Abstract
RNA polymerase (Pol) I–transcribed ribosomal genes of budding yeast exist as a tandem array (about 150 repeats) with transcription units separated by spacer sequences. Half of these rDNAs are inactivated by repressive chromatin structure, whereas the rest exist in an open conformation transcribed by closely spaced Pol I elongation complexes. Whereas previous studies have suggested that active rDNA is devoid of nucleosomal structure, we demonstrate that active rDNA has nucleosomal structure, according to chromatin immunoprecipitation and biochemical fractionation. Using a yeast strain with reduced numbers of all actively transcribed rDNA repeats, we show that rDNA exists in a dynamic chromatin structure of unphased nucleosomes. Furthermore, it is associated with chromatin-remodeling enzymes Chd1p, Isw1p and Isw2p, whose inactivation causes defects in transcription termination. We suggest that Pol I transcription, like that of Pol II, may be modulated by specific chromatin structures.
Eukaryotic genomes are packaged into chromatin structures that act to fold DNA into the limited three-dimensional space of the nucleus. For silent parts of the genome, extensive higher-order folding of nucleosomal DNA into complex loop structures occurs, partly accounting for gene repression1. In contrast, transcribed genes are known to adopt a more open nucleosomal structure2. These core nucleosomes provide for an important level of gene regulation, in part through their specific displacement or remodeling3,4.
For genes transcribed by RNA polymerase II, tight regulation is achieved by the selective remodeling of nucleosomes, which provides access for the transcription apparatus during all stages of transcription2,5. At initiation, nucleosomes must be precisely repositioned to allow transcription factor binding to promoter and enhancer sequences followed by Pol II recruitment to set up the preinitiation complex. Elongation then commences, requiring the transient displacement of each successive nucleosome along the gene template. Eventually, the end of the gene is reached, and this too may be marked by specific nucleosomal arrangements6. The transcription cycle for genes transcribed by Pol II is consequently highly complex, requiring multiple chromatin-modifying and chromatin-remodeling enzymes. In Saccharomyces cerevisiae, a range of chromatin-remodeling enzymes are known to be crucial for Pol II transcription, including SWI/SNF and Isw1p remodeling factors at initiation and Isw1p and Chd1p for elongation and termination of transcription6–9. In addition to these ATP-dependent nucleosome ‘moving machines’, other elongation factors seem to facilitate transient nucleosome displacement, allowing Pol II passage along the gene10. These include FACT and Spt6p, as well as additional elongation factors, some of which covalently modify both histone tails (methylation and acetylation) and the heptad repeat of Pol II’s C-terminal domain (Ser2 and Ser5 phosphorylation)11.
Chromatin structure also regulates the transcription of ribosomal genes, which are arranged as tandem repeats of identical gene and spacer sequences. In S. cerevisiae, a single array of 150 repeats generates the full complement of ribosomal RNAs12. Unlike in higher eukaryotes, each repeat also contains a single 5S RNA gene transcribed by Pol III. Transcription of the remaining rRNAs is achieved by the dedicated Pol I, which synthesizes a 35S precursor (pre)-rRNA that is processed into mature 18S, 5.8S and 25S rRNAs. In total, rRNA synthesis comprises over 60% of all nuclear transcripts12.
Notably, about half of the rDNA repeats are transcriptionally silent at any one time13,14. This rDNA silencing is mediated by a chromatin-remodeling complex called NoRC in higher eukaryotes15,16. NoRC and a corepressor, SIN3, are recruited to Pol I promoters through interactions with the transcription factor TTF1 (Reb1p in S. cerevisiae)17. This leads to remodeling and covalent modification of nucleosomes over the rDNA repeat, so that Pol I is blocked from transcription initiation and elongation. Deacetylation and methylation of histone tails, as well as DNA methylation, all combine to maintain this repressed state. The remaining active rDNAs are transcribed at high efficiency, with an estimated 50 Pol I complexes engaged per gene14.
The high density of elongating Pol I along the transcribed rDNA sequence would seem incompatible with nucleosome structures. Consequently, the view has long been held that active rDNA genes are essentially devoid of chromatin structure. However, it has also been noted not only that the number of chromatin-repressed rDNA repeats may vary but also that the efficiency of Pol I transcription initiation may alter on active repeats18, changing the loading of Pol I complexes across the rDNA gene. Both these mechanisms exist to precisely regulate the required abundances of rRNA under different growth conditions and thereby reduce energetic waste. Evidence that inactive rDNA repeats are selectively repressed by chromatin structure comes from studies using psoralen cross-linking techniques. Actively transcribed rDNA is readily cross-linked by psoralen, indicating an open, possibly nucleosome-free DNA conformation, whereas inactive rDNA is resistant to psoralen cross-linking, indicating that it is packaged into a tight nucleosomal structure13,19. In contrast, earlier micrococcal nuclease (MNase) digestion studies suggested that active rDNAs have at least some regular chromatin structure20, and formaldehyde and UV cross-linking studies have also indicated their association with core histones21,22. Notably, a more recent study has shown that partial depletion of histone H3 causes inhibition of Pol I transcription23. However, as both H3 and H4 are known components of the Pol I initiation factor UAF24, partial H3 depletion may affect Pol I transcription by inhibiting transcription initiation rather than removing a required nucleosome structure.
We decided to revisit the question of whether Pol I transcription occurs on a chromatin template in yeast and investigate the possibility that chromatin-remodeling activities control Pol I transcription in a way that is similar to their role in Pol II transcription. We used a yeast strain having a reduced number of rDNA repeats, all of which are transcriptionally active at any given time14. Using chromatin immunoprecipitation experiments as well as biochemical purification of nucleosomes, we demonstrate that Pol I does indeed transcribe a chromatin template. Furthermore, we show that the chromatin-remodeling enzymes Chd1p, Isw1p and Isw2p are required for correct transcription termination.
Results
Pol I transcription on rDNA associated with histones
Experimental evidence for and against a role of chromatin in the regulation of Pol I transcription has been described13,19–23. We therefore re-evaluated this issue by performing chromatin immunoprecipitation (ChIP) analysis coupled with quantification by real-time PCR to determine whether nucleosome components histone H2B and histone H3 are associated with active and inactive rDNA repeats. We used primer pairs spanning the entire rDNA repeat (from upstream of the promoter to the 5S gene downstream of the site of termination) and control primers at the Pol II–transcribed CYC1, UTR1 and ISY1 loci (Fig. 1a and Methods). To first check the specificity of our assay, we used an antibody to HA with a strain having HA-tagged Pol I25,26. As expected, Pol I was reproducibly associated with the entire rDNA transcription unit, with a profile reflecting that seen in transcription run-on (TRO) analysis27. The specificity of these results was further confirmed by the absence of Pol I from the Pol II–transcribed CYC1, UTR1 and ISY1 loci (Fig. 1b). We next used ChIP analysis to look for histones H2B and H3 on rDNA. We made use of a strain that has only 42 rDNA repeats (NOY886). In this strain, all repeats are actively transcribed at all times, which allows differentiation between active and inactive rDNA repeats14,28. This low repeat number is maintained stably by the strain (data not shown and ref. 28).
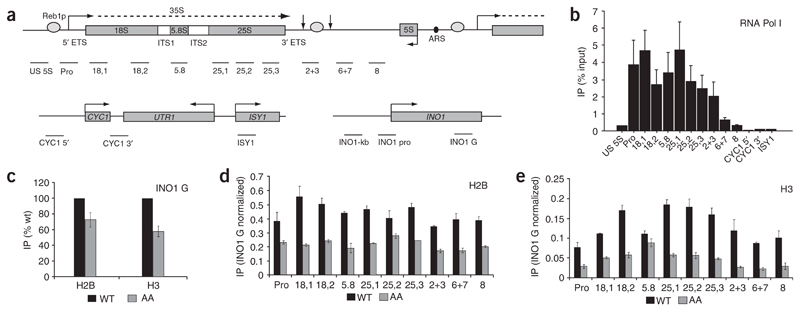
Figure 1. Histones H2B and H3 are present across active rDNA repeats.
(a) Diagram of the rDNA repeat (top) and the two Pol II–transcribed loci (one with CYCY1 UTR1 ISY1 and one with INO1; bottom). The primary rRNA transcript is indicated by a dashed line. The external (ETS) and internal (ITS) transcribed sequences are shown. Vertical arrows to the left and right of the Reb1p- binding site denote the +93 primary termination site and the +250 fail-safe termination site, respectively. Black oval denotes the autonomously replicating sequence (ARS). Horizontal bars denote the locations of regions amplified by real-time PCR. Exact locations and sizes of PCR products are detailed in Supplementary Methods and Supplementary Table 1. (b) Profile of Pol I across Pol I– and Pol II–transcribed loci, determined by ChIP analysis. Signals from the primers US 5S to 8 show Pol I across the rDNA repeat. High signals are evident over the promoter, throughout the genic region and over the termination region (2+3), as expected. The Pol II–transcribed locus produces background signals, confirming absence of Pol I and thus specificity of the ChIP analysis. Quantification of all real-time PCR data was carried out as detailed in Methods. All ChIP analyses were repeated (n = 2–4) and average data sets are presented. Error bars show s.d. throughout. (c) ChIP analysis of the Pol II–transcribed INO1 locus reveals that abundances of histones H3 and H2B are decreased in the AA strain as compared with WT. Histones H2B (d) and H3 (e) are associated with actively transcribed rDNA. Comparison of an AA strain with a WT strain reveals that histones H2B and H3 remain associated with rDNA during transcription. Values shown are normalized to the abundance of H2B (d) or H3 (e) in the Pol II–transcribed locus (INO1 G) in c. According to the ratio of H2B or H3 in the AA strain to that in the WT strain across all amplified regions, AA repeats contribute, on average, 40% of the histone H2B and 25% of the histone H3 detected in the WT strain.
Polyclonal antibodies against H2B and H3 were used to first compare the levels of these core histones on a Pol II–transcribed gene sequence, INO1. Comparing ChIP signals between the all-active (AA) strain and its isogenic wild-type (WT) strain (Fig. 1c), we note that for this INO1 gene primer pair, as well as for other tested Pol II genes (data not shown), the H2B and H3 signals are reduced 50%–75% in AA as compared with WT chromatin preparations. This difference may reflect altered sensitivity of chromatin isolated from the AA strain to the ChIP procedure. We then performed ChIP analysis on AA and WT chromatin with the full range of Pol I gene primers, using the H2B (Fig. 1d) and H3 (Fig. 1e) antibodies. These measurements were repeated multiple times, allowing accurate error bars (s.d.) to be established, and are presented as a ratio between the observed Pol II signal (INO1 G primers) and the full set of Pol I primers. Note that the quantitative ChIP analyses presented in this study are corrected for the different rDNA-repeat copy numbers in AA and WT strains (see Methods). Both the H2B and H3 ChIP analyses show the clear and reproducible presence of these core histones across the whole Pol I locus in the AA strain, with an average of 40% of the abundance seen in the WT chromatin for H2B and 25% for H3. Previous studies have predicted that histone H3 would be found over the rDNA promoter, as histones H3 and H4 are components of UAF, a member of the Pol I initiation complex24. However, our results suggest that histone H3, together with H2B, is associated with the entire actively transcribed rDNA repeat. Consequently, we predict that a reduced abundance of nucleosomes is present on genes actively transcribed by Pol I, in spite of their high Pol I occupancy.
Actively transcribed rDNA has a nucleosomal structure
To further characterize the potential chromatin structure associated with Pol I–transcribed rDNA, we performed limit MNase digestion on WT and AA yeast chromatin (Fig. 2). DNAs purified from these MNase digestion reactions were fractionated on agarose gels as shown, and the mononucleosome fraction (indicated by arrowhead) was isolated by gel extraction (Fig. 2b). Increasing amounts of micrococcal nuclease resulted in only a slight increase in the mononucleosome fraction, supporting the assumption that these were limit digestions. Note the near-complete degradation of naked DNA in the DNA control lanes. PCR analysis (Fig. 2c) of the mononucleosome DNA fractions was carried out using primer pairs that span a single nucleosome repeat (150 base pairs (bp)), corresponding to different parts of the Pol I transcription unit (Fig. 2a). PCR signals were detected for both WT and AA rDNA that partly accumulate over the 3′ end of the Pol I transcription unit (−2 primers). We showed, by varying the PCR cycle number (data not shown), that the signals obtained (Fig. 2c) reflect a logarithmic amplification stage, providing semiquantitative results. Notably, very little signal (other than background, primer-dimer signal) was detectable with primers 2+3, corresponding to the Reb1p-binding site. It is likely that Reb1p blocks nucleosome formation at this position in both WT and AA rDNA repeats. The AA mononucleosome DNA gave consistently lower PCR signals. This is in part due to the lower copy number of rDNA in this strain. Even so, it is clear that a proportion of the AA strain is nucleosomal, especially in the termination region (−2 primers).
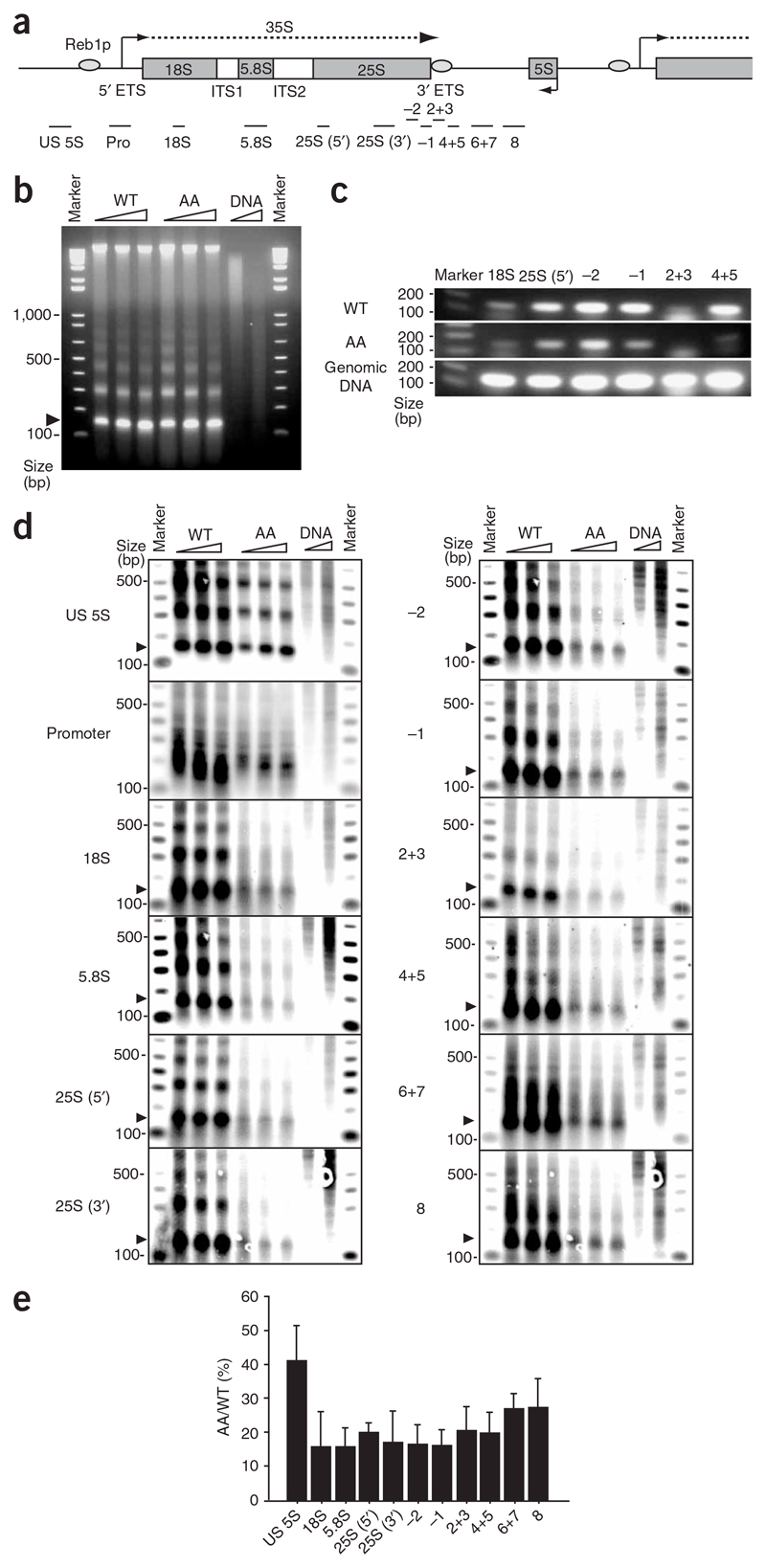
Figure 2. Biochemical analysis of Pol I–associated nucleosomes.
(a) Location of the probes used in nucleosome PCR and in Southern blot analysis. rDNA repeat map is shown as in Figure 1a, with probes indicated by horizontal bars. Exact positions of probes are described in Supplementary Methods and Supplementary Table 2. (b) Agarose gel fractionation of DNA isolated from MNase-digested chromatin (300, 600 and 900 units ml−1 for 3.5 min; triangles above gels indicate increasing concentration) of AA and isogenic WT strains. ‘DNA’ lanes are naked DNA control digested by MNase (10 units ml−1 for 30 s and 90 s). Arrowhead indicates DNA derived from mononucleosome. (c) Nucleosome PCR. Gel-purified mononucleosome DNA of WT and AA strains was PCR-amplified. Locations of primer sets are shown in a. Control PCR (bottom) was carried out with yeast genomic DNA. (d) Nucleosome structure exists on active 35S rDNA gene. Shown are Southern blot analyses of agarose gels like that in b, with the 12 probes indicated. 32P-labeled marker DNA is shown. (e) Relative values of mononucleosome-band intensity of AA versus WT strain. Ratios of mononucleosome signals between AA and WT strains averaged from each MNase concentration are shown with s.d.
To extend these results using an alternative assay, we carried out Southern blot analysis on the chromatin DNA fractions (Fig. 2b) using a range of 32P-labeled DNA probes spanning the whole rDNA repeat (Fig. 2a). The quality of these data can be assessed by the specific variations in labeled products observed (Fig. 2d). For the promoter probe, we note that a complex pattern of products was detected, with major bands between 150–300 bp, for both WT and AA chromatin. We predict that this promoter profile reflects stable Pol I initiation complexes, which are distinct from the nucleosome patterns observed with the other DNA probes. For the spacer probe (US 5S), clear mono-, di- and trinucleosome bands are apparent for both WT and AA DNAs. Furthermore, the ratio of AA to WT signal is about 40%, matching the reduced number of rDNA repeats in the AA strain. Thus, in this region of the spacer sequence, the AA strain is fully nucleosomal. In contrast, for the transcribed regions of the rDNA repeat probed for in this experiment, the level of nucleosomal signal was proportionally lower in the AA strain (about 15% of WT; see Fig. 2e). Furthermore, in general, only mononucleosome bands were clearly detectible with the AA chromatin digests, suggesting that the regular arrays of nucleosomes observed in the WT and AA spacer region are disrupted. These data indicate that although nucleosomes are detectible in transcribed regions, they are highly unstable, presumably reflecting their continuous displacement by the passage of Pol I elongation complexes. Notably, probe 2+3 (corresponding to the Reb1p terminator binding site) again gave lower overall signals, consistent with the PCR analysis described above (Fig. 2c).
In summary, the results obtained from these biochemical analyses of rDNA chromatin in WT and AA strains demonstrates that Pol I–transcribed rDNA repeats have nucleosomal structure. However, it is apparent that the regular nucleosomal structure associated with inactive rDNA or spacer sequence is substantially disrupted in the Pol I transcription unit, suggesting that nucleosomes exist in a highly dynamic state, presumably in a continuous exchange with elongating Pol I. These results are consistent with the levels of H2B and H3 ChIP signals seen across rDNA (Fig. 1), supporting a lower nucleosome level than seen in the inactive rDNA genes.
Chd1p and Isw1p chromatin remodelers on active rDNA
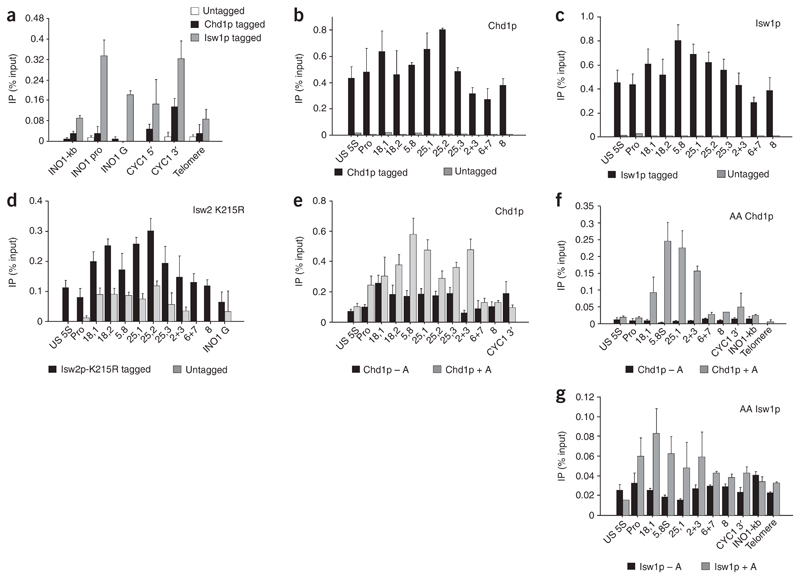
The data presented above describes the presence of nucleosome components across active rDNA repeats. Furthermore, we suggest that this chromatin structure is likely to be highly dynamic to allow passage of multiple elongating Pol I complexes across rDNA. This situation implies the close involvement of chromatin-remodeling factors during Pol I transcription. We have previously shown that such factors are required for Pol II transcription termination6. In addition, different forms of Isw1p have been shown to be crucial in the initiation and elongation stages of Pol II transcription29. To test for the association of chromatin-remodeling factors with rDNA, we carried out ChIP analysis using Myc antibody on strains carrying Myc-tagged Chd1p30 and Isw1p31. Again, we first tested Pol II–transcribed genes (see Fig. 1a) and showed that Isw1p is detectible in both intergenic and genic regions31, whereas Chd1p is predominantly associated with the 3′ end of the tested CYC1 gene (Fig. 3a), as previously described6. Lower signals were detected with the telomere primers, providing a negative control for this analysis. We then performed ChIP analysis on rDNA in the Myc-Chd1p and Myc-Isw1p strains and found that both remodeling enzymes were reproducibly present at appreciable abundances across the whole rDNA repeat (Fig. 3b,c). Note the absence of signals with the untagged strain, confirming the specificity of our ChIP analysis. We also performed ChIP analysis on a strain containing Flag-tagged Isw2p with a further mutation in the ATPase catalytic domain (Fig. 3d). This allows the use of Flag antibody to detect Isw2p on chromatin, which is otherwise hard to detect owing to the rapid turnover of this remodeling enzyme on chromatin32. We note that the positive Isw2p signal is only a few times greater than the untagged background signal, reflecting the lower occupancy of this chromatin-remodeling enzyme compared with Chd1p and Isw1p abundances on rDNA chromatin.
Figure 3. Chromatin-remodeling factors are associated with the entire rDNA repeat.
(a) ChIP analysis of Chd1p and Isw1p over Pol II gene loci (INO1 and CYC1) and telomere DNA showing accumulation of signal from CYC1 3′ probe6. For details of ChIP PCR primers used, see Figure 1a. (b) ChIP analysis of Chd1p across the rDNA repeat in the Chd1p-Myc strain compared with an untagged isogenic control strain. Chd1p is found across the entire repeat, with higher levels seen over genic regions (18,1 to 25,2). Appreciable abundances of Chd1p are also maintained over the termination region and into the spacer region. (c) ChIP analysis of Isw1p across the rDNA repeat in an Isw1p-Myc strain compared with an untagged isogenic control strain. Isw1p is associated with the entire rDNA repeat. (d) ChIP analysis of Isw2-K215R protein across the rDNA repeat in an Isw2-K215R–Flag strain compared with an untagged isogenic control strain. Isw2p-K215R is also associated with the entire rDNA repeat. (e) RNase A treatment (+A) enhances the Chd1p ChIP signals over the transcribed region, indicating that Chd1p does not interact with rRNA. The increase is probably due to enhanced antibody binding upon clearance of RNA, indicating that Chd1p interacts with actively transcribed repeats. (f,g) Chd1p and Isw1p are associated with active rDNA repeats. RNase A treatment before ChIP enhances signals, especially in Chd1-tagged AA strains, over the transcribed region and early-termination region.
The association of Chd1p with the rDNA repeat could occur through rDNA or rRNA, as it has been shown that chromodomains can bind RNA33 and Chd1p has two such domains, though these are different from the MOF chromodomain implicated in RNA interactions. We therefore used a modified ChIP assay that includes RNase A treatment before immunoprecipitation34. If Chd1p associates with the repeats via rRNA, a drop in ChIP signal is expected, whereas signals should remain the same if the association occurs via the rDNA. To our surprise, the addition of RNase A caused the ChIP signals to selectively increase over the transcribed part of the rDNA repeat (regions Pro to 2+3), whereas the signals remained almost unchanged upstream of the promoter (US 5S) and over the untranscribed spacer (regions 6+7 and 8). In particular, region 2+3 shows a seven-fold increase in Chd1p signal after RNase A treatment (Fig. 3e). These data can be explained by considering the availability of the epitope for interaction with antibody. The high transcription rate of rDNA genes generates a large amount of rRNA, which could interfere with the interaction of Myc-tagged Chd1p with anti-Myc. When rRNA is degraded by RNase A, the epitope becomes more accessible to the antibody, leading to an increase in signal. This hypothesis is consistent with the large increase in signal upon RNase A treatment over regions Pro to 2+3 while the signals do not increase over regions 6+7 and 8. This is presumably because the polymerase has terminated and the rRNA has been released. Notably, the increase in signal seen upon RNase A treatment also indicates that Chd1p is associated with the active repeats and that these signals are not simply due to Chd1p binding to inactive repeats.
Our ChIP analysis showing that Chd1p, Isw1p and Isw2p are associated with rDNA (Fig. 3b–e) implies a role for these enzymes in rDNA expression. However, the data fall short of proving their direct involvement in active rDNA transcription. We therefore constructed new AA yeast strains with either the Chd1p or Isw1p HA-tagged. We then compared the abundances of these two chromatin remodeling enzymes with or without RNase treatment across the rDNA sequence (Fig. 3f,g). For Chd1p, we observed strong RNase A–dependent signals across the Pol I transcription unit, with much lower signals in the spacer sequence. Our data therefore demonstrate that Chd1p binds specifically to the rDNA transcription unit and its detection is largely masked by nascent rRNA. Similar results were obtained for Isw1p, where again higher Isw1p occupancy was observed over the Pol I transcription unit when chromatin was treated with RNase. The specificity and selective association of these chromatin-remodeling enzymes implies that both Chd1p and Isw1p have direct roles in rDNA transcription.
Remodeling factors are required for Pol I termination
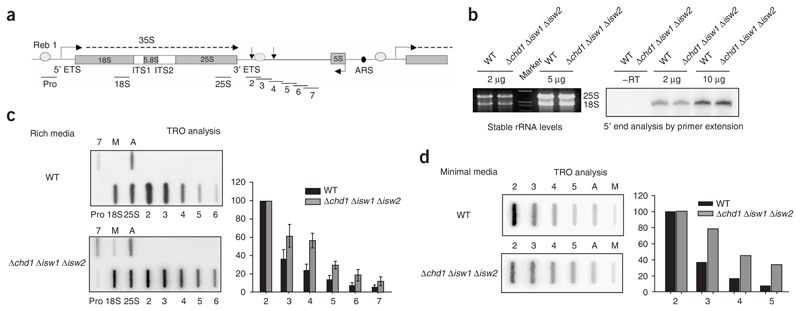
The presence of chromatin and chromatin-remodeling factors at active rDNA repeats suggests that these factors help to regulate efficient transcription. We therefore analyzed rRNA transcription in a yeast strain with all three chromatin remodeling enzymes deleted (Δchd1 Δisw1 Δisw2; Fig. 4). This triple-mutant strain, though viable, has a severe slow-growth phenotype. We initially measured the steady-state abundance of rRNA produced by the triple-mutant strain, as compared with its isogenic WT strain. Using equalized cell density, we observed unchanged rRNA abundance and unchanged abundance of pre-rRNA 5′ termini, as determined by primer extension (Fig. 4b). These data support the idea that transcription initiation and accumulation of stable rRNA are unaffected by lack of the deleted chromatin-remodeling enzymes.
Figure 4. Chromatin remodeling factors Chd1p, Isw1p and Isw2p modulate Pol I’s transcription termination profile.
(a) Diagram of rDNA repeat as in Figure 1a. Black horizontal bars denote M13 TRO probes. Locations and sizes of probes have been described27. (b) Profiles of stable rRNA (left) and the 5′ end of the 35S rRNA (right) are the same between the WT and the Δchd1 Δisw1 Δisw2 strain. RNA was isolated from each strain, and amounts indicated were analyzed by direct agarose gel fractionation (ethidium bromide stain) or by primer extension. (c,d) Loss of Chd1p, Isw1p and Isw2p causes a distortion in Pol I transcription termination profile, with more read-through after the +93 termination site. Representative TRO hybridization profiles are presented for the WT and the Δchd1 Δisw1 Δisw2 strain grown in either rich media (c) or minimal media (d). ‘A’ denotes the actin positive control. The negative control (M) is an M13 DNA lacking rDNA sequence. Error bars in c show s.d. of four experiments.
We next performed nascent-transcription analysis on the triple-mutant remodeling strain by TRO analysis. This procedure is carried out by incubating detergent-permeabilized yeast cells in a buffer containing [α-32P]UTP, which allows cells to continue transcribing, with polymerases ‘running on’ between 50 and 100 nucleotides. Labeled RNA is then hybridized to strand-specific DNA probes spanning the rDNA repeat unit, and the signal obtained determines the density of transcriptionally engaged polymerase present on the gene and the profile of nascent transcription across the gene35. TRO analysis was carried out using probes specific to the known Pol I termination region spanning from downstream of the 25S gene to the Pol III–transcribed 5S gene (Fig. 4a), as described27. Control promoter, 18S and 25S probes were also included in this analysis.
Comparison of the TRO profiles between the WT and triple-mutant strains revealed a clear Pol I termination defect in the triple-mutant strain (Fig. 4c). In particular, WT TRO signals beyond the normal 3′ end of the transcription unit27 (detected by probe 2) drop to low levels (90% reduction by probe 5). In contrast with the triple-mutant strain, about two-fold higher TRO signals persist into the spacer sequence. These data are based on an average of four independent experiments. Furthermore, TRO analysis of both strains grown in minimal medium reveals a similar, though slightly more pronounced, termination defect (Fig. 4d). We also note that, especially for the WT TRO, signals were substantially lower over the 18S- and 25S-probed regions than over the region outside the mature rRNA sequence but still within the transcription unit (probe 2). Titrating lower amounts of TRO-labeled RNA gave more equal 18S, 25S and probe 2 signals (Supplementary Figure 1 online). This equal distribution of TRO signals reflects the more equal abundances of Pol I detected by ChIP across rDNA (Fig. 1b). In addition, it has been suggested that mature rRNA present at high abundance in yeast cells may compete with the 32P-labeled TRO RNA and thus cause an underrepresentation of signals from the 18S and 25S probes. Consequently, we normalized our TRO data by fixing the probe 2 TRO signal as 100%. We also performed TRO analysis on yeast strains deleted for Chd1p, Isw1p and Isw2p singly or pairwise. In each case, little difference from the WT TRO profile was observed (data not shown). Apparently, these factors act redundantly at the rDNA locus, as has previously been observed for the Pol II–transcribed GAL10-7 locus6. The requirement of chromatin-remodeling enzymes for proper Pol I termination may reflect the presence of, in particular, Chd1p up to but not beyond the normal termination region.
Discussion
Although chromatin-remodeling factors function at all stages of Pol II transcription6,8,37, their role in Pol I transcription is unknown. Previous studies have suggested that rDNA is devoid of nucleosomes during active transcription by Pol I13,19, consistent with the high density of Pol I elongation complexes across actively transcribed rDNA genes. However, these data rely on the observed accessibility of the DNA cross-linking reagent psoralen. Transcribed rDNA has been found to be accessible in these studies, whereas untranscribed rDNA has not. Although it is clear from these analyses that transcribed rDNA has a more open chromatin structure, it remains possible that dynamic nucleosomes, as described here, would not preclude psoralen accessibility38. Our previous study showing similarities between transcription by Pol I and Pol II27 led us to reinvestigate the possibility that some form of chromatin structure exists in actively transcribed ribosomal genes and that this structure is maintained by chromatin-remodeling factors.
Active rDNA is associated with dynamic chromatin
Our ChIP analysis on a yeast strain (NOY886) having reduced numbers of rDNA repeats, of which a large proportion are active, implies the close association of histones with actively transcribed rDNA genes. As WT yeast has 150 repeats, half of which are inactive and known to be packaged into repressive chromatin structure13,14, we expected to see association of histones H2B and H3 with rDNA in WT strains. However, we also found that histones H2B and H3 are associated with the entire active rDNA repeat. The amount of histone H2B on active repeats is ~40% of the total histone H2B seen in the WT strain, and the amount of histone H3 on active repeats is ~25% of the total H3 in WT. Even though these data suggest that histones are a major component of actively transcribed rDNA chromatin, they do not necessarily imply a regular nucleosomal structure. Furthermore, the difference in the abundances of H2B and H3 is notable. Canonical nucleosomes contain equal amounts of all four core histones. The unequal signals seen here may, therefore, suggest that the chromatin present on active repeats is dynamic and unlikely to form canonical nucleosomes. However, the presence of both histones H2B and H3 argues against the possibility that the chromatin environment seen on active rDNA repeats represents a form of ‘prenucleosome’ containing only histones H3 and H4, the two histones deposited first during nucleosome assembly39. The presence of both histones H2B and H3 suggests that all four core histones may be present on active rDNA repeats. To confirm this, studies should be carried out using antibodies to histones H2A and H4. Available antibodies were not suitable for such ChIP analysis.
To further characterize the nature of actively transcribed rDNA chromatin, we carried out biochemical purification of chromatin nucleosomal DNA from both WT and AA strains. This DNA fraction was then probed with rDNA sequences, by both PCR and Southern blotting. Our results confirm the presence of nucleosome-like structures over active rDNA, but at reduced levels, consistent with a dynamic chromatin structure. Simple PCR analysis (Fig. 2c) shows that there are some mononucleosome structures in AA rDNA. However, more notably, the Southern blot analysis shows that, whereas the spacer sequence in AA chromatin is fully occupied with nucleosomes, the transcribed regions of the AA rDNA repeats have three-fold less nucleosomes than the amount expected for full occupancy (Fig. 2d,e). Furthermore, very little signal other than that of the mononucleosomes is apparent for most transcribed rDNA probes, suggesting that an ordered array of nucleosomes is absent. The fact that the promoter probe in both WT and AA strains gives a non-nucleosomal but consistent hybridization pattern suggests that the promoter is fully occupied with Pol I initiation factor (UAF) whether or not the rDNA repeat is active.
Overall, our data on the nature of chromatin associated with actively transcribed rDNA indicate that it is comprised of dynamic nucleosomes as well as a fraction of non-nucleosomal histones (at least H3 and H2B). It seems plausible that elongating Pol I encounters dynamic nucleosomal structures during transcription and that this structure is remodeled, just as occurs with genes transcribed by Pol II.
Chromatin remodelers on rDNA dictate normal transcription
The presence of dynamic chromatin structure across active rDNA suggests that chromatin-remodeling factors are involved in Pol I transcription. To address this issue, we carried out ChIP analysis on strains having tagged Chd1p, Isw1p and Isw2p (Fig. 3). We found that all three remodeling enzymes are detectable across the rDNA repeat at comparable levels. Notably, HA-Chd1p and HA-Isw1p AA yeast strains also gave clear signals for these remodeling enzymes across the Pol I transcription unit. As Chd1p and Isw1p have been implicated in elongation and termination of transcription by Pol II6,29,37, their presence across the Pol I transcription unit on active rDNA suggests that they are probably involved in productive Pol I transcription. Treatment of Chd1p and Isw1p ChIP samples with RNase led to a strong increase in signal, especially for Chd1p, indicating that rRNA impedes the interaction of the antibody. Finally, we investigated whether chromatin-remodeling factors have an active role during Pol I transcription by studying the Pol I nascent-transcription profile in WT and mutant strains. We found that loss of Chd1p, Isw1p and Isw2p causes a clear Pol I termination defect.
The presence of Chd1p and Isw1p across active rDNA suggests that they have a role in the Pol I elongation process. Even so, deletion of their genes does not detectibly reduce rRNA levels. However, the slow-growth phenotype of this mutant strain may reflect Pol I transcription deficiency. In contrast, the chromatin-remodeling deletion strain does show a clear Pol I termination defect. This may indicate that normal Pol I termination requires a chromatin structure set up by these remodeling enzymes, and their absence thus perturbs the normal termination process.
Conclusions
Our data demonstrate that histones and chromatin-remodeling factors are associated with active rDNA and suggest that a chromatin environment is present during active transcription. Our results, together with the reported detrimental effect of histone H3 depletion on Pol I transcription23, suggest that modifications in the chromatin environment of active rDNA can function to recruit chromatin-remodeling factors, which in turn are required for efficient transcription by Pol I. The results presented in this study therefore modify the current view of rDNA chromatin structure. Although inactive rDNA repeat units are associated with repressive chromatin structures, active repeats are also associated with a dynamic chromatin structure that may act to facilitate efficient rRNA synthesis.
Methods
Yeast strains
The following strains were used in this study: AS14 (Matα ade2 ura3-52 trp1Δ63 TRP1 his3Δ200 leu2Δ1 lys2-801 RPA14-3HA), NOY886 (Matα rpa135Δ::LEU2 ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 fob1Δ::HIS3 pNOY117 (CEN RPA135 TRP1); rDNA copy number ~42), NOY1051 (same as NOY886, but rDNA copy number is ~140), YHT149 (Mata 6MYC::6HIS::CHD1::TRP1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1 pep4Δ:: HIS3 prb1-Δ1.6R), Isw1-Myc (Matα 13Myc::KanMX6::ISW1 ura3-52 trp1-289 leu2-3 1his3Δ1 MAL2-8CSUC2), YTT166 (Mata ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5), YTT227 (same as YTT166, but chd1::TRP1 isw1::ADE2 isw2::LEU2), YTT1401 (same as YTT166, but Mata ste2::HygB ste3:: KanMX), YTT2164 (same as YTT166, but Mata ste2::HPH ste3::NAT isw2-K215R-3FLAG-KanMX), AA-Chd1 (same as NOY886, but CHD1-3HA::KanMX) and AA-Isw1 (same as NOY886, but ISW1-3HA::KanMX). All S. cerevisiae media and growth conditions are standard. All strains were grown in YPD medium at 30 °C, except for the TRO analysis that used minimal medium (Fig. 4d).
Chromatin immunoprecipitation
Strains used for ChIP analysis were AS14, NOY1051, NOY886, YHT149, Isw1-Myc, YTT1401, YTT2164, AA-Chd1 and AA-Isw1. Analysis was carried out as described29 with the following modifications: the cross-linking time was 6 min and proteinase K treatment was carried out for 1 h before reversal of cross-links. Cross-links were reversed overnight. DNA was sonicated using a bioruptor and purified using Qiagen minicolumns. The locations of the PCR products are shown in Figure 1a and their primer sequences and coordinates are shown in Supplementary Methods and Supplementary Table 1 online.
All real-time PCR reactions were done with the Corbett Rotorgene system. Input DNA was diluted 1:20, and immunoprecipitations (IPs) and ‘no antibody’ controls (NOs) were diluted 1:5. All samples were run in triplicate to ensure accuracy of the data. PCR of 45 cycles was done using the Qiagen QuantiTect SYBR Green PCR Master mix. Primers were used at 0.5 μM for each experiment. Relevant fluorescent intensities were calculated by the equation:
All values obtained for IP signals and NO signals were multiplied by 5 and all input signals were multiplied by 20 before the values were input into the equation, to account for the dilutions noted above. Division of all IP values by input values takes copy number into account, allowing the comparison of data from multiple- and single-copy loci.
The protocol for RNase A treatment was adapted from previous studies34. After addition of IP buffer, two sets each of input, IP and NO aliquots were made. RNase A was added to one set. All tubes were incubated at room temperature for 30 min. The following amounts of RNase A were added: input, 6 μl; IP and NO, 19 μl. RNase A/T1 cocktail was purchased from Ambion (catalog number 2286). Antibodies for the ChIP analysis were anti-HA clone F7 (Santa Cruz Biotechnology.), anti–histone H2B (see Acknowledgments), anti–histone H3 ab1791 (Abcam), anti-Flag M2 F3165 (Sigma-Aldrich) and anti-Myc clone 9E10 (Sigma-Aldrich).
Biochemical chromatin analysis
Strains used were NOY1051 and NOY886. Analysis was carried out as described31. Briefly, samples of detergent-permeabilized yeast spharoplasts (equivalent to 4.0 × 108) were incubated with MNase (300, 600 and 900 units ml−1) for 3.5 min at 37 °C. DNA was purified and separated on 1.5% (w/v) agarose gels and blotted onto nylon membranes. MNase digests of deproteinized DNA were also included as ‘naked DNA’ controls, together with size marker DNA. Blots (Fig. 2d) were hybridized with specific primer-labeled 32P probes and marker DNA and washed at high stringency. Probes were derived from DNA fragments amplified by PCR from yeast genomic DNA. The sequence and location of probes are shown in Figure 2a, Supplementary Methods and Supplementary Table 2 online.
Mononucleosome DNA was purified from above agarose gels using standard procedures. PCR reactions were done in 22 cycles with primers spaced 150 bp apart over the same DNA sequence as in the Southern blot probes. Yeast genomic DNA was also used as a template for control.
Primer extension
Strains used were YTT166 and YTT227. Analysis was done as described13, with modification. Briefly, RNA was prepared with the hot phenol method from 25 ml culture (A600 = 0.4–0.6), growing exponentially in YPD medium. The primer used for the analysis was 5′-ACACGCTGTATAGAGACTAGGC-3′, which hybridizes to 35S precursor rRNA 130 nucleotides downstream of the Pol I start site. End-labeled primer (0.2 pmol) was allowed to hybridize to each RNA sample for 1 h at 60 °C. After the mixture had been slowly cooled to room temperature, reverse transcription was carried out for 1 h at 37 °C. After extraction with phenol, chloroform and isoamyl alcohol and precipitation with ammonium acetate, the samples were electrophoresed in an 8% (w/v) polyacrylamide gel.
Transcription run-on analysis
Strains used were YTT166 and YTT227. TRO analysis and rDNA probes were both as described27,35 and locations of the probes used are shown in Figure 4a. Filers were quantified by phosphor-imaging. Values plotted are relative to probe 2 signal (set as 100%) to allow for direct comparison between WT and triple-mutant strains.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
We thank members of N.J.P.’s laboratory for useful discussions. We are grateful to A. Johnson (University of California, San Francisco), J. Mellor (University of Oxford), M. Nomura (McGill University), M. Riva (Centre National de la Recherche Scientifique, Campus de Gif sur Yvette) and T. Tsukiyama (Fred Hutchinson Cancer Center) for providing the strains used in this work. We thank D. Bentley (University of Colorado, School of Medicine) for providing the H2B antibody. H.S.J. was supported by a Wellcome Trust Prize Studentship and J.K. by a fellowship from the Japan Heart Foundation. This work was supported by a Wellcome Trust Programme grant to N.J.P.
Footnotes
Competing Interests Statement
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/ reprints and permissions
References
- 1.Wolffe AP. Chromatin: Structure and Function. 3rd edn. Academic Press; San Diego: 1998. [Google Scholar]
- 2.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 3.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr Opin Genet Dev. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Langst G, Becker PB. Nucleosome remodeling: one mechanism, many phenomena? Biochim Biophys Acta. 2004;1677:58–63. doi: 10.1016/j.bbaexp.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Krebs JE, Peterson CL. Understanding “active” chromatin: a historical perspective of chromatin remodeling. Crit Rev Eukaryot Gene Expr. 2000;10:1–12. [PubMed] [Google Scholar]
- 6.Alen C, et al. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 7.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 8.Mellor J, Morillon A. ISWI complexes in Saccharomyces cerevisiae. Biochim Biophys Acta. 2004;1677:100–112. doi: 10.1016/j.bbaexp.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Sif S. ATP-dependent nucleosome remodeling complexes: enzymes tailored to deal with chromatin. J Cell Biochem. 2004;91:1087–1098. doi: 10.1002/jcb.20005. [DOI] [PubMed] [Google Scholar]
- 10.Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 11.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 13.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 14.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol. 2005;25:2539–2546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strohner R, et al. Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol Cell Biol. 2004;24:1791–1798. doi: 10.1128/MCB.24.4.1791-1798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex. NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss T. At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohr D. Chromatin structure differs between coding and upstream flanking sequences of the yeast 35S ribosomal genes. Biochemistry. 1983;22:927–934. doi: 10.1021/bi00273a034. [DOI] [PubMed] [Google Scholar]
- 21.Belikov SV, Dzherbashyajan AR, Preobrazhenskaya OV, Karpov VL, Mirzabekov AD. Chromatin structure of Drosophila melanogaster ribosomal genes. FEBS Lett. 1990;273:205–207. doi: 10.1016/0014-5793(90)81085-3. [DOI] [PubMed] [Google Scholar]
- 22.Dimitrov SI, et al. Binding of histones to Xenopus laevis ribosomal genes with different levels of expression. Eur J Biochem. 1992;204:977–981. doi: 10.1111/j.1432-1033.1992.tb16719.x. [DOI] [PubMed] [Google Scholar]
- 23.Tongaonkar P, et al. Histones are required for transcription of yeast rRNA genes by RNA polymerase I. Proc Natl Acad Sci USA. 2005;102:10129–10134. doi: 10.1073/pnas.0504563102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keener J, Dodd JA, Lalo D, Nomura M. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc Natl Acad Sci USA. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bischler N, et al. Localization of the yeast RNA polymerase I-specific subunits. EMBO J. 2002;21:4136–4144. doi: 10.1093/emboj/cdf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peyroche G, et al. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prescott EM, et al. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc Natl Acad Sci USA. 2004;101:6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morillon A, et al. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- 30.Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15:619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodelling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilised templates. Mol Cell Biol. 2001;21:2098–2106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 34.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birse CE, Lee BA, Hansen K, Proudfoot NJ. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 1997;16:3633–3643. doi: 10.1093/emboj/16.12.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang WH, Morrow BE, Ju Q, Warner JR, Reeder RH. A model for transcription termination by RNA polymerase I. Cell. 1994;79:527–534. doi: 10.1016/0092-8674(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 37.Simic R, et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, Rothblum LI, Chen C. Ribosomal chromatin organisation. Biochem Cell Biol. 2006;84:444–449. doi: 10.1139/o06-089. [DOI] [PubMed] [Google Scholar]
- 39.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.