Abstract
We examined the intensity and anisotropy decays of DNA labeled with two ruthenium metal-ligand complexes, [Ru(bpy)2(dppz)]2+ and [Ru(phe)2(dppz)]2+. Both complexes display high emission anisotropies in the absence of rotational diffusion, making them suitable probes for rotational motions. When bound to DNA, these complexes display decay times as long as 294 ns, providing long-lived probes of DNA dynamics. The decay times of both complexes were rather insensitive to dissolved oxygen. We examined anisotropy decays of these complexes bound to B-form DNA. The anisotropy decays revealed correlation times near 10, 50, and several hundred nanoseconds, suggesting that these probes are sensitive to a wide range of DNA motions. The use of metal-ligand complexes should allow resolution of both the torsional and bending motions of DNA, the latter of which has been mostly inaccessible using shorter-lived fluorescent probes bound to DNA.
Keywords: Metal-ligand complexes, luminescent probes, DNA, ruthenium, (II) complexes, DNA intercalator
INTRODUCTION
Time-resolved fluorescence has been widely used to study the solution dynamics of DNA.(1–8) Essentially all these studies have been performed using fluorophores such as acridine, 2-aminopurine, and ethidium bromide, which display decay times shorter than 30 ns. However, extensive theoretical studies of DNA have predicted a wide range of times for the torsional and bending motion of DNA. (9–12) While nanosecond probes can reveal the faster torsional motions of DNA, it is well-known that the 10−7- to 10−5-s bending motions of DNA are difficult to resolve using fluorophores with nanosecond decay times.(13, 14)
We recently described a new class of long-lived fluorescence probes which display useful anisotropy properties. We found that molecules of the [Ru(bpy)3]2+5 class, but which contain one suitable nonidentical organic ligand, display high anisotropy in the absence of rotational diffusion.(15–17) The observation of usefully high anisotropies for molecules such as [Ru(bpy)3]2+ was surprising in that one expects such symmetrical molecules to display low anisotropy. This is not an obvious result because some studies of these compounds have suggested that the emission is depolarized by energy transfer between the organic diimine ligands.(18–20) Our studies have demonstrated high time 0 anisotropies, and that the anisotropies were sensitive to the rates of rotational diffusion.(15–17) We have used such metal-ligand complexes to measure rotational correlation times longer than 5 μs(17) and for fluorescence polarization immunoassays of high molecular weight species.(16) Hence, measurement of the anisotropy decays of macromolecules labeled with these probes extends the time scale of anisotropy decay measurements to much longer times or slower motions than can be observed using nanosecond decay time fluorophores.
We now describe the use of luminescent metal-ligand complexes to study the hydrodynamic properties of DNA. We used complexes of the type shown in Scheme I, where one of the ligands is dppz, and the other can be bpy or phe. The use of such complexes as DNA probes has been reported by Barton and co-workers.(21–23) The main advantage of these probes is that they display little or no emission in water but are highly fluorescent when bound to DNA. This property is apparently due to the dppz ligand. When dissolved in solution interactions with the distal nitrogens on dppz with water appear to quench the emission of the complex, an effect which is no longer present when bound to DNA.(22) We now show that the metal-ligand complexes shown in Scheme I display anisotropies as high as 0.2 when excited in the last absorption band near 500 nm. The long decay times of these probes when bound to DNA, and their favorable anisotropy properties, allowed us to detect rotational motions as long as several hundred nanoseconds. Since it is known that the intensity decay times of such complexes can range from several nanoseconds to 100 μs, (24, 25) metal-ligand complexes should be useful for studies of DNA motions in a wide range of cellular environments.
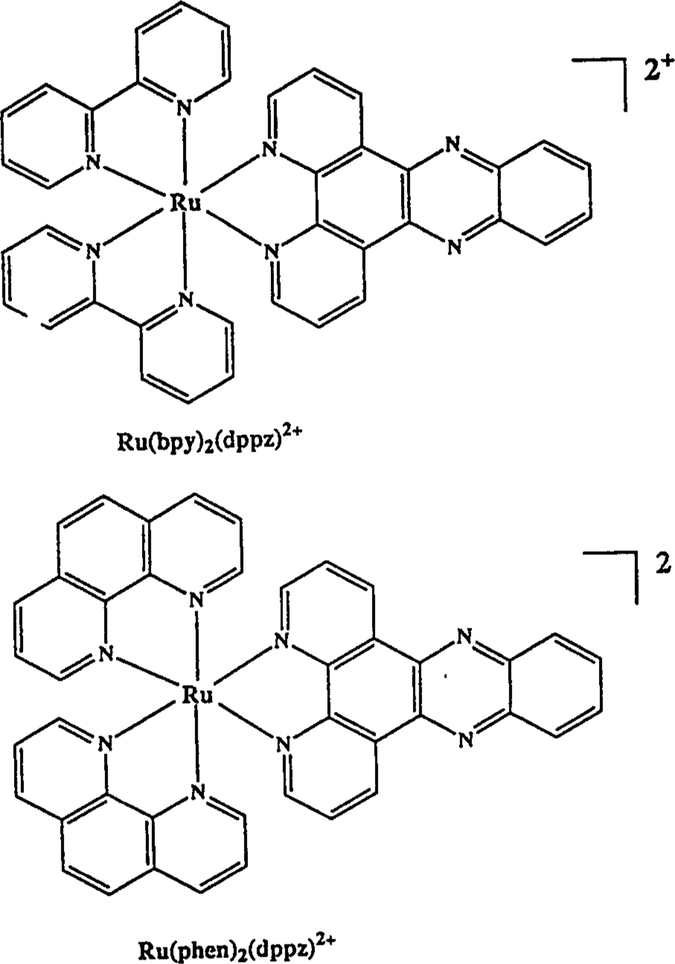
Scheme I.
Structures of [Ru(bpy)2(dppz)]2+ and [Ru(phen)2(dppz)]2+.
MATERIALS AND METHODS
Materials
RuCl3 hydrate, 2,2’-bipyridine (bpy) (99%), 1,10-phenanthroline (phe) (99+%), and ammonium hexafluorophosphate (99.99%) were obtained from Aldrich and used as received; o-phenylenediamine (Aldrich, 98%) was recrystallized from water before use. Water was deionized with a Barnsted nanopure system. Acetonitrile was dried over molecular sieves and distilled from CaH2. All solvents and other chemicals were of at least reagent-grade quality and were used without further purification. NMR spectra were obtained on a Bruker AMX 300 MHz spectrometer with all chemical shifts referenced to TMS. Time-resolved luminescence measurements on the complexes in dry, argon-saturated CH3CN were performed as previously described.(26)
Synthesis
Ru(bpy)2(Cl)2 and Ru(phe)2C12 were prepared through the same method using RuC13 and the appropriate diimine.(27) 1,10-Phenanthroline-5,6-dione was prepared according to a literature procedure(28) and was subsequently recrystallized from MeOH. Dipyrido[3,2-a:2’,3-c]phenazine (dppz) was synthesized by reaction of 1,10-phenanthroline-5,6-dione with 1.1 equivalents of o-phenylenediamine in refluxing EtOH.(29) 1H NMR of dppz in (CD3)2SO was in agreement with the literature spectrum.(30)
[Ru(L-L)2(dppz)(PF6)2. Either Ru(bpy)2(Cl)2 (15.8 mg, 4.35 × 10−5 mol) or Ru(phe)2(C1)2 (22 mg, 3.87 × l0−5 mol) was suspended in a minimal volume of MeOH (10 mL) with 1.1 equivalents of dppz. The solution was refluxed with stirring for 2.5 hr, then cooled to room temperature. This red solution was filtered to remove solid impurities, followed by the addition of a concentrated aqueous solution of NH4PF6. The precipitate was collected by vacuum filtration on a medium frit and was washed with cold MeOH followed by ether. Typical yields ranged between 70 and 85%. 1H NMR spectra of both Ru(bpy)2(dppz)(PF6)2 and Ru(phe)2(dppz)(PF6)2 in (CD3)2CO and CD2Cl2 were in agreement with literature spectra.(30, 31) UV-vis absorbance spectra of the complexes were also consistent with the literature.(30, 31)
These compounds were nonfluorescent in water, which is consistent with the absence of [Ru(bpy)3]2+ or [Ru(phe)3]2+ impurities. The lifetimes of the bpy and phe complexes in very dry acetonitrile and without O2 measured by time-resolved emission were single exponentials with decay times of 750 and 787 ns, respectively. Barton and co-workers(22) reported smaller values, which could be due to dissolved oxygen or water in the solvent used for their measurements.
Calf thymus DNA was obtained from Sigma Chemical Co. and was used without further purification and without sonication. DNA was dissolved in 5 mM tris (pH 7.0) containing 50 mM NaC1. The concentration of metal-ligand complex and DNA base pairs in these solutions was 6 and 200 μM, resulting in one probe molecule per 33 base pairs. For measurement of the excitation anisotropy spectra 5 μM bpy or phe complex was dissolved in 100% glycerol and the emission was observed through a monochromater at 670 nm.
Emission spectra were recorded on a SLM AB2 spectrofluorometer. Time-resolved data were obtained by time-correlated single-photon counting (TCSPC).(32) The light source was the frequency-doubled output of a Pridine 1 dye laser, cavity-dumped at 0.5 or 1 MHz and frequency-doubled to 365 nm. For TCSPC measurements the emission was isolated using a long pass filter transmitting above 520 nm. The detector was a Hamamatsu R2809 red-sensitive microchannel plate PMT.
The intensity decays were analyzed by nonlinear least-squares using software provided by IBH, Inc., in terms of a multiexponential decay model,
| (1) |
where αi and the preexponential factors associated with the decay time (τi). The fractional intensity (fi) of each decay time component to the steady-state emission can be obtained from
| (2) |
The anisotropy decay data were also analyzed with software from IBH, Inc., using the multicorrelation time model
| (3) |
where ri are the amplitudes associated with the ith correlation time (θi).
RESULTS
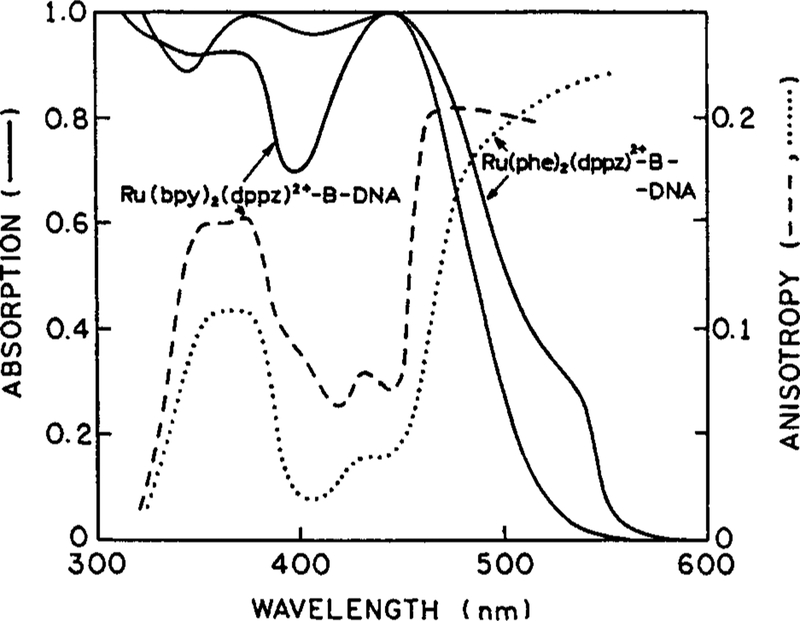
The absorption and anisotropy spectra of [Ru(bpy)2(dppz)]2+ and [Ru(phe)2(dppz)]2+ are shown in Fig. 1. The spectral properties of these complexes are similar, except the phe complex absorbs at somewhat longer wavelengths. In the absence of rotational motions (−60°C in glycerol), the excitation anisotropy spectra of the bpy and phe complexes both display maxima near 365 and 500 nm. These high values of the anisotropy indicate that the excitation is localized on one of the organic ligands, and not randomized among these ligands. Since the dppz ligand intercalates into DNA, and the emission intensity is sensitive to intercalation, it seems likely that the excited state is a charge transfer state between the Ru and the dppz.
Fig. 1.
Absorption and excitation anisotropy spectra of the two MLC DNA probes. Absorption spectra are in aqueous solution, bound to DNA. Anisotropy spectra are in glycerol at −60°C.
The anisotropy spectra for the bpy and dppz complexes are rather favorable for measurements of time-resolved anisotropies. One can select a range of excitation wavelengths, from 330 to 350 nm and from 480 to 550 nm. Such wavelengths can be obtained from a variety of pulsed lasers.
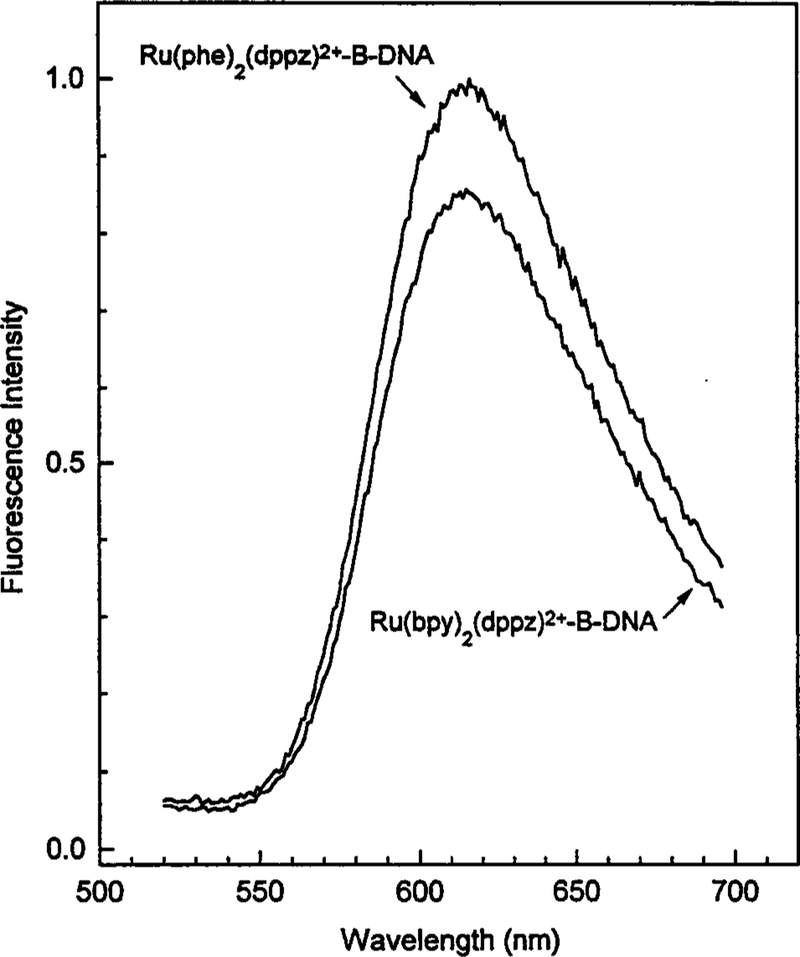
The emission spectra of the bpy and pile complexes bound to calf thymus DNA are shown in Fig. 2. In aqueous solution the probes are nearly undetectable by the emission. In the presence of DNA the luminescence of these probes is remarkably enhanced, an effect attributed to intercalation of the dppz ligand into double-helical DNA.(33, 34) Hence probe emission is observed only from the DNA-bound form, without contributions from free probe in solution. In the intensity-normalized emission spectra (not shown), one notices that the emission of the phe complex is shifted several nanometers to longer wavelengths, compared to the bpy complex.
Fig. 2.
Emission spectra of [Ru(bpy)2(dppz)]2+ and [Ru(phe)2(dppz)]2+ bound to B-DNA.
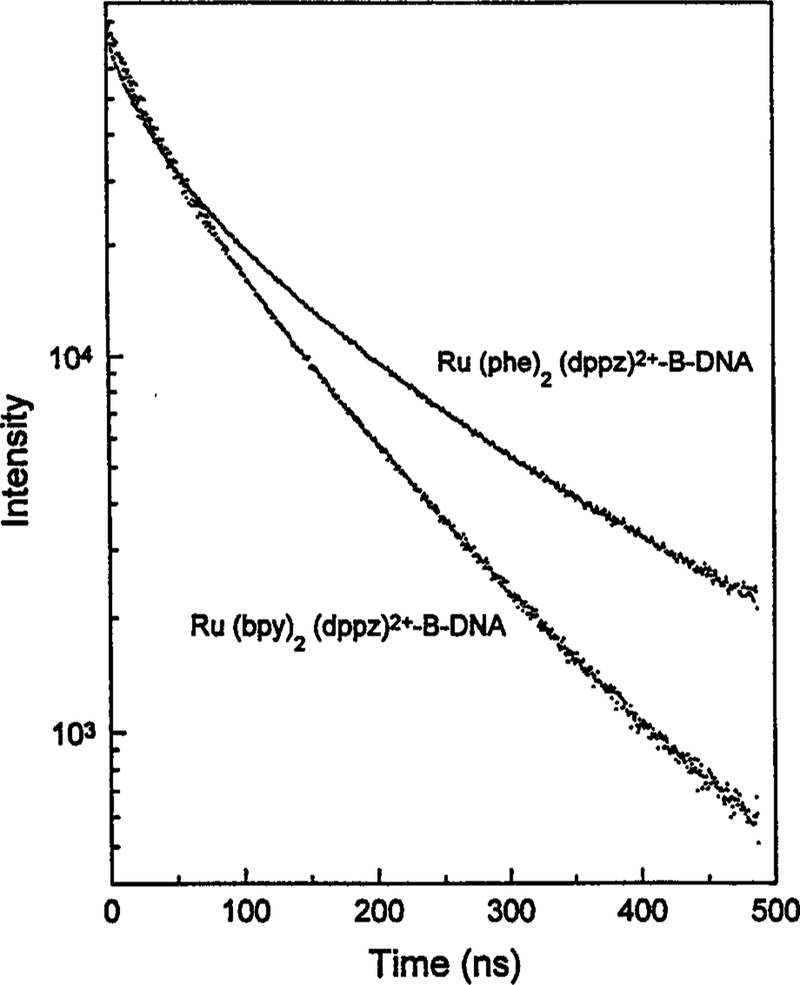
The time-resolved intensity decays of the bpy and phe complexes bound to calf thymus DNA are shown in Fig. 3. The intensity decays are best fit by triple-exponential decays for both the bpy and the phe complexes (Table I). Use of the double-exponential model yielded reduced values which were four- to sevenfold larger than for the triple-exponential fit. Such a large difference in indicates that at least three decay times are needed to explain the data. However, the intensity decay could be still more complex, but the additional complexity not resolvable with the present experimental data. The mean lifetime (τ) of the phe complex bound to DNA (τ = 145 ns) is about 50% longer than that of the bpy complex (τ = 99 ns). This difference in decay time is readily visible in the intensity decay (Fig. 3) and suggests that the phe complex is the better choice for examining DNA motions on the submicrosecond timescale.
Fig. 3.
Intensity decays of [Ru(bpy)2(dppz)]2+ on [Ru(phe)2(dppz)]2+ bound to B-form DNA.
Table I.
Intensity Decays of [Ru(bpy)2(dppz)]2+ and [Ru(phe)2(dppz)]2+ Bound to B-DNAa
| Lifetime | % rel. amplitudeb | ||||||
|---|---|---|---|---|---|---|---|
| Probea | τ1 (ns) |
τ2 (ns) |
τ3 (ns) |
f1 | f2 | f3 | |
| [Ru(bpy)2(dppz)]2+ | |||||||
| 78.97 | — | — | 100 | — | — | 68 | |
| 34.28 | 107.74 | — | 22 | 78 | — | 7 | |
| 22.28 | 67.09 | 149.90 | 9 | 48 | 41 | 0.94 | |
| [Ru(phe)2(dppz)]2+ | |||||||
| 138.41 | — | — | 100 | — | — | 239 | |
| 42.03 | 191.59 | — | 23 | 77 | — | 4.27 | |
| 24.14 | 89.37 | 250.40 | 8 | 37 | 54 | 1.02 | |
The concentration of DNA was 2 × 10−4 M in base pairs, and the concentration of the metal-ligand complex was 6 μM, for a base pair-to-probe ratio of 33:1. The buffer was 50 mM NaCl, 5 mM Tris, pH 7.0, at 20°C.
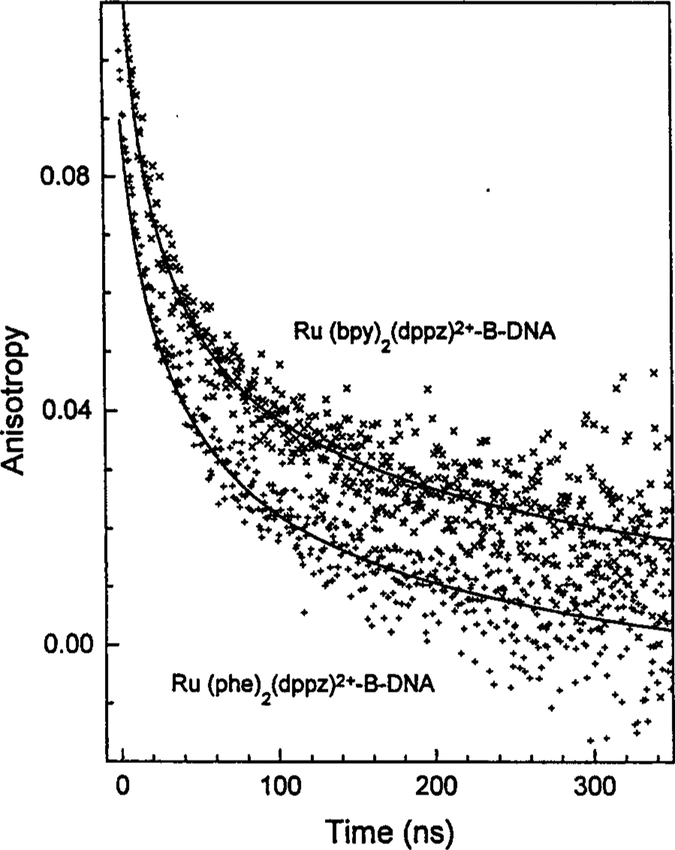
Time-resolved anisotropy decays of the DNA-bound phe and bpy complexs are shown in Fig. 4. The anisotropy decays are not characterized by a single correlation time, and fitting the data requires up to three correlation times (Table II). Once again, the values for the double-exponential fit indicate that the decay is more complex than a two-correlation time model. Similar correlation times and amplitudes were observed with both complexes (Table II). The correlation times range from 22 to 250 ns, and the complexity of the recovered anisotropy decay indicates that these probes will be useful for quantifying a range of DNA motions. At present we feel that the frequency-domain method will be more effective than TCSPC for quantifying the anisotropy decays of these long-lived complexes. However, our present instrumentation did not permit frequency-domain measurements on these long-lived complexes. Importantly, the results shown in Fig. 4 indicate that rotational motions as long as 300 ns are readily measured using these metal-ligand probes.
Fig. 4.
Anisotropy decays of [Ru(bpy)2(dppz)]2+ and [Ru(phe)2(dppz)]2+ bound to B-DNA.
Table II.
Anisotropy Decay of [Ru(bpy)2(dppz)]2+ and [Ru(phen)2(dppz)]2+ Bound to B-DNAa
| Correlation time | Anisotropy | ||||||
|---|---|---|---|---|---|---|---|
| Probe | θ1 (ns) |
θ2 (ns) |
θ3 (ns) |
r1 | r2 | r3 | |
| [Ru(bpy)2(dppz)]2+ | |||||||
| 45.17 | — | — | 0.084 | — | — | 2.50 | |
| 12.30 | 106.87 | — | 0.05 | 0.057 | — | 1.39 | |
| 6.06 | 32.73 | 243.90 | 0.03 | 0.05 | 0.04 | 1.35 | |
| [Ru(phe)2(dppz)]2+ | |||||||
| 59.52 | — | — | 0.075 | — | — | 1.35 | |
| 19.28 | 144.28 | — | 0.041 | 0.050 | — | 0.989 | |
| 8.74 | 37.66 | 224.58 | 0.017 | 0.0396 | 0.040 | 0.982 | |
See Table I, footnote a, for experimental conditions.
We examined the effects of dissolved oxygen on the intensity decays of the complexes bound to DNA. These results are presented as mean decay times in the absence and presence of oxygen (Table III). These values can be used to calculate the oxygen biomolecular quenching contact kq using
| (4) |
where τ0 and τ are the decay times in the absence and presence of oxygen, respectively, and [O2] is the oxygen concentration. When equilibrated with 1 atm of oxygen, the concentration in water is 0.001275 M.(35) For fluorophores in aqueous solution, in the absence of steric barriers to contact with oxygen, kq is near 1 × 1010 M−1 s−1.(35) Hence, the probes when bound to DNA are shielded by a factor of 20- to 40-fold from contact with oxygen. This degree of shielding is comparable to the 30-fold shielding of ethidium bromide from oxygen when bound to DNA.(35) Ethidium bromide is known to intercalate between the DNA base pairs. Hence, the inaccessibility of these MLC probes to oxygen quenching supports the hypothesis that the excited state is localized between the dppz and the Ru and that the dppz ligand is intercalated into the DNA helix.
Table III.
Mean Decay Times of the Metal-Ligand Complexes Bound to B-DNA in the Absence and Presence of Oxygen, and the Oxygen Biomolecular Quenching Constants
| Probe | τ0 (ns)a | τ (ns) | kq (M−1 s−1)b |
|---|---|---|---|
| [Ru(bpy)2(dppz)]2+ | 99.4 | 97.1 | 2.4 × 108 |
| [Ru(phe)2(dppz)]2+ | 175 | 159 | 4.5 × 108 |
Calculated using τ0 or τ = Σfi τi.
Calculated using Eq. (4) with an oxygen concentration of 1.275 × 10−3 M.
DISCUSSION
The long-lived probes of DNA dynamics can have numerous applications for biophysical studies of DNA. This probe can be used to study DNA in a wide range of environments, as has been done for DNA labeled with ethidium bromide. This probe, which displays a decay time near 30 ns, has been used to study DNA in solution, (36) in chromatin, (37) and in bacteriophage.(38) The long lifetime of the metal-ligand complexes may provide additional insights into the effect of the local environment on DNA dynamics. Additionally, one can imagine the use of global analysis(39) with short- and long-lived probes to provide increased resolution of DNA dynamics.
The use of long-lived probes may also provide a means to measure site-to-site motions of DNA. It is known that site-to-site diffusion coefficients in proteins can be determined from measurements of time-resolved fluorescence resonance energy transfer(40) However, using nanosecond decay time probes, such measurements are limited to rather rapid rates of diffusion, in excess of 10−7 cm2/s. Much slower site-to-site motions should be measured with these long-lived complexes.
In closing, the use of metal-ligand complexes as DNA probes is in its infancy, and additional experimentation is needed to realize the potential of these probes.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institute of Health (RR-08119 and GM-35154), with support for instrumentation from Grants RR-07510 and RR-10416.
Footnotes
Dedicated to Professor Robert F. Steiner upon his retirement.
Abbreviations used: bpy, 2,2’-bipyridyl; bpy complex, [Ru(bpy)2(dppz)]2+; DNA, calf thymus DNA; dppz, dipyrido[3,2-a’,2’,3’,-c]phenazine; EB, ethidium bromide; MLC, metal-ligand complexes; phe, 1,10-phenanthroline; phe complex, [Ru(phe)2(dppz)]2+; TCSPC, time-correlated single-photon counting.
REFERENCES
- 1.Millar DP, Robbins RJ, and Zewail AH (1980) Proc. Natl. Acad. Sci. USA 77(10), 5593–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millar DP, Robbins RJ, and Zewail AH (1982) J. Chem. Phys 76(4), 2080–2094. [Google Scholar]
- 3.Magde D, Zappala M, Knox WH, and Nordlund TM (1983) J. Phys. Chem 87, 3286–3288. [Google Scholar]
- 4.Genest D, Wahl Ph., Erard M, Champagne M, and Daune M (1982) Biochimie 64, 419–427. [DOI] [PubMed] [Google Scholar]
- 5.Georghiou S (1977) Photochem. Photobiol 26, 59–68. [DOI] [PubMed] [Google Scholar]
- 6.Steiner RF and Kubota Y (1983) in Steiner RF (Ed.), Excited States of Biopolymers, Plenum Press, New York, pp. 203–254. [Google Scholar]
- 7.Xu D, Evans KO, and Norlund TM (1994) SPIE Proc. 2137, 661–672. [Google Scholar]
- 8.Guest CR, Hoehstrasser RA, Sowers LC, and Millar DP (1991) Biochemistry 30, 3271–3279. [DOI] [PubMed] [Google Scholar]
- 9.Allison SA and Schurr JM (1979) Chem. Phys 41, 35–59. [Google Scholar]
- 10.Barkley MD and Zimm BH (1979) J. Chem. Phys 70(60), 2991–3007. [Google Scholar]
- 11.Garcia de la Toree J, Navarro S, Lopez Martinez MC, Diaz FG, and Lopez Cascales JJ (1994) Biophys. J 67, 530–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia de la Torre J, Navarro S, and Lopez Martinez MC (1994) Biophys. J 66, 1573–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schurr JM, Fujimoto BS, Wu P, and Song L (1992) in Lakowicz JR (Ed.), Topics in Fluorescence Spectroscopy, Vol. 3: Biochemical Applications, Plenum, New York, pp. 137–229. [Google Scholar]
- 14.Millar DP, Ho KM, and Aroney MJ (1988) Biochemistry 27, 8599–8606. [DOI] [PubMed] [Google Scholar]
- 15.Terpetschnig E, Szmacinski H, Malak H, and Lakowicz JR (1995) Biophys. J 68, 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terpetschnig E, Szmacinski H, and Lakowicz JR (1995) Anal. Biochem 227, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szmacinski H, Terpetschnig E, and Lakowicz JR (1996) Biophys. Chem 62, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson J and Krausz E (1987) Inorg. Chem 26, 1383–1386. [Google Scholar]
- 19.Pogge JL and Kelley DF (1995) Chem. Phys. Lett 238, 16–24. [Google Scholar]
- 20.Riesen H, Gao Y, and Krausz E (1994) Chem. Phys. Len 228, 610–615. [Google Scholar]
- 21.Barton JK (1986) Science 233, 727–734. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins Y, Friedman AE, Turro NJ, and Barton JK (1992) Biochemistry 31, 10809–10816. [DOI] [PubMed] [Google Scholar]
- 23.Murphy CJ, Arkin MR, Ghatlia ND, Bossmann S, Turro NJ, and Barton JK (1994) Proc. Natl. Acad. Sci. USA 91, 5315–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demas JN and DeGraff BA (1992) Makromol. Chem. Macromol. Symp 59, 35–51. [Google Scholar]
- 25.Reitz GA, Demas JN, DeGraff BA, and Stephens EM (1988) J. Am. Chem. Soc 110, 5051–5059. [Google Scholar]
- 26.Castellano FN, Heimer TA, Tandhasetti MT, and Meyer GJ (1994) Chem. Mater 6, 1041–1048. [Google Scholar]
- 27.Sullivan BP, Salmon DJ, and Meyer TJ (1978) Inorg. Chem 17, 3334–3341. [Google Scholar]
- 28.Yamada M, Tanaka Y, Yoshimoto Y, Kuroda S, and Shimao I (1992) Bull. Chem. Soc. Ipn 65, 1006–1011. [Google Scholar]
- 29.Dickeson JE and Summers LA (1970) Aust. J. Chem 23, 1023–1027. [Google Scholar]
- 30.Amouyal E, Homsi A, Chambron J-C, and Sanvage J-P (1990) J. Chem. Soc. Dalton Trans 1841–1845. [Google Scholar]
- 31.Hartshorn RM and Barton JK (1992) J. Am. Chem. Soc 114, 5919–5925. [Google Scholar]
- 32.Birch DJS and Imhof RE (1991) in Lakowicz JR (Ed.), Topics in Fluorescence Spectroscopy, Vol. 1. Techniques, Plenum, New York, pp. 1–45. [Google Scholar]
- 33.Friedman AE, Chambron J-C, Sauvage J-P, Turro NJ, and Barton JK (1990) J. Am. Chem. Soc 112, 4960–4962. [Google Scholar]
- 34.Hartshorn RM and Barton JK (1992) J. Am. Chem. Soc 114, 5919–5925. [Google Scholar]
- 35.Lakowicz JR and Weber G (1973) Biochemistry 12, 4161–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashikawa I, Kinostia K, and Ikegami A (1984) Biochim. Biophys. Acta 782, 87–93. [DOI] [PubMed] [Google Scholar]
- 37.Ashikawa I, Kinosita K, Ikegami A, Nishimura Y, Tsuboi M, Watanabe K, Iso K, and Nakano T (1983) Biochemistry 22, 6018–6026. [DOI] [PubMed] [Google Scholar]
- 38.Ashikawa I, Furono T, Kinosita K, Ikegami A, Takahashi H, and Akutsu H (1984) J. Biol. Chem 259(13), 8338–8344. [PubMed] [Google Scholar]
- 39.Beechem JM, Gratton E, Ameloot M, Knutson JR, and Brand L (1991) in Lakowicz JR (Ed.), Topics in Fluorescence Spectroscopy, Vol. 2. Principles, Plenum, New York, pp. 241–305. [Google Scholar]
- 40.Lakowicz JR, Gryczynski I, Kuśba J, Wiczk W, Szmacinski H, and Johnson ML (1994) Photochem. Photobiol 59, 16–29. [DOI] [PubMed] [Google Scholar]