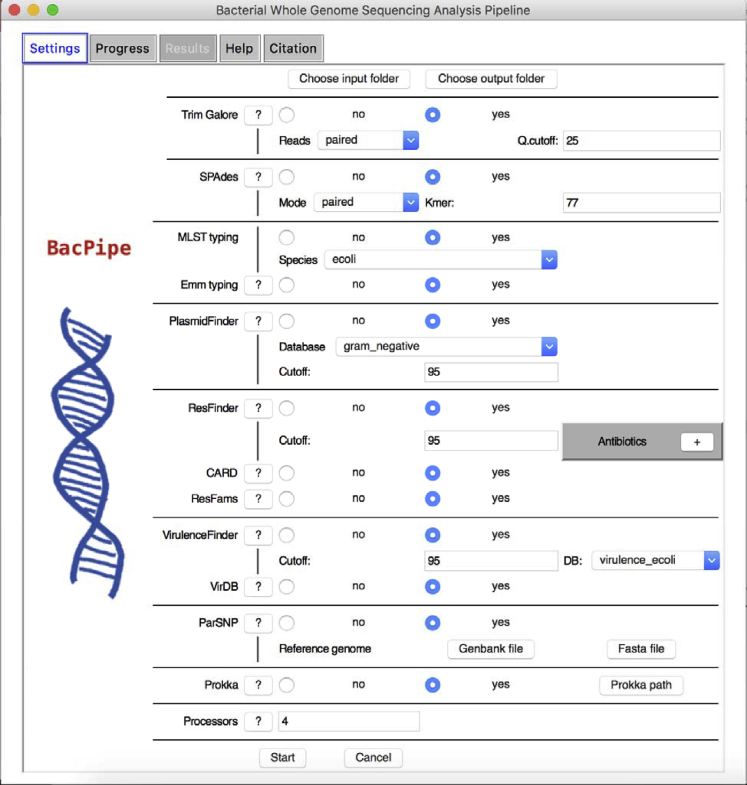
Figure 2.
Snapshot of BacPipe
BacPipe graphical user interface (GUI). See also Figure S1.
Results and Discussion
BacPipe Implementation and Running Time on Small Number of Strains (as a Function of Genome Size and Sequencing Coverage)
To demonstrate the impact of bacterial genome sizes on the computational time required to obtain results with BacPipe, we used three pathogen genomes that vary considerably in size, Streptococcus pyogenes (∼1.8 Mb), Escherichia coli (∼5.2 Mb), and Pseudomonas aeruginosa (∼6.8 Mb). Whole genome sequences of each pathogen were normalized to the same fold-coverage to demonstrate an increase in computational time as a function of genome size. These internal isolates were sequenced from our in-house MiSeq, and as the P. aeruginosa PAO1 had 70-fold coverage, we randomly selected reads from the other two strains resulting in the same coverage. Expectedly, computational time increased with increasing genome size totalling 9, 25, and 41 min for S. pyogenes, E. coli, and P. aeruginosa, respectively (Figure 3A). Among all the tools employed in the pipeline, as expected, genome assembly (SPAdes) was found to be the most computationally intensive, taking on average 36% of the total running time. Also, we assessed the added value of parallelizing the post-assembly tools (PlasmidFinder, ResFinder, VirulenceFinder, MLST, and emm typing) and post-annotation tools (ResFams, VirDB, and CARD search). Parallelizing these tools resulted in a reduction of time-to-result (computational time) by 56%, 29%, and 25% for the three pathogen sequences, respectively (data not shown). Additionally, to emphasize the increase in the computational time due to higher coverage, we subsampled the E. coli sequences at 50-, 70-, 100-, and 120-fold-coverage. The required computational time for 50-, 70-, 100-, 120-fold-coverages were 21, 25, 28 and 30 min, respectively (Figure 3B). For this benchmark, we used a MacBook Pro, 2.5 GHz, quad-core i7 with 16 GB RAMS (DDR3), 4 cores, and SSD hard drive.
Figure 3.
BacPipe Running Time
Impact of different genome sizes at equal sequencing coverage (70-fold) on the computational time taken for each analysis step in BacPipe (A). Impact of varying sequencing coverage of an E. coli genome on the computational time taken for each analysis step in BacPipe (B).
BacPipe Implementation and Running Time on Publicly Available Bacterial Genomes at Large Scale (EBI-SELECTA Framework)
Within the SELECTA framework, BacPipe was used to analyze 4,139 paired-end publicly available WGS sequence reads for the bacterial genomes listed in Table S1. An example of an analysis result can be found here (https://www.ebi.ac.uk/ena/data/view/ERZ799760). This implementation demonstrated the potential of BacPipe in processing a large number of runs on a short timescale (Figure 4).
Figure 4.
Large Scale Validation of BacPipe
BacPipe running time (on average 50 min/run) over 4,000 paired-end sequence reads of bacterial genomes. This process was performed on the EBI high-performance computing platform is an EBI shared facility made up of 130 nodes with 130Gb of RAM each and 2 core per node with 40 CPUs (See also Figures S2 and S3 and Table S1).
Validation of BacPipe's Functionality Using Prior Published Data
We challenged BacPipe with various bacterial genomes including those with higher GC content and multiple repeat regions (M. tuberculosis). Mainly, five previously published and analyzed WGS datasets (raw reads or assembled contigs) from hospital outbreaks caused by MRSA and carbapenem-resistant K. pneumoniae (Snitkin et al., 2012), a 3-year long in-hospital transmission study of C. difficile (Jia et al., 2016), a community-based surveillance and transmission study of M. tuberculosis (Kohl et al., 2014), and finally a foodborne outbreak caused by S. enterica (Taylor et al., 2015) were utilized. We attempted to recreate the same analyses as reported in the respective publications to demonstrate the “one-stop” analysis with BacPipe.
Outbreak Dynamics of MRSA in an Academic Hospital of Paramaribo, Republic of Suriname
The recent work of Sabat et al reported an investigation of an MRSA outbreak at the Academic Hospital Paramaribo (AZP), Suriname from April to May 2013. The outbreak included 12 patients and one healthcare worker/nurse at the AZP totaling 24 isolates that were used to investigate phylogenetic relatedness and transmission (Sabat et al., 2017). In this study, isolates were sequenced on the MiSeq (V3 kit), and downstream analysis were done using commercial software SeqMan NGen and SeqMan Pro (DNASTAR Inc., USA). Annotation was done using NCBI prokaryotic genome annotation pipeline (PGAP) (Tatusova et al., 2016), and MLST, acquired resistance genes, and SNP analyses were performed using the CGE (http://genomicepidemiology.org/) tools. The data are available under the Bioproject accession number PRJNA312385.
We analyzed all raw reads belonging to 24 isolates and 63 plasmids from this study using BacPipe and produced same results. Firstly, we constructed an SNP-based phylogenetic tree similar to that of Sabat et al. (Sabat et al., 2017), consisting of six distinct clusters (A–F) and one singleton (SUR7) (Figures 5A and 5B). Secondly, the pipeline assessed the MLST of all isolates as ST8, as reported, and confirmed the loss of splD and splE genes (representing important virulence factors) from Cluster F (Data S1). Similar to what was reported, antibiotic resistance patterns of all isolates showed the presence of dfrG trimethoprim resistance, with exception to ClusterE, whereas the ermC gene, conferring resistance to clindamycin, was identified in all isolates of ClusterF and two of ClusterA isolates. For the remainder, it was possible to confirm identical resistance profiles found in all isolates to the previously reported ones including blaZ, mecA, ermC, aphA3, str, msrA, and mphC genes. Thus, similar to the conclusions of Sabat et al. (Sabat et al., 2017), we also identified utilizing BacPipe, a heterogeneous population structure, during this outbreak driven by the different body sites of the same patient or existence of direct transmission between patients. Additionally, virulence factors ssp, atl, efb, and esa were also detected in all analyzed strains by the VFDB database in BacPipe (Chen et al., 2016) (Data S1).
Figure 5.
Comparison of Phylogenetic Analysis
Phylogenetic maximum likelihood tree generated from core-genome SNPs generated through BacPipe and visualized by TreeView tool (A) and from Sabat et al. (Sabat et al., 2017) (B). The scale bar indicates the evolutionary distance between the sequences determined by 0.1 substitutions per nucleotide at the variable positions. See also Data S1.
Tracking a Hospital Outbreak of Carbapenem-Resistant Klebsiella pneumoniae
Snitkin et al described a carbapenem-resistant K. pneumoniae (CRE) outbreak in 2011 at the US National Institutes of Health Clinical Center that affected 18 patients of whom 11 died (Snitkin et al., 2012). The first patient colonized with CRE was placed under contact isolation and treated, yet after three weeks of her discharge, one new case of colonization or active infection was detected every week at the center totalling up to 17 patients. To answer the central question whether patient 1 had initiated the outbreak and if so, how was she linked to the other affected patients, CRE isolated from the 18 patients' samples were analyzed by WGS Roche/454 XLR instrument (Roche Life Sciences). Assembly and annotation was done using gsAssembler and NCBI PGAP, respectively (Snitkin et al., 2012). The data is available under Bioproject accession number PRJNA73841.
The 18 strain sequences were processed through BacPipe. As reported in the study, all 18 CRE belonged to the epidemic ST258 clone and harbored blaKPC-3. SNP-based phylogenetic construction showed two large clusters and a third cluster consisting only of patient 8 and demonstrated that patient 1 was not only linked to the outbreak but also that three independent transmissions of genetically distinct isolates occurred from patient 1 to other patients (Figures 6A and 6B).
Figure 6.
Comparison of Phylogenetic Analysis
Phylogenetic maximum likelihood tree generated through BacPipe and visualized by TreeView tool (A). Putative map of K. pneumoniae transmission during outbreak reproduced from Snitkin et al. (Snitkin et al., 2012). Nodes represent patients, and arrows indicate a transmission event directly or indirectly from one patient to another (B). See also Data S1.
Additional antibiotic resistance genes such as blaSHV, blaTEM, blaOXA, fosA, mphA, catA, oqxA, oqxB, sul1, dfrA12, and aadA2 were also identified in the isolates as were plasmid types IncFII(K), IncFIB (pQil), IncFIB (K), and ColRNAI and a virulence gene, cii in a single process rather than multi-stage analysis as in publication (Data S1).
Tracing Nosocomial Transmission of Clostridium difficile Ribotype 027 in a Chinese Hospital, 2012–2014
In the study by Jia et al. 2016 (Jia et al., 2016), a rare case of C. difficile bloodstream infection (CDBI) was identified. Consequently, all cases or strains that had emerged from the same ward during the past three years were retrospectively analyzed by WGS. Of the 75 patients presenting with diarrhea, C. difficile was isolated from 20 patients, including the case with CDBI. Isolates were sequenced on the HiSeq platform, reads were mapped to R20291 (NAP1/BI/027, ST1) reference strain using the REALPHY tool (Bertels et al., 2014), and the phylogenetic tree was reconstructed by the BEAST tool, whereas the genomic SNP differences between strains were detected using SOAP2 (Li et al., 2009). The data are available under Bioproject accession number PRJNA271048.
BacPipe analysis was able to reproduce the MLST results, where the isolates were characterized into five STs: ST1 (11 patients), ST2 (2 patients), ST8 (2 patients), ST37 (2 patients), and, and ST81 (3 patients) (Data S1). From the SNP-based phylogenetic analysis, we confirmed the finding of Jia et al. of a clear separation between isolates of different STs and that all ST1 isolates were monoclonal (Figures 7A and 7B).
Figure 7.
Comparison of Phylogenetic Analysis
Phylogenetic maximum likelihood tree of C. difficile generated through BacPipe and visualized by TreeView tool (A) and tree reconstructed from multimapping files via Bayesian evolutionary analysis by BEAST from Jia et al. (Jia et al., 2016) (B). See also Data S1.
Additional data not reported in this study but generated through BacPipe were as follows: aac(6′)-aph(2″) gene conferring aminoglycoside resistance and erm(B) conferring macrolide resistance were identified in all isolates belonging to ST1, ST38, and ST81, whereas tet(M) conferring tetracycline resistance was identified in isolates belonging to ST37 and ST81. Additionally, for all isolates belonging to ST1, we were able to identify rep1 plasmid, which was not detected in the other non-ST1 isolates (Data S1).
Whole-Genome-Based Surveillance of Mycobacterium tuberculosis
Kohl et al. trace an M. tuberculosis complex (MTBC) longitudinal outbreak compromising 26 isolates (between 2001 and 2010) showing identical IS 6110 DNA fingerprint and spoligotype patterns. These underwent WGS using MiSeq (Illumina), reads were mapped to the H37Rv reference genome using the exact alignment program SARUMAN, and SNPs were extracted from the mapped reads by customized Perl scripts (Kohl et al., 2014). Raw reads are available on Bioproject accession number PRJEB6276.
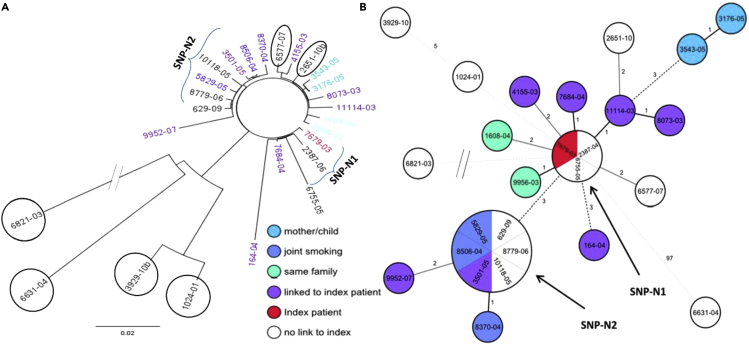
Using BacPipe, we confirmed that 22 isolates were grouped into one major cluster while four were outliers. Within the primary cluster, we also confirmed two sub-groups compromising of three and six strains, SNP-N1 and SNP-N2, respectively (Figures 8A and 8B).
Figure 8.
Comparison of Phylogenetic Analysis
Phylogenetic maximum likelihood tree of M. tuberculosis core-genome SNPs generated through BacPipe and visualized by TreeView tool (A) and a minimum spanning tree of concatenated sequences of the 322 SNPs of the same data from Kohl et al (Kohl et al., 2014) (B). See also Data S1.
Additional data not reported in this study but generated through BacPipe were an assessment of antibiotic resistance where all isolates harbored aac(2′)-Ic and subclass B1 beta-lactamase genes conferring aminoglycoside and beta-lactam resistance, respectively (Supplemental Information).
Characterization of Foodborne Outbreaks of Salmonella enterica Serovar Enteritidis with Whole Genome Sequencing for Surveillance and Outbreak Detection
Taylor et al. described the application of whole genome sequencing for the detection of S. enterica serovar Enteritidis outbreaks from isolates previously characterized by PFGE in Minnesota and Ohio between 2001 and 2014 (Taylor et al., 2015). The cohort contained 28 isolates from seven epidemiologically confirmed foodborne outbreaks and 27 non-epidemiologically linked sporadic isolates that were assessed by WGS (MiSeq). Reads were mapped to the reference genome using BWA-MEM tools, which were later sorted and de-duplicated by the Picard tool. The variant call file (VCF) was produced with BCF tool, and the maximum-likelihood phylogenetic tree was calculated with PhyML. Raw reads are available on accession number: PRJNA237212.
BacPipe was able to retrieve the same phylogenetic tree, confirming that all isolates within the same outbreak were closely related (ranging from 2–7 isolates per outbreak) (Figure 9A). We derived the same conclusion as the original study, that the serovar Enteritidis shows little genetic diversity in the host over time, from investigating isolates MDH-2014-00222, MDH-2014-00223, MDH-2014-00225, and MDH-2014-00228 that were isolated from an individual over a five-week period (see outbreak 2 in Figure 9B).
Figure 9.
Comparison of Phylogenetic Analysis
Maximum-likelihood tree of S. enteritidis produced by SNP analysis showing outbreak clusters and time frame (month[s] and year) and the State from where each isolate originated. The phylogenetic analysis generated through BacPipe and visualized by TreeView tool (A) and tree reproduced from Taylor et al. (Taylor et al., 2015) (B). See also Data S1.
Additional data not reported in this study but generated through BacPipe were the assignment of all strains as ST11 and identification of IncFIB(S) and IncFII(S) plasmids within all isolates, with exception of MDH-2014-00232, MDH-2014-00245, and outbreak 6/7 isolates—where no plasmids were detected—and MDH-2014-00215 and MDH-2014-00247 isolates—where IncHI1B, Incl1, IncHI1A, and IncFIA(HI1) plasmids were identified. Similarly, from MDH-2014-00215 and MDH-2014-00247, antibiotic resistance genes such as blaTEM-1B, catA1, sul1, tet(B), and dfrA7 were identified, whereas none were found in the remaining isolates (Data S1).
Conclusion
Here we have presented BacPipe, a bacterial whole genome sequencing analysis pipeline and demonstrated its robustness in handling diverse genomes of clinically important pathogens characterized by different sizes, GC content, and presence of repeat regions that are challenging for downstream data analysis. Along with being comprehensive and modular, BacPipe has the advantage of being a fast-on account of parallel computing, and requiring computationally low-resource as pipeline functionality does not require an internet connection or high-end computers. This would also allow the analysis to be performed locally, which is highly advantageous to hospitals that are mandated to comply with data protection guidelines.
A graphical interface makes it very user-friendly; a user can specify the tools to allow visualization of the included in the analysis and adjust the database/parameters from a drop-down list or buttons. BacPipe can be run either with raw reads from various sequencing platforms or can pick-up the analysis from any step throughout the workflow giving tremendous flexibility. The open-source nature and GNU license would allow more expert users to modify and adapt the software to their preferences.
The endpoint of the analysis provides various levels of details, from an overview comparing the results across all analyzed samples, to an Excel file with all of the compiled tool results, to very detailed folders dedicated for each tool output and log files. Additionally, the output of BacPipe can easily be used to study the pan-genome and perform comparative genome analysis, define gene acquisition/loss through horizontal gene transfer, and perform functional analysis through the KEGG ortholog database. Finally, although prior publications have utilized numerous heterogeneous tools to delineate hospital or community-based pathogen transmission, we demonstrated that the collection of tools within BacPipe could reproduce the entire analyses as a “one-stop” platform in less than an hour. Future development of BacPipe entails expansion of tools to enable identification of prophages, IS (insertion sequence) elements, CRISPR-Cas elements, and, depending upon their open-access status, whole-/core-/pan-genome MLST schemes. We believe this fully automated pipeline will help to overcome one of the primary barriers to analyzing and interpreting WGS data, facilitating applications for routine patient care in hospitals and public health and infection control monitoring.
Limitations of the Study
Although Bacpipe is a fully automated pipeline with a user-friendly GUI that requires minimal user intervention, the heavy reliance on open-access data resources makes it imperative that these are well-curated, up-to-date, and comprehensive.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The study and M.B. were supported by European Union Horizon 2020 Research and Innovation Programme: Compare (COllaborative Management Platform for detection and Analyses of (Re-) emerging and foodborne outbreaks in Europe: Grant No. 643476). B.B.X. was supported by the Innovative Medicines Initiative Joint Undertaking under grant agreement n° 115523; COMBACTE (Combatting Bacterial Resistance in Europe, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies in kind contribution). J.A.C. and B.R.G. were partially funded by BacGenTrack (Tubitak/0004/2014); Fundação para a Ciência e a Tecnologia (FCT)/Scientific and Technological Research Council of Turkey(TUBITAK); and Oneida project (LISBOA-01-0145-Feder-016417) “Fundos Europeus Estruturais E De Investimento” (FEEI) from “Programa operacional Regional LisBOA2020” and FCT National Funds. Fundacao para a ciencia e a technologia (PSFRH/BD/101448/2014) Ph.D grant to B.R.G.
Author Contributions
This work was conceptualized by S.M.K. The study was designed by S.M.K. and B.B.X. The pipeline was developed and validated by M.M., B.B.X., M.B., and C.L. B.R.G. and J.A.C. contributed to the dockerization of the platform. B.T.F.A. and P.H. integrated and validated tool at EBI-SELECTA. The manuscript was drafted by B.B.X., M.M., S.K.S., G.C., H.G., and S.M.K. and was reviewed by all authors.
Declaration of Interests
None declared.
Published: January 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.100769.
Data and Code Availability
BacPipe can be obtained through https://hub.docker.com/r/mahmed/bacpipe (docker image) https://github.com/wholeGenomeSequencingAnalysisPipeline/BacPipe (Github approach).
Supplemental Information
References
- Afgan E., Sloggett C., Goonasekera N., Makunin I., Benson D., Crowe M., Gladman S., Kowsar Y., Pheasant M., Horst R. Genomics virtual laboratory: a practical bioinformatics workbench for the cloud. PLoS One. 2015;10:e0140829. doi: 10.1371/journal.pone.0140829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgün M., Bayrak A.O., Ozer B., Sağıroğlu M.Ş. Privacy preserving processing of genomic data: a survey. J. Biomed. Inform. 2015;56:103–111. doi: 10.1016/j.jbi.2015.05.022. [DOI] [PubMed] [Google Scholar]
- Arnold C. Outbreak breakthrough: using whole-genome sequencing to control hospital infection. Environ. Health Perspect. 2015;123:A281–A286. doi: 10.1289/ehp.123-A281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertels F., Silander O.K., Pachkov M., Rainey P.B., van Nimwegen E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014;31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zheng D., Liu B., Yang J., Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccuru G., Orsini M., Pinna A., Sbardellati A., Soranzo N., Travaglione A., Uva P., Zanetti G., Fotia G. Orione, a web-based framework for NGS analysis in microbiology. Bioinformatics. 2014;30:1928–1929. doi: 10.1093/bioinformatics/btu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg R.H., Bathoorn E., Chlebowicz M.A., Couto N., Ferdous M., García-Cobos S., Kooistra-Smid A.M.D., Raangs E.C., Rosema S., Veloo A.C.M. Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol. 2017;243:16–24. doi: 10.1016/j.jbiotec.2016.12.022. [DOI] [PubMed] [Google Scholar]
- Didelot X., Bowden R., Wilson D.J., Peto T.E.A., Crook D.W. Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 2012;13:601. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Du P., Yang H., Zhang Y., Wang J., Zhang W., Han G., Han N., Yao Z., Wang H. Nosocomial transmission of Clostridium difficile ribotype 027 in a Chinese hospital, 2012–2014, traced by whole genome sequencing. BMC Genomics. 2016;17:405. doi: 10.1186/s12864-016-2708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen K.G., Scheutz F., Lund O., Hasman H., Kaas R.S., Nielsen E.M., Aarestrup F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl T.A., Diel R., Harmsen D., Rothgänger J., Walter K.M., Merker M., Weniger T., Niemann S. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J. Clin. Microbiol. 2014;52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koser C.U., Ellington M.J., Cartwright E.J., Gillespie S.H., Brown N.M., Farrington M., Holden M.T., Dougan G., Bentley S.D., Parkhill J. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012;8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong J.C., McCallum N., Sintchenko V., Howden B.P. Whole genome sequencing in clinical and public health microbiology. Pathology. 2015;47:199–210. doi: 10.1097/PAT.0000000000000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Lüth S., Kleta S., Al Dahouk S. Whole genome sequencing as a typing tool for foodborne pathogens like Listeria monocytogenes – the way towards global harmonisation and data exchange. Trends Food Sci. Technology. 2018;73:67–75. [Google Scholar]
- Moran-Gilad J. Whole genome sequencing (WGS) for food-borne pathogen surveillance and control - taking the pulse. Euro Surveill. 2017;22:30547. doi: 10.2807/1560-7917.ES.2017.22.23.30547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir P., Li S., Lou S., Wang D., Spakowicz D.J., Salichos L., Zhang J., Weinstock G.M., Isaacs F., Rozowsky J. The real cost of sequencing: scaling computation to keep pace with data generation. Genome Biol. 2016;17:53. doi: 10.1186/s13059-016-0917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punina N.V., Makridakis N.M., Remnev M.A., Topunov A.F. Whole-genome sequencing targets drug-resistant bacterial infections. Hum. Genomics. 2015;9:19. doi: 10.1186/s40246-015-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat A.J., Hermelijn S.M., Akkerboom V., Juliana A., Degener J.E., Grundmann H., Friedrich A.W. Complete-genome sequencing elucidates outbreak dynamics of CA-MRSA USA300 (ST8-spa t008) in an academic hospital of Paramaribo, Republic of Suriname. Scientific Rep. 2017;7:41050. doi: 10.1038/srep41050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snitkin E.S., Zelazny A.M., Thomas P.J., Stock F., Henderson D.K., Palmore T.N., Segre J.A. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.J., Lappi V., Wolfgang W.J., Lapierre P., Palumbo M.J., Medus C., Boxrud D. Characterization of foodborne outbreaks of Salmonella enterica serovar enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J. Clin. Microbiol. 2015;53:3334–3340. doi: 10.1128/JCM.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M.C.F., Ahrenfeldt J., Cisneros J.L.B., Jurtz V., Larsen M.V., Hasman H., Aarestrup F.M., Lund O. A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS One. 2016;11:e0157718. doi: 10.1371/journal.pone.0157718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BacPipe can be obtained through https://hub.docker.com/r/mahmed/bacpipe (docker image) https://github.com/wholeGenomeSequencingAnalysisPipeline/BacPipe (Github approach).