Abstract
Human immunodeficiency virus Type 1 (HIV-1) is the major cause of acquired immune deficiency syndrome (AIDS). In 2014, it was estimated that 1.2 million people died from AIDS-related illnesses. RNA interference-based therapy to block HIV replication is a field that, as of now, is without any FDA-approved drugs available for clinical use. In this chapter we describe a protocol for testing and utilizing a new approach that relies on reassociation of RNA–DNA hybrids activating RNAi and blocking HIV replication in human cells.
Keywords: RNA–DNA hybrids, RNA interference, HIV-1, Hybrids reassociation, Dicer substrate RNA, RNA nanotechnology
1. Introduction
RNA-based therapy is a new frontier in cancer research, creating an intriguing alternative to small molecule based treatments [1]. In a posttranscriptional, sequence-specific gene silencing process known as RNA interference [2, 3], longer double-stranded RNAs or self-folded, single-stranded RNAs are processed by an enzyme known as Dicer into shorter duplexes, called small interfering RNA (siRNA), or micro RNAs (miRNAs) [4–7]. The processed duplexes are then loaded onto an RNA-induced silencing complex (RISC) which uses, based on the thermodynamic asymmetry, one of the RNA strands as a guide to target the messenger RNA (mRNA) sequence [8, 9]. The targeted mRNA is then cleaved, thereby preventing it from undergoing translation [10], effectively curtailing the corresponding protein production. This method of cell regulation has a myriad of possible applications in the biomedical field.
RNA-based therapy provides an advantage over traditional pharmacological approaches by potentially being able to effectively target any protein in the diseased cells. However, there are still obstacles in making RNA therapy a viable clinical treatment [7]. First, RNAs are quickly degraded in the bloodstream by nucleases, which provide a natural defense against foreign nucleic acids. Secondly, analyzing the pharmacodynamics of RNA-based therapies is difficult because adding functionalities, such as fluorescent probes for visualization, to the RNA strands has the potential to alter the RNA’s activity. Lastly, ensuring conditional activation of the RNAi in the targeted diseased cells alone may be essential for minimizing collateral damage in healthy cells. The recently developed technique that is based on the reassociation of RNA–DNA hybrids [11] is a novel approach that may offer some solutions to the problems encountered in the field of nucleic acid-based therapeutic technologies.
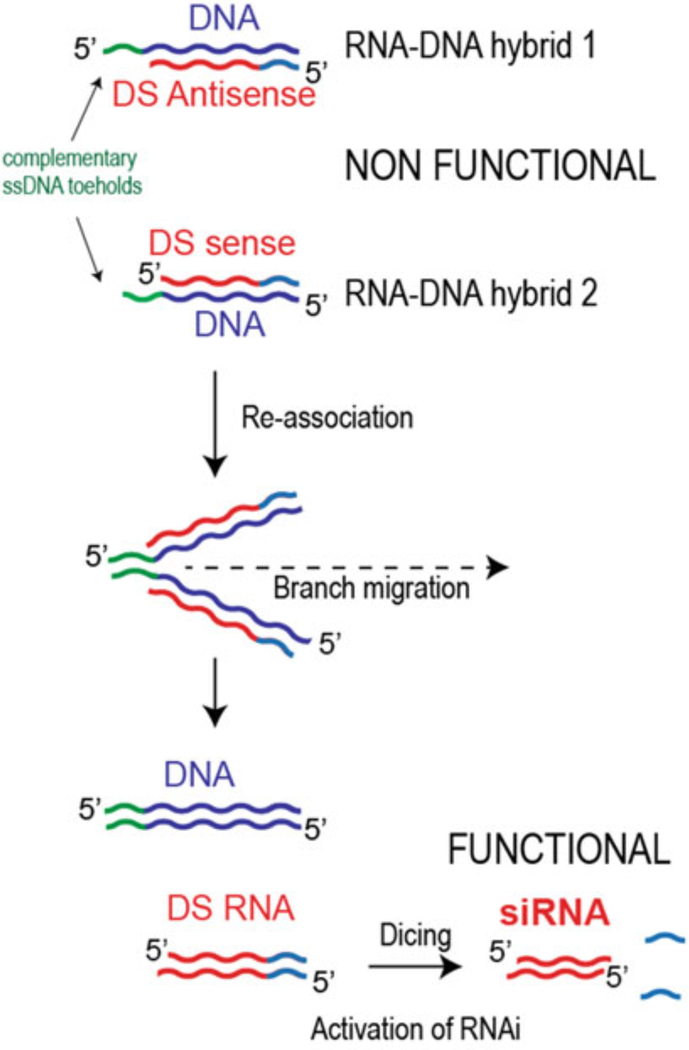
The concept behind the hybrids is similar to split-protein systems [12] which involve the division of functional proteins into nonfunctional pieces that under certain conditions reassociate, restoring the original function. This permits tighter control over the functionality of the proteins, as well as an increase in molecule detectability. The RNA–DNA hybrids are used to split the RNAi triggers into two pieces, by having one strand of the Dicer substrate RNA [13] (DS RNA) in each of the two hybrids, and letting the DNA serve as the other half of each duplex. RNA–DNA hybrids cannot be processed by Dicer and as such are inactive. When cognate hybrids are present in the same cell, the complementary sequences on the hanging ends of their DNA strands, known as toeholds, recognize each other and initiate the interactions. The DNA strands of hybrids then reassociate into the longer DNA duplexes, releasing, through branch migration, the RNA fragments which assemble into the functional DS RNAs, thus activating RNAi (Fig. 1). The use of RNA toeholds was also shown to be functional [14]. It is important to mention that the use of conventional 21-mer siRNAs in place of DS RNAs for RNA/DNA hybrids does not entirely deactivate the individual hybrids and some gene silencing is still observed for the siRNA antisense-carrying hybrids [15]. Novel computational algorithms were later developed to follow, in silico, the process of hybrid formation and reassociation, thereby automating hybrid design [16].
Fig. 1.
Schematic illustration of the reassociation of RNA–DNA hybrids through the ssDNA toehold interactions that results in release of Dicer substrate RNAs and further RNAi activation through dicing
The simultaneous release of multiple DS RNAs (up to seven at once) on reassociation of two hybrids has been previously demonstrated [15]. However, the resulting long DNA duplexes showed some immunostimulatory effects. To avoid this, while still simultaneously activating multiple DS RNAs, previously characterized RNA-, RNA/DNA-, and DNA-based nanoparticles, such as nanorings [17–19] and nanocubes [20, 21], were decorated with multiple nonfunctional RNA–DNA hybrids. During the addition of cognate hybrids, the activation of nanoparticles and triggering of specific RNAi were promoted [22, 23]. Additional functionalities such as fluorescent dyes and targeting agents can be added onto both the 5′- and 3′-ends of the DNA strands to allow a more detailed analysis of the treatment effects and delivery rates without interfering with the RNA function. The RNA–DNA hybrid approach allows for greater control over RNA interference activation, while allowing the direct visualization of intracellular reassociation in real time. RNA–DNA hybrids are also more stable in human blood compared to conventional RNAs [11]. The RNA–DNA hybrid approach also allows for simultaneous activation of other split functionalities such as, for example, FRET and functional aptamers [15, 24]. Recently, the use of modified RNAs, in place of DNA strands, has been shown to offer certain advantages [25].
According to UNAIDS, it is estimated that more than 37 million people are currently living with HIV, and the number of people with HIV continues to increase. Although effective treatment against HIV is available, the virus continues to evolve and acquire drug resistance mutations, becoming resistant to available therapies. Therefore, it is important to continue research for new drugs as well as new targets. Blocking HIV replication using RNAi is a field well studied using cell culture assays. Several regions within the HIV genome have been tested in order to find potential effective targets. Berkhout’s group has worked extensively in this field, and to test the hypothesis that RNA–DNA hybrids could inhibit HIV replication we selected two HIV targets, viral protease (PR) and surface envelope glycoprotein (gp120), validated by their published work [26].
2. Materials
All solutions should be prepared using double-deionized ultrapure water (18 MΩ at 25 °C) and biological grade reagents. It is recommended to have all reagents freshly made and filtered. Once prepared, all reagents and buffers can be stored at room temperature (unless indicated otherwise). All waste disposal regulations should be followed when disposing of waste materials. RNAs, DNAs, and fluorescently labeled DNAs for hybrid duplexes can be purchased from commercial vendors such as Integrated DNA Technologies, Inc. or others. In the case of the fluorescently labeled DNAs, we recommend the use of an additional linker of two nucleotides (e.g., TT) prior to the fluorescent tags. Different software tools such as, for example, IDT OligoAnalizer or Scripps Oligo Extinction Coefficient Calculator can be used to determine the extinction coefficients for each oligo.
The following equipment is required for the described experiments: vortex mixer, SpeedVac concentrator, horizontal gel electrophoresis, PCR thermocycler, water bath, ChemiDoc XS, fume hood, CO2 incubator, biosafety cabinet type II A2, microfuge centrifuge, oven, Shaker, Personal Molecular Imager™ (PMI™) System and correspondent software and Geiger counter.
2.1. Preparation and Analysis of RNA–DNA Hybrids
10× TBE: 0.89 M Tris, 0.89 M boric acid, 20 mM EDTA, pH ~8.3.
10× TB: 0.89 M Tris, 0.89 M boric acid, pH ~8.3.
Eluting buffer: 10 mM Tris pH 7.5, 300 mM NaCl, 0.5 mM EDTA.
5× assembly buffer: 5×TB (pH 8.3), 10 mM Mg(OAc)2.
Denaturing gel polyacrylamide gel electrophoresis (PAGE): 8 M urea, 8% acrylamide with 19:1 acrylamide–bis-acrylamide ratio, 1× TBE.
Nondenaturing native PAGE: 8% acrylamide with 19:1 acrylamide–bis-acrylamide ratio, 1× TB, 2 mM Mg(OAc)2.
2.2. Cell Culture and Transfection Reagents
Plasmid: Full-length HIV-1 molecular clone pNL4–3 [27] (Although pNL4–3 is not a harmful reagent, it is still a recombinant DNA that can generate full length virus, and therefore should be handled with caution according to NIH Guidelines for recombinant or synthetic nucleic acids).
RNAi inducers: PR RNA–DNA hybrids, Env RNA–DNA hybrids, PR DS RNA, Env DS RNA (5, 10, and 20 nM final concentrations) (see Subheading 3—Hybrid Design).
Transfection reagent: Lipofectamine 2000™ (Invitrogen).
Medium: Dulbecco’s modified Eagle’s medium (DMEM) High Glucose supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine and 5% fetal bovine serum (FBS), Opti-MEM-I reduced serum, Roswell Park Memorial Institute 1640 Medium lacking the amino acids methionine (Met) and cysteine (Cys) (RPMI 1640 Cys−/Met−), supplemented or not with 2% (vol/vol) dialyzed (dFBS).
2.3. Metabolic Labeling
Protein Labeling Mix: [35S]Met/Cys (a PerkinElmer mixture containing both 35S-L-methionine and 35S-L-cysteine).
Cell lysis buffer: 300 mM NaCl, 50 mM Tris–HCl (pH 7.5), 0.5% Triton X-100 (TX-100), 10 mM iodoacetamide, and two tablets of cOmplete protease inhibitor cocktail tablets per 100 mL of solution.
Triton wash buffer: 300 mM NaCl, 50 mM Tris–HCl (pH 7.5), 0.5% Triton X-100.
Protein A Agarose for Immunoprecipitation (ThermoFisher).
Bovine serum albumin (BSA).
Antibody: HIV-IgG (pool of purified serum from HIV-1-infected patient obtained from AIDS Research and Reference Reagent Program. Although the reagent is not considered to be biohazardous, it should be handled with caution).
SDS/DOC wash buffer: 300 mM NaCl, 50 mM Tris–HCl (pH 7.5), 0.1% SDS, 0.1% deoxycholic acid.
2× sample buffer: 2× Laemmli buffer: 120 mM Tris–HCl (pH 6.8), 4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.02% bromophenol blue.
2.4. Radioimmunoprecipitation SDS-PAGE Gel
-
Acrylamide–AcrylAide lower gel: 29% acrylamide–1% AcrylAide gel reagent for SDS-PAGE, 1.5 M Tris–HCl, (pH 8.7), 0.4% SDS, 0.5 mL 2% ammonium persulfate (APS), 40 μL N,N,N′, N′-tetramethylethylenediamine (TEMED).
Important: AcrylAide Cross-Linker can be obtained as a special order to Lonza, Cat No. 51015.
Stacking upper gel: 30% acrylamide–0.8% bis-acrylamide solution for SDS-PAGE, 0.5 M Tris–HCl, pH 6.8, 0.4% SDS, 0.2 mL 2% APS, 20 μL TEMED.
Gel bond paper.
Tris–Glycine–SDS running buffer: 25 mM Tris, 192 mM glycine, 1% (w/v) SDS.
Fixative: 40% (vol/vol) methanol, 10% (vol/vol) acetic acid in dH2O.
Enhancer solution: 1 M salicylic acid with 2% glycerol (vol/vol).
Storage Phosphor screen for gel exposure.
Radiacwash decontaminant to cleanup isotope contamination.
3. Experimental Procedure
Hybrid design
General designing strategy:
Identify target gene and find corresponding siRNAs (21/21 nts) based on the published literature.
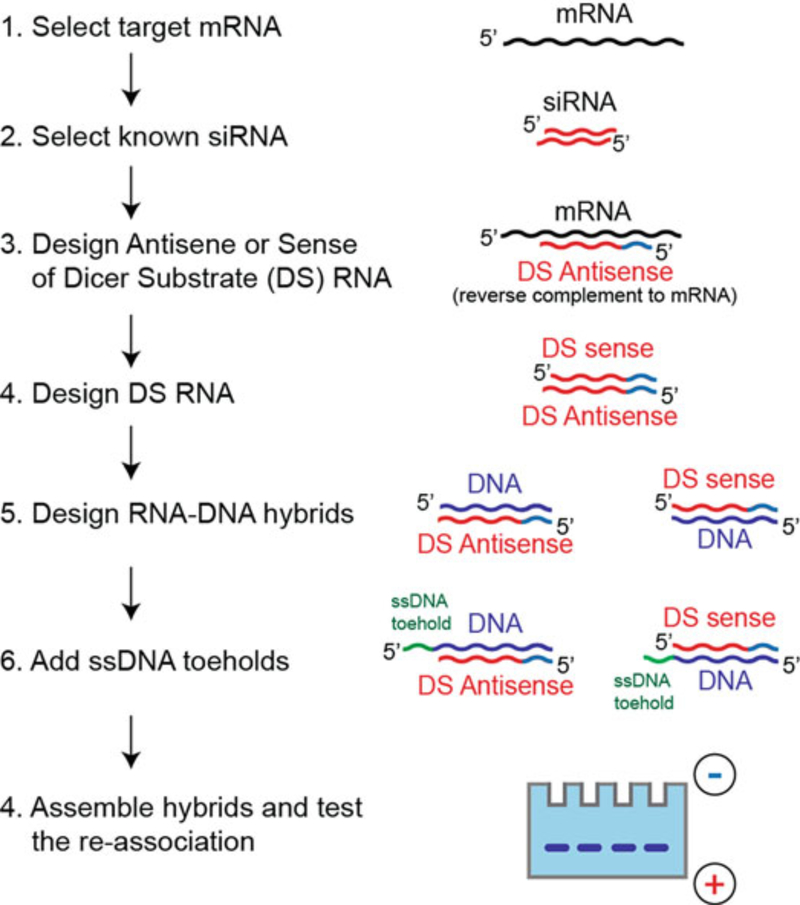
Using the sequences of known siRNAs, design asymmetric (25/27 nts) Dicer substrate RNAs (DS siRNAs) through elongation of the siRNA sense strands with corresponding sequences identified and extracted from mRNAs, as shown in Fig. 2.
Design DNA strands by taking the reverse complements of individual DS RNA strands.
Each of the DNA strands in the hybrids is decorated with ssDNA toehold. The toeholds in the hybrids are complementary to each other for hybridization. The standard toehold sequence [11] that has been used in multiple designs is as follows: 5′-GGA-GACCGTGAC-hybrid (and its complement hybrid -GTCACGGTCTCC-3′), and can be routinely used for any new hybrid designs.
Fig. 2.
Main steps required for RNA–DNA hybrids design and characterization: First target mRNA should be identified, then Dicer substrate (DS) RNA should be designed by elongating the known siRNA sequences, and the corresponding DNA strands with ssDNA toeholds should be added to form the hybrids. The resulting hybrids can be assembled and their reassociation confirmed by gel electrophoretic techniques
Hybrid design for HIV targets:
-
Identify gene locations where previous studies have shown that siRNAs targeting that sequence inhibit HIV replication.
Important: a single nucleotide difference, upstream or down-stream of the siRNA sequence can abrogate the ability to block HIV replication.
Use a sequence location where multiple siRNAs are tested.
Two viral targets are used, viral protease (PR) and surface envelope glycoprotein (gp120). More precisely, positions 2332 to 2356 for PR and 7642 to 7665, according to the virus strain NL43. The sequence accession number AX032749 can be obtained from NCBI GenBank. The following DS RNAs and DNAs for RNA–DNA hybrids are designed as described above.
Hybrids designed against Protease:
DS sense
5′-pGAGCAGAUGAUACAGUAUUAGAAGA
DNA that binds DS sense
5′-TCTTCTAATACTGTATCATCTGCTCCTGTCACGGTC TCC
DS antisense
5′-UCUUCUAAUACUGUAUCAUCUGCUCCU
DNA that binds DS antisense
5′-GGAGACCGTGACAGGAGCAGATGATACAGTATTAG AAGA
Hybrids designed against Envelope:
DS sense
5′-GGACAAUUGGAGAAGUGAAUUAUAUU
DNA that binds DS sense:
5′-AATATAATTCACTTCTCCAATTGTCCGTCACGGTCT CC
DS antisense
5′-pUAUAAUUCACUUCUCCAAUUGUCC
DNA that binds DS antisense
5′-GGAGACCGTGACGGACAATTGGAGAAGTGAATTAT
ATT
3.1. Purification of DNAs and RNAs with Urea Gels
Gel electrophoresis
It’s recommended to purify all the purchased DNAs and RNAs prior to further experiments with RNA–DNA hybrids. To purify oligos, prepare a denaturing gel with TBE. Polymerization is initiated by mixing APS solution and TEMED.
-
Run a vertical gel with spacer thickness of 0.75 mm, TBE is used as a running buffer solution (see Notes 1–4).
Important: Use a syringe to wash the loading wells several times with running buffer prior to loading your samples.
Separate the glass plates and wrap the gel in Saran Wrap.
Visualize the nucleotide bands on TLC plate using a UV lamp at short wavelength (254 nm).
Outline the nucleic acid bands and cut out the DNA and RNA bands.
Recover the DNA and RNA strands by incubating the gel pieces overnight at 4 °C in eluting buffer.
Recover as much supernatant as possible, without collecting gel, and place into new tube.
Add equal volumes of 100% ethanol to each tube containing eluted oligos.
Place oligos in a −20 °C freezer for 3 h or overnight.
Centrifuge for 30 min at 11,000 × g and 4 °C.
Discard supernatant.
Rinse with of 90% ethanol and repeat steps 10 and 11.
Discard supernatant and dry the oligos in the vacuum concentrator.
-
Suspend dry pellets in water.
Important: Resuspended oligos should be always kept on ice or frozen. If frozen, oligos must be vortex-mixed for 5 sec prior to each use.
Measure the absorbance at 260 nm (A260) for each purified oligo using a UV spectrometer.
Based on the sequence, determine the extinction coefficients for each oligo using IDT OligoAnalyzer tools (https://www.idtdna.com/calc/analyzer).
Use the Beer–Lambert law to calculate the concentration of purified oligos:
A = a × b × c, where A is the measured absorbance at 260 nm; a—extinction coefficient; b—path length; c—concentration.
3.2. Assembly of RNA–DNA Hybrids
Mix cognate RNA and DNA strands of designed hybrids at equimolar concentrations in water.
-
Incubate the mixture in a heat block at 95 °C for 2 min to melt all hydrogen bonds.
Important: Do not incubate the mixture longer, as it can promote the degradation of RNAs.
Snap-cool the mixture by rapidly transferring it to the heating block set at 30 °C. As an alternative, snap-cooling to RT can be used.
Add 5× assembly buffer (to the final 1×) and incubate for 20 min.
Run quality control experiments using native polyacrylamide gel electrophoresis (native PAGE) [15]. For visualization, use total staining with ethidium bromide. As an alternative, assemblies containing one fluorescently labeled DNA strand (e.g., with Alexa 488) together with nonlabeled RNA can be directly visualized on the gels using ChemiDoc MP system (see Notes 5–7).
3.3. Native PAGE Experiments for RNA–DNA Assemblies and Visualization of Reassociation
Mix two cognate hybrids at equimolar concentrations and incubate for 30 min at 37 °C.
Prepare a sequencing gel for vertical electrophoresis for high-resolution native PAGE, i.e., 10% (19:1) acrylamide, 1× assembly buffer.
-
For vertical gel with the spacer thickness of 0.75 mm, load samples in individual lanes of a gel (5 μL per lane), perform electrophoresis for 2 h at 20 W at 4 °C.
Important: Use a syringe to wash the loading wells several times with running buffer before loading.
For EtBr-stained gels, a ChemiDoc MP can be used for visualization.
3.4. HIV-1 Inhibition Experiments
Cotransfection of HIV and siRNAs
HeLa cells are cultured with DMEM High Glucose, and are maintained at 37 °C in 5% CO2 incubator humid.
Day 1: Plate 120,000 HeLa cells per well in a 12-well plate.
- Day 2: Cotransfect 800 μg of pNL4–3 molecular clone [27] plus 5, 10, and 20 nM (final concentrations) of RNA–DNA hybrids, or pNL43 plus DS RNAs as positive controls. For knockdown, negative control RNA–DNA hybrids and DS RNAs targeting other genes (e.g., eGFP) can be used. Virus control is measured only with pNL43 transfection and Mock only transfection reagent is used. Use 1 μL of Lipofectamine 2000 (L2K) per well. For RNA–DNA hybrids, incubate DS sense and DS antisense containing hybrids in separate tubes using half of the volume of L2K, and 400 μg of pNL4–3 for each tube.
-
(a)Add to each tube Opti-MEM I (to dilute L2K and nucleic acids, to a maximum final volume of 50 μL).
-
(b)Add RNA–DNA hybrids (containing sense or antisense) and DS RNAs. For virus control do not add RNA–DNA hybrids or DS RNAs.
-
(c)Add 400 or 800 μg of pNL4–3.
-
(d)Add 0.5 or 1 μL of L2K.
-
(e)Incubate for 30 min.
-
(f)Discard old DMEM and add 450 μL of fresh DMEM without penicillin and streptomycin to each well from plated cells.
-
(g)Add dropwise the complexes of L2K/pNL4–3; L2K/pNL4–3/RNA–DNA hybrid sense+L2K/pNL4–3/RNA–DNA hybrid antisense; or L2K/pNL4–3/DS RNA.
-
(h)Discard the media 5 h post transfection.
-
(i)Add 1 mL of DMEM 5% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine.
-
(j)Incubate for 2 days before metabolic labeling (Day 4).
-
(a)
Metabolic Labeling
During the metabolic labeling assay culture cells with RPMI 1640 Medium lacking the amino acids methionine (Met) and cysteine (Cys), supplemented or not with 2% (vol/vol) dFBS [28].
Discard the DMEM media.
Starve the cells with 1 mL of RPMI 1640 Cys−/Met− and incubate for 30 min at 37 °C.
Discard starvation media.
Add 1 mL of RPMI 1640 Cys−/Met− supplemented with 2% dFBS and 250 μCi[35S] Met/Cys (a mixture containing both 35S-L-methionine and 35S-L-cysteine) (see Notes 8–11).
-
Incubate at 37 °C for 4 h.
Important: Handling isotopes requires special training and approval. All solid and liquid waste should be discarded separately following the rules of your institution. If any questions rise, consult the radiation safety program at your Institution.
Dispose the supernatant if you will not measure the virus production.
Lyse the cells using 250 μL of Cell lysis buffer at room temperature.
Immunoprecipitation
Important: Preparation of cell lysates and all subsequent steps use screw-cap tubes with O-rings (Sarstedt) to reduce aerosol production and contamination of equipment with isotopes.
- Preclearance of cell-associated material.
-
(a)Spin the cell lysate at 11,000 × g for 5 min using a microfuge.
-
(b)Transfer 50 μL of the cell lysate supernatant to a new tube.
-
(c)Add 500 μL of Triton wash buffer.
-
(d)Mix with 50 μL Protein A agarose beads/tube and 50 μL 1% BSA/tube.
-
(e)Incubate in low rotation at 4 °C for 1 h.
-
(f)Spin samples for 1 min at 11,000 × g using a microfuge.
-
(g)Collect 550 μL from the supernatant, avoiding disturbing the pellet (This is the precleared lysate).
-
(a)
- Couple HIV-IgG with protein A agarose.
-
(a)Prepare a mixture of 50 μL protein A agarose beads + 30 μL 1% BSA + 1 μL of HIV-1 IgG in 1 mL of Phosphate-buffered saline (PBS) per sample.
-
(b)Incubate in low rotation at 4 °C for 1 h.
-
(c)Spin beads now coupled with HIV-IgG for 5 min at 500 × g using a microfuge.
-
(d)Wash beads 2× with 500 μL of PBS, repeating the spinning from step c.Important: Instead, the coupling of the beads and the washing steps can be performed in a pool by mixing enough solution to all samples in 15 mL tubes (500 μL/per sample). Then before the last spin transfer to a tube and waste the supernatant.
-
(e)After the last wash aspirate all the PBS leaving the beads coupled with HIV-IgG.
-
(f)Add the precleared lysate to the tube with the beads pellet.
-
(g)Immunoprecipitate the samples by incubating in slow rotation at 4 °C overnight.
-
(a)
- Washing the beads complexed with HIV-IgG and HIV proteins from the cell lysate (see Note 12)
-
(a)Spin samples for 1 min at 14,000 RPM.
-
(b)Wash beads 2× with 1 mL of Triton wash buffer room temperature (RT) (Aspirate the supernatant, and inverting the tube 2× after addition of 1 mL of Triton wash buffer, then repeat the spin for 1 min at 11,000 × g).
-
(c)Add 1 mL of SDS/DOC wash buffer to the beads pellet.
-
(d)Incubate at RT for 5 min.
-
(e)Spin samples for 1 min at 11,000 × g.
-
(f)Aspirate the SDS/DOC wash buffer.
-
(g)Resuspend the beads in 100 μL 2× Sample buffer.
-
(h)Vortex briefly.
-
(i)Incubate in dry heating block at 99 °C for 5 min.
-
(j)Spin samples for 1 min at 11,000 × g.
-
(k)Transfer 80 μL of the samples supernatant to a new tube.
-
(a)
Radioimmunoprecipitation SDS-PAGE
- Gel preparation
-
(a)Acrylamide–AcrylAide lower gel.
-
(b)Stacking upper gel.
-
(c)Add gel bond paper to the smaller glass, with the hydrophobic side facing the glass.
-
(e)Once the gel is casted and polymerized load the samples.
-
(a)
Run the gel using 1× Tris–glycine–SDS buffer.
Gently remove the gel bond paper with the gel attached.
Fix the gel: Add the gel to a container with gel fixative solution.
Incubate by slow shaking at room temperature for 2 h.
Discard the gel fixative.
Add Enhancer solution to the container.
Incubate in slow shaking at RT for 2 h.
Place the gel on paper towel.
Incubate in dry 100 °C oven until the gel dries (see Note 13).
Place the gel in an X-ray film cassette.
-
Add a phosphorimage screen for quantification.
Important: Face the gel to the white side of the phosphorimage screen. The time of exposure will depend on the signal. We recommend the first exposure to be overnight, or use the Geiger counter for measuring signal intensity.
Read the screen in any Phosphorimager (Fig. 3).
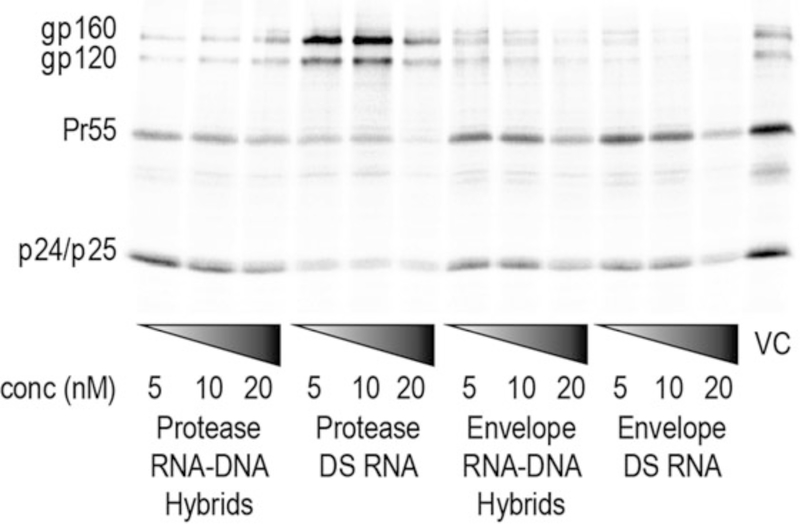
Fig. 3.
Inhibition of HIV-1 expression and production by intracellular reassociation of RNA–DNA hybrids and activation of RNAi. HeLa cells were transfected with pNL4–3, without (VC virus control) and with RNA–DNA hybrids or DS RNAs. At 48 h posttransfection, HeLa cells were metabolically labeled with [35S] MetCys for 4 h. Cell lysates were radioimmunoprecipitated. Positions of envelope glycoprotein precursor (gp160), and surface glycoprotein (gp120); Pr55Gag (Pr55), CA/CA-SP1 (p24/p25). (Original figure from Afonin et al., Nat Nano 2013)
Quantifying HIV proteins inside the cell
Important: Use the software from the Phosphorimager used to quantify the bands of interest. Always subtract the background value from each band. Since we target PR, and PR is located in the full length mRNA, we quantify the expression of Gag. Although the siRNA targeting Envelope binds to the Envelope mRNA, it also binds the full-length mRNA.
-
14.
Quantification of total cell-associated Gag Pr55Gag (Pr55), matrix-capsid (p41), and capsid/capsid-SP1 (p24/p25) (Pr55 + p41 + p25 + p24).
-
15.
Plot a column graph on Excel normalizing the relative amount of Gag Pr55 + p41 + p25 + p24 from at least three repetitions.
4. Notes
Avoid overloading samples to prevent less efficient separation in native PAGE gel.
Prior to pipetting, mix well to avoid unequal concentrations.
Prerun the native PAGE gel as a calibration and quality control step to remove magnesium ions.
After loading of the samples, run the gel at a lower voltage to allow all nucleic acid strands of all lengths to begin separation at the same point.
In order to obtain the best result for strand association, make sure each DNA–RNA duplex is in exactly equal concentrations.
Cool tubes before opening after incubating at 95 °C to avoid loss of the volatilized sample so that the concentration remains unchanged.
For native PAGE visualization use total staining with ethidium bromide. As an alternative, assemblies containing fluorescence labeled RNA/DNA strands together with nonlabeled RNA/DNAs can be visualized using a gel imaging system such as ChemiDoc.
Use the Geiger counter to monitor any contamination with 35S during the work with radioisotopes.
Check your gloves with the Geiger counter before disposing of them.
Radiacwash decontaminant to cleanup isotope contamination works better than regular soap.
For more difficult isotope contamination use household bleach, then rinse with ethanol 70%.
When removing or aspirating supernatants, avoid disturbing the pellet.
The gel will change from clear to opaque white. Do not over-dry to avoid breaking of the gel. Use an oven connected to a vacuum line for shorter incubation time.
References
- 1.Bramsen JB, Kjems J (2012) Development of therapeutic-grade small interfering RNAs by chemical engineering. Front Genet 3:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811 [DOI] [PubMed] [Google Scholar]
- 3.Parrish S, Fleenor J, Xu S et al. (2000) Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol Cell 6(5):1077–1087 [DOI] [PubMed] [Google Scholar]
- 4.Lund E, Dahlberg JE (2006) Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol 71:59–66 [DOI] [PubMed] [Google Scholar]
- 5.Ji X (2008) The mechanism of RNase III action: how dicer dices. Curr Top Microbiol Immunol 320:99–116 [DOI] [PubMed] [Google Scholar]
- 6.Macrae IJ, Zhou K, Li F et al. (2006) Structural basis for double-stranded RNA processing by Dicer. Science 311(5758):195–198. doi: 10.1126/science.1121638 [DOI] [PubMed] [Google Scholar]
- 7.Dao BN, Viard M, Martins AN et al. (2015) Triggering RNAi with multifunctional RNA nanoparticles and their delivery. DNA RNA Nanotechnol 1(1):27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarz DS, Hutvagner G, Du T et al. (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115(2):199–208 [DOI] [PubMed] [Google Scholar]
- 9.Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115(2):209–216 [DOI] [PubMed] [Google Scholar]
- 10.Pratt AJ, MacRae IJ (2009) The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem 284(27):17897–17901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afonin KA, Viard M, Martins AN et al. (2013) Activation of different split functionalities on re-association of RNA-DNA hybrids. Nat Nanotechnol 8(4):296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shekhawat SS, Ghosh I (2011) Split-protein systems: beyond binary protein-protein interactions. Curr Opin Chem Biol 15(6):789–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose SD, Kim DH, Amarzguioui M et al. (2005) Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res 33(13):4140–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afonin KA, Viard M, Tedbury P et al. (2016) The use of minimal RNA toeholds to trigger the activation of multiple functionalities. Nano Lett 16(3):1746–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afonin KA, Desai R, Viard M et al. (2014) Co-transcriptional production of RNA-DNA hybrids for simultaneous release of multiple split functionalities. Nucleic Acids Res 42 (3):2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afonin KA, Bindewald E, Kireeva M et al. (2015) Computational and experimental studies of reassociating RNA/DNA hybrids containing split functionalities. Methods Enzymol 553:313–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afonin KA, Grabow WW, Walker FM et al. (2011) Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 6 (12):2022–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grabow WW, Zakrevsky P, Afonin KA et al. (2011) Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett 11(2):878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yingling YG, Shapiro BA (2007) Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett 7 (8):2328–2334 [DOI] [PubMed] [Google Scholar]
- 20.Afonin KA, Bindewald E, Yaghoubian AJ et al. (2010) In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 5 (9):676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afonin KA, Kasprzak W, Bindewald E et al. (2014) Computational and experimental characterization of RNA cubic nanoscaffolds. Methods 67(2):256–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afonin KA, Viard M, Kagiampakis I et al. (2015) Triggering of RNA interference with RNA-RNA, RNA-DNA, and DNA-RNA nanoparticles. ACS Nano 9(1):251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afonin KA, Viard M, Koyfman AY et al. (2014) Multifunctional RNA nanoparticles. Nano Lett 14(10):5662–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers TA, Andrews GE, Jaeger L et al. (2015) Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer. ACS Synth Biol 4(2):162–166 [DOI] [PubMed] [Google Scholar]
- 25.Groves B, Chen YJ, Zurla C et al. (2016) Computing in mammalian cells with nucleic acid strand exchange. Nat Nanotechnol 11 (3):287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Low JT, Knoepfel SA, Watts JM et al. (2012) SHAPE-directed discovery of potent shRNA inhibitors of HIV-1. Mol Ther 20(4):820–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adachi A, Gendelman HE, Koenig S et al. (1986) Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59 (2):284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waheed AA, Ono A, Freed EO (2009) Methods for the study of HIV-1 assembly. Methods Mol Biol 485:163–184 [DOI] [PMC free article] [PubMed] [Google Scholar]