Abstract
The applications of cell patterning are widespread due to the high-throughput testing and different resolutions offered by these platforms. Cell patterning has aided in deconvoluting in vivo experiments to better characterize cellular mechanisms and increase therapeutic output. Here, we present a technique for engineering an artificial surface via surface chemistry to form large-scale arrays of cells within a microchannel by employing microstamping. By changing the approach in surface chemistry, H1568 cells were patterned hydrodynamically using immunoaffinity, and neutrophils were patterned through self-assembly via chemotaxis. The high patterning efficiencies (93% for hydrodynamic patterning and 68% for self-assembled patterning) and the lack of secondary adhesion demonstrate the reproducibility of the platform. The interaction between H1568 and neutrophils was visualized and quantified to determine the capability of the platform to encourage cell-cell interaction. With the introduction of H1568 cells into the self-assembled patterning platform, a significant hindrance in the neutrophils' ability to swarm was observed, indicating the important roles of inflammatory mediators within the nonsmall cell lung cancer tumor microenvironment.
INTRODUCTION
Cellular heterogeneity has presented a consistent challenge in characterizing cellular behavior and formulating therapeutics to combat diseases.1–3 Spatially organizing cells in vitro can remove the complexities associated with animal studies4,5 by either extrapolating observations to larger populations6 or by highlighting the importance of variability between cells.7 Although most patterning methods lack the third-dimension that more accurately models the in vivo microenvironment,8 two-dimensional patterns can be investigated temporally with little convolution.6,9 To combat the loss of in vivo imitation, cocultured cells10 or extracellular matrix materials11,12 have been included within two-dimensional formulations. Cell patterning can also be performed at different resolutions such as single-cell patterning13,14 or carefully segregated colonies15 to understand single-cell mechanisms or population dynamics. Since micropatterning allows for the high-throughput testing of several experimental conditions, this technique can be used to measure real-time cytotoxicity,16 genotoxicity,17 and the genetic alterations behind drug resistance,18 leading to more efficient drug screening platforms that can increase the output of therapeutic discovery.19
Robotics have been used to create arrays of mammalian cells;20 however, with the recent advancements in biomicroelectromechanical system (BioMEMS) fabrication, BioMEMS have become alternate, cost-effective candidates that offer spatial confinement conducive to cell patterning.21 The dimensions on the scale of a cell promote the patterning of single cells and allow for definite microarrays.6 Developments with BioMEMS have been made to physically pattern cells using electrical, optical, acoustic, and magnetic impulses.21 However, these techniques can overheat the cell,22 photobleach,23 limit the variability in patterning,6,24 or require cellular manipulation.25 The use of surface chemistry could avoid these external forces that are either damaging to the cell or offer limited patterning capacity. However, the current methods of chemical attachment require large changes in temperature,26 UV radiation,27,28 or the removal of monolayers using plasma or electrochemistry,29 which may still be harmful to the cells. Using substrates that are readily found within the body and that can attract cells naturally may bypass these obstacles while maintaining the microenvironment. For example, nonsmall cell lung cancer (NSCLC) cells, such as the H1568 cell line, overexpress epithelial markers, which can be used to anchor the cells onto a microfluidic device coated with antibodies against epithelial cells.30 On the other hand, neutrophils, whose main role lies within the innate immune response,31 have been patterned dynamically into large-scale arrays through swarming.32–34 Neutrophil swarming within these arrays occurs in four successive steps. Scouting is the first step, which is when free-flowing neutrophils come in contact with pathogen-associated molecular patterns leading them to activate and adhere to their target.33–35 Upon activation, the neutrophil releases chemotactic mediators that recruit more neutrophils to the site of infection in the next step referred to as the growth phase.33,34,36,37 The third step is the equilibrium phase, where the migration dwindles into a constant swarm size, followed by the resolution of the infection.33,34,38,39
Here, we engineered a large-scale array of cells within a microchannel via microstamping, allowing facile surface chemistry within only regions of interest (ROI). The cells were patterned by exploiting the inherent attributes present within H1568 cells and neutrophils by changing the approach in surface chemistry. The H1568 cells were patterned hydrodynamically at an efficiency of 92.9 ± 6.22% by tethering the cells to antibodies against epithelial cells. The neutrophils were patterned through self-assembly at an efficiency of 68.5 ± 15.9% by employing the neutrophils' ability to swarm to localized infection points simulated by Escherichia coli bioparticles. The addition of the microchannel decreased the amount of secondary cellular adhesion experienced without the need of nonbiofouling molecules or self-assembling monolayers.28,29,40 The self-assembled cell-patterning platform was validated for cell-cell communication by introducing H1568 cells. A significant decrease in the neutrophils' capability to swarm was observed (ANOVA, p < 0.0001 for all three donors). Furthermore, the pooled-average efficiency in the self-assembled pattern dropped from 68.5 ± 15.9% to 22.4 ± 11.9% (t-test, p < 0.0001) in the presence of H1568 cells.
MATERIALS AND METHODS
Materials
For hydrodynamic patterning, gelatin (Sigma-Aldrich, St. Louis, MO) was patterned into a microarray. The gelatin microarray was coated with streptavidin-coated polystyrene nanoparticles (Spherotech, Lake Forest, IL). The biotinylation of anti-EGFR (epithelial growth factor receptor) (ImClone Systems, New York City, NY), anti-EpCAM (epithelial cellular adhesion molecule) (R&D Systems, Minneapolis, MN), and anti-HER2 (human epithelial growth factor receptor 2) (R&D Systems, Minneapolis, MN) was performed with a biotinylation kit from Life Technologies (Carlsbad, CA) allowing for the attachment of the antibodies onto the microarray. For self-assembled patterning, Zetag® 8185 (BASF, Florham Park, NJ) was patterned into a microarray. The Zetag microarray was coated with E. coli (K-12) bioparticles conjugated with Alexa Flour 594 (Invitrogen, Carlsbad, CA). The H1568 cells were stained with CellTracker™ Red CMPTX (Invitrogen, Carlsbad, CA) or CellTracker Green CMFDA (Invitrogen, Carlsbad, CA), and the neutrophils were stained with Hoechst 33342 (Life Technologies, Carlsbad, CA). The polydimethylsiloxane (PDMS) microchannel and the microstamp were fabricated with the SYLGARD™ 184 Silicone Elastomer Base (Krayden, Denver, CO) and with the SYLGARD 184 Silicone Elastomer Curing Agent (Krayden, Denver, CO).
Microfabrication of PDMS devices
Standard photolithography was used to fabricate silicon masters of the microfluidic channel and the microstamp. Briefly, the photoresist SU-8 2025 (MicroChem, Westborough, MA) was added onto a 4 in. silicon wafer. A chrome mask was used to pattern the photoresist with UV light. The excess of the photoresist was removed using the SU-8 Developer (MicroChem, Westborough, MA). The silicone elastomer base was mixed with the silicone elastomer curing agent at a ratio of 10:1. The mixture was added to the silicon master, placed under vacuum, and later allowed to cure overnight at 65 °C.
Cell preparation
The H1568 cell line was cultured in RPMI-1640 (Thermo Fisher Scientific, Waltham, MA) containing 10% (v/v) fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA) and 1% (v/v) penicillin-streptomycin (Thermo Fisher Scientific, Waltham, MA). CellTracker Red or CellTracker Green was used to stain the cytoplasm of cells. Neutrophils were isolated from fresh blood collected from healthy donors. Blood specimens were obtained after informed volunteer’s consent according to the institutional review board (IRB) protocol No. 2018H0268 reviewed by the Biomedical Sciences Committee at The Ohio State University. Neutrophils were isolated using an Enrichment Kit from STEMCELL Technologies (Vancouver, Canada), and their nuclei were stained with Hoechst 33342. The enrichment kit works on a negative selection principle. An antibody cocktail with specificity for the cellular components of human blood aside from neutrophils was added to the buffy coat from fresh blood. The antibodies were then conjugated onto magnetic particles and separated via a magnetic field. Isolated neutrophils were resuspended in IMDM (Thermo Fisher Scientific, Waltham, MA) with 20% (v/v) FBS.
Microstamping
The working solution (ink) was spread over a clean glass slide, which was then spin-coated to uniformly distribute the solution. The microstamp was then placed face down on the inked glass slide with a 95-g weight to imprint the ink onto the microstamp. After 20 min, the microstamp was removed and placed face down onto an ozone-treated glass slide with the 95-g weight for 10 min. The ink used for hydrodynamic patterning was a 0.5% (w/v) gelatin solution with poly-l-lysine-FITC (Sigma-Aldrich, St. Louis, MO). The ink used for self-assembled patterning was 1.6 mg/ml of Zetag.
Particle patterning and microfluidic channel bonding
For hydrodynamic patterning, 0.005% (w/v) streptavidin-coated polystyrene nanoparticles were physically adsorbed onto the gelatin pattern by allowing them to sit overnight on a rocker at 4 °C. Similarly, for self-assembled patterning, E. coli (K-12) bioparticles were attached to the Zetag pattern via electrostatic binding for 20–30 min before washing. The microfluidic channel and the patterned glass slide were oxygen-plasma treated for 15 s to reduce damage to the surface chemistry (Fig. 1 in the supplementary material). Then, they were irreversibly bonded together using a hot plate at 45 °C for 20 min.
Antibody functionalization
Devices used for hydrodynamic patterning were functionalized with antibodies. Anti-EGFR, anti-EpCAM, and anti-HER2 were biotinylated using a biotinylation kit. 10 μg/ml of the biotinylated antibody was added to a 1% (v/v) bovine serum albumin (BSA) solution (Sigma-Aldrich, St. Louis, MO). The device was primed with liquid, and then the antibodies were added and incubated overnight at 4 °C.
Hydrodynamic cell patterning
The stained H1568 cells were concentrated to approximately 2 million cells/ml and filtered to remove cell aggregates. A blocking solution with 0.05% (v/v) polyoxyethylenesorbitan monolaurate (Sigma-Aldrich, St. Louis, MO) in 3% (v/v) BSA was loaded into the device and allowed to sit for an hour before removing with the phosphate saline buffer (PBS). The H1568 cells were put on a rocker and fed through the device at a rate of 1 ml/h for 1 h via gravity-driven flow (Fig. 2 in the supplementary material). The cells were then washed by injecting PBS through the outlet.
Self-assembled cell patterning
The stained neutrophils were concentrated to 20 million cells/ml and were added to the device via injection after wetting with 100% ethanol and subsequently with PBS. After injection, the inlet and outlet of the device were pinched shut to inhibit unwanted flow. The cells were then washed by injecting PBS through the outlet. When introducing the mixture of both cells, the ratio of H1568 cells to neutrophils was 1:20, where the same concentration of neutrophils was maintained. This mixture was injected into the device, similar to how the neutrophils alone were added.
Live-cell imaging and analysis
The cells were imaged using a Nikon® Ti2 microscope immediately after introducing them into the microchannel. The surrounding environment was set to 37 °C and 5% CO2 by a cage incubator. The H1568 cells and the patterned substrates were imaged for brightfield and fluorescence at magnifications of 10× and 20×. The migration of neutrophils was visualized for brightfield and fluorescence for 2.5 h, employing automated time-lapse imaging at a magnification of 10×. The migration of neutrophils was quantified by measuring the normalized fluorescent intensity at 405 nm within regions of interest (ROI). The ROI was defined by nine 75 μm diameter circles surrounding randomly selected points chosen via a random number generator within the array. The efficiency of the hydrodynamic cell patterning was measured by the number of occupied patterns per number of total patterns within a nine-by-nine array for three different arrays and reproduced by three experiments. The efficiencies for the self-assembled cell patterning were measured employing the same method and reproduced across three healthy donors. The histograms were generated by counting the number of cells inside 100 points chosen at random within the array for the three different trials.
RESULTS
Microstamping for large-scale patterning within a microchannel
Microstamping was used to generate large-scale arrays of cells following two patterning approaches: hydrodynamically or through self-assembly (Fig. 1). These different forms of cellular attachment were governed by simple changes in the surface chemistry that exploit the characteristics specific to the cell in use. For example, when patterning the cells hydrodynamically, antibodies specific to NSCLC were used to attach the NSCLC cells through immunoaffinity. On the other hand, when patterning the cells through self-assembly, E. coli bioparticles were used to attract the neutrophils employing positive chemotaxis. These patterning methods could be extended to other cell types so long as the proper antibodies and chemoattractants are selected, the latter of which requires cells that are highly chemotactic. A microfluidic channel with 3D herringbone patterns was chosen, since its flow regime maximizes the contact cells have with the glass surface to which the channel is attached, increasing the probability of cellular adhesion.41 Therefore, using the microchannel on a chemically modified surface in distinct positions would maximize the contact time of the cells with the pattern, leading to better patterning efficiency and increased contact between different cell types or stimuli.
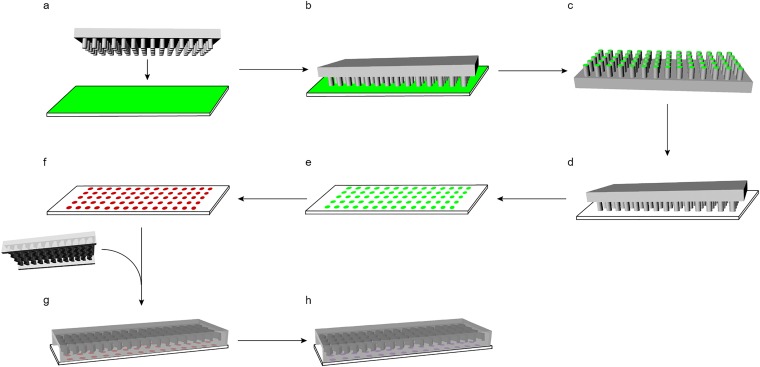
FIG. 1.
Platform preparation using microstamping. (a) A PDMS microstamp is fabricated by photolithography with 30 μm diameter columns spaced 150 μm center-to-center and a clean glass slide is spin-coated with gelatin for hydrodynamic patterning or Zetag for self-assembled patterning. (b) The microstamp and the glass slide are placed in conjunction. (c) The ink is transferred onto the microstamp. (d) A plasma-treated glass slide and the inked stamp are placed in conjunction. (e) The ink is transferred to the plasma-treated glass slide yielding a microarray. (f) Particles particular to the form of patterning (streptavidin-polystyrene nanoparticles for hydrodynamic patterning or E. coli bioparticles for self-assembled patterning) are deposited onto the substrate. (g) The microchannel with a length of 40 mm, width of 2.5 mm, and a total height of 90 μm (with a 50-μm distance from the glass surface to the herringbone design) was irreversibly bound to the particle-patterned slide via oxygen-plasma treatment. (h) For hydrodynamic patterning, biotinylated antibodies are added with flow through the microchannel.
Hydrodynamic cell patterning
An antibody cocktail against epithelial markers including EGFR, EpCAM, and HER2 was used to capture the cells within the microchannel with a high efficiency, by accounting for the heterogeneous expression levels of surface antigens on cancer cells.42 The antibodies were connected by employing the strong affinity biotin has for streptavidin.43 First, the gelatin was stamped onto the device, where the streptavidin-polystyrene nanoparticles were physically adsorbed. Second, the biotinylated antibodies were attached after the adhesion of the microchannel. By running the cells at a low flow rate of 1 ml/h and a high cell density of 2 million cells/ml, the cells were able to attach to the tethered antibodies, which were only available on the gelatin pattern.
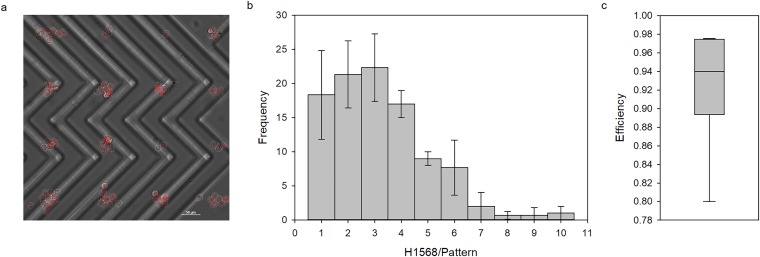
The flow present within the microchannel restricted the cells from adhering onto the nonpatterned glass surface, which allowed the free-flowing cells to be easily washed with PBS, yielding a clean pattern [Fig. 2(a)]. Without flow, the cells adhered to the glass even after washing vigorously amounting to a random, nondefined pattern [Fig. 3(a) in the supplementary material]. The 30 μm diameter substrate pattern allowed for the adhesion of single cells as can be seen by the distribution that is skewed to the left [Fig. 2(b)]. The average number of cells per pattern was 3.23 ± 1.84 cells per pattern. The clear, distinct pattern as well as the high efficiency of 92.9 ± 6.22% calculated by the number of filled patterns per total number of gelatin patterns [Fig. 2(c)] render this patterning method as a promising technique.
FIG. 2.
Hydrodynamic cell patterning. (a) H1568 cells stained with CellTracker (red) at a concentration of approximately 2 million cells/ml are patterned onto the gelatin microdomain via immunoaffinity capture. (b) The distribution for the number of H1568 cells on each pattern (n = 100) with three different trials (error bars indicate the standard deviation) shows an average pattern size of 3.23 ± 1.84 cells/pattern. (c) The efficiency of the patterning method using immunoaffinity is 92.9 ± 6.2%.
Self-assembled cell patterning
Taking advantage of the cellular communication between neutrophils, the patterned substrate mimicked a bacterial localized infection point by attaching E. coli bioparticles to Zetag via electrostatic interaction. Therefore, by exploiting this chemotactic behavior, the cells swarmed toward the bacterial bioparticles, forming a pattern through self-assembly following the four steps of neutrophil swarming. The neutrophils were added within the device at a concentration of 20 million cells/ml, where the flow was restricted by closing the inlet and the outlet of the device. A slight movement was observed that did not overpower the ability of the neutrophils to swarm.
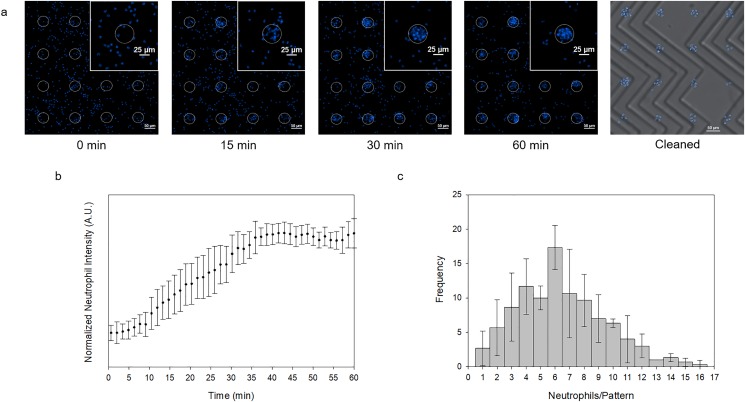
The neutrophils swarmed to the nearest bacterial target and formed a clear pattern. Similar to the hydrodynamic patterning, the slight presence of flow hindered the unwanted adhesion of cells outside of the pattern when rinsed with PBS [Fig. 3(a)]. The clean pattern can also be attributed to the flow regime within the microchannel during the cleaning step. When introduced to a platform without flow, secondary adhesion was observed [Fig. 3(b) in the supplementary material]. The formation of the pattern began at around five minutes and was fully formed after approximately 40 min. After the full formation of the pattern, the size would neither increase nor decrease shown by the asymptotic behavior of the normalized neutrophil intensity indicating the equilibrium phase [Fig. 3(b)]. Although the substrate microarray was of the same size as that in the hydrodynamic patterning platform (30 μm diameter), the average pattern size was approximately twice as large with respect to the hydrodynamic patterns at 6.46 ± 3.02 cells per pattern [Fig. 3(c)]. Given that the distribution is not supported by a Poisson statistic (Pearson χ2-test, p < 0.0001), the larger pattern sizes may be a result of the patterning mechanism as opposed to only the cell concentration and the cell size. Since the patterning was dictated by the migration of the cells to a single target, the cells often clumped together and adhered to each other, which may have contributed to larger pattern sizes.
FIG. 3.
Self-assembled cell patterning. (a) Neutrophils stained with Hoechst (blue) seeded at approximately 4000 cells/mm2 swarm toward the Zetag pattern indicated by the white, dotted circles over 60 min. The cleaned product is shown to the right. (b) The normalized neutrophil intensity measured by a normalized fluorescent intensity at 405 nm is graphed vs time, quantitatively representing Fig. 3(a). Three trials were performed (error bars indicate the standard deviation). (c) The distribution for the number of neutrophils on each pattern (n = 100) with three different trials (error bars indicate the standard deviation) shows an average pattern size of 6.46 ± 3.02 cells/pattern.
Neutrophil and cancer cell interaction
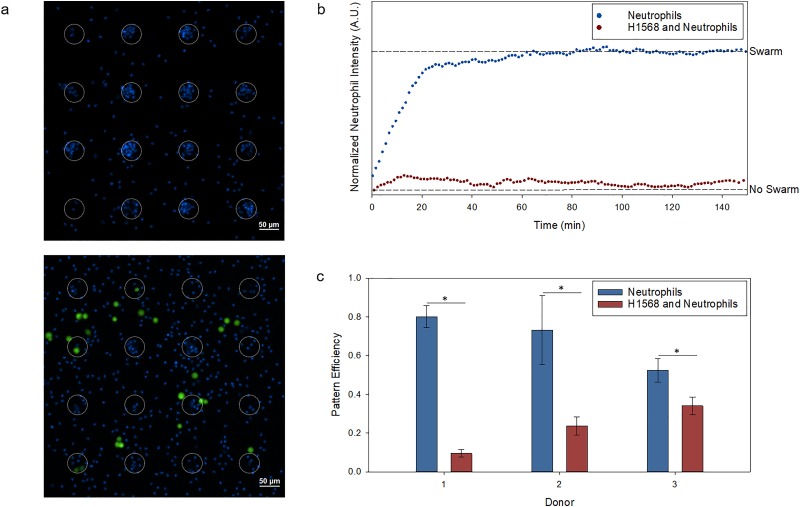
H1568 cells were added to the neutrophil self-assembly platform at a ratio of 1:20, respectively. In the presence of H1568 cells, neutrophils lacked the ability to swarm enough to form distinct patterns [Fig. 4(a)]. Although there was swarming when H1568 cells were present, the neutrophils' ability to swarm and form a pattern was drastically hindered [Fig. 4(b)]. This statistically significant outcome was observed for neutrophils derived from three healthy donors (Fig. 4 in the supplementary material). A metric indicative of the patterning capacity can be defined by normalizing the fluorescent intensity at 405 nm surrounding the E. coli bioparticles after the swarming had ceased (60 min), where a value of zero describes free-flowing neutrophils without a distinct pattern and a value of one describes the maximum swarm observed over time. With the introduction of H1568, the swarming capacity decreased from 0.966 ± 0.0131 to 0.0459 ± 0.0156 for donor 1 (ANOVA, p < 0.0001), 0.940 ± 0.0342 to 0.527 ± 0.0279 for donor 2 (ANOVA, p < 0.0001), and 0.910 ± 0.0456 to 0.429 ± 0.0265 for donor 3 (ANOVA, p < 0.0001).
FIG. 4.
Cellular interaction between H1568 and neutrophils. (a) Neutrophils stained with Hoechst (blue) seeded at approximately 4000 cells/mm2 form stable swarms after 60 min, where the white, dotted circles represent the Zetag pattern (top). The introduction of approximately 200 cells/mm2 H1568 cells stained with CellTracker (green) demonstrates a lack of pattern formation (bottom). (b) The average normalized neutrophil intensity (n = 9) measured by a normalized fluorescent intensity at 405 nm is graphed vs time, showing a significant difference in swarming capabilities between solely neutrophils (blue) and neutrophils and H1568 cells (red) [ANOVA, p < 0.0001 for all three donors (Fig. 4 in the supplementary material)]. (c) The pattern efficiency, measured by the number of swarms per Zetag pattern, shows a significant difference (*t-test, n = 3, p < 0.05) across all three donors (the error bars indicate the standard deviation).
To further quantify the loss in neutrophil function during pattern formation, the pattern efficiency was calculated by the number of swarms per the total number of E. coli bioparticle patterns. Again, the efficiencies across every donor were significantly higher for the platforms that did not have H1568 cells present and highlighted the heterogeneity between the immune responses of the different donors [Fig. 4(c)]. The efficiencies decreased from 80.0 ± 5.66% to 9.51 ± 1.95% for donor 1 (t-test, p < 0.0001), 73.1 ± 17.8% to 23.7 ± 4.70% for donor 2 (t-test, p = 9.70 × 10−3), and 52.4 ± 6.13% to 34.0 ± 4.58% for donor 3 (t-test, p = 0.0141). These results imply that there is a loss in the ability for neutrophils to initiate the swarming response within the NSCLC tumor microenvironment.
DISCUSSION AND CONCLUSIONS
The applications for cell patterning are diverse and ubiquitous, ranging from assembling subpopulations of cells to mimic the cellular arrangement observed in tissues44 to measuring the firing properties and signal propagation of neurons.13,14 To tailor the needs of the different practices, a cell-patterning technique that can be easily tuned is appealing. The tunability for cell patterning for this platform was demonstrated by showing two different mechanisms of patterning, simply by changing the substrate that is microstamped and the successive layers formed using surface chemistry. Patterning via immunoaffinity used the hydrodynamic flow present within the microchannel, whereas patterning using chemotaxis exploited the cellular communication cells innately possess to pattern with self-assembly. The patterning efficiency for hydrodynamic patterning was 92.9 ± 6.22% and a pooled average of 68.5 ± 15.9% for self-assembled patterning, rendering this patterning method as a potential candidate for a wide array of applications.
In vitro platforms remove the complexity associated with single cellular mechanisms present in cancer progression whose nature is vastly heterogeneous and is often convoluted within animal models.1,4,45 The immune cell that makes the largest presence in NSCLC is the neutrophil, so understanding the interaction of the two could lead to better immune-based therapeutics.46 To demonstrate the ability of this platform to track cell-cell communication, the interaction between NSCLC cells and activated neutrophils was investigated. The neutrophils were activated artificially by the E. coli bioparticles, simulating an inflammatory response.47 With the introduction of NSCLC cells, neutrophils lost their ability to swarm. Tumor-associated neutrophils (TANs) have been shown to adopt different phenotypes that are either antitumorigenic or protumorigenic as a result of intercellular communication within the cancer microenvironment.48,49 The platform for self-assembled patterning encouraged the cell signaling between neutrophils and NSCLC cells insofar as the NSCLC cells interfered with the natural inflammatory response of the neutrophils. Understanding the signaling molecules that are responsible for this apparent protumorigenic behavior could aid in formulating immune-based therapeutics against NSCLC.50
Although the immobilization of antibodies to capture flowing cancer cells has been well reported,41,42,51,52 the novelty behind this platform is its ability to capture cells in specific surface locations. Engineering the surface within a microchannel greatly reduced the secondary adhesion of cells outside the patterned region, leading to greater specificity in patterning and high efficiencies for both hydrodynamic and self-assembled patterning. Other methods require nonbiofouling molecules or self-assembling monolayers to combat unspecific adhesion,28,29,40 which could also add to unwanted stimulation. This platform only requires chemical modifications in areas where cellular attachment is desired, forming an inert microarray that can enhance cellular communication and high-throughput screening.53
SUPPLEMENTARY MATERIAL
See the supplementary material for the effects of plasma treatment on the surface chemistry (Fig. 1), the schematic for the gravity-driven flow for hydrodynamic patterning (Fig. 2), the unwanted cellular adhesion when patterning outside a microchannel (Fig. 3), and the statistical decrease in neutrophil swarming with the introduction of H1568 cells (Fig. 4).
ACKNOWLEDGMENTS
We thank the volunteers who participated in this study and kindly donated their blood. This work was partially supported by a grant from the National Institutes of Health (NIH) (No. 1UG3TR002884) with additional funding from the William G. Lowrie Department of Chemical and Biomolecular Engineering and the Comprehensive Cancer Center at The Ohio State University.
REFERENCES
- 1.Bhatia S., Frangioni J. V., Hoffman R. M., Iafrate A. J., and Polyak K., “The challenges posed by cancer heterogeneity,” Nat. Biotechnol. 30, 604–610 (2012). 10.1038/nbt.2294 [DOI] [PubMed] [Google Scholar]
- 2.Altschuler S. J. and Wu L. F., “Cellular heterogeneity: Do differences make a difference?,” Cell 141, 559–563 (2010). 10.1016/j.cell.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldman S. L. et al. , “The impact of heterogeneity on single-cell sequencing,” Front. Genet. 10, 8 (2019). 10.3389/fgene.2019.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao S. S., Kondapaneni R. V., and Narkhede A. A., “Bioengineered models to study tumor dormancy,” J. Biol. Eng. 13, 3 (2019). 10.1186/s13036-018-0137-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones C. N. et al. , “Microfluidic assay for precise measurements of mouse, rat, and human neutrophil chemotaxis in whole-blood droplets,” J. Leukoc. Biol. 100, 241–247 (2016). 10.1189/jlb.5ta0715-310rr [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins D. J. et al. , “Two-dimensional single-cell patterning with one cell per well driven by surface acoustic waves,” Nat. Commun. 6, 8686 (2015). 10.1038/ncomms9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park J. A. et al. , “Freeform micropatterning of living cells into cell culture medium using direct inkjet printing,” Sci. Rep. 7, 14610 (2017). 10.1038/s41598-017-14726-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong X. and Mills K. L., “Large-scale patterning of single cells and cell clusters in hydrogels,” Sci. Rep. 8, 3849 (2018). 10.1038/s41598-018-21989-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X. et al. , “Controlled cell patterning on bioactive surfaces with special wettability,” J. Bionic Eng. 14, 440–447 (2017). 10.1016/S1672-6529(16)60409-2 [DOI] [Google Scholar]
- 10.Zhao L. et al. , “Tape-assisted photolithographic-free microfluidic chip cell patterning for tumor metastasis study,” Anal. Chem 90, 15 (2018). 10.1021/acs.analchem.7b03225 [DOI] [PubMed] [Google Scholar]
- 11.Fernandes T. G., Diogo M. M., Clark D. S., Dordick J. S., and Cabral J. M. S., “High-throughput cellular microarray platforms: Applications in drug discovery, toxicology and stem cell research,” Trends Biotechnol. 27, 342–349 (2009). 10.1016/j.tibtech.2009.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaim C. J., Teng D., Chien S., and Bhatia S. N., “Combinatorial signaling microenvironments for studying stem cell fate,” Stem Cells Dev. 17, 29–39 (2008). 10.1089/scd.2007.0085 [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto H., Hayakawa T., Netoff T. I., and Hirano-Iwata A., “A single-cell based hybrid neuronal network configured by integration of cell micropatterning and dynamic patch-clamp,” Appl. Phys. Lett. 113, 133703 (2018). 10.1063/1.5049487 [DOI] [Google Scholar]
- 14.Yamamoto H. et al. , “Unidirectional signal propagation in primary neurons micropatterned at a single-cell resolution,” Appl. Phys. Lett. 109, 043703 (2016). 10.1063/1.4959836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin X. and Riedel-Kruse I. H., “Biofilm lithography enables high-resolution cell patterning via optogenetic adhesin expression,” Proc. Natl. Acad. Sci. U.S.A. 115, 3698–3703 (2018). 10.1073/pnas.1720676115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiao S. C. et al. , “Real time assays for quantifying cytotoxicity with single cell resolution,” PLoS One 8, e66739 (2013). 10.1371/journal.pone.0066739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao Y., An J., and Ma L., “Single cell array based assay for in vitro genotoxicity study of nanomaterials,” Anal. Chem. 85, 4107–4112 (2013). 10.1021/ac400242w [DOI] [PubMed] [Google Scholar]
- 18.Zhou H., Zhao L., and Zhang X., “In-channel printing-device opening assay for micropatterning multiple cells and gene analysis,” Anal. Chem. 87, 2048–2053 (2015). 10.1021/ac504823s [DOI] [PubMed] [Google Scholar]
- 19.Castel D., Pitaval A., Debily M.-A., and Gidrol X., “Cell microarrays in drug discovery,” Drug Discov. Today 11, 616–622 (2006). 10.1016/j.drudis.2006.05.015 [DOI] [PubMed] [Google Scholar]
- 20.Woodruff K., Fidalgo L. M., Gobaa S., Lutolf M. P., and Maerkl S. J., “Live mammalian cell arrays,” Nat. Methods 10, 550–552 (2013). 10.1038/nmeth.2473 [DOI] [PubMed] [Google Scholar]
- 21.Nilsson J., Evander M., Hammarström B., and Laurell T., “Review of cell and particle trapping in microfluidic systems,” Anal. Chim. Acta 649, 141–157 (2009). 10.1016/j.aca.2009.07.017 [DOI] [PubMed] [Google Scholar]
- 22.Voldman J., “Electrical forces for microscale cell manipulation,” Annu. Rev. Biomed. Eng. 8, 425–454 (2006). 10.1146/annurev.bioeng.8.061505.095739 [DOI] [PubMed] [Google Scholar]
- 23.Jeffries G. D. M. et al. , “Using polarization-shaped optical vortex traps for single-cell nanosurgery,” Nano Lett. 7, 415–420 (2007). 10.1021/nl0626784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marx J., “Cancer’s Bulwark against immune attack: MDS cells,” Science 319, 154–156 (2008). 10.1126/science.319.5860.154 [DOI] [PubMed] [Google Scholar]
- 25.Marx V., “Biophysics: Using sound to move cells,” Nat. Methods 12, 41 (2015). 10.1038/nmeth.3218 [DOI] [PubMed] [Google Scholar]
- 26.Bearinger J. P. et al. , “Chemical tethering of motile bacteria to silicon surfaces,” Biotechniques 46, 209–216 (2009). 10.2144/000113073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi J. S., Kim D. H., and Seo T. S., “Facile endothelial cell micropatterning induced by reactive oxygen species on polydimethylsiloxane substrates,” Biomaterials 84, 315–322 (2016). 10.1016/j.biomaterials.2016.01.041 [DOI] [PubMed] [Google Scholar]
- 28.Jang K. et al. , “Surface modification by 2-methacryloyloxyethyl phosphorylcholine coupled to a photolabile linker for cell micropatterning,” Biomaterials 30, 1413–1420 (2009). 10.1016/j.biomaterials.2008.11.013 [DOI] [PubMed] [Google Scholar]
- 29.Shah S. S. et al. , “Micropatterning of proteins and mammalian cells on indium tin oxide,” ACS Appl. Mater. Interfaces 1, 2592–2601 (2009). 10.1021/am900508m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maheswaran S. et al. , “Detection of mutations in lung cancer,” New Engl. J. Med. 359, 366–377 (2008). 10.1056/NEJMoa0800668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosales C., “Neutrophil: A cell with many roles in inflammation or several cell types?,” Front. Physiol. 9, 113 (2018). 10.3389/fphys.2018.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J. J. et al. , “Large-scale patterning of living colloids for dynamic studies of neutrophil-microbe interactions,” Lab Chip 18, 1514 (2018). 10.1039/c8LC00228B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walters N., Nguyen L. T. H., Zhang J., Shankaran A., and Reátegui E., “Extracellular vesicles as mediators of in vitro neutrophil swarming on a large-scale microparticle array,” Lab Chip 19, 2874 (2019). 10.1039/C9LC00483A [DOI] [PubMed] [Google Scholar]
- 34.Reátegui E. et al. , “Microscale arrays for the profiling of start and stop signals coordinating human-neutrophil swarming,” Nat. Biomed. Eng. 1, 0094 (2017). 10.1038/s41551-017-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolaczkowska E. and Kubes P., “Neutrophil recruitment and function in health and inflammation,” Nat. Rev. Immunol. 13, 159–175 (2013). 10.1038/nri3399 [DOI] [PubMed] [Google Scholar]
- 36.Mayadas T. N., Cullere X., and Lowell C. A., “The multifaceted functions of neutrophils,” Annu. Rev. Pathol. Mech. Dis. 9, 181–218 (2014). 10.1146/annurev-pathol-020712-164023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Oliveira S., Rosowski E. E., and Huttenlocher A., “Neutrophil migration in infection and wound repair: Going forward in reverse,” Nat. Rev. Immunol. 16, 378–391 (2016). 10.1038/nri.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lämmermann T., “In the eye of the neutrophil swarm-navigation signals that bring neutrophils together in inflamed and infected tissues,” J. Leukoc. Biol. 100, 55–63 (2016). 10.1189/jlb.1mr0915-403 [DOI] [PubMed] [Google Scholar]
- 39.Kienle K. and Lämmermann T., “Neutrophil swarming: An essential process of the neutrophil tissue response,” Immunol. Rev. 273, 76–93 (2016). 10.1111/imr.12458 [DOI] [PubMed] [Google Scholar]
- 40.Rozkiewicz D. I. et al. , “Covalent microcontact printing of proteins for cell patterning,” Chemistry 12, 6290–6297 (2006). 10.1002/chem.200501554 [DOI] [PubMed] [Google Scholar]
- 41.Stott S. L. et al. , “Isolation of circulating tumor cells using a microvortex-generating herringbone-chip,” Proc. Natl. Acad. Sci. U.S.A. 107, 18392–18397 (2010). 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reátegui E. et al. , “Tunable nanostructured coating for the capture and selective release of viable circulating tumor cells,” Adv. Mater. 27, 1593–1599 (2015). 10.1002/adma.201404677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chivers C. E., Koner A. L., Lowe E. D., and Howarth M., “How the biotin-streptavidin interaction was made even stronger: Investigation via crystallography and a chimaeric tetramer,” Biochem. J. 435, 55–63 (2011). 10.1042/BJ20101593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guillotin B. and Guillemot F., “Cell patterning technologies for organotypic tissue fabrication,” Trends Biotechnol. 29, 183–190 (2011). 10.1016/j.tibtech.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 45.Kar S., Molla M. D. S., Katti D. R., and Katti K. S., “Tissue-engineered nanoclay based 3D in vitro breast cancer model for studying breast cancer metastasis to bone,” J. Tissue Eng. Regen. Med. 13, 119–130 (2018). 10.1002/term.2773 [DOI] [PubMed] [Google Scholar]
- 46.Kargl J. et al. , “Neutrophils dominate the immune cell composition in non-small cell lung cancer,” Nat. Commun. 8, 14381 (2017). 10.1038/ncomms14381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coussens L. M. and Werb Z., “Inflammation and cancer,” Nature 420, 860–867 (2002). 10.1038/nature01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fridlender Z. G. et al. , “Polarization of tumor-associated neutrophil phenotype by TGF-β: ‘N1’ versus ‘N2’ TAN,” Cancer Cell 16, 183–194 (2009). 10.1016/j.ccr.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sagiv J. Y. et al. , “Phenotypic diversity and plasticity in circulating neutrophil subpopulations in cancer,” Cell Rep. 10, 562–573 (2015). 10.1016/j.celrep.2014.12.039 [DOI] [PubMed] [Google Scholar]
- 50.Eser PÖ and Jänne P. A., “TGFβ pathway inhibition in the treatment of non-small cell lung cancer,” Pharmacol. Ther. 184, 112–130 (2018). 10.1016/j.pharmthera.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 51.Li W. et al. , “Biodegradable nano-films for capture and non-invasive release of circulating tumor cells,” Biomaterials 65, 93–102 (2015). 10.1016/j.biomaterials.2015.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park M. H. et al. , “Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip,” J. Am. Chem. Soc. 139, 2741–2749 (2017). 10.1021/jacs.6b12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seo J. et al. , “High-throughput approaches for screening and analysis of cell behaviors,” Biomaterials 153, 85–101 (2018). 10.1016/j.biomaterials.2017.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See the supplementary material for the effects of plasma treatment on the surface chemistry (Fig. 1), the schematic for the gravity-driven flow for hydrodynamic patterning (Fig. 2), the unwanted cellular adhesion when patterning outside a microchannel (Fig. 3), and the statistical decrease in neutrophil swarming with the introduction of H1568 cells (Fig. 4).