Abstract
Background
Exercise is widely recognised for its health enhancing benefits. Despite this, an overproduction of reactive oxygen and nitrogen species (RONS), outstripping antioxidant defence mechanisms, can lead to a state of (chronic) oxidative stress. DNA is a vulnerable target of RONS attack and, if left unrepaired, DNA damage may cause genetic instability.
Objective
This meta-analysis aimed to systematically investigate and assess the overall effect of studies reporting DNA damage following acute aerobic exercise.
Methods
Web of Science, PubMed, MEDLINE, EMBASE, and Scopus were searched until April 2019. Outcomes included (1) multiple time-points (TPs) of measuring DNA damage post-exercise, (2) two different quantification methods (comet assay and 8-oxo-2′-deoxyguanosine; 8-OHdG), and (3) protocols of high intensity (≥ 75% of maximum rate of oxygen consumption; VO2-max) and long distance (≥ 42 km).
Results
Literature search identified 4316 non-duplicate records of which 35 studies were included in the meta-analysis. The evidence was strong, showcasing an increase in DNA damage immediately following acute aerobic exercise with a large-effect size at TP 0 (0 h) (SMD = 0.875; 95% CI 0.5, 1.25; p < 0.05). When comparing between comet assay and 8-OHdG at TP 0, a significant difference was observed only when using the comet assay. Finally, when isolating protocols of long-distance and high-intensity exercise, increased DNA damage was only observed in the latter. (SMD = 0.48; 95% CI − 0.16, 1.03; p = 0.15 and SMD = 1.18; 95% CI 0.71, 1.65; p < 0.05 respectively).
Conclusions
A substantial increase in DNA damage occurs immediately following acute aerobic exercise. This increase remains significant between 2 h and 1 day, but not within 5–28 days post-exercise. Such an increase was not observed in protocols of a long-distance. The relationship between exercise and DNA damage may be explained through the hormesis theory, which is somewhat one-dimensional, and thus limited. The hormesis theory describes how exercise modulates any advantageous or harmful effects mediated through RONS, by increasing DNA oxidation between the two end-points of the curve: physical inactivity and overtraining. We propose a more intricate approach to explain this relationship: a multi-dimensional model, to develop a better understanding of the complexity of the relationship between DNA integrity and exercise.
Electronic supplementary material
The online version of this article (10.1007/s40279-019-01181-y) contains supplementary material, which is available to authorized users.
Key Points
| Acute exercise can damage single-stranded DNA, and DNA repair likely occurs within at least 3 days. |
| Multiple factors affect the extent of exercise-induced DNA damage and its repair. |
| An elaborate, multi-dimensional approach should be considered to fully understand the complex relationship between exercise, reactive oxygen and nitrogen species, and DNA damage. |
Introduction
Exercise is widely regarded as a primary conduit to a proficient state of health, and there is now ample evidence from both observational studies and randomised trials to postulate that regular exercise is a contributing factor in the prevention of cardiovascular disease, cancer, diabetes, and other chronic conditions, as well as reducing the risk of all-cause mortality [1, 2].
Despite this paradigm, multiple studies have established a link between strenuous and/or exhaustive exercise, and the increased formation of reactive oxygen and nitrogen species (RONS) [3]. RONS are generated endogenously in most aerobic organisms by an incomplete reduction of oxygen, and mainly via the mitochondrial electron transport chain during cellular respiration [4]. It is currently well understood that between 0.12 and 2% of the oxygen utilised by mitochondria during normal respiration is not converted to water (tetravalent reduction), but instead is reduced to the superoxide anion (O2·−), which can subsequently be reduced to hydrogen peroxide (H2O2) and further to the more potent hydroxyl free radical (OH·) [5, 6]. However, of note, the percentage estimation of total oxygen consumption in mitochondrial RONS production refers primarily to the in vitro-based experiments performed by Chance and colleagues [7]; as such, the production of O2− in vivo may indeed be much less [8–10].
RONS are often implicated in complex molecular mechanisms designed to explain the process of human ageing and associated chronic disease states [11]. Molecules such as lipid, protein, and DNA are known vulnerable targets of RONS attack, and therefore, can be oxidatively modified [12]. Oxidative free radical attack and subsequent damage to DNA in particular, is of prime biomedical importance and interest, as if left unrepaired, significant DNA alterations (e.g., chromosomal rearrangement, base damage, and strand breaks) may lead to rapid ageing, mutagenesis, and ultimately carcinogenesis [13–15]. Paradoxically, although excessive RONS production may be implicated in the pathology of numerous diseases [16], when produced in moderate/low amounts (i.e., not inducing a state of oxidative stress defined as an ‘imbalance between oxidants and antioxidants in favor of the oxidants, leading to a disruption of redox signaling and control and/or molecular damage’) [17], they act as key intracellular signalling molecules regulating a host of physiological and biological processes [11, 18]. RONS are generated in skeletal muscle, and play a key role in skeletal muscle adaptation to aerobic exercise training [19, 20]. In vitro work has shown myotubes exposed to hydrogen peroxide (exogenous), which increases the expression of peroxisome proliferator-activated receptor (PPAR-γ), and peroxisome-gamma co-activator-1 alpha (PGC-1α), whereas further exposure to N-acetylcysteine, an antioxidant, impeded its activity [21]. PGC-1α, which is induced by AMP kinase (AMPK), is a signalling pathway involved in adaptation to endurance exercise leading to mitochondrial biogenesis [18]. Similarly, in vivo work has demonstrated that antioxidant supplementation can hinder essential training adaptation mechanisms in humans. A study administrating 1 g of vitamin C per day during 8 weeks of training (3 days/week at 65–80% VO2-max; 5% increase every 2 weeks) resulted in a decreased expression of PGC-1α and mitochondrial transcription factor A, both of which are key transcription factors involved in mitochondrial biogenesis [22].
Mechanistically, there are several ways free radicals can be generated during exercise. While exercising, the energy requirements in the body greatly increase, leading to a substantially higher rate of oxygen uptake up to 15-fold, and in active muscle, the oxygen flux may increase to approximately 100-fold compared to resting values [23, 24]. The primary radical species produced by the contracting skeletal muscle are O2·− and nitric oxide (NO) [25]. When electron transfer occurs normally through the electron mitochondrial transport chain to reduce oxygen to water, approximately 1–3% of all electrons are leaked resulting in the formation of O2·− by adding one electron to molecular oxygen [13, 26]. Apart from the mitochondria, there are enzymatic sources that contribute substantially to free radical production such as nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, the enzyme which catalyses the one electron reduction of molecular oxygen (reaction 1) upon the activation of phagocytosis [13, 27].
Central to these mechanisms is the generation of superoxide anion (O2·−; one-electron reduction), and subsequently, produced through the superoxide dismutases (SODs), hydrogen peroxide (H2O2; two-electron reduction) [13]. Following the production of H2O2, the hydroxyl radical (OH·; three-electron reduction) can be produced in the presence of transition metal catalysts, through the Haber–Weiss Fenton reaction (reaction 2) [13, 26]. O2·− also reacts with NO (reaction 3) to produce peroxynitrite (ONOO-), a highly reactive molecule, that can damage DNA and nitrate proteins [26]:
As RONS accumulate in the cell, from either metabolic signalling (NADPH) pathways or external sources, they are balanced by scavenging antioxidant systems [28]. Under these balanced conditions, RONS are used as signalling molecules or, under unbalanced conditions, can interact with Fe2+ through Fenton chemistry, as mentioned, and cause DNA damage due to hydroxyl radicals (OH·), which in turn can be attenuated by DNA repair mechanisms. In the case of over-accumulation of such DNA damage and insufficient repair, it is conceivable to suggest that rapidly dividing cells may promote a mutational profile leading to disease. However, per their signalling role, RONS and DNA damage can trigger physiological programmed cell death (apoptosis) by activating p53 to prevent mutagenesis/carcinogenesis [28]. Therefore, it is important to differentiate whether cell death is caused by oxidative stress (i.e., DNA damage), which can be avoided (scavenging systems and DNA repair mechanisms), or programmed cell death via RONS signalling which could be advantageous when the cell becomes compromised, as a result of DNA damage [28]. RONS are therefore, important molecules involved in the fate of the cell’s destiny as they regulate crucial processes such as growth, differentiation, and cell death [29]. Once DNA is damaged, it is normally repaired by mechanisms such as base excision repair (BER), nucleotide excision repair (NER), or through a process of homologous recombination (HR) or nonhomologous end joining (NHEJ); the type of DNA repair will depend on the mechanism and the extent of the damage [30] (Electronic Supplementary Material Figure S1).
Exercise represents an intriguing model to examine the dynamic role of RONS from both a physiological and pathological perspective. Evidence suggests that only exhaustive (long-distance) and/or strenuous exercise (high-intensity maximal exercise, marathons, triathlons, and overtraining) can induce detrimental DNA alterations, if left unrepaired [18, 31]. However, during low or moderate intensity and distance exercise, RONS may serve to act as signalling molecules responsible for the initiation of exercise and skeletal muscle adaptation [19, 31, 32], as often conceptualised through the hormesis theory.
The aim of this work is to systematically investigate data reporting DNA damage following acute aerobic exercise, and perform a meta-analysis to examine the overall effect from these studies. There are discrepancies regarding exercise intensity and that it necessarily needs to be very exhaustive/strenuous to cause oxidative damage and/or stress, and this review will aim to elucidate this. Furthermore, the possible physiological and/or pathological consequences of exercise-induced DNA damage need to be considered in relation to the exercising individual in line with a new proposed multi-dimensional model. This is the first meta-analysis aimed to investigate the relationship between DNA damage and exercise.
Methods
Search Strategy
According to the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) guidelines [33], a detailed search was conducted to identify all relevant studies (including a range of publication from 1900 to April 2019) across the following five databases: Web of Science, PubMed, MEDLINE, EMBASE, and Scopus. Searching was limited to articles published in English and the filter “in humans” was applied on PubMed, MEDLINE, and EMBASE.
Inclusion/Exclusion Criteria
All published studies were checked for the following criteria: (1) the study was a full report published in a peer-reviewed journal; (2) the study assessed humans; and (3) the keyword combination referred to the following terms (used in all possible combinations): exercise, exercis*, exercise training, endurance, exhaustive, exercise-induced, acute exercis*, physical activity, DNA, nucleoid DNA, deoxyribonucleic acid, 8-hydroxy-2′-deoxyguanosine, 8-hydroxy-2-deoxyguanosine, 8-oxo-7,8-dihydro-2′-deoxyguanosine, 8-oxo-7,8-dihydro-2-deoxyguanosine, 8-hydroxy-2-deoxy guanosine, 8-hydroxydeoxyguanosine, 8-oxoguanine, 8-hydroxyguanosine, 8-oxo-2-deoxyguanosine, 8-OHdG, 8OHdG, 8-OH-dG, 8-OHG, 8-oxo-dG, 8-oxodG, 8-oxo-dG, 8-oxo-G, damage, oxidative damage, oxidative stress. Note that for the purposes of this review, we used the term DNA damage to encompass DNA single-strand breakage and nucleotide base oxidation.
One investigator initially reviewed records generated from all databases and applied the inclusion/exclusion criteria to identify eligible studies for inclusion; these were then agreed with at least three of the authors. The inclusion/exclusion criteria are shown in Table 1. To note, acute exercise was defined as aerobic exercise performed over a short period of time, but could also extend up to 1–3 days of a marathon event. To minimise the limitation of various biological samples, studies utilising urine, red blood, and muscle cells were also excluded. Please see Electronic Supplementary Material Table S1 for information and details of excluded studies.
Table 1.
Inclusion/exclusion criteria
| Criteria | Include | Exclude |
|---|---|---|
| Participants | Humans | Animals |
| Age group | 18–70 | Children, elderly |
| Exercise protocol | Acute, aerobic | Anaerobic, training (chronic) |
| Sample | White blood cells (leukocytes, lymphocytes, PBMC) | Urine, red blood cells (erythrocytes), muscle |
| Outcome measure | SBs (%), tail damage (%), tail length, tail moment, 8-OHdG | Double SBs |
SBs strand breaks, 8-OHdG 8-hydroxy-2’–deoxyguanosine, PBMC peripheral blood mononuclear cells
Data Extraction
A general extraction form was used, once the number of included studies was finalised. Characteristics of the participants (sample size, age, and sex), the exercise protocol (distance and intensity), assayed biomarkers, and methods of DNA quantification used were extracted by one investigator. The outcome measure, DNA damage, was expressed using multiple descriptors, and with regard to the comet assay, these were: DNA in the tail (%); DNA migration (μm) (otherwise known as tail length); tail moment (also known as olive tail moment) which is the product of tail (%) and tail length [34]. The biomarker used was 8-OHdG. Due to variations in the analytical approach, high-performance liquid chromatography (HPLC) or enzyme-linked immunosorbent assay (ELISA), 8-OHdG (pg/ml), and 8-OHdG/105 dG are also reported. The tail DNA (%), DNA migration (μm) or tail length and tail moment correspond to the comet assay and 8-OHdG (ng/ml) or (pg/ml) and 8-OHdG/105 dG to HPLC or ELISA methods. In reference to the comet assay, where multiple image descriptors were reported by one study, the authors used tail (%), as this is regarded as the most sensitive descriptor/parameter compared to tail moment or length [35]. Data were collected as means and standard deviation (SD) or standard error of the mean (SEM). Graph digitizer software (DigitizeIt, Braunschweig, Germany) and WebPlotDigitizer (Web Plot Digitizer, V.4.2. Texas, USA: Ankit Rohatgi, 2019) were used to obtain data from studies where data were only presented in a figure format. In two studies [36, 37], data were not extractable and, therefore, not included in the meta-analysis.
Numerous studies included heterogeneous groups of participants: trained or untrained, young or old, sport-specific volunteers (such as swimmers, rowers, and runners), physically active and sedentary participants, and a few studies compared men and women. Furthermore, three studies [38–40] used more than one parameter to quantify DNA damage. Finally, some studies measured DNA damage at only one time-point (TP), while other studies included multiple post-exercise measures of DNA damage following exercise. Table 2 details corresponding TPs for each investigation.
Table 2.
Individual time-points (TP) of measures of DNA damage after exercise for each investigation
| Study | TP 0 (0 h) | TP 1 (15 min–1 h) | TP 2 (2 h) | TP 3 (3 h) | TP 4 (4–6 h) | TP 5 (1 day) | TP 6 (2 days) | TP 7 (3 days) | TP 8 (4 days) | TP 9 (5 days) | TP 10 (6–7 days) | TP 11 (14–28 days) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bloomer et al. [59] | 0 h | |||||||||||
| Briviba et al. [49] | 20 min | |||||||||||
| Davison et al. [65] | 0 h | 1 h | ||||||||||
| Fogarty et al. [67] | 0 h | |||||||||||
| Fogarty et al. [66] | 0 h | |||||||||||
| Harms-Ringdahl et al. [50] | 1 h | |||||||||||
| Hartmann et al. [70] | 0 h | 6 h | 1d | 2d | 3d | 4d | ||||||
| Hartmann et al. [68] | 1d | |||||||||||
| Hartmann et al. [69] | 0 h | 1d | 2d | 3d | 4d | 5d | ||||||
| Inoue et al. [56] | 0 h | |||||||||||
| Itoh et al. [60] | 0 h | 1 h | 1d | |||||||||
| Kim et al. [79] | 0 h | 3d | ||||||||||
| Liu et al. [38] | 0 h | |||||||||||
| Mastaloudis et al. [51] | 0 h | 1d | 2d | 3d | 4d | 5d | 6d | |||||
| Meihua et al. [58] | 0 h | |||||||||||
| Møller et al. [34] | 0 h | 1d | 2d | |||||||||
| Niess et al. [39] | 15 min | 1d | ||||||||||
| Niess et al. [71] | 1 h | |||||||||||
| Paik et al. [72] | 0 h | 1 h | ||||||||||
| Peters et al. [73] | 0 h | 3 h | ||||||||||
| Pittaluga et al. [53] | 30 min | 1d | ||||||||||
| Revan [61] | 0 h | 30 min | ||||||||||
| Ryu et al. [40] | 0 h | 3d | ||||||||||
| Sacheck et al. [62] | 1d | |||||||||||
| Sardas et al. [55] | 1d | |||||||||||
| Saritaş et al. [63] | 0 h | 1d | ||||||||||
| Sato et al. [64] | 0 h | 1 h | 1d | 2d | ||||||||
| Shi et al. [57] | 0 h | 3 h | 1d | |||||||||
| Tanimura et al. [74] | 0 h | 1 h | 2 h | 3 h | 4 h | |||||||
| Tanimura et al. [54] | 1d | |||||||||||
| Tsai et al. [75] | 0 h | 1d | 3d | 7d | 14d | |||||||
| Turner et al. [76] | 0 h | 1d | 7d | 28d | ||||||||
| Wagner et al. [77] | 20 min | 1d | 5d | 19d | ||||||||
| Williamson et al. [80] | 0 h | |||||||||||
| Zhang et al. [78] | 2 h | 6 h | 1d |
Data Analysis
The primary outcome was defined as DNA oxidative damage before and following exercise at TP 0 (0 h) grouped by method of DNA damage quantification (1) comet assay and (2) 8-OHdG. Secondary outcomes included: (3) high intensity (≥ 75% of maximum rate of oxygen consumption; VO2-max) and (4) long distance (≥ 42 km) as different exercise protocols were measured, and finally (5), DNA damage at further time-points 1–11 (ranging from 15 min to 28 days).
Quality Assessment
To assess the quality of included studies, the risk of bias was assessed by one investigator using the 12 criteria (rating: yes, no, and unsure) recommended by the Cochrane Back Review Group (Table 3) [41]. The criteria assess risk of bias using the five following categories: selection bias; performance bias; attrition bias; reporting bias, and detection bias. However, due to the inherent difficulties in blinding participants to exercise treatments, 7 of the 12 criteria were not applicable, and as such not included. These were: adequate method of randomization; allocation concealment; outcome assessor blinding; participant and provider blinding; similarity or not of co-interventions; and intention-to-treat analysis. In contrast, two additional sources of bias, smoking and training status, were included as criteria given their potential to influence exercise responses. Following these modifications, the maximum score that studies could gather was seven, with the lowest scores indicating high risk of bias and higher scores indicating lower risk of bias. To establish a clearer overall assessment of bias, a high-, moderate-, and low-risk scale was developed according to how studies scored. Therefore, the following ranges were developed: 1–3 = high risk, 4–5 = moderate, and 6–7 = low risk.
Table 3.
Quality assessment for risk of bias of the included studies using the criteria recommended by the Cochrane back review group
| Study | Selection bias | Performance bias | Attrition bias | Reporting bias | Study-specific | Total score (maximum 7) | ||
|---|---|---|---|---|---|---|---|---|
| Similar baseline characteristics | Acceptable compliance | Acceptable and described drop-out rate | No selective outcome reporting | Similar timing of outcome assessment | Training status reported | Smoking status reported | ||
| Asami et al. [36]a | Unsure | Yes | No | Yes | Yes | No | Yes | 4 |
| Bloomer et al. [59] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Briviba et al. [49] | No | Yes | Yes | Yes | Yes | No | No | 4 |
| Davison et al. [65] | Yes | Yes | Yes | Yes | Yes | No | Yes | 6 |
| Fogarty et al. [67] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Fogarty et al. [66] | Yes | Yes | Yes | Yes | Yes | No | No | 5 |
| Harms-Ringdahl et al. [50] | No | Yes | Yes | Yes | Yes | No | Yes | 5 |
| Hartmann et al. [70] | No | Yes | Yes | Yes | Yes | Yes | Yes | 6 |
| Hartmann et al. [68] | Yes | Yes | Yes | Yes | Yes | No | Yes | 6 |
| Hartmann et al. [69] | No | Yes | Yes | Yes | Yes | Yes | Yes | 6 |
| Inoue et al. [56] | No | Yes | Yes | Yes | Yes | Yes | Yes | 6 |
| Itoh et al. [60] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Kim et al. [79] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Liu et al. [38] | Yes | Yes | Yes | Yes | Yes | No | Yes | 6 |
| Mastaloudis et al. [51] | No | Yes | Yes | Yes | Yes | No | Yes | 5 |
| Meihua et al. [58] | Yes | Yes | Yes | Yes | Yes | No | Yes | 6 |
| Møller et al. [34] | No | Yes | Yes | Yes | Yes | No | No | 4 |
| Niess et al. [39] | Yes | Yes | Yes | Yes | Yes | Yes | No | 6 |
| Niess et al. [71] | Yes | Yes | Yes | Yes | Yes | Yes | No | 6 |
| Orlando et al. [48]a | Yes | Yes | Yes | No | Yes | Yes | No | 5 |
| Paik et al. [72] | Yes | Yes | Yes | Yes | Yes | No | No | 5 |
| Peters et al. [73] | Yes | No | No | Yes | Yes | Yes | No | 4 |
| Pittaluga et al. [53] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Revan [61] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Roh et al. [37]a | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Ryu et al. [40] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Sacheck et al. [62] | No | Yes | Yes | Yes | Yes | Yes | Yes | 6 |
| Sardas et al. [55] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Saritaş et al. [63] | Yes | Yes | Yes | Yes | Yes | No | Yes | 6 |
| Sato et al. [64] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Shi et al. [57] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Tanimura et al. [74] | Yes | Yes | Yes | Yes | Yes | No | Yes | 6 |
| Tanimura et al. [54] | No | Yes | Yes | Yes | Yes | Yes | Yes | 6 |
| Tsai et al. [75] | Yes | Yes | Yes | Yes | Yes | No | No | 5 |
| Turner et al. [76] | Yes | No | No | Yes | Yes | No | Yes | 4 |
| Wagner et al. [77] | Yes | Yes | Yes | Yes | Yes | Yes | No | 6 |
| Williamson et al. [80] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Zhang et al. [78] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
aNot included in meta-analyses
Statistical Analysis
Assessment of effect size Meta-analyses were calculated using Comprehensive Meta-Analysis (Version 3.3.070, NJ:USA: Biostat, Inc). A random-effects model was used, since it assumes statistical heterogeneity among studies and that studies represent a random sample of effect sizes that could have been observed [42, 43]. Standardised mean differences (SMD) adjusted with Hedges’ g and 95% confidence intervals (CI) were calculated as the difference in means before and after exercise divided by the pooled standard deviation [43]. Where studies did not report standard deviations, these were calculated from standard errors [42]. The SMD measure was used to express effect size, the magnitude of which was calculated using Cohen’s categories: (1) small: SMD = 0.2–0.5, (2) medium: SMD = 0.5–0.8, and (3) large: SMD > 0.8 [44, 45]. A positive SMD measure was considered to show increased DNA damage after exercise compared to rest, whereas a negative SMD measure would show greater DNA damage at rest in comparison to after exercise. The overall effect was assessed using Z scores with a set significance level of p < 0.05.
Assessment of heterogeneity The Chi-square Cochran’s Q test and the I2 statistic were used for the assessment of statistical heterogeneity among studies. The Chi-square test assesses whether the observed differences in results are compatible with chance alone and a p value ≤ 0.10 was considered to display significant heterogeneity [42]. Furthermore, the I2 statistic was used to quantify inconsistency across studies, with (1) I2 = 0–30% showing no heterogeneity, (2) I2 = 30–49% showing moderate heterogeneity, (3) I2 = 50–74% showing substantial heterogeneity, and (4) I2 = 5–100% showing considerable heterogeneity [42].
Subgroup and sensitivity analysis Subgroup analyses were performed for multiple time-points of DNA damage quantification after exercise grouped by different methodologies in DNA quantification (comet assay versus 8-OHdG), and according to the exercise protocol: high intensity (≥ 75% VO2-max) versus long distance (≥ 42 km). To assess the robustness of the significant outcome data, sensitivity analysis was planned by excluding studies with high risk of bias.
Publication bias Publication bias was assessed, when at least ten studies were included in the meta-analyses, by visually analysing funnel plots. In general, asymmetrical funnel plots were considered to indicate high risk of publication bias, while symmetrical funnels plots were considered to indicate low risk [46].
Results
Literature Search
The number of articles identified from all electronic database searches and the selection process is shown in Fig. 1. Four thousand four hundred and twenty records (4420) were retrieved in the database search, one hundred and four (104) of which were duplicates. Four thousand one hundred and forty-one articles (4141) were excluded after title screening, leaving one hundred and seventy-five (175) records for abstract screening. One hundred and thirteen (113) records were excluded after abstract screening and sixty-two (62) full-text articles were assessed for eligibility. Twenty-three (23) full-text articles were excluded due to various reasons (detailed in Electronic Supplementary Material Table S1). The most common reason for study exclusion was the exercise protocol not consisting of acute and aerobic exercise. Thirty-nine studies (39) were included in the qualitative analysis, out of which one (1) was excluded due to same sample size [47], one had unpublished data [48], and two (2) due to non-extractable data [36, 37], and therefore, thirty five (35) were included in the quantitative analysis.
Fig. 1.
PRISMA flow diagram displaying the electronic search and selection process
Study Characteristics
The characteristics of each study including participants, exercise protocol, sample source, biomarker, quantification technique, and results are presented in Table 4.
Table 4.
Characteristics of the included studies and relevant outcomes
| Study | Participants | Exercise protocol | Sample | Biomarker | Technique | Findings |
|---|---|---|---|---|---|---|
| Møller et al. [34] | 12 Healthy subjects (7 males and 5 females); MA ± SD = 26.1 ± 4.9 y/o | Maximal bicycle exercise (VO2-max test) under normal conditions and at 4559 m (high-altitude hypoxia) for 3 days | Lymphocytes | SBs,FPG-s s, ENDO III-s s | Comet assay | Increased SBs at hypoxia on all 3 days; ENDO III-s s increased on day 3 at hypoxia; FPG-s s remained unchanged |
| Fogarty et al. [66] | 10 Healthy males; MA ± SD = 23 ± 4 y/o | Treadmill test to exhaustion | Lymphocytes | Tail DNA (%) | Comet assay | DNA damage increased 63% (p < 0.05) after exercise compared to rest |
| Sardas et al. [55] | 12 Male rowers (MA ± SD = 22 ± 3.8 y/o) and 11 PE students (MA ± SD = 21.8 ± 3.8 y/o) | Three-staged exercise test on a rowing (rowers) and bicycle ergometer (PE students) | Lymphocytes | Tail DNA (%) | Comet assay | DNA damage was increased 24 h post exercise compared to pre-exercise in both rowers and PE students but overall pre- and post-exercise damage was significantly higher in rowers compared to PE students |
| Reichhold et al. [47]a | 28 Male triathletes; MA ± SD = 32.7 ± 6.3 y/o | Ironman triathlon (3.8 km swim, 180 km cycle, 42 km run) | Lymphocytes | Tail DNA (%) | Comet assay |
The % of SBs decreased significantly immediately post-race, then increased 1 d post-race and declined again 5 d after the race Between days 5 and 19 post-race the levels of strand breaks decreased further below initial levels |
| Hartmann et al. [70] | 2 Males (20 and 30 y/o) and 1 female (32 y/o) | Treadmill test to exhaustion | White blood cells | DNA migration (μm) | Comet assay | No increase in DNA migration was seen immediately after the test but a significant increase in DNA migration was found after 6, 24 and 48 h. |
| Liu et al. [38] | 8 Males athletes; MA ± SD = 21.35 ± 1.04 | Treadmill exercise to exhaustion | Peripheral blood cells | Tail length; Olive tail moment | Comet assay | Compared to before exercise, both tail length and olive tail moment significantly rose after exercise (p < 0.001) |
| Inoue et al. [56] | 9 Swimmers (MA ± SD = 20.1 ± 1.2 y/o) and 9 distance runners (MA ± SD = 20.9 ± 2.1 y/o) | 1500 m for 90 min (swimmers) and 15 km distance running for 70 min (runners) | Lymphocytes | 8-OHdG | HPLC | 8-OHdG decreased significantly after swimming, but no significant change was observed after running |
| Sacheck et al. [62] | 8 Young (MA ± SD = 25.4 ± 2.2 y/o) and 8 older (MA ± SD = 69.3 ± 3.5 y/o) | Downhill running on a treadmill for 45 min at 75% VO2-max | Leukocytes | 8-OHdG | HPLC | No change in 8-OHdG 24 h post-exercise |
| Revan [61] | 14 Healthy males; MA ± SEM = 22.3 ± 0.5 y/o | Incremental exercise test to exhaustion on a cycle ergometer | Plasma | 8-OHdG | Cayman ELISA Kit | No significant difference in 8-OHdG levels before and after exercise |
| Tanimura et al. [54] | 8 Young untrained (MA ± SD = 23.8 ± 3.2 y/o) and 8 endurance-trained (MA ± SD = 21.1 ± 3.7 y/o) men | Cycling at 75% VO2-max 1 h daily for three consecutive days | Lymphocytes | Tail DNA (%) | Comet assay | DNA damage at day 4 was significantly greater than that at day 1 in both groups |
| Saritaş et al. [63] | 22 Healthy trained men; MA ± SEM = 21.45 ± 0.43 y/o | 12 min run test | Serum | 8-OHdG | ELISA Kit | 8–OHdG levels were not different between the time periods (before exercise, immediately after exercise and 24 h after the exercise) (p > 0.05) |
| Mastaloudis et al. [51] | 5 Males (MA ± SEM = 39 ± 8 y/o) and 5 females (MA ± SEM = 35 ± 4 y/o) | 50 km ultramarathon | Leukocytes | Tail DNA (%) | Comet assay | On average, in both placebo and experimental groups, DNA damage increased at mid-race but returned to baseline values by the end of the race |
| Harms-Ringdahl et al. [50] | 15 Healthy untrained males and females (7 females and 8 males; MA ± SD = 30.5 ± 6.9 y/o | 20 min cycling at 80% of maximum heart rate | Serum | 8-OHdG | ELISA Kit | Levels of 8-OHdG increased 42% after the exercise |
| Hartmann et al. [69] | 3 Male and 3 female athletes (aged between 21-33 y/o) | Short-distance triathlon competition (1.5 km swimming, 40 km cycling, 10 km running) | Lymphocytes | Tail moment (DNA migration) | Comet assay | Increase in DNA migration was seen at 24 h post-exercise, whereas at 48 h the values were lower compared to 24 h but higher than the pre-exercise values. At 72 h, the maximum increase in DNA migration was found and baseline values were still elevated after 120 h |
| Fogarty et al. [67] | 12 Healthy males; MA ± SD = 23 ± 4 y/o) | Bike test at 40,70 and 100% of VO2-max | Leukocytes | Tail length | Comet assay | Increase in DNA damage observed at moderate (70% VO2max) and high-intensity exercise (100% VO2max) compared to rest |
| Tanimura et al. [74] | 15 Healthy sedentary males; MA ± SD = 23.7 ± 1.1 y/o | Cycling at 75% VO2-max for 1 h | Lymphocytes | Tail DNA (%) | Comet assay | There was increased DNA damage at 3 h post-exercise compared to pre-exercise (p < 0.05) |
| Paik et al. [72] | 10 Healthy males; MA ± SD = 25.6 ± 0.8 y/o | Treadmill run to exhaustion at 80% VO2-max | Lymphocytes | Tail DNA (%), tail length, tail moment | Comet assay | There was increased DNA damage measured just prior to the termination of exercise compared to pre-exercise (p < 0.05) |
| Ryu et al. [40] | 30 Male runners, 10 in each group; 10 km group (MA ± SD = 36.5 ± 10.9 y/o), 21 km group (MA ± SD = 45.0 ± 7.8 y/o), 45 km group (MA ± SD = 37.9 ± 13.6 y/o) | 3 different marathon distances: 10 km,21 km and 45 km | Lymphocytes | Tail DNA (%), tail length, tail moment | Comet assay | Compared to rest and recovery, tail moment was significantly higher in all groups. Also, at 45 km tail moment was found to be higher a post-exercise compared to the 10 km and 21 km (p < 0.05). No differences in DNA in tail (%) or tail length was observed |
| Itoh et al. [60] | 8 Untrained males; MA ± SD = 21.8 ± 2.1 y/o | 10 km run | Plasma | 8-OHdG | ELISA kit | Decreased plasma 8-OHdG levels both immediately and 1 h after the 10-km run compared to resting values (p < 0.05) |
| Tsai et al. [75] | 14 Male runners (median age 21, range 20–24 y/o) | 42 km marathon race | Peripheral blood mononuclear cells | Tail DNA (%), FPG-s s, ENDO III-s s | Comet assay | SBs increased on day 1; increased levels of FPG-s s were observed immediately and 24 h (day 1) after the race; ENDO III-s s levels also increased immediately post-exercise and on day 7 |
| Bloomer et al. [59] | 11 & 6 Aerobically trained men and women; MA ± SD = 23.3 ± 5.2 y/o | Treadmill run for 30 min at 80% VO2-max | Plasma | 8-OHdG | ELISA Kit | No significant difference was observed in 8-OHdG levels before and after exercise |
| Turner et al. [76] | 9 Healthy men; MA ± SD = 46.1 ± 5.3 y/o | 233 km ultraendurance race | Peripheral blood mononuclear cells | Tail DNA (%), FPG | Comet assay | Increased SBs immediately and 24 h after the race, compared to baseline (p < 0.01); an increase in FPG-dependent oxidative DNA damage was also observed immediately after the race (p < 0.05) |
| Peters et al. [73] | 8 Male athletes; MA ± SD = 34.2 ± 2.44 y/o | 2.5-h treadmill run at 75% VO2-max | Lymphocytes | Tail length/DNA migration (μm) | Comet assay | There was no significant increase in DNA strand breaks before and after the exercise (p > 0.05) |
| Zhang et al. [78] | 11 Healthy male students aged between 18–20 y/o | Bicycle exercise to exhaustion | White blood cells | Tail length/DNA migration (mm) | Comet assay | Significantly increased DNA migration at 6 h and 24 h compared to pre-exercise (p < 0.01) |
| Wagner et al. [77] | 42 Male athletes; MA ± SD = 35.3 ± 7 y/o | Ironman triathlon (3.8-km swimming, 180 km cycling and 42.2-km running) | Lymphocytes | SBs, Tail DNA (%), ENDO III-s s, FPG-s s | Comet assay | SBs decreased post-race; DNA migration increased 1 day post-race due to SBs (p < 0.01), then 5 days post-race returned to pre-race levels and decreased further to below the initial levels 19 days post-race (p < 0.01) |
| Sato et al. [64] | 15 Male subjects aged 19–29 y/o (7 physically active and 8 sedentary) | 50% VO2-max of cycle ergometer exercise for 30 min | Leukocytes | 8-OHdG | HPLC | No change in physically active subjects but decreased in sedentary subjects |
| Hartmann et al. [68] | 8 Healthy men (29-34 y/o) | Treadmill run to exhaustion | White blood cells | Tail moment | Comet assay | Increase in DNA damage was seen 24 h after the run (mean increase = 35.3 ± 8.3%) |
| Shi et al. [57] | 5 Healthy males (aged 22–38 y/o) | Exercise on a cycle ergometer at 50% VO2-max for 10.5 ± 1.3 min | Leukocytes | 8-OHdG | HPLC | After aerobic exercise, no significant change in leukocyte 8OHdG level was seen. (However, a significant increase was detected in samples taken 24 h after anaerobic exercise) |
| Davison et al. [65] | 7 Healthy males; MA ± SD = 22.3 ± 4.1 y/o | Treadmill test to exhaustion | Peripheral blood mononuclear cells | Tail moment | Comet assay | An increase in DNA damage was observed after exercise |
| Asami et al. [36]a | 23 Healthy males aged 19-50 y/o (10 untrained and 13 trained) | Maximal cycling exercise | Leukocytes | 8-OHdG | HPLC | A significant decrease in 8-OHdG levels was observed in the untrained subjects only (p < 0.05) |
| Briviba et al. [49] | 10 Subjects for half-marathon (5 males and 5 females; MA ± SD = 43 ± 9 y/o) and 12 subjects for marathon (2 males and 10 females; MA ± SD = 45 ± 10 y/o) | Half-marathon (21.1 km) and a marathon (42.195 km) | Lymphocytes | Tail DNA (%), FPG-s s and ENDO III-s s | Comet assay | No significant changes in the levels of DNA strand breaks in lymphocytes after either race. However, a significant difference was found in the % of ENDO III s in the tail after both races (p < 0.05), whereas the % of FPG-s s was slightly increased but not significantly (p > 0.05) |
| Pittaluga et al. [53] | 7 Females; MA ± SD = 68.1 ± 2.7 y/o | Exhaustive bout on a cycle ergometer | Serum | 8-OHdG | HPLC | No significant difference in 8-OHdG levels before and after exercise |
| Niess et al. [39] | 5 UT and 6 TR | Treadmill run test to exhaustion | Leukocytes | Tail moment/DNA migration(μm) | Comet assay | An increase in DNA migration from 2.31 ± 0.20 (TR) and 2.22 ± 0.16 (UT) at rest to 2.65 ± 0.30 (TR) and 3.00 ± 0.41 tail moment (UT) was observed 24 h after exercise |
| Niess et al. [71] | 12 Male runners; MA ± SD = 27.3 ± 4.1 y/o | Half-marathon (21.1 km) | Leukocytes | DNA migration (μm) | Comet assay | DNA migration rose significantly 24 h after the race, compared to rest (p < 0.01) |
| Williamson et al. [80] | 10 Recreationally males; MA ± SD = 22 ± 2 y/o | Treadmill test to exhaustion | Peripheral blood mononuclear cells | Tail DNA (%) | Comet assay | Tail intensity was increased by 18.2% post-exercise (p < 0.05) |
| Orlando et al. [48]a | 21 Rugby male athletes; MA ± SD = 26 ± 5 y/o | 40 min run at 85% of maximum heart rate | Peripheral blood mononuclear cells | Tail DNA (%) | Comet assay | No significances of DNA damage were observed after the exercise bout |
| Meihua et al. [58] | 10 Male athletes; MA ± SD = 21.1 ± 1.13 y/o | VO2-max (Bruce protocol) | Peripheral blood cells and plasma | DNA damage index and 8-OHdG | Comet assay and HPLC | Exercise increased DNA damage index as measured by comet assay; plasma 8-OHdG levels also increased following exhaustive exercise (p < 0.01) |
| Roh et al. [37]a | 10 Male college athletes; MA ± SD = 18.8 ± 0.8 y/o | 1 h run at 75% of heart rate reserve | Lymphocytes | Tail DNA (%) | Comet assay | DNA tail (%) increased following exercise (p < 0.05) |
| Kim et al. [79] | 11 Amateur male triathletes; MA ± SD = 37.9 ± 6.2 y/o | 2 triathlon races (O2 and Olympic courses) | Lymphocytes | Tail DNA (%) | Comet assay | In the Olympic course, DNA tail intensity (%) increased after match and decreased after 3 and 6 days of recovery; In the O2 course, tail (%) decreased after match, increased after 3 days, and decreased after 6 days of recovery (p < 0.01) |
SBs strand breaks, FPG-s s formamidopyrimidine glycosylase-sensitive sites, ENDO III-s s endonuclease III-sensitive sites, 8-OHdG 8-hydroxy-2′–deoxyguanosine, MA mean age, SD standard deviation, HPLC high-performance liquid chromatography, ELISA enzyme-linked immunosorbent assay, PE physical education, VO2-max maximum rate of oxygen consumption; untrained, UT trained, TR, y/o years old
anot included in meta-analyses
Participants Participant age ranged from 18 to 70 years old. Five studies included both male and female participants [49–53]. Three studies included groups of untrained and trained subjects [36, 39, 54]; one study [55] used rowers and physical education students, while another [56] used swimmers and runners. Finally, two studies used volunteers participating in multiple running distances [40, 49].
Biomarkers/Analytical Techniques With regard to the biomarker and the techniques used to quantify DNA damage, 12 studies used 8-OHdG [36, 50, 53, 56–64] with either HPLC or ELISA. A total of 27 studies used tail DNA (%) or strand breaks, tail length, and tail moment with the comet assay technique [37–40, 48, 49, 51, 52, 54, 55, 65–80].
Exercise Protocol There was variation in the chosen exercise protocols, most often involving treadmill exercise and cycling whilst employing different exercise intensities (ranged from 40 to 100% VO2-max). Eight studies included marathons, half-marathons, or ultra-marathons [40, 49, 51, 69, 71, 75, 76, 79], and three studies [47, 77, 79] involved a triathlon as part of the exercise protocol.
Quality assessment in individual studies No study scored in the high-risk bias range (1–3), 11 studies scored in the moderate-risk range (4–5), and the remaining 27 studies scored in the low-risk range (6–7).
Analysis of Overall Effects
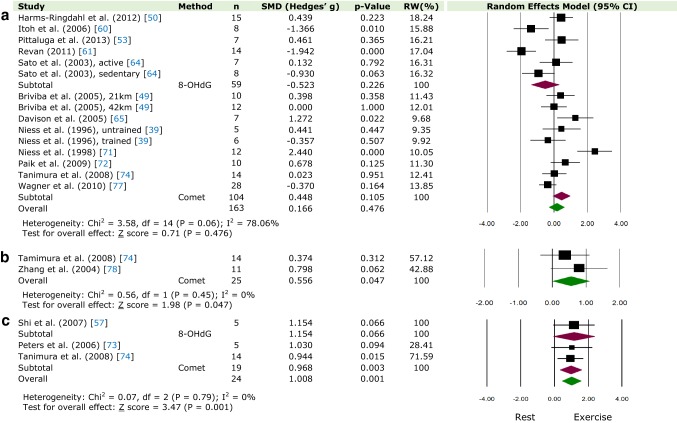
In summary, as seen in Table 5, a significant increase in DNA damage following exercise was observed at time-points 0 (0 h), 2 (2 h), 3 (3 h), 4 (4–6 h), 5 (1 days), and 7 (3 days). No significant differences were found at time-points 1 (15 min–1 h), 6 (2 days), 8 (4 days), 9 (5 days), 10 (6–7 days), and 11 (14–28 days).
Table 5.
Summary of results from all meta-analyses
| Time-point | N (sample size) | Effect (Hedges’ g) | p value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 8-OHdG | Comet | Overall | 8-OHdG | Comet | Overall | 8-OHdG | Comet | Overall | |
| 0 (0 h) | 109 | 203 | 312 | 0.150 | 1.140 | 0.875 | 0.684 | 0.000* | 0.000* |
| 1 (15 min–1 h) | 59 | 104 | 163 | − 0.523 | 0.448 | 0.166 | 0.226 | 0.105 | 0.476 |
| 2 (2 h) | – | 25 | – | – | 0.556 | – | – | 0.047* | – |
| 3 (3 h) | 5 | 19 | 24 | 1.154 | 0.968 | 1.008 | 0.066 | 0.003* | 0.001* |
| 4 (4–6 h) | – | 28 | – | – | 1.480 | – | – | 0.010* | – |
| 5 (1 day) | 73 | 148 | 221 | 0.141 | 2.468 | 1.113 | 0.720 | 0.000* | 0.000* |
| 6 (2 days) | 15 | 28 | 43 | − 1.208 | 0.338 | 0.231 | 0.509 | 0.497 | 0.630 |
| 7 (3 days) | – | 85 | – | – | 0.627 | – | – | 0.022* | – |
| 8 (4 days) | – | 19 | – | – | 0.691 | – | – | 0.380 | – |
| 9 (5 days) | – | 44 | – | – | 0.315 | – | – | 0.548 | – |
| 10 (6–7 days) | – | 33 | – | – | 0.359 | – | – | 0.491 | – |
| 11 (14–28 days) | – | 51 | – | – | 0.105 | – | – | 0.801 | – |
| High intensitya | 33 | 69 | 102 | 0.718 | 1.571 | 1.179 | 0.044* | 0.000* | 0.000* |
| Long distanceb | – | 89 | – | – | 0.437 | – | – | 0.151 | – |
a≥ 75% VO2-max at time-point 0 (0 h) and 5 (1 day)
b≥ 42 km at time-point 0 (0 h) and 1 (15 min–1 h)
– not applicable
*significance at p < 0.05
Overall Effect of DNA Damage After Exercise at TP 0
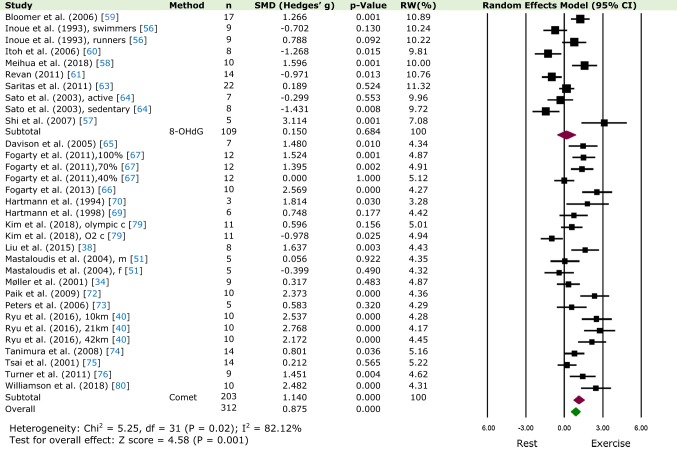
For DNA damage after exercise at TP 0 (0 h), data were available from 24 studies, with a total number of 312 participants. As seen in Fig. 2, compared to rest, there was a significant increase in DNA damage after exercise (SMD = 0.875; 95% CI 0.5, 1.25; p < 0.05). Heterogeneity among studies was found to be considerable (χ2 = 5.25, p = 0.02, I2 = 82.12%).
Fig. 2.
Relative weight (RW) standardised mean difference (SMD) and 95% CI (Hedges’ g adjusted) of DNA damage compared between rest and after an exercise bout at time-point 0 (0 h). Values for individual trials and pooled data (random model) are shown and grouped by method of quantification. c course, m males, f females
Comet Assay vs 8-OHdG at TP 0
Similarly, as shown in Fig. 2, for studies utilizing only the comet assay with 203 participants, DNA damage remained significantly higher after exercise at TP 0 (0 h) compared to rest (SMD = 1.14; 95% CI 0.7, 1.58; p < 0.05). Moreover, although the number of studies using the 8-OHdG biomarker was considerably less, with 109 participants, no change in DNA damage compared to rest using this assay was observed (please see Fig. 2; SMD = 0.15; 95% CI − 0.58, 0.88; p = 0.68).
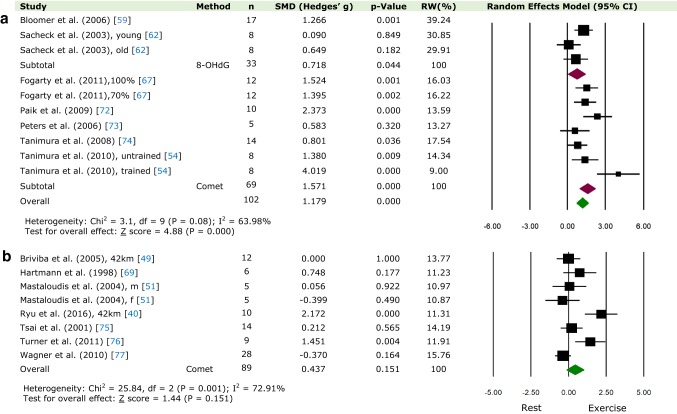
High-Intensity Exercise (≥ 75% VO2-max)
DNA damage was increased after high-intensity exercise (≥ 75% VO2-max), measured at time-point 0 (0 h) and 5 (1 day) (Fig. 3a; SMD = 1.18; 95% CI 0.71, 1.65; p < 0.05; heterogeneity: χ2 = 3.1, p = 0.08, I2 = 63.98%).
Fig. 3.
Relative weight (RW) standardised mean difference (SMD) and 95% CI (Hedges’ g adjusted) of DNA damage compared between rest and after an exercise bout at a high-intensity exercise (≥ 75% VO2-max) at time-point 0 (0 h) and 5 (1 day) and b long-distance exercise (≥ 42 km) at time-point 0 (0 h) and 1 (15 min–1 h). Values for individual trials and pooled data (random model) are shown and grouped by method of quantification. m males, f females
Long-Distance Exercise (≥ 42 km)
As shown in Fig. 3b, DNA damage was not significantly higher after long- distance (≥ 42 km) exercise at time-point 0 (0 h) and 1 (15 min–1 h) (SMD = 0.48; 95% CI − 0.16, 1.03; p = 0.15; heterogeneity: χ2 = 25.84, p = 0.001, I2 = 72.91%).
Summary of Findings
Using data from 35 studies and 312 participants, this paper quantitatively demonstrates, for the first time, that DNA damage increases immediately following acute aerobic exercise (Fig. 2). Based on C4ohen’s classification, the effect on DNA damage was large (> 0.8). No significant differences were seen at 1 h (Fig. 4a); however, increased DNA damage was observed from 2 h to 1 day following exercise (Fig. 4b, c; Electronic Supplementary Material Figure S2a, S2b). Similarly, no DNA damage was observed 2 days following exercise (Electronic Supplementary Material Figure S3a) but significantly increased 3 days post-exercise (Electronic Supplementary Material Figure S3b). Furthermore, when comparing the two methods of DNA damage (comet assay and 8-OHdG), a significant difference was observed only in studies using the comet assay, at time-point 0 h, 3 h and 1 day, again with a large effect size. No significant differences were observed 5–28 days post-exercise (Electronic Supplementary Material Figure S4a, S4b and S4c). Finally, when isolating protocols of high intensity (≥ 75% VO2-max) and long-distance (≥ 42 km), greater DNA damage following exercise was observed only in the former (Fig. 3a, b). However, it should be noted that no long-distance study in our analysis used 8-OHdG as a biomarker for oxidative damage, whereas, in the high-intensity protocols, a mixture of both methods was utilized. As has been suggested [51, 77], DNA damage measured after long-distance exercise (7–10 h race) may not be detected due to the activation of repair mechanisms and increased clearance of damaged cells initiated during the race, which would otherwise not be observed when measured after a shorter exercise protocol. In addition, these processes could be further enhanced due to the intake of antioxidants ingested during the race.
Fig. 4.
Relative weight (RW) standardised mean difference (SMD) and 95% CI (Hedges’ g adjusted) of DNA damage compared between rest and after an exercise bout at a time-point 1 (15 m–1 h), b time-point 2 (2 h), and c time-point 3 (3 h). Values for individual trials and pooled data (random model) are shown and grouped by method of quantification
Discussion
The main purpose of this meta-analysis was to examine the effect of acute exercise on DNA damage. These results suggest that exhaustive exercise leads to increased DNA damage. Acute aerobic exercise, regardless of intensity, seems to produce sufficient stimulus for a greater production of RONS which may evoke damaging effects to DNA. After longer distance events, such as triathlons and ultramarathons, added protection against DNA damage may be offered through the initiation of repair systems and adequate antioxidant intake from food/drinks consumed during such events; however, more studies are needed to confirm this.
Mechanisms of Free Radical Production During Exercise
Previous work from our laboratory has shown a 63% increase in DNA damage following exercise as well as a greater production of H2O2 as a function of exercise, compared to rest, indicating a possible mechanism of exercise-induced DNA damage through the increased production of H2O2 [66]. Moreover, the activation of inflammatory cells such as neutrophils and lymphocytes during exercise, due to muscle tissue damage, can further enhance superoxide production, which can cause direct damage to DNA [75, 80]. In addition, catecholamines released during exercise can be autoxidized and lead to the production of non-radicals such as H2O2 [81]. Finally, during high-intensity aerobic exercise, tissue ischaemia occurs, resulting in an increased number of hydrogen ions which can in turn react with superoxide anions to produce further RONS [82].
Free Radical-Induced Damage to DNA/Repair
Although O2·− and NO are the primary radical species produced by contracting skeletal muscle, these do not directly cause damage to DNA [14]. Instead, OH· reacts with the different components of DNA, such as DNA bases and the deoxyribose sugar, causing damage either by hydrogen addition or abstraction, producing multiple products, as well as single- and/or double-strand breaks, tandem lesions, and DNA protein cross-links [14, 83]. Among the four DNA bases, guanine has the least reduction potential, and acts as an excellent electron donor and is the most prone to oxidation by OH· [83]. For this reason, the product 8-OHdG is the most popular biomarker of DNA damage in urine and blood samples [12]. Furthermore, compared to guanine, adenine has a greater reduction potential and is not oxidized to the same extent [83]. Just as with guanine, OH· reacts with adenine by adding a hydrogen molecule to its double bonds at specific locations but in a slightly different distribution to that of guanine [83]. The base excision repair pathway is normally activated to repair DNA damage, and this occurs following the activation of a number of enzymes such as DNA glycosylase-1 [84], endonuclease phosphodiesterase, and DNA polymerase [85]. Repair to DNA is almost always controlled by a number of factors such as availability of said enzymes and others such as p53 and RAS [86, 87].
Hormesis Theory
The relationship between exercise, RONS, and DNA damage has been explained in the context of the hormesis theory (displayed in Fig. 5) [18]. In toxicology, hormesis refers to an environmental agent’s beneficial effect on a cell or organism at low doses that is otherwise harmful at high doses, creating a bell-shaped curve [88]. In this instance, exercise acts as the stimulus and the subsequent effects of exercise-induced RONS (physiological or pathological) are determined by the dose. Being physically inactive is a major risk factor for numerous chronic diseases and physiological disorders such as cancer, type 2 diabetes, cardiovascular disease, metabolic syndrome, hypertension, and obesity [89–91]. In 2000, physical inactivity in combination with poor diet was the second leading cause of death after tobacco in the US, contributing to 16.6% of total US deaths [92].
Fig. 5.
The relationship between exercise and DNA oxidation and its effects explained by the hormesis curve (one-dimensional model). RONS reactive oxygen and nitrogen species
Physical inactivity represents one end-point of the hormesis curve, while overtraining and strenuous unaccustomed/unindividualized exercise represents the opposite end-point; both result in a higher risk of disease and decreased physiological function and are mediated by elevated RONS production and oxidative stress [93]. Regular exercise can lead to adaption through up-regulation of molecular and cellular pathways, redox signalling, and antioxidant repair systems, resulting in the enhanced capacity of the organism to overcome greater stress [11, 93]. In addition, exercise training can further extend that adaptive response by ‘stretching’ the capacity to tolerate even higher levels of RONS [93]. Yet, when RONS production outstrips antioxidant defence mechanisms and there is insufficient repair of DNA double-strand breaks, this can cause chromosome instability and gene mutation can occur [15, 94]. However, it is unclear where the threshold limit exists between the beneficial effects of regular exercise and the point of overtraining associated with higher oxidative DNA damage and insufficient repair. This makes the concept of hormesis definitive but narrow. Defining this point is inherently complex due to the heterogeneous variations across individuals based on sex, age, fitness, and exercise intensity and distance. However, along with these, there are even more complex factors (discussed in Sect. 4.5) that influence the degree of the damage and, therefore, the overall effect of the beneficial adaptations and the harmful effects of the two end-points that should be considered.
One-Dimensional vs Multi-dimensional Model
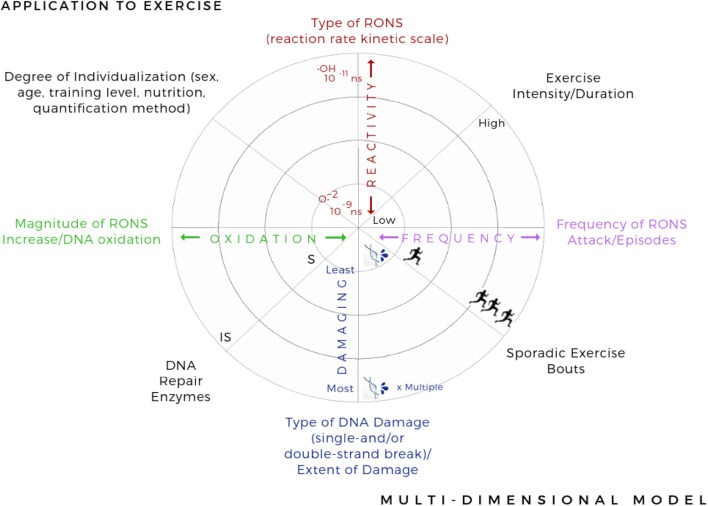
A role for RONS in exercise-mediated adaptations and responses is evident [95]. The concept of hormesis can allow us, to some extent, to understand how the relationship between exercise and DNA oxidation can fit into a bell-shaped curve. However, it only considers levels of RONS/DNA oxidation, rendering it somewhat one-dimensional. While this may be an important factor in explaining the fundamental adaptive responses to exercise, when investigating the extent of DNA oxidative damage, there are multiple factors to consider. We propose three basic factors (instead of only articulating RONS/DNA oxidation) in a more intricate and adaptable multi-dimensional model, visualised in a radar chart starting from the centre (least damaging) to the edge of the circle (most damaging) in a linear scale manner, as shown in Fig. 6. Thus, this proposed multi-dimensional model would consist of the following four factors: (1) type of RONS, ranging from the least reactive (such as O2·−) to the most reactive radical (such as OH·); (2) frequency of RONS attacks/episodes, ranging from one to multiple episodes; (3) type/extent of DNA damage, either single- or double-strand breaks, ranging from the least to the most damaging effect; and (4) magnitude of RONS/DNA oxidation, ranging from lowest to maximum levels of DNA oxidation/RONS increase. When applying this multi-dimensional model to the exercise stimulus, there are four further specific factors to consider: (5) exercise intensity/ distance; (6) exercise frequency; (7) sufficiency of DNA repair enzymes; and (8) degree of individualization (sex, age, training level, and nutrition quantification method) (Fig. 6).
Fig. 6.
Multi-dimensional model showing multiple factors to be considered when assessing the degree of oxidative damage when applied to the exercise model. IS insufficient, RONS reactive oxygen and nitrogen species, S sufficient
As exercise occurs, adaptive mechanisms are stimulated and these lead to the accentuation of antioxidant enzymes, as a result of training adaptation [96]. However, if multiple individual sporadic bouts of acute, but not regular, exercise occur (effect of overtraining and/or excessive exercise resulting from very high-intensity and/or long-distance exercise), without sufficient rest periods in between, the repair systems most likely fail due to higher oxidative stress resulting from enhanced RONS production [96]. Successful adaptations are thus unlikely and detrimental health outcomes may occur as a consequence. In contrast, individual bouts of exercise with complete recovery in between could revoke any oxidative stress via the antioxidant enzymes which are upregulated within the muscle as a function of training, suggesting that exercise itself can exert an antioxidant effect [97, 98]. In turn, this supports the now established theory that RONS production is in fact a necessary step to stimulate the adaption of the skeletal muscle in response to exercise [19]. Furthermore, the severity of the damage and whether genome stability is being compromised or not depends on the type of damage/oxidation that has occurred to the DNA—base oxidation, single- or double-strand breaks.
Cumulatively, these factors can affect the degree of DNA oxidative damage by causing the least to the most amount of DNA damage and, in turn, possibly creating (individual) different individual thresholds for the end-points of physical inactivity and overtraining and the way the hormesis effect unfolds as a bell-shaped curve. Obviously, a combination of reaching the higher end of the scale in all factors (towards the circumference of the circle) would result in the most harmful kind of oxidative DNA alteration, compared to the lower end of the scale (towards the centre of circle) where the outcome may be less harmful.
This meta-analysis suggests that aerobic exercise leads to an increase in oxidative DNA damage as measured by the comet assay. It is important to elucidate what this means in relation to health outcomes. The literature collectively suggests that a single acute bout of exercise (even of high intensity/long distance) is not likely able to cause any long-term and significantly harmful effects as explained under the hormesis theory. Ironman triathlon studies have shown that well-trained athletes show a large decline in DNA damage post-race. For instance, Mastaloudis et al. reported that DNA damage decreased below baseline levels 2 days after a 50-km ultramarathon [51]. Moreover, an 8% decrease below baseline was observed after 6 days. Similarly, Wagner et al. showed that DNA damage after an Ironman triathlon returned to baseline 5 days after the event, suggesting non-persistent DNA damage [77]. This can be attributed to the up-regulation of repair mechanisms and endogenous antioxidative networks, which indicates that endurance training can enhance the body’s ability to prevent and repair DNA damage, largely by increasing its antioxidant defenses [77]. The non-trained cohorts may not have the added antioxidant protection as a function of adaptive training. Master endurance athletes are also shown to have longer telomere length (TL), a marker of biological age, than non-athlete age-matched controls [99, 100]. Telomeres are responsible for stopping cell division by activating DNA damage recognition systems [101]. TL shortening is attenuated by long-term endurance training, and thus, reduced antioxidant activity and accumulation of RONS may contribute to TL debilitation [100]. Taking this into account, along with the use of different experimental designs, fitness levels, and methods of DNA damage detection at various post exercise TPs, all these factors may affect the degree of the damage and the extent to which it is (efficiently) repaired.
In summary, both the advantageous and harmful effects of exercise-associated adaptations and the two end-points, physical inactivity and overtraining, are caused by non-exposure or repeated exposure to the stimulus (inactivity or repeated exercise bouts) combined with a varying degree of DNA oxidative damage. Whether or not physiological or pathological consequences occur, and to what extent, may depend on all factors mentioned in the multi-dimensional model.
Strengths and Limitations
This is the first meta-analysis available on DNA damage and exercise. DNA damage was distinguished while performing sensitivity analysis of two of the most common methods of quantification found across studies, the comet assay, and 8-OHdG. The overall risk bias was low, since studies scored well in the quality assessment table. Finally, PRISMA guidelines [33] and Cochrane collaboration recommendations were followed [42].
Some limitations have been identified in the included studies. A number of study data had to be manually extracted from figures due to data not being presented in the text. However, the degree of error should be minimal due to the high accuracy of the software used. Moreover, the sample size for the two quantification methods was not equal, and while this is expected given the variety of study methodologies used is nonetheless noteworthy. This may be the main reason why only studies utilising the comet assay showed significantly greater DNA damage following exercise as opposed to 8-OHdG. However, this could also result due to interlaboratory differences. In 2005, the European Standards Committee on Oxidative DNA Damage found no association between levels of oxidative DNA damage in a sample of 88 healthy males measured by the comet assay and 8-OHdG by HPLC methods in six different laboratories [102]. Therefore, the validity and comparability of different methods of oxidative DNA damage across laboratories may be questioned. Similarly, the number of studies/sample size at all time-points and in the subgroup analysis (high-intensity and long-distance studies) varied, and could explain the difference between observed significance and non-significance between the two protocols.
The authors chose to focus solely on studies that have quantified DNA damage assayed from blood as these represent the most frequently measured in the literature. Nevertheless, we acknowledge that DNA damage can also be determined in urine and muscle. Studies measuring DNA damage following exercise in tissues/specimens other than white blood cells (e.g., muscle and urine) support our data demonstrating that exercise induces DNA damage. Previous work from our laboratory shows an 86% increase, compared to rest, in muscle 8-OHdG concentration following 100 isolated and continuous maximal knee contractions [103]. Moreover, during a 4-day race, urinary 8-OHdG of five super-marathon runners was monitored; where after day 1 (93 km) 8-OHdG increased, on day 2 (120 km), no further increase occurred, while on days 3 and 4 (56 and 59 km, respectively), there was a decrease in 8-OHdG suggesting the likelihood of exercise adaptation and upregulation of antioxidant systems [104]. Similarly, after 8 days of running (30 ± 3 km/day) at a training camp, 8-OHdG measured from urine increased significantly by 26% [105]. Another investigation showed that, after 1 h of cycling at 70% of maximal O2 uptake, urinary 8-OHdG was elevated, and this increase remained significant 1 day post-exercise [106].
Furthermore, training status was not distinguished across studies and was only taken into account as to whether it was reported or not in the literature in the quality assessment of this review. There were a few studies using marathons and/or triathlons as the exercise protocol, but most of the investigations did not report the training status of participants. This is important as trained athletes may be less susceptible to oxidative stress due to their enhanced expression of antioxidant enzymes and up-regulation of repair systems, acquired from previous training [77]. Across studies, the time of post-exercise measurement ranged from immediately post-exercise to 28 days following exercise (Table 2). However, in most studies, DNA damage was measured immediately post-exercise. Although this was further investigated by analysis of subsequent time-points, a significant increase at some of those time-points may not have been found, due to a smaller sample size.
Future Research
A relatively new biomarker has been used recently, the γ-H2AX, to assess DNA double-strand breaks in cancer research [94]. This assay is considered a sensitive method of measuring DNA damage, due to its ability to detect very low levels of double-strand breaks, which the comet assay could not otherwise detect [107]. Lippi et al. reported an increase in DNA injury, associated with running distance and intensity, with γ-H2AX foci analysis in lymphocytes. Amateur runners completed a 5-km, 10-km, 21-km, and 42-km running trial on four separate occasions. The authors observed a small increase in γ-H2AX foci after both 5 km and 10 km of running, a larger increase after 21 km, and an even larger increase after 42 km, indicating a dose-dependent relationship of DNA damage with distance and intensity [108]. This method could represent a salient methodological approach for future research to better address the complexity of exercise and DNA damage. Similarly, although challenging, incorporating direct free radical detection in parallel studies may yield more robust results and sensitive data. Finally, the role of antioxidant supplementation and its potential effects on DNA damage following exercise could be the next focal point of future meta-analyses. As a final practical aspect of performing subsequent meta-analyses, future authors are recommended to include all numeric values in text for easier extraction.
Conclusions
This systematic review and meta-analysis demonstrates a large increase in DNA damage immediately following an acute exercise bout as well as after 2 h and up to 1 day post-exercise, while such an increase was not evident between 5 and 28 days. Furthermore, only studies using the comet assay showed significance, compared to 8-OHdG. The analysis further showed that high-intensity exercise results in an increase in DNA damage, suggesting that greater DNA damage maybe be positively associated with increasing exercise intensity in a dose-dependent manner, while no significance was observed in the long-distance studies, possibly due to the initiation of repair systems during such events. However, due to limitations discussed and the paucity of evidence for most secondary outcomes, findings should be viewed with a degree of caution. Although an increase in DNA damage occurs after exercise, this is not necessarily a negative outcome per se. Such damage is most likely repaired within 3 days, or likely even sooner, as the long-distance studies may suggest, and thus may be transitory and should not confer any long-term adverse health outcomes on the individual or athlete. However, this will differ across individuals due to variation in individual thresholds, since there are multiple factors to consider as explained (but not limited to) in the multi-dimensional model. The hormesis curve describes, in a somewhat one-dimensional manner, how exercise modulates any advantageous or harmful effects through RONS by increasing DNA oxidation between the two-end points of the curve, physical inactivity and overtraining. Finally, the proposed multi-dimensional model may allow for a better understanding of the complex and multi-factorial relationship between DNA damage and exercise.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author Contributions
DVT performed the data search, selection, and analyses. DVT, CM, MGN, and GWD all contributed to writing, discussion, and final approval of all results and the manuscript.
Data Availability
The data presented in this systematic review are available in the associated studies, and references are provided.
Compliance with Ethical Standards
Funding
No sources of funding were used to assist in the preparation of this article.
Conflict of interest
The authors Despoina V. Tryfidou, Conor McClean, Michalis G. Nikolaidis, and Gareth W. Davison declare they have no conflicts of interest relevant to the content of this review.
Footnotes
The original version of this article was revised due to a Figures 2 and 3 update.
Change history
10/1/2019
Page 17, Fig. 2
References
- 1.Finaud J, Biologie L. Oxidative stress relationship with exercise and training. Sports Med. 2006;36:327–358. doi: 10.2165/00007256-200636040-00004. [DOI] [PubMed] [Google Scholar]
- 2.Warburton DER, Nicol CW, Bredin SSD. Prescribing exercise as preventive therapy. CMAJ. 2006;174:961–974. doi: 10.1503/cmaj.1040750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguiló A, Tauler P, Fuentespina E, Tur JA, Córdova A, Pons A. Antioxidant response to oxidative stress induced by smoking. Physiol Behav. 2005;84:1–7. doi: 10.1016/j.physbeh.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 4.Ray PD, Huang B-W, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Packer L, Cadenas E, Davies KJA. Free radicals and exercise: an introduction. Free Radic Biol Med. 2008;44:123–125. doi: 10.1016/j.freeradbiomed.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 6.Urso ML, Clarkson PM. Oxidative stress, exercise, and antioxidant supplementation. Toxicology. 2003;189:41–54. doi: 10.1016/s0300-483x(03)00151-3. [DOI] [PubMed] [Google Scholar]
- 7.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 8.Boveris A, Oshino N, Chance B. The cellular production of hydrogen peroxide. Biochem J. 1972;128:617–630. doi: 10.1042/bj1280617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown GC, Borutaite V. There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion. 2012;12:1–4. doi: 10.1016/j.mito.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boccatonda A, Tripaldi R, Davì G, Santilli F. Oxidative stress modulation through habitual physical activity. Curr Pharm Des. 2016;22:3648–3680. doi: 10.2174/1381612822666160413123806. [DOI] [PubMed] [Google Scholar]
- 12.Reichhold S, Neubauer O, Bulmer AC, Knasmüller S, Wagner KH. Endurance exercise and DNA stability: is there a link to duration and intensity? Mutat Res. 2009;682:28–38. doi: 10.1016/j.mrrev.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5:9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobley JN, Margaritelis NV, Morton JP, Close GL, Nikolaidis MG, Malone JK. The basic chemistry of exercise-induced DNA oxidation: Oxidative damage, redox signaling, and their interplay. Front Physiol. 2015;6:1–8. doi: 10.3389/fphys.2015.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davison GW. Exercise and oxidative damage in nucleoid DNA quantified using single cell gel electrophoresis: present and future application. Front Physiol. 2016;7:1–7. doi: 10.3389/fphys.2016.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesta D, Roden M. The Janus head of oxidative stress in metabolic diseases and during physical exercise. Curr Diab Rep. 2017;17:41. doi: 10.1007/s11892-017-0867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sies H, Jones DP. Oxidative stress. 2. Amsterdam: Elsevier; 2007. [Google Scholar]
- 18.Radák Z, Chung HY, Koltai E, Taylor AW, Goto S. Exercise, oxidative stress and hormesis. Ageing Res Rev. 2008;7:34–42. doi: 10.1016/j.arr.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Powers SK, Duarte J, Kavazis AN, Talbert EE. Reactive oxygen species are signalling molecules for skeletal muscle adaptation. Exp Physiol. 2010;95:1–9. doi: 10.1113/expphysiol.2009.050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemes R, Koltai E, Taylor AW, Suzuki K, Radák Z. Reactive oxygen and nitrogen species regulate key metabolic, anabolic, and catabolic pathways in skeletal muscle. Antioxidants. 2018;7:85. doi: 10.3390/antiox7070085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irrcher I, Ljubicic V, Hood DA. Interactions between ROS and AMP kinase activity in the regulation of PGC-1alpha transcription in skeletal muscle cells. Am J Physiol Cell Physiol. 2009;296:116–123. doi: 10.1152/ajpcell.00267.2007. [DOI] [PubMed] [Google Scholar]
- 22.Gomez-Cavrera M-C, Domenech E, Romagnoli M, Arduini A, Borras C, Pallardo FV, et al. Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance. Am J Clin Nutr. 2008;87:142–149. doi: 10.1093/ajcn/87.1.142. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee AK, Mandal A, Chanda D, Chakraborti S. Oxidant, antioxidant and physical exercise. Mol Cell Biochem. 2003;253:307–312. doi: 10.1023/a:1026032404105. [DOI] [PubMed] [Google Scholar]
- 24.Cooper CE, Vollaard NBJ, Choueiri T, Wilson MT. Exercise, free radicals and oxidative stress. Biochem Soc Trans. 2001;30:280. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 25.Jackson MJ, Vasilaki A, McArdle A. Cellular mechanisms underlying oxidative stress in human exercise. Free Radic Biol Med. 2016;98:13–17. doi: 10.1016/j.freeradbiomed.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Powers SP, Jackson MJ. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol Rev. 2008;88:1243–1276. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner JE, Bosch JA, Aldred S. Measurement of exercise-induced oxidative stress in lymphocytes. Biochem Soc Trans. 2011;39:1299–1304. doi: 10.1042/BST0391299. [DOI] [PubMed] [Google Scholar]
- 28.Mittler R. ROS are good. Trends Plant Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Nocella C, Cammisotto V, Pigozzi F, Borrione P, Fossati C, Amico AD, et al. Systems : Short- and long-term implications for athletes’ health. Nutrients. 2019;11:E1353. doi: 10.3390/nu11061353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher-Wellman K, Bloomer RJ. Acute exercise and oxidative stress: a 30 year history. Dyn Med. 2009;8:1–25. doi: 10.1186/1476-5918-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachdev S, Davies KJA. Production, detection, and adaptive responses to free radicals in exercise. Free Radic Biol Med. 2008;44:215–223. doi: 10.1016/j.freeradbiomed.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Moher David, Liberati Alessandro, Tetzlaff Jennifer, Altman Douglas G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Medicine. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Møller P, Loft S, Ersson C, Koppen G, Dusinska M, Collins A. On the search for an intelligible comet assay descriptor. Front Genet. 2014;5:3–6. doi: 10.3389/fgene.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins A, Brunborg G. Comet assay interest group, frequently asked questions. 2017. http://cometassay.com/index_files/Page345.htm.
- 36.Asami S, Hirano T, Yamaguchi R, Itoh H, Kasai H. Reduction of 8-hydroxyguanine in human leukocyte DNA by physical exercise. Free Radic Res. 1998;29:581–584. doi: 10.1080/10715769800300621. [DOI] [PubMed] [Google Scholar]
- 37.Roh H, Cho S, So W, Paik I, Suh S. Effects of different fluid replacements on serum HSP70 and lymphocyte DNA damage in college athletes during exercise at high ambient temperatures. J Sport Heal Sci. 2016;5:448–455. doi: 10.1016/j.jshs.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu D, Liang S, An L. Effect of allicin on peripheral blood cell DNA damage in human body after exhaustive exercise. Carpath J Food Sci Technol. 2015;7:86–92. [Google Scholar]
- 39.Niess AM, Hartmann A, Grünert-Fuchs M, Poch B, Speit G. DNA damage after exhaustive treadmill running in trained and untrained men. Int J Sports Med. 1996;17:397–403. doi: 10.1055/s-2007-972868. [DOI] [PubMed] [Google Scholar]
- 40.Ryu JH, Paik IY, Woo JH, Shin KO, Cho SY, Roh HT. Impact of different running distances on muscle and lymphocyte DNA damage in amateur marathon runners. J Phys Ther Sci. 2016;28:450–455. doi: 10.1589/jpts.28.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furlan AD, Pennick V, Bombardier C, van Tulder M, Group EBCBR Updated method guidelines for systematic reviews in the Cochrane back review group. Spine (Phila Pa 1976) 2009;2009(34):1929–1941. doi: 10.1097/BRS.0b013e3181b1c99f. [DOI] [PubMed] [Google Scholar]
- 42.Higgins JP, Green S. Cochrane handbook for systematic reviews of interventions: The Cochrane Collaboration. 2011. http://handbook.cochrane.org/.
- 43.Borenstein M, Hedges LV, Higgins JP, Rothstein HR. Introduction to meta-analysis. 1. Hoboken: Wiley; 2009. [Google Scholar]
- 44.Cohen J. Statistical power analysis for the behavioral sciences. USA: Lawrence Erlbaum Associates; 1988. [Google Scholar]
- 45.Faraone SV. Interpreting estimates of treatment effects: implications for managed care. P T. 2008;33:700–711. [PMC free article] [PubMed] [Google Scholar]
- 46.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reichhold S, Neubauer O, Hoelzl C, Stadlmayr B, Valentini J, Ferk F, et al. DNA damage in response to an Ironman triathlon. Free Radic Res. 2009;43:753–760. doi: 10.1080/10715760903040628. [DOI] [PubMed] [Google Scholar]
- 48.Orlando P, Silvestri S, Galeazzi R, Antonicelli R, Cirilli I, Bacchetti T, et al. Effect of ubiquinol supplementation on biochemical and oxidative stress indexes after intense exercise in young athletes. Commun Free Radic Res. 2018;23:136–145. doi: 10.1080/13510002.2018.1472924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Briviba K, Watzl B, Nickel K, Kulling S, Bös K, Haertel S, et al. A half-marathon and a marathon run induce oxidative DNA damage, reduce antioxidant capacity to protect DNA against damage and modify immune function in hobby runners. Redox Rep. 2005;10:325–331. doi: 10.1179/135100005X83716. [DOI] [PubMed] [Google Scholar]
- 50.Harms-Ringdahl M, Jenssen D, Haghdoost S. Tomato juice intake suppressed serum concentration of 8-oxodG after extensive physical activity. Nutr J. 2012;11:1–5. doi: 10.1186/1475-2891-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mastaloudis A, Yu TW, O’Donnell RP, Frei B, Dashwood RH, Traber MG. Endurance exercise results in DNA damage as detected by the comet assay. Free Radic Biol Med. 2004;36:966–975. doi: 10.1016/j.freeradbiomed.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Møller P, Loft S, Lundby C, Olsen NV. Acute hypoxia and hypoxic exercise induce DNA strand breaks and oxidative DNA damage in humans. FASEB J. 2001;15:1181–1186. doi: 10.1096/fj.00-0703com. [DOI] [PubMed] [Google Scholar]
- 53.Pittaluga M, Sgadari A, Tavazzi B, Fantini C, Sabatini S, Ceci R, et al. Exercise-induced oxidative stress in elderly subjects: The effect of red orange supplementation on the biochemical and cellular response to a single bout of intense physical activity. Free Radic Res. 2013;47:202–211. doi: 10.3109/10715762.2012.761696. [DOI] [PubMed] [Google Scholar]
- 54.Tanimura Y, Shimizu K, Tanabe K, Kono I, Ajisaka R. Effects of three consecutive days exercise on lymphocyte DNA damage in young men. Eur J Appl Physiol. 2010;110:307–314. doi: 10.1007/s00421-010-1499-2. [DOI] [PubMed] [Google Scholar]
- 55.Sardas S, Omurtag GZ, Monteiro IFC, Beyoglu D, Tozan-Beceren A, Topsakal N, et al. Assessment of DNA damage and protective role of vitamin E supplements after exhaustive exercise by Comet Assay in athletes. J Clin Toxicol. 2012;S:1. [Google Scholar]
- 56.Inoue T, Mu Z, Sumikawa K, Adachi K, Okochi T. Effect of physical exercise on the content of 8-hydroxydeoxyguanosine in nuclear DNA prepared from human lymphocytes. Jpn J Cancer Res. 1993;84(7):720–725. doi: 10.1111/j.1349-7006.1993.tb02035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi M, Wang X, Yamanaka T, Ogita F, Nakatani K, Takeuchi T. Effects of anaerobic exercise and aerobic exercise on biomarkers of oxidative stress. Environ Health Prev Med. 2007;12:202–208. doi: 10.1265/ehpm.12.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meihua S, Duoduo Y, Shuilian Z. Preventive effect of cranberry consumption against dna damage after exhaustive exercise in athlete men. Acta Medica Mediterr. 2018;34:499–506. [Google Scholar]
- 59.Bloomer RJ, Goldfarb AH, McKenzie MJ. Oxidative stress response to aerobic exercise: comparison of antioxidant supplements. Med Sci Sports Exerc. 2006;38:1098–1105. doi: 10.1249/01.mss.0000222839.51144.3e. [DOI] [PubMed] [Google Scholar]
- 60.Itoh H, Ohkuwa T, Yamazaki Y, Shimoda T, Takahashi H, Shimaoka K. Influence of endurance running on plasma 8-hydroxy-deoxyguanosine levels in humans. Jpn J Phys Fit Sports Med. 2006;55:S241–S246. [Google Scholar]
- 61.Revan S. Effects of acute high-intensity aerobic and anaerobic exercise on oxidative damage to lipids, proteins and DNA in untrained subjects. Afr J Pharm Pharmacol. 2011;5:1321–1326. [Google Scholar]
- 62.Sacheck JM, Milbury PE, Cannon JG, Roubenoff R, Blumberg JB. Effect of vitamin E and eccentric exercise on selected biomarkers of oxidative stress in young and elderly men. Free Radic Biol Med. 2003;34:1575–1588. doi: 10.1016/s0891-5849(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 63.Saritaş N, Uyanik F, Hamurcu Z, Çoksevim B. Effects of acute twelve minute run test on oxidative stress and antioxidant enzyme activities. Afr J Pharm Pharmacol. 2011;5:1218–1222. [Google Scholar]
- 64.Sato Y, Nanri H, Ohta M, Kasai H, Ikeda M. Increase of human MTH1 and decrease of 8-hydroxydeoxyguanosine in leukocyte DNA by acute and chronic exercise in healthy male subjects. Biochem Biophys Res Commun. 2003;305:333–338. doi: 10.1016/s0006-291x(03)00774-5. [DOI] [PubMed] [Google Scholar]
- 65.Davison GW, Hughes CM, Bell RA. Exercise and mononuclear cell DNA damage: the effects of antioxidant supplementation. Int J Sport Nutr Exerc Metab. 2005;15:480–492. doi: 10.1123/ijsnem.15.5.480. [DOI] [PubMed] [Google Scholar]
- 66.Fogarty MC, Hughes CM, Burke G, Brown JC, Davison GW. Acute and chronic watercress supplementation attenuates exercise-induced peripheral mononuclear cell DNA damage and lipid peroxidation. Br J Nutr. 2013;109:293–301. doi: 10.1017/S0007114512000992. [DOI] [PubMed] [Google Scholar]
- 67.Fogarty MC, Hughes CM, Burke G, Brown JC, Trinick TR, Duly E, et al. Exercise-induced lipid peroxidation: implications for deoxyribonucleic acid damage and systemic free radical generation. Environ Mol Mutagen. 2011;52:35–42. doi: 10.1002/em.20572. [DOI] [PubMed] [Google Scholar]
- 68.Hartmann A, Niess AM, Grünert-Fuchs M, Poch B, Speit G. Vitamin E prevents exercise-induced DNA damage. Mutat Res Lett. 1995;346:195–202. doi: 10.1016/0165-7992(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 69.Hartmann A, Pfuhler S, Dennog C, Germadnik D, Pilger A, Speit G. Exercise-induced DNA effects in human leukocytes are not accompanied by increased formation of 8-hydroxy-2′-deoxyguanosine or induction of micronuclei. Free Radic Biol Med. 1998;24:245–251. doi: 10.1016/s0891-5849(97)00249-9. [DOI] [PubMed] [Google Scholar]
- 70.Hartmann A, Plappert U, Raddatz K, Griinert-Fuchs M, Speit G. Does physical activity induce DNA damage? Mutagenesis. 1994;9:269–272. doi: 10.1093/mutage/9.3.269. [DOI] [PubMed] [Google Scholar]
- 71.Niess AM, Baumann M, Roecker K, Horstmann T, Mayer F, Dickhuth H-H. Effects of intensive endurance exercise on DNA damage in leucocytes. J Sports Med Phys Fitness. 1998;38:111–115. [PubMed] [Google Scholar]
- 72.Paik IY, Jeong MH, Jin HE, Kim YI, Suh AR, Cho SY, et al. Fluid replacement following dehydration reduces oxidative stress during recovery. Biochem Biophys Res Commun. 2009;383:103–107. doi: 10.1016/j.bbrc.2009.03.135. [DOI] [PubMed] [Google Scholar]
- 73.Peters EM, Van Eden M, Tyler N, Ramautar A, Chuturgoon AA. Prolonged exercise does not cause lymphocyte DNA damage or increased apoptosis in well-trained endurance athletes. Eur J Appl Physiol. 2006;98:124–131. doi: 10.1007/s00421-006-0227-4. [DOI] [PubMed] [Google Scholar]
- 74.Tanimura Y, Shimizu K, Tanabe K, Otsuki T, Yamauchi R, Matsubara Y, et al. Exercise-induced oxidative DNA damage and lymphocytopenia in sedentary young males. Med Sci Sports Exerc. 2008;40:1455–1462. doi: 10.1249/MSS.0b013e31817242cf. [DOI] [PubMed] [Google Scholar]
- 75.Tsai K, Hsu TG, Hsu KM, Cheng H, Liu TY, Hsu CF, et al. Oxidative DNA damage in human peripheral leukocytes induced by massive aerobic exercise. Free Radic Biol Med. 2001;31:1465–1472. doi: 10.1016/s0891-5849(01)00729-8. [DOI] [PubMed] [Google Scholar]
- 76.Turner JE, Hodges NJ, Bosch JA, Aldred S. Prolonged depletion of antioxidant capacity after ultraendurance exercise. Med Sci Sport Exerc. 2011;43:1770–1776. doi: 10.1249/MSS.0b013e31821240bb. [DOI] [PubMed] [Google Scholar]
- 77.Wagner KH, Reichhold S, Hölzl C, Knasmüller S, Nics L, Meisel M, et al. Well-trained, healthy triathletes experience no adverse health risks regarding oxidative stress and DNA damage by participating in an ultra-endurance event. Toxicology. 2010;278:211–216. doi: 10.1016/j.tox.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 78.Zhang M, Izumi I, Kagamimori S, Sokejima S, Yamagami T, Liu Z, et al. Role of taurine supplementation to prevent exercise-induced oxidative stress in healthy young men. Amino Acids. 2004;26:203–207. doi: 10.1007/s00726-003-0002-3. [DOI] [PubMed] [Google Scholar]
- 79.Kim D, Paik I, Cho S, Woo J, Bae J, Roh H. Effects of long-term aerobic exercise on the antioxidant system and lymphocyte dna damage by triathlon distance. J Mens health. 2018;14:42–55. [Google Scholar]
- 80.Williamson J, Hughes C, Davison G. Exogenous plant-based nutraceutical supplementation and peripheral cell mononuclear DNA damage following high intensity exercise. Antioxidants. 2018;7:70. doi: 10.3390/antiox7050070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Danese E, Lippi G, Sanchis-Gomar F, Brocco G, Rizzo M, Banach M, et al. Physical exercise and DNA injury: good or evil? Adv Clin Chem. 2017;81:193–230. doi: 10.1016/bs.acc.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 82.Jenkins RR. Exercise, oxidative stress, and antioxidants : a review. Int J Sport Nutr. 1993;3:356–375. doi: 10.1123/ijsn.3.4.356. [DOI] [PubMed] [Google Scholar]
- 83.Dizdaroglu M, Jaruga P. Mechanisms of free radical-induced damage to DNA. Free Radic Res. 2012;46:382–419. doi: 10.3109/10715762.2011.653969. [DOI] [PubMed] [Google Scholar]
- 84.German P, Szaniszlo P, Hajas G, Radák Z, Bacsi A, Hazra TK, et al. Activation of cellular signaling by 8-oxoguanine DNA glycosylase-1-initiated DNA base excision repair. DNA Repair (Amst). 2014;12:856–863. doi: 10.1016/j.dnarep.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carter RJ, Parsons JL. Base excision repair, a pathway regulated by posttranslational modifications. Mol Cell Biol. 2016;36:1426–1437. doi: 10.1128/MCB.00030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seo YR, Jung HJ. The potential roles of p53 tum or suppressor in nucleotide excision repair (NER) and base excision repair (BER) Exp Mol Med. 2004;36:505–509. doi: 10.1038/emm.2004.64. [DOI] [PubMed] [Google Scholar]
- 87.Boldogh I, Hajas G, Aguilera-aguirre L, Hegde ML, Radák Z, Bacsi A, et al. Activation of Ras GTPases by OGG1 complexed with 8oxoG. J Biol Chemisrty. 2012;287:20760–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mattson MP. Hormesis defined. Ageing Res Rev. 2008;7:1–7. doi: 10.1016/j.arr.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Booth FW, Roberts CK, Layne MJ. Lack of exercise is a major cause of chronic diseases. Compr Physiol. 2012;2:1143–1211. doi: 10.1002/cphy.c110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Knight JA. Review physical inactivity: associated diseases and disorders. Ann Clin Lab Sci. 2012;42:320–337. [PubMed] [Google Scholar]
- 91.Martin CK, Church TS, Thompson AM, Earnest CP, Blair SN. Exercise dose and quality of life: results of a randomised controlled trial. Arch Intern Med. 2009;169:269–278. doi: 10.1001/archinternmed.2008.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 93.Radák Z, Ishihara K, Tekus E, Varga C, Posa A, Balogh L, et al. Exercise, oxidants, and antioxidants change the shape of the bell-shaped hormesis curve. Redox Biol. 2017;12:285–290. doi: 10.1016/j.redox.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327:123–133. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Margaritelis NV, Cobley JN, Paschalis V, Veskoukis AS, Theodorou AA, Kyparos A, et al. Principles for integrating reactive species into in vivo biological processes: examples from exercise physiology. Cell Signal. 2016;28:256–271. doi: 10.1016/j.cellsig.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 96.Wagner KH, Reichhold S, Neubauer O. Impact of endurance and ultraendurance exercise on DNA damage. Ann N Y Acad Sci. 2011;1229:115–123. doi: 10.1111/j.1749-6632.2011.06106.x. [DOI] [PubMed] [Google Scholar]
- 97.de Sousa CV, Sales MM, Rosa TS, Lewis JE, de Andrade RV, Simões HG. The antioxidant effect of exercise: a systematic review and meta-analysis. Sports Med. 2017;47:277–293. doi: 10.1007/s40279-016-0566-1. [DOI] [PubMed] [Google Scholar]
- 98.Gomez-Cabrera MC, Domenech E, Viña J. Moderate exercise is an antioxidant: Upregulation of antioxidant genes by training. Free Radic Biol Med. 2008;44:126–131. doi: 10.1016/j.freeradbiomed.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 99.Simoes HG, Sousa CV, Rosa S, Aguiar S, Deus LA, Carine E, et al. Longer telomere length in elite master sprinters : relationship to performance and body composition authors. Int J Sport Med. 2017;38:1111–1116. doi: 10.1055/s-0043-120345. [DOI] [PubMed] [Google Scholar]
- 100.Sousa CV, Aguiar SS, Santos PA, Barbosa LP, Knechtle B, Nikolaidis PT, et al. Telomere length and redox balance in master endurance runners : the role of nitric oxide. Exp Gerontol. 2019;117:113–118. doi: 10.1016/j.exger.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 101.Corbett N, Alda M. On telomeres long and short. J Psychiatry Neurosci. 2015;40:3–4. doi: 10.1503/jpn.140347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gedik CM, Collins A, Loft S, Poulsen H, Riis B, Cadet J, et al. Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 103.Fogarty MC, Devito G, Hughes CM, Burke G, Brown JC, Mceneny J, et al. Effects of α-lipoic acid on mtDNA damage after isolated muscle contractions. Med Sci Sport Exerc. 2013;45:1469–1477. doi: 10.1249/MSS.0b013e31828bf31e. [DOI] [PubMed] [Google Scholar]
- 104.Radák Z, Pucsuk J, Boros S, Josfai L, Taylor AW. Changes in urine 8-hydroxydeoxyguanosine levels of super-marathon runners during a four-day race period. Life Sci. 2000;66:1763–1767. doi: 10.1016/s0024-3205(00)00499-9. [DOI] [PubMed] [Google Scholar]
- 105.Okamura K, Doi T, Hamada K, Masao S, Yoshioka Y, Mitsuzono R, et al. Effect of repeated exercise on urinary 8-hydroxy- deoxyguanosine excretion in humans. Free Radic Res. 1997;26:507–514. doi: 10.3109/10715769709097821. [DOI] [PubMed] [Google Scholar]
- 106.Orhan H, van Holland B, Krab B, Moeken J, Vermeulen NPE, Hollander P, et al. Evaluation of a multi-parameter biomarker set for oxidative damage in man : increased urinary excretion of lipid, protein and DNA oxidation products after one hour of exercise. Free Radic Res. 2004;38:1269–1279. doi: 10.1080/10715760400013763. [DOI] [PubMed] [Google Scholar]
- 107.Heylmann D, Kaina B. The γH2AX DNA damage assay from a drop of blood. Sci Rep. Nature Publishing Group. 2016;6:1–9. doi: 10.1038/srep22682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lippi G, Buonocore R, Tarperi C, Montagnana M, Festa L, Danese E, et al. DNA injury is acutely enhanced in response to increasing bulks of aerobic physical exercise. Clin Chim Acta. 2016;460:146–151. doi: 10.1016/j.cca.2016.06.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this systematic review are available in the associated studies, and references are provided.