In this study, Wu et al. set out to understand the regulation of linkage-specific ubiquitin signaling at sites of DNA damage. Using a ubiquitin binding domain protein array screen, they identify Cezanne and Cezanne2 as two new components in the complex ubiquitin landscape at DNA damage sites for regulating recruitment of DNA repair and signaling proteins. They further show that Cezanne serves as a “reader” of the Lys63-linkage localizing to DNA damage sites and an “eraser” of the Lys11-linkage ubiquitination, indicating a crosstalk between linkage-specific ubiquitination at DNA damage sites.
Keywords: 53BP1, Abraxas, BRCA1, Cezanne, DNA damage response, DNA double-strand breaks, DUB, Lys11-linkage ubiquitin, Lys63-linkage ubiquitin, Rap80
Abstract
The establishment of polyubiquitin conjugates with distinct linkages play important roles in the DNA damage response. Much remains unknown about the regulation of linkage-specific ubiquitin signaling at sites of DNA damage. Here we reveal that Cezanne (also known as Otud7B) deubiquitinating enzyme promotes the recruitment of Rap80/BRCA1-A complex by binding to Lys63-polyubiquitin and targeting Lys11-polyubiquitin. Using a ubiquitin binding domain protein array screen, we identify that the UBA domains of Cezanne and Cezanne2 (also known as Otud7A) selectively bind to Lys63-linked polyubiquitin. Increased Lys11-linkage ubiquitination due to lack of Cezanne DUB activity compromises the recruitment of Rap80/BRCA1-A. Cezanne2 interacts with Cezanne, facilitating Cezanne in the recruitment of Rap80/BRCA1-A, Rad18, and 53BP1, in cellular resistance to ionizing radiation and DNA repair. Our work presents a model that Cezanne serves as a “reader” of the Lys63-linkage polyubiquitin at DNA damage sites and an “eraser” of the Lys11-linkage ubiquitination, indicating a crosstalk between linkage-specific ubiquitination at DNA damage sites.
Posttranslational modification of proteins by ubiquitination regulates multiple cellular processes in a dynamic and reversible manner. Ubiquitination occurs by covalent attachment of a ubiquitin molecule to substrate lysine residue mediated by E1-activating, E2-conjugating, and E3 ligase enzymes; deubiquitination removes ubiquitin moiety by deubiquitinating enzymes (DUB). Ubiquitin can be added to a target protein as mono- (monoubiquitination) or poly- (polyubiquitination) conjugates. Since additional ubiquitin molecules can be tethered to any of the seven lysine residues (Lys6, 11, 27, 29, 33, 48, and 63) or through the N-terminal methionine residue in the first ubiquitin molecule, polyubiquitin chains form with distinct linkages (Pickart 2001). Linkage-specific ubiquitin chains adopt distinct topologies and often result in diverse functional outcomes in cells, manifesting the magnitude of complexity of ubiquitination (Komander and Rape 2012).
Cellular response to DNA damage involves a sophisticated ATM/ATR-mediated sensory network—the DNA damage response (DDR), which senses damage and signals to initiate DNA repair, activates the cell cycle checkpoint, regulates transcription, or triggers apoptosis or senescence if the damage is beyond repair. The DNA double-strand break (DSB) is one of the most cytotoxic lesions experienced by cells that, if not repaired, leads to genomic instability. Growing evidence over the last decade has illustrated a ubiquitin landscape at DSBs, demonstrating that ubiquitin modification on damaged chromatin plays important roles in the signaling and repair of DSBs (Jackson and Durocher 2013; Elia et al. 2015). Upon induction of DSBs and subsequent activation of ATM, phosphorylation of histone H2AX directly recruits MDC1, which then leads to the recruitment of a ubiquitin E3 ligase RNF8 (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007). RNF8 and RNF168 E3 ligases, together with a Lys63 (K63)-linkage-specific E2-conjugating enzyme, Ubc13, modify chromatin in the vicinity of DSBs, generating K63-linked ubiquitin conjugates on damaged chromatin including histones H2A/H2AX for the recruitment of DNA repair proteins like 53BP1 and BRCA1 (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007; Doil et al. 2009; Stewart et al. 2009; Mattiroli et al. 2012). The UIM domains of Rap80, a component of the Abraxas/BRCA1-A complex, bind to K63-linked chains recruiting Rap80 and the BRCA1-A complex to DNA damage sites (Kim et al. 2007; Sobhian et al. 2007; Wang and Elledge 2007; Wang et al. 2007, 2009; Wang 2012). In addition to Rap80 UIMs, the UMI-MIU1 and MIU2 domains of RNF168 and the UBZ domain of Rad18 also recognize K63-polyubiquitin at DSBs to facilitate the recruitment of these proteins (Huang et al. 2009; Pinato et al. 2011; Panier et al. 2012). It remains largely unclear whether there are additional ubiquitin binding motifs that can specifically recognize K63-linked ubiquitin conjugates at DSBs.
While K63-linked ubiquitination plays a key regulatory role in the DDR-signaling pathway in recruiting DDR factors, other linkage-specific ubiquitinations have also been indicated in the DDR (Morris and Solomon 2004; Feng and Chen 2012; Jackson and Durocher 2013; Elia et al. 2015; Gatti et al. 2015; Paul and Wang 2017). Recently, we have shown that RNF8 functions with a K11-linkage-specific ubiquitin E2 enzyme Ube2S catalyzing K11-ubiqutin conjugation on damaged chromatin (Paul and Wang 2017). The K11-linked ubiquitin conjugation plays a critical role in inhibiting transcription at sites of damage and is essential for cellular resistance to ionizing radiation (IR). Although RNF8 appears to be involved in catalyzing both K11- and K63-linked ubiquitination, it remains unknown whether there is crosstalk between K11- and K63-linkage ubiquitination.
Cezanne and Cezanne2 are members of the ovarian tumor (OTU) subfamily DUBs (Komander et al. 2009). Both Cezanne and Cezanne2 contain a UBA domain at the N terminus followed by an OTU DUB domain. It has been shown that Cezanne is a DUB that prefers to cleave K11-linked ubiquitin conjugates through its OTU domain (Bremm et al. 2010), playing a role in NF-κB signaling, mTORC signaling and other cellular functions (Hu et al. 2013, 2016; Bremm et al. 2014; Wang et al. 2017; Bonacci et al. 2018). Cezanne2 is a less well characterized paralog of Cezanne. In response to DSBs, Cezanne antagonizes the activity of RNF8 and Ube2S in K11-linkage ubiquitin modification of chromatin-bound proteins, including H2A/H2AX, on damaged chromatin (Paul and Wang 2017). While knockdown of RNF8 decreased the K11-linkage ubiquitin modification, depletion of Cezanne in RNF8-deficient cells significantly reversed the decrease of K11-linkage ubiquitination, indicating that RNF8/Ube2S and Cezanne function in the same pathway to modify K11-linked ubiquitin conjugation in response to DSBs. The role of Cezanne and Cezanne2 in the DDR is still largely unknown.
In this study, we identify Cezanne and Cezanne2 as “readers” and “erasers” of linkage-specific ubiquitin “code” at DNA damage sites facilitating recruitment of DNA repair factors. We developed a ubiquitin binding domain microarray screen for K63-linked ubiquitin chain-specific binders; we found that Cezanne and Cezanne2 UBA domains selectively bind to the K63-, but not K48- or K11-linked chain. Cezanne promotes Rap80/Abraxas/BRCA1 recruitment to DNA damage sites through its UBA domain binding to K63-conjugates and its DUB activity disassembling K11-linked polyubiquitin. The binding of Rap80 UIMs to the K63-linked chain is largely decreased when a K63/K11-mixed chain is formed. Cezanne2 interacts with Cezanne, playing a facilitating role in Rap80/Abraxas/BRCA1, Rad18, and 53BP1 recruitment, DNA repair, and cellular resistance to IR.
Results
Ubiquitin binding domain microarray screen identifies Cezanne and Cezanne2 UBA domain as selective K63-linked ubiquitin chain binding domains
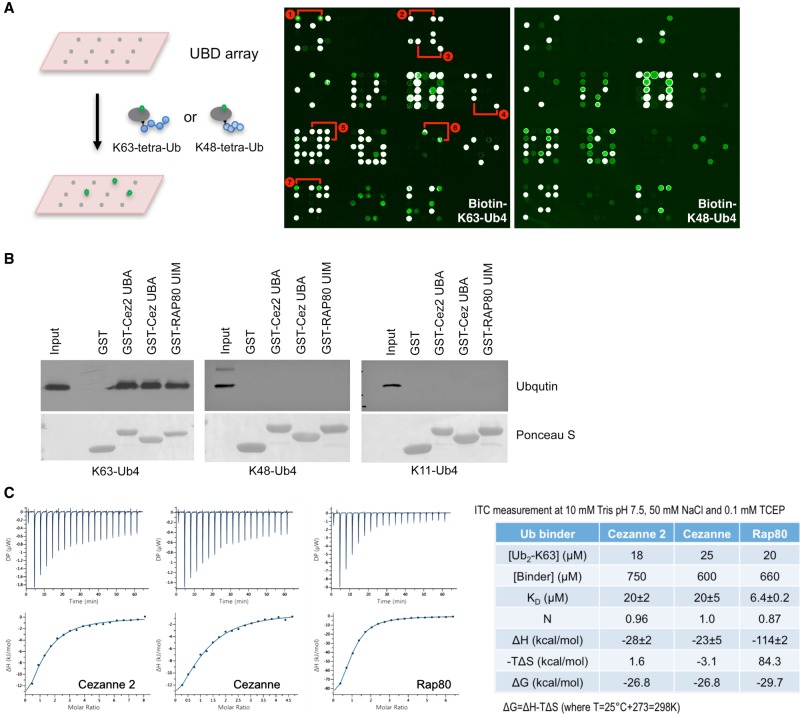
To identify proteins that selectively interact with K63-linked ubiquitin chains, we have generated protein arrays consisting of a library of recombinant ubiquitin binding domains (UBDs) and performed a screen (Fig. 1A). The UBD library contains 148 GST fusion proteins that harbor a number of UBDs, including UIM, UBA, CUE, GAT, VHS, PAZ, PFU, UBM, UBZ, and JAB/MPN domains (Supplemental Fig. S1). We carried out the screen by probing the array with K63- or K48-linked tetra-ubiquitin chains that were directly labeled with streptavidin-Cy3 on a biotin moiety (at position 20 of the first ubiquitin molecule). In addition to known K63-linked ubiquitin binding domains Rap80 UIMs (Kim et al. 2007; Sobhian et al. 2007; Wang and Elledge 2007; Wang et al. 2007), RNF168 MIU2 (Doil et al. 2009; Stewart et al. 2009), and ASCC2 CUE (Brickner et al. 2017), we have identified several new K63-linked polyubiquitin binders including Cezanne UBA, Cezanne2 UBA, TDRD3 UBA, and TOM1L2 VHS. Similar results were obtained when we used labeled K63-di-ubiquitin as a probe (Supplemental Fig. S2A). A screen using untagged K63-tetra-ubiquitin followed by anti-ubiquitin antibody and FITC-labeled secondary antibody also identified Cezanne and Cezanne2 UBA as selective “readers” of the K63-linked chain (Supplemental Fig. S2B). We confirmed the binding of the UBA domain of Cezanne and Cezanne2 to K63-, but not K48- or K11-tetra-ubiquitin in the in vitro pull-down assay using purified recombinant GST-tagged protein fragments and linkage-specific ubiquitin chains (Fig. 1B). Analyzing the UBA domain from Cezanne or Cezanne2 binding to K63-di-Ub in vitro using isothermal titration calorimetry (ITC) showed that both of the UBA domains bind to K63-di-Ub, with a binding affinity (KD = 20 ± 2 and 20 ± 5 μM), approximately threefold lower than that of the UIMs of Rap80 (KD = 6.4 ± 0.2 μM) (Fig. 1C). Thus, Cezanne and Cezanne2 UBA domains specifically bind to the K63-linked ubiquitin chain.
Figure 1.
Identification of Cezanne and Cezanne2 UBA domain as a K63-linked ubiquitin binding motif. (A) A protein array screen identifies the K63-ubiquitin binding motif. Cy3-streptavidin-labeled biotin-K63- and K48- tetra-ubiquitin were used to probe the UBD arrays. The positions of selective K63-linked ubiquitin chain binding protein (domain) are marked: 1-ASCC2 (CUE); 2-Cezanne (UBA); 3-Cezanne2 (UBA); 4-TDRD3 (UBA); 5-RAP80 (UIMs); 6-TOM1L2 (VHS); and 7-RNF168 (MIU2). (B) In vitro pull-down assay using purified recombinant GST-Cezanne UBA, Cezanne2 UBA, or Rap80 UIMs incubated with K63-, K48-, or K11-tetra-ubiquitin chains. Anti-ubiquitin antibody was used for the western blot. (C) ITC analysis of Cezanne, Cezanne2 UBA, or Rap80 UIMs binding to K63-di-Ub.
Rap80/Abraxas/BRCA1 recruitment is diminished in Cezanne-deficient cells
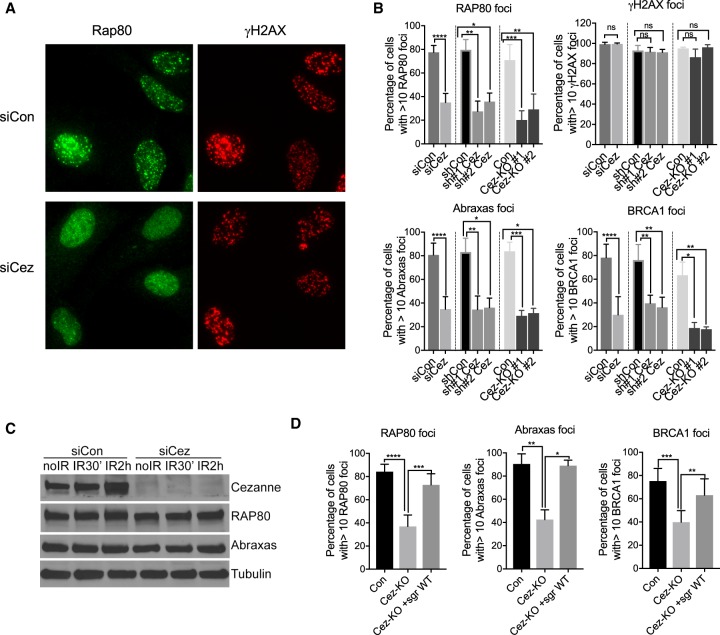
We found that in cells depleted of Cezanne by siRNAs, IR-induced foci (IRIF) of Rap80 were significantly decreased (Fig. 2A,B). The Rap80 protein level, however, did not change in the knockdown cells (Fig. 2C), indicating that the diminished Rap80 IRIF was not due to decreased levels of Rap80 protein. Rap80 is a component of the BRCA1-A complex and is essential for the recruitment of the BRCA1-A complex in which Abraxas serves as an adaptor protein mediating the binding of Rap80 to BRCA1 (Wang and Elledge 2007; Wang et al. 2009). Consistent with a role of Rap80 in the recruitment of the BRCA1-A complex to DNA damage sites (Kim et al. 2007; Sobhian et al. 2007; Wang et al. 2007), depletion of Cezanne also led to diminished IRIF of Abraxas and BRCA1 (Fig. 2B; Supplemental Fig. S3). Cezanne shRNAs knockdown cells or Cezanne knockout (KO) cells generated by CRISPR-Cas9 showed similar defects in IRIF of Rap80, Abraxas, and BRCA1 (Fig. 2B; Supplemental Fig. S3). Cezanne deficiency, however, did not result in much change in γH2AX foci (Fig. 2A,B), indicating that Cezanne is downstream from γH2AX in the hierarchy of recruitment of Rap80 and BRCA1-A complex. Importantly, the deficiency of Cezanne KO in recruitment of Rap80, Abraxas, and BRCA1 can be rescued by complementation of wild-type (WT) Cezanne gene expression (Fig. 2D). Thus, Cezanne plays a key role in regulating the recruitment of Rap80 and the BRCA1-A complex to DSBs.
Figure 2.
Cezanne regulates Rap80/Abraxas/BRCA1 recruitment to DNA damage sites. (A) Representative images of decreased Rap80 IRIF in Cezanne siRNA-treated U2OS cells. Cells were treated with 10 Gy IR followed by 2 h incubation before fixation and staining. (B) Quantification of Rap80, Abraxas, BRCA1 IRIF in Cezanne siRNAs-, shRNAs-treated cells, or Cezanne KO (Cez-KO#1 and KO#2) cells. Student's t-test (con versus siCez) or nonparametric Kruskal–Wallis ANOVA (con versus shCez, con versus KO) was used for statistical analysis. (C) Western blots of lysates from siRNAs-treated U2OS cells untreated (no IR), or 30 min and 2 h post-IR treatment. (D) Expression of Cezanne WT gene rescues the deficiency of Cezanne KO cells in IRIF of Rap80, Abraxas, and BRCA1. Nonparametric Kruskal–Wallis ANOVA was used for statistics analysis.
UBA domain and DUB activity are required for Cezanne's role in regulating recruitment of Rap80/Abraxas/BRCA1
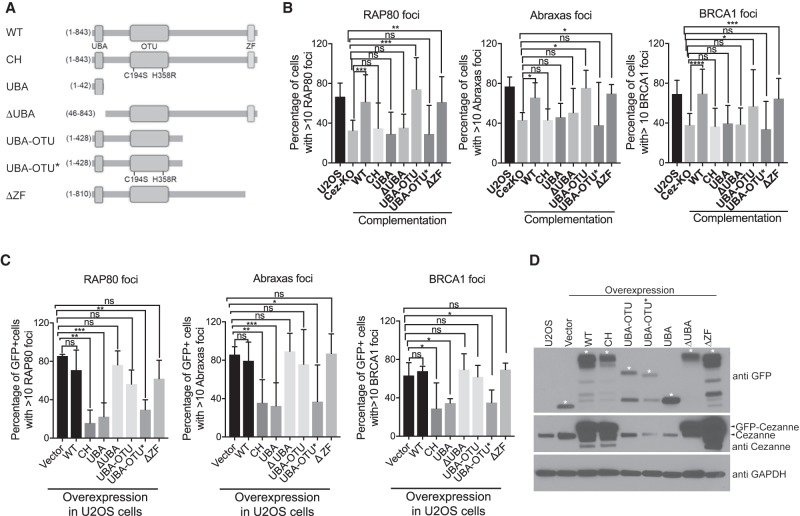
Cezanne is an OTU family DUB, containing a UBA domain followed by an OTU DUB domain at the N terminus and a zinc-finger domain at the C terminus (Bremm et al. 2010; Mevissen et al. 2013, 2016). By generating and analyzing Cezanne mutants, we found that the UBA domain and the DUB activity are essential for Cezanne to regulate Rap80 recruitment (Fig. 3; Supplemental Fig. S4). In Cezanne KO cells, we examined whether the defect of Rap80, Abraxas, and BRCA1 IRIF formation can be rescued when Cezanne mutants were re-introduced for expression. Compared to WT, expression of UBA-only or a UBA domain deletion mutant (ΔUBA) failed to rescue the defect. The GFP-tagged or HA-tagged DUB inactive mutant (CH) that previously was shown to abolish the DUB activity (Hu et al. 2013; Mevissen et al. 2016) is defective in rescuing Rap80, Abraxas, and BRCA1 IRIF in Cezanne KO cells (Fig. 3B; Supplemental Fig. S4B). Whereas the UBA domain by itself does not rescue RAP80, Abraxas, and BRCA1 IRIF, a truncated N terminus of Cezanne that contains both the UBA domain and the DUB domain (UBA-OTU), but not a DUB inactivated version (UBA-OUT*), rescues the phenotype (Fig. 3B). The C-terminal zinc-finger domain had minimal effect on the IRIF of Rap80, as the mutant without this domain (ΔZF) rescues the phenotype (Fig. 3B). These data indicate that the UBA domain and the DUB activity together are sufficient for regulating Rap80 recruitment.
Figure 3.
Cezanne UBA domain and DUB activity are essential for Rap80, Abraxas, and BRCA1 recruitment. (A) A diagram of Cezanne WT and mutants. (B) UBA and DUB activity of Cezanne are essential for Rap80, Abraxas, and BRCA1 IRIF. Cezanne KO cells were complemented with expression of GFP-tagged Cezanne WT or mutants. Nonparametric Kruskal–Wallis ANOVA was used for statistics analysis. (C) Overexpression of Cezanne DUB mutants decreases Rap80/Abraxas/BRCA1 recruitment. GFP-tagged Cezanne WT or mutants were expressed in U2OS cells. Nonparametric Kruskal–Wallis ANOVA was used for statistics analysis. (D) Western blot showing expression of GFP-tagged Cezanne WT and mutants in cells. * marks expression of indicated protein fragments.
Since Cezanne DUB activity is required for Rap80 recruitment, we investigated whether expression of Cezanne mutants lacking the DUB activity interferes with the endogenous Cezanne activity at sites of damage. When overexpressed in cells, GFP-tagged DUB inactive CH mutant of Cezanne plays a dominant-negative effect on the recruitment of Rap80 (Fig. 3C,D; Supplemental Fig. S5). In addition, although overexpression of the N-terminal half of Cezanne, UBA-OUT, does not appear to have a significant effect on Rap80 IRIF, expression of a DUB-inactive version of this truncated protein (UBA-OUT*) greatly reduced Rap80 IRIF. So did overexpression of the UBA domain-only mutant that lacks the DUB domain (Fig. 3C,D; Supplemental Fig. S5). Overexpression of HA-tagged Cezanne CH mutant also dominant-negatively decreased Rap80 IRIF (Supplemental Fig. S5). Thus, lack of DUB activity of Cezanne at DSBs can lead to decreased recruitment of Rap80, Abraxas and BRCA1.
Cezanne deficiency increases K11-ubiquitin conjugation but does not affect K63- conjugation at DNA damage sites
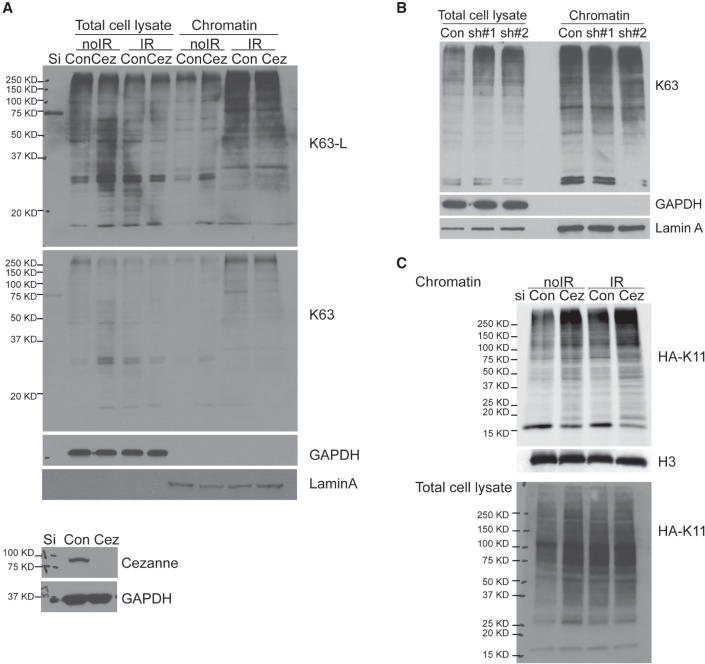
The recruitment of Rap80 to DNA damage sites depends on its UIMs binding to the K63-linked ubiquitin chain (Kim et al. 2007; Sobhian et al. 2007; Wang et al. 2007). To determine whether Cezanne deficiency affects the K63-linked ubiquitin conjugation at sites of damage, we examined K63-linked ubiquitin levels on damaged chromatin using a K63-linkage-specific antibody (Supplemental Fig. S6A,B). Cezanne knockdown did not lead to a decrease in K63-linked ubiquitin levels on damaged chromatin from cells treated with IR (Fig. 4A,B; Supplemental Fig. S6C), indicating that the failure of Rap80 recruitment to DNA damage sites is not due to decreased K63-linked polyubiquitin conjugation in Cezanne-deficient cells. Consistently, immunofluorescence (IF) staining using a K63-specific antibody also did not show a significant change of K63 IRIF (Supplemental Fig. S6D). Although overexpression of UBA-only or the DUB inactive CH mutant of Cezanne in WT cells resulted in decreased Rap80 recruitment (Fig. 3C), it did not lead to decreased levels of K63-linked polyubiquitin on damaged chromatin (Supplemental Fig. S6E). Thus, Cezanne DUB activity at DNA damage sites is not likely directly involved in affecting K63-linked ubiquitin conjugation. Instead, in the absence of Cezanne, HA K11-linked ubiquitin (an HA-tagged ubiquitin K11 mutant in which all of the lysine residues except lysine 11 are mutated to arginine) conjugation increased, consistent with a role of Cezanne DUB activity in antagonizing K11-conjugation on damaged chromatin (Fig. 4C; Paul and Wang 2017).
Figure 4.
Cezanne-deficiency does not affect K63-polyubiquitin levels on damaged chromatin. (A,B) K63-ubiquitin levels on damaged chromatin are not affected in Cezanne siRNA (A) or shRNAs (B) treated U2OS cells. Cells were treated with 10 Gy IR, collected at 2 h after incubation. Western blots were carried out with antibodies to K63-linkage-specific ubiquitin, GAPDH, or Lamin A. (C) HA K11-ubiquitin levels on damaged chromatin in cells is increased. Cells were transfected with HA K11, treated with 10 Gy IR, followed by 2 h incubation. Western blot was carried out with antibodies to HA.
Cezanne K11-DUB activity is required for regulating Rap80 recruitment
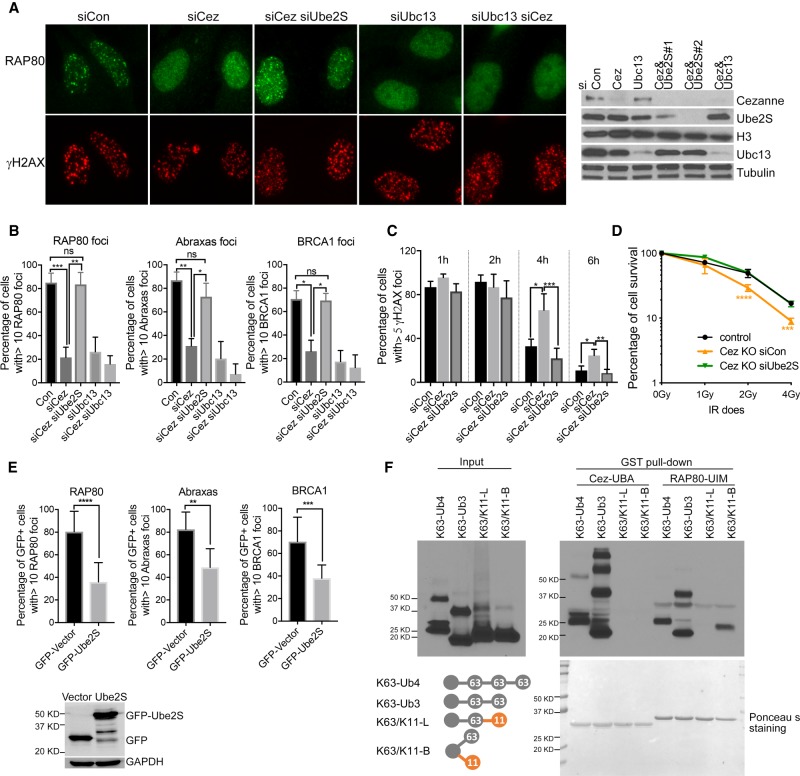
Cezanne is a K11-linkage-specific DUB (Bremm et al. 2010; Mevissen et al. 2016). In the absence of Cezanne, increased K11-ubiquitin conjugation may interfere with the binding of Rap80 UIMs to K63-ubiquitin conjugates, thus compromising the recruitment of Rap80. If this is the case, decreasing the K11-linked ubiquitin conjugation at sites of damage may rescue the recruitment of Rap80 in Cezanne-deficient cells. Indeed, when we knocked down Ube2S, an E2 conjugating enzyme for K11-ubiquitin conjugation (Baboshina and Haas 1996; Garnett et al. 2009; Williamson et al. 2009; Paul and Wang 2017), in Cezanne KO cells, Rap80 IRIF, as well as Abraxas and BRCA1 IRIF, were rescued (Fig. 5A,B; Supplemental Fig. S7A). As a control, knockdown of Ubc13, an E2 for K63-ubiquitin conjugation (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007), in Cezanne KO cells did not rescue the defect in Rap80/Abraxas/BRCA1 recruitment (Fig. 5A,B). In addition, the DNA repair defect in Cezanne-deficient cells marked by a delayed clearance of γH2AX staining of cells treated with 2 Gy IR was rescued when Ube2S was knocked down (Fig. 5C). The increased cellular sensitivity to IR in Cezanne KO cells was also rescued when Ube2S was knocked down (Fig. 5D). On the other hand, overexpression of GFP-tagged Ube2S in cells led to decreased recruitment of Rap80/Abraxas/BRCA1 (Fig. 5E; Supplemental Fig. S7B), consistent with the idea that increased K11-conjugation compromises recruitment of Rap80. Together, these data indicate that the K11-linkage-specific DUB activity of Cezanne removes K11-ubiquitin conjugates at DSBs to promote Rap80 recruitment.
Figure 5.
Cezanne promotes Rap80/Abraxas/BRCA1 recruitment through its K11-linkage DUB activity. (A,B) Depletion of Ube2S in Cezanne knockdown cell rescues Rap80 IRIF. Cells were treated with 10 Gy IR, followed by 2 h incubation before fixation and staining. Representative images (A) and quantification of Rap80, Abraxas, and BRCA1 IRIF (B) are shown. Nonparametric Kruskal–Wallis ANOVA was used for statistical analysis. (C) Double depletion of Ube2S and Cezanne rescues the repair deficiency of Cezanne-depleted cells. U2OS cells treated with indicated siRNAs were treated with 2Gy IR, fixed, and stained with γ-H2AX antibody at indicated times. The percentage of γ-H2AX foci positive cells were quantified. Nonparametric Kruskal–Wallis ANOVA was used for statistical analysis. (D) Depletion of Ube2S rescues the IR sensitivity of Cezanne KO cells detected by colony formation assay. Student's t-test was performed to compare Cez KO treated with siCon (orange) or siUbe2S (green) at indicated IR doses. “Control” (black) represents cells established from transfection with the construct without sgRNA. (E) Overexpression of GFP-tagged Ube2S in U2OS cells decreases Rap80/Abraxas/BRCA1 IRIF. Student's t-test was used for statistical analysis. (F) In vitro GST-pull-down assay using purified recombinant GST-Cezanne UBA or Rap80-UIMs incubated with K63-linked tetra-ubiquitin (K63-Ub4), tri-ubiquitin (K63-Ub3), mixed K63/K11-linkage tri-ubiquitin (K63/K11-L), or branched K63/K11- tri-ubiquitin (K63/K11-B).
Decreased binding of Rap80 UIMs to mixed K63/K11-chain
To determine the possibility that increased K11-conjugation compromises Rap80 IRIF by “corrupting” the K63-chain that recruits Rap80 recruitment, we tested in vitro the binding of Rap80 UIMs to a K63-chain or K63- and K11-mixed or branched chain by carrying out a pull-down assay using purified recombinant Cezanne UBA, Rap80 UIMs, and synthesized ubiquitin chains with different linkages. We found that Rap80 UIMs binding to mixed K63- and K11-tri-Ub (K63/K11-L) was largely abolished, indicating that mixed K63/K11-chain formation is detrimental to the binding of UIMs. Whereas Rap80 UIMs showed much weaker binding to branched K63- and K11-tri-Ub (K63/K11-B) when compared to the binding to K63-Ub3, Cezanne UBA did not appear to bind to either mixed or branched K63/K11 chains (Fig. 5F). These data suggest that Cezanne binds to K63-chains at DNA damage sites to prevent K63/K11-mixed chain formation to promote Rap80 recruitment.
Cezanne localizes to DNA damage sites regulating recruitment of DNA repair factors
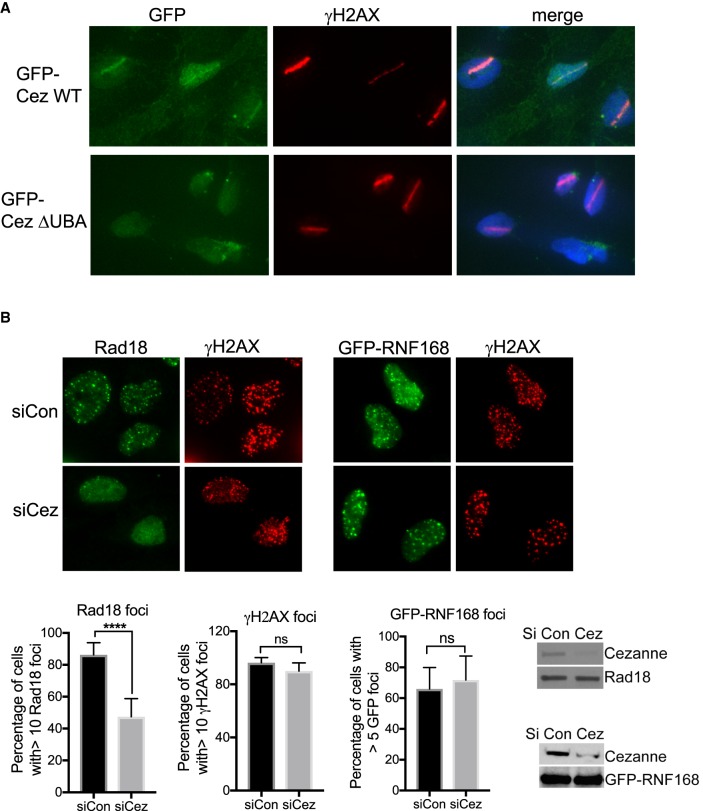
We then tested whether Cezanne localizes to DNA damage sites through its UBA domain. In cells microirradiated with a laser, GFP-tagged Cezanne accumulated at laser-induced DNA damage, and lack of the UBA domain (ΔUBA mutant) abolished the recruitment of Cezanne to DNA damage sites, indicating that Cezanne localization to DNA damage sites depends on the UBA domain (Fig. 6A).
Figure 6.
Cezanne localizes to DNA damage sites and promotes Rad18 IRIF. (A) GFP-Cezanne, but not the Cezanne mutant lacking the UBA domain, accumulates to laser-induced DNA damage. U2OS cells stably expressing GFP-Cezanne WT or ΔUBA treated with laser microirradiation were fixed and stained. (B) Cezanne deficiency leads to decreased Rad18 IRIF. U2OS or U2OS stably expressing GFP-RNF168 cells were treated with control or Cezanne siRNA, irradiated with 10 Gy IR, followed by 2 h incubation before fixation and staining. Student's t-test was used for statistical analysis.
It has been shown that ubiquitin K63-conjugation is critical for several additional DNA repair factors, including Rad168, Rad18, and 53BP1 (Huang et al. 2009; Pinato et al. 2011; Panier et al. 2012). We found that knockdown of Cezanne led to a decrease in Rad18 IRIF, but not RNF168 or 53BP1 IRIF (Fig. 6B; Supplemental Fig. S8), indicating that the K63-ubiquitin conjugation-dependent recruitments of DNA repair proteins are differentially regulated by Cezanne.
Cezanne2 interacts with Cezanne, promoting recruitment of Rap80/Abraxas and 53BP1 and DNA repair
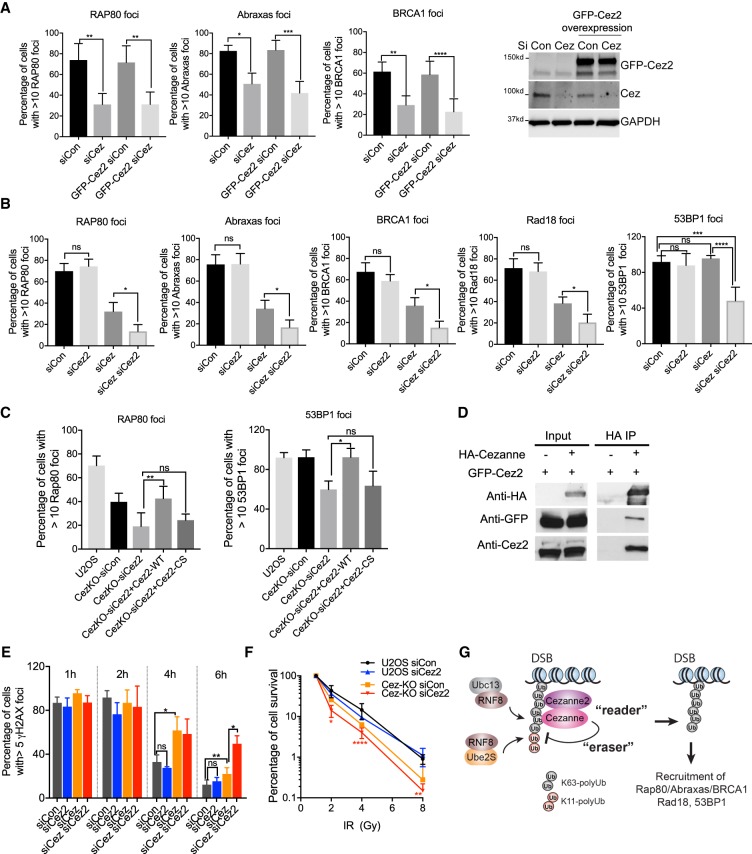
Cezanne2 is a paralog of Cezanne with a UBA domain which also binds to the K63-linked polyubiquitin chain (Fig. 1) followed by a DUB domain (Supplemental Fig. S8A); however, its function is not clear. We tested whether Cezanne2 functions redundantly with Cezanne as a backup of the Cezanne gene. We found that overexpression of Cezanne2 does not rescue the defect of Cezanne-depleted cells (Fig. 7A), indicating that Cezanne2 does not simply function as a backup of Cezanne.
Figure 7.
Cezanne and Cezanne2 play critical roles in recruitment of Rap80/Abraxas/BRCA1, Rad18 and 53BP1, cellular resistance to IR, and DNA damage repair. (A) Overexpression of GFP-Cezanne2 does not rescue Cezanne-deficiency in recruitment of Rap80/Abraxas/BRCA1. U2OS or U2OS cells overexpressing GFP-Cezanne2 were transfected with control or Cezanne siRNA, treated with 10 Gy IR, and incubated for 2 h before fixation and staining. Nonparametric Kruskal–Wallis ANOVA was used for statistical analysis. (B) Cezanne and Cezanne2 promote recruitment of Rap80/Abraxas/BRCA1, Rad18, and 53BP1. Cells were treated with 10 Gy IR, followed by 2 h incubation. Nonparametric Kruskal–Wallis ANOVA was used for statistical analysis. (C) Cezanne2 DUB activity is required for its role in promoting recruitment of Rap80 and 53BP1. Cezanne KO cells were transfected with Cezanne2 siRNA and complemented with expression of siRNA-resistant Cezanne2 WT or the DUB mutant (Cez2-CS) construct. Forty-eight hours after transfection, cells were treated with 10 Gy IR and incubated for 2 h. Nonparametric Kruskal–Wallis ANOVA was used for statistical analysis. (D) Cezanne interacts with Cezanne2. 293T cells were cotransfected with GFP-Cezanne2 and vector or HA-Cezanne expression plasmids. Total lysates were used for immunoprecipitation. (E) Cezanne and Cezanne2 are required for efficient DNA repair. Cells were treated with 2 Gy IR, fixed, and stained with γ-H2AX antibody at indicated times. Nonparametric Kruskal–Wallis ANOVA was used for statistics analysis. (F) Colony survival assay for cells treated with indicated doses of IR. Two-way ANOVA statistical analysis was performed to compare Cezanne-deficient (Cez-KO siCon, orange) with Cezanne- and Cezanne2-double deficient (Cez-KO siCez2, red) cells. (G) A proposed model showing that Cezanne and Cezanne2 promote recruitment of Rap80/Abraxas/BRCA1, Rad18, and 53BP1 through binding to K63-linked ubiquitin (“reader”) antagonizing K11-linked ubiquitin conjugation (“eraser”) at DNA damage sites.
Unlike knockdown of Cezanne, depletion of Cezanne2 alone had a minimal effect on Rap80, Abraxas, or BRCA1 IRIF (Fig. 7B; Supplemental Fig. S8C). However, siRNAs depletion of both Cezanne and Cezanne2 led to a greater decrease of Rap80/Abraxas/BRCA1 IRIF when compared to Cezanne-deficient cells (Fig. 7B). Similarly, when Cezanne2 is depleted in Cezanne-KO cells, the recruitment of Rap80/Abraxas/BRCA1 is further decreased (Supplemental Fig. S8). Double depletion of Cezanne and Cezanne2 also led to a further decrease in Rad18 IRIF but had a minimal effect on RNF168 recruitment (Fig. 7B; Supplemental Fig. S8). Thus, Cezanne2 facilitates Cezanne in regulating Rap80/Abraxas/BRCA1 and Rad18 recruitment.
Whereas knockdown of either Cezanne or Cezanne2 alone did not result in much change in 53BP1 IRIF (Fig. 7B; Supplemental Fig. S8), depletion of both genes by siRNAs, however, led to a significant decrease in 53BP1 IRIF (Fig. 7B). Similarly, although Cezanne KO cells did not display a deficiency in 53BP IRIF, knocking down Cezanne2 in Cezanne KO cells resulted in a defect in 53BP1 recruitment (Supplemental Fig. S8G). These data indicate that both Cezanne and Cezanne2 are required to recruit 53BP1 to DNA damage sites.
The defects caused by the deficiency of Cezanne2 in recruiting Rap80 and 53BP1 can be rescued by expression of Cezanne2 WT but not the DUB mutant (Cez2-CS) (Fig. 7C), indicating that the DUB activity of Cezanne2 is required for its function. In addition, Cezanne appears to interact with Cezanne2 in a co-IP experiment using cells co-expressing HA-Cezanne and GFP-Cezanne2 (Fig. 7D). Thus, it is likely that Cezanne and Cezanne2 form a complex and that the DUB activity of both of these two proteins is required for promoting Rap80/Abraxas/BRCA1, Rad18, and 53BP1 recruitment.
Due to the role of Cezanne and Cezanne2 in the recruitment of the Rap80/BRCA1-A complex, Rad18, and 53BP1, we examined their roles in DNA damage repair and cellular resistance to IR. Using the γH2AX clearance assay to examine repair efficiency in cells treated with 2 Gy IR, we found that the deficiency of Cezanne-depleted cells was further enhanced when both Cezanne and Cezanne2 were knocked down (Fig. 7E). Thus, Cezanne2 assists Cezanne in the repair of DNA damage in response to IR. When tested for the role of Cezanne and Cezanne2 in HR and NHEJ using reporter assays, it is clear that Cezanne is required for HR, whereas its role in NHEJ is not apparent (Supplemental Fig. S9). Knockdown of Cezanne2 appears to have a minimal role in HR or NHEJ (Supplemental Fig. S9). We then examined the resection of DSB ends using phosphorylated RPA32pS4/8 as a surrogate marker. We found that RPA32pS4/8 is not affected by siRNAs to Cezanne, Cezanne2, or combined (Supplemental Fig. S9), suggesting that Cezanne is not directly involved in DSB end resection responding to IR. Since Cezanne and Cezanne2 promote recruitment of multiple DNA repair factors, their roles in HR or NHEJ are likely to be complex. In a colony survival assay, Cezanne-siRNA-treated cells displayed increased sensitivity to IR, and lack of Cezanne2 enhanced the sensitivity of Cezanne-siRNA-treated cells to IR, consistent with their roles in the recruitment of Rap80, Rad18, and 53BP1 (Fig. 7F; Supplemental Fig. S9). Together, these data indicate that Cezanne and Cezanne2 play critical roles in DNA repair and cellular resistance to IR.
Discussion
Our study identifies Cezanne and Cezanne2 as two new components in the complex ubiquitin landscape at DNA damage sites for regulating recruitment of DNA repair and signaling proteins, including Rap80/Abraxas/BRCA1, Rad18, and 53BP1. We present a model that Cezanne serves as a “reader” of the K63-linkage and “eraser” of the K11-linkage ubiquitination for the regulation of DNA damage response (Fig. 7G), introducing an example that there is crosstalk between linkage-specific ubiquitination at DNA damage sites.
Cezanne as a K63-“reader” and a K11-“eraser” of the “ubiquitin code” at DSBs
The K63-linked ubiquitin conjugation plays a major role in regulating DNA repair and signaling (Jackson and Durocher 2013; Panier and Durocher 2013). Several known K63-chain binding proteins in the response to DNA damage were identified from our screen, including Rap80 UIMs (Kim et al. 2007; Sobhian et al. 2007; Wang et al. 2007), the RNF168 MIU2 domain, (Doil et al. 2009; Pinato et al. 2009, 2011; Stewart et al. 2009; Panier et al. 2012), and the ASCC2 CUE domain which was recently shown to mediate the recruitment of ASCC2 to alkylation damage sites (Brickner et al. 2017). It remains to be seen whether the other domains identified in our screen also bind to K63-polyubiquitin at DSBs or are involved in DNA damage responses. Although the “reader” list for the K63-chain at DSBs may still not be complete, our study adds Cezanne and Cezanne2 as two new “readers” of the K63-chain ubiquitin code for the regulation of DNA damage response.
The fact that Cezanne and Cezanne2 contain both a UBA domain and an OTU DUB domain predicts that they can be “reader” and “eraser” of the “ubiquitin code.” Since neither the UBA deletion mutant nor DUB mutant rescues the defect of Cezanne KO cells in Rap80 IRIF formation, it indicates that both the “reader” and “eraser” activity are essential for the role of Cezanne in promoting Rap80 recruitment. Cezanne “reads” the K63-chain (Fig. 1) and localizes to DNA damage sites through the UBA domain (Fig. 6A). It is noted that the affinity of the Cezanne and Cezanne2 UBA binding to di-K63-Ub is less than that of the Rap80 UIMs in vitro (Fig. 1); thus, Cezanne and Cezanne2 are less likely to compete with Rap80 in binding to K63-linked conjugates at DSBs. Indeed, exogenously overexpressed wild-type Cezanne or the Cezanne N-terminal fragment containing the UBA and intact DUB domain do not appear to compromise Rap80 IRIF (Fig. 3C). While the K63-“reader” activity localizes Cezanne to DSBs, the DUB activity of Cezanne is required for Cezanne's role in promoting Rap80 recruitment. Mutation of the residues that abolish the DUB activity compromises the role of Cezanne in the recruitment of Rap80 (Fig. 3). Moreover, Cezanne mutants lacking DUB activity, when overexpressed in wild-type cells, play a dominant negative role in regulating Rap80 recruitment to DSBs (Fig. 3).
Cezanne and Cezanne2 are the only DUBs known to be specific for K11-linked polyubiquitin (Bremm et al. 2010, 2014; Mevissen et al. 2013). Depleting Cezanne leads to increased K11-linked ubiquitin conjugation on chromatin (Fig. 4C). This is consistent with our previous finding that Cezanne antagonizes RNF8 and Ube2S-dependent K11-conjugation on damaged chromatin (Paul and Wang 2017). Recently, however, Cezanne was also found to possess a K63-DUB activity toward modifying proteins with K63-chains in NF-kB activation or mTORC2/AKT signaling (Enesa et al. 2008; Wang et al. 2017). Such a Cezanne K63-DUB activity is unlikely to be occurring at DNA damage sites since we did not observe a significant increase of K63-linked polyubiquitin modification on damaged chromatin in the absence of Cezanne (Fig. 4). Thus, although it is possible that Cezanne may display differential DUB activities processing K11- or K63-ubiquitin chains conjugated by different E2s or E3s, it is unlikely that Cezanne is directly involved in processing K63-ubiquitin chains on damaged chromatin.
Our results indicate that overly increased K11-ubiquitin conjugation impairs the recruitment of DNA repair factors, and the DUB activity of Cezanne that “erases” the K11-linked ubiquitin chain plays a critical role in the DDR. Knocking down Ube2S in Cezanne -deficient cells, which rebalances the K11-conjugation, restores the recruitment of Rap80 and rescues the deficiency of Cezanne-depleted cells in DNA repair and cellular resistance to IR (Fig. 5). On the other hand, an enforced increase of K11-ubiquitination by overexpression of Ube2S in cells results in impaired Rap80/Abraxas/BRCA1 recruitment similar to that caused by depletion of Cezanne. Why does increased K11-conjugation have a negative effect on Rap80 recruitment? We show that, in vitro, the binding of Rap80 UIMs to the K63/K11-mixed chain is largely abolished (Fig. 5F). Thus, it is likely that elevated K11-conjugation due to lack of Cezanne DUB activity increases the formation of a K63/K11-mixed chain at sites of damage, which impairs the binding of Rap80 UIMs and thus the recruitment of Rap80.
Together, our study paints a model that Cezanne promotes Rap80 recruitment through its UBA domain binding to the K63-linked ubiquitin chain and its DUB antagonizing K11-linked ubiquitin conjugation (Fig. 7G).
Crosstalk between K11- and K63-linked polyubiquitin modification
The “ubiquitin code” at sites of DNA damage is complex. Multiple forms of linkage-specific ubiquitin conjugation have been implicated, including K6, K11, K27, K33, K48, and K63 (Morris and Solomon 2004; Feng and Chen 2012; Jackson and Durocher 2013; Elia et al. 2015; Gatti et al. 2015; Paul and Wang 2017). Little is known about whether there is coordination between these different linkages. Our study provides an example that there is crosstalk between K63- and K11-linked ubiquitination, suggesting that linkage-specific ubiquitination at DSBs is likely coordinated.
The K63-linked chains are key to the recruitment of BRCA1 and 53BP1, and the K11-linked chains are implicated in regulating inhibition of transcription (Paul and Wang 2017). The crosstalk likely fine-tunes the linkage-specific ubiquitination for coordination of these events in the DNA damage response. Cezanne K11-DUB activity may be important to generate a chromatin environment at DSBs that favors K63-conjugation by antagonizing K11-conjugation to allow for efficient recruitment of DNA repair factors. Considering that both K63- and K11-conjugation are catalyzed by the same E3 ligase RNF8, the crosstalk carried out by Cezanne and Cezanne2 to coordinate the linkage-specific ubiquitination is likely to be particularly important. Since it has been shown that Cezanne K11-DUB activity not only functions toward K11-chains but also extends to K11-linkage within K11/K63- and K11/Lys48-branched chains (Mevissen et al. 2016), it is likely that at least one of the functions of Cezanne is to prevent K11-conjugation on K63-chains forming mixed K63/K11-chains to protect the “purity” of K63-chains. Such a crosstalk is essential in promoting K63-ubiquitin chain-dependent recruitment of Rap80, Rad18, and 53BP1.
Roles of Cezanne and Cezanne2 in promoting Rap80, Rad18, and 53BP1 recruitment and DNA repair
Our data indicate that both Cezanne and Cezanne2 play critical roles in the DDR promoting recruitment of DNA repair factors, in DNA repair, and in cellular resistance to IR. Although Cezanne2 shares sequence and protein domain similarity with Cezanne, it does not replace the function of Cezanne (Fig. 7A). Unlike Cezanne, Cezanne2 depletion alone does not have a major effect on Rap80/Abraxas/BRCA1 or Rad18 recruitment. Cezanne2 deficiency, however, greatly increases the defects of Rap80/Abraxas/BRCA1 and Rad18 foci formation in Cezanne-deficient cells, indicating that in the regulation of Rap80/BRCA1-A complex and Rad18 recruitment, Cezanne plays a major role and Cezanne2 plays a facilitating role. The regulatory role of Cezanne and Cezanne2 in 53BP1 recruitment is somewhat different. Knockdown of either Cezanne or Cezanne2 does not affect 53BP1 foci; only when both are depleted, the recruitment of 53BP1 is compromised (Fig. 7). It indicates that both Cezanne and Cezanne2 are required for regulation of 53BP1 foci formation. The different roles of Cezanne and Cezanne2 in promoting Rap80 or Rad18 and 53BP1 suggest that the K63-dependent recruitments of DNA repair proteins are differentially regulated by Cezanne and Cezanne2. This is consistent with the difference in mechanisms of recruitment of Rap80 or Rad18 and 53BP1. Rap80 or Rad18 are respectively recruited to DSBs through the UIMs of Rap80 or the UBZ of Rad18 specifically interacting with K63-linked polyubiquitin (Kim et al. 2007; Sobhian et al. 2007; Wang et al. 2007). 53BP1 recruitment, however, depends on its Tudor domain binding to dimethylated histone H4 lysine 20 and its ubiquitination-dependent recruitment (UDR) motif interacting with histone H2A K15 monoubiqutinated by RNF168 (Fradet-Turcotte et al. 2013; Wilson et al. 2016). Since Cezanne and Cezanne2 have little effect on the recruitment of RNF168, they are likely downstream from RNF168 in the recruitment of Rap80, Rad18, or 53BP1. In addition, Cezanne2 interacts with Cezanne (Fig. 7D), and the DUB activity of Cezanne2 is important for its function (Fig. 7B). Thus, Cezanne and Cezanne2 likely form a complex, with both of the DUB activities required for promoting recruitment of DNA repair factors.
In summary, our work demonstrates that polyubiquitin modification at DSBs is a highly orchestrated process that involves crosstalk between different linkage ubiquitin chains, a mechanism that likely applies to other cellular processes that involve linkage-specific ubiquitination.
Materials and methods
UBD array screen
Purified recombinant GST-fused ubiquitin binding domains were arrayed onto glass slides. The current version of the library contains 148 GST fusion proteins that harbor members of protein domain families known to interact with ubiquitin, including UIMs, UBA, CUE, GAT, VHS, PAZ, PFU, UBM, UBZ, and JAB/MPN domains. We carried out the screen by probing the array with K63-linked or K48-linked tetra-Ub chains that were directly labeled with streptavidin-Cy3 on a biotin moiety (at position 20 of the first Ub molecule). The S20 site of the first ubiquitin was mutated to C and biotinylated. This position was then used for Cy3-Streptavidin labeling.
Isothermal titration calorimetry (ITC) of Rap80 UIMs, Cezanne UBA, and Cezanne2 UBA with K63-diUb
DNA fragments encoding Rap80 UIMs (60–144 amino acids), Cezanne UBA (1–66 aa), and Cezanne2 UBA (1–88 aa) were cloned into pGEX-4T-1. The Gst-tagged proteins were expressed in Escherichia coli BL21 (DE3) cells induced with 0.1 mM isopropyl β-D-thiogalactoside overnight. Cell pellets were lysed in 20 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, 0.5 mM tris(2-carboxyethyl) phosphine (TCEP) with sonication. Lysate was cleared with centrifugation followed by 8 mL glutathione Sepharose (GE Healthcare) and extensive washes. Thrombin protease was added, mixed into the beads, and allowed to cut the fusion overnight at 4°C in the column. Flow-through containing the ubiquitin binding domains was collected, concentrated, and loaded onto a Hitrap Superdex 16/60 S200 preparative sizing column (GE Healthcare). Selected fractions from the sizing column profile were collected, flash frozen, and stored at −80°C. Purified K63-diUB in PBS buffer at pH 7.4 was purchased from Lifesensors, Inc. Buffer exchange to buffers (10 mM Tris pH 7.2 and 50 mM NaCl, 0.1 mM TCEP) was completed either by running through a Superdex 200 Increase 10/300 GL analytical sizing column (GE Healthcare) or rounds of concentration/dilution using an Amicon Ultra 0.5 mL Centrifugal 3K cutoff concentrator (Millipore) for all proteins. The K63-diUB was placed in the cell, and the respective ubiquitin binding domains in the syringe at the approximate concentrations (18 μM for Cezanne2 UBA, 25 μM for Cezanne UBA, and 20 μM for Rap80 UIMs) were measured using a ThermoScientific Nanodrop One either at 280 nm (Cezanne and Cezanne2) or at peptide bond wavelength (Rap80 and K63-diUb), as these proteins had no tryptophan. Experiments were done at 25°C using the MicroCal PEAQ-ITC automated system (Malvern Instrument Ltd). Binding constants (KD) were calculated by fitting the data using the MicroCal PEAQ-ITC Analysis ITC software (Malvern).
In vitro GST pull-down assay
Ten micrograms of purified recombinant GST-RAP80 UIMs, Cezanne UBA, and Cezanne2 UBA bound to Glutathione Sepharose 4B beads was mixed with 100 ng K63-, K48- or K11-linked tetra-ubiquitin chain, K63-linked Ub3, K11/K63-mixed Ub3, or K11/K63-branched Ub3 in 400 μL NETN butter, and incubated at 4°C with rocking for 3 h. After three washes with NETN buffer, beads were boiled in 5× SDS sample loading buffer and loaded to a protein gel for western blot analysis.
Synthesis of tri-ubiquitin chain
K11/K63-linked mixed (linear) and branched tri-ubiquitins were assembled from ubiquitin monomers containing chain terminating mutations (K11R&K63R, K63R, and K11R&D77 for the former and K11R&K63R and D77 for the latter) in a controlled stepwise manner using linkage-specific E2 enzymes Ube2s (for K11) and Ubc13/MMS2 (for K63) following the strategy described in Castañeda et al. (2013) and Nakasone et al. (2013). K63-linked tri-ubiquitin was assembled from WT ubiquitin using Ubc13/MMS2; the trimer species was separated from the reactants and other products using cation-exchange and size-exclusion chromatography.
Cell lines, cell culture, and antibodies
The human U2OS cell line was grown in McCoy's 5A with L-glutamine medium (Cellgro, Corning) supplemented with 10% FBS (GenDEPOT) and 1% penicillin/streptomycin (Gibco). The 293T cell line was grown in DMEM (Cellgro, Corning) with 4.5 g/L glucose, L-glutamine, and sodium pyruvate medium supplemented with 10% FBS and 1% penicillin/streptomycin. Antibodies used are: Cezanne (Santa Cruz, sc-514402), RAP80 (Bethyl Laboratories, A300-763A), Abraxas (homemade), HA (Cell Signaling Technology, 3724s, 2367s), GFP (Invitrogen, A11122, A11120), K63 (EMD Millipore, 05-1308), ubiquitin (Santa Cruz, sc-8017), Ubc13 (Zymed, 37-1100), Ube2S (Cell Signaling Technology, 11878s), Lamin A (Sigma, L1293), GAPDH (Invitrogen, MA5-15738), BRCA1 (Santa Cruz, sc-6954), 53BP1 (Upstate, 05-726), γH2AX (Upstate, 05-636 JBW103), Rad18 (Abcam, ab188235), and pRPA32 (Bethyl, A300-245).
Plasmid, siRNA, and shRNA
The pENTR-Cezanne was purchased from the MDACC shRNA and ORFeome Core at MD Anderson and was inserted into a MSCV-GFP retroviral expression vector or pCDNA3-HA expression vectors by LR recombination. SiRNAs targeting Cezanne, Cezanne2, Ube2S, and Ubc13, and negative control siRNA were purchased from Invitrogen. Cezanne, Ube2S, Ubc13, and negative control siRNA sequences were published previously (Paul and Wang 2017). Cezanne2 siRNA sequences are: 5′-AAAUCCUCGCUGUACACGC-3′, and 5′-UCAUCAUGGUAUAGAGAGC-3′. SiRNAs were transfected into cells using lipofectamine RNAiMAX reagent (Invitrogen). Lentiviral Cezanne and Cezanne 2 shRNA plasmids were purchased from the MDACC shRNA and ORFeome Core. The shRNA sequences for Cezanne are: 5′-TGAGCAAGGACAAAGACGT-3′ and 5′-ATTCTGCTGTGTCTGCTGC-3′. The shRNA sequences for Cezanne2 are: 5′-TGATCATAGGCCAGAACCA-3′ and 5′-GTATCTGCCACAACAACGA-3′.
Generation of Cezanne KO cells
The CRISPR-Cas9 system was used to generate Cezanne KO U2OS cells. Briefly, U2OS cells were infected by pLentiCRISPR lentivirus carrying Cas9 and sgRNA targeting Cezanne, which expresses Cas9 endonuclease and the targeting sequences: 5′-TCAGATTTTGTCCGTTCCAC-3′. Cells were subjected to puromycin selection. Single colonies were selected and expanded for a screen for Cezanne KO clones.
Construction of Cezanne mutants
Cezanne mutants were generated using a QuikChange II site-directed mutagenesis kit (Agilent Technologies) in the pENTR-Cezanne plasmid and then were inserted into the retroviral expression vector MSCV-GFP or pCNDA3-HA expression vector by LR recombination. Primers used for mutagenesis are: CH 5′-CTGGAGATGGGAACAGCCTCCTGCATGCA-3′ and 5′-TGCATGCAGGAGGCTGTTCCCATCTCCAG-3′ (for C194S mutation), and 5′-GCCTATGATCAGGCCCGCTTTTCTGCACTCGTG-3′ and 5′-CACGAGTGCAGAAAAGCGGGCCTGATCATAGGC-3′ (for H358R mutation), UBA 5′-TTCCAGCATGGACTTGACGCTACTATTCAAAATCACTGAGGGCGGCATTC-3′ and 5′-GAATGCCGCCCTCAGTGATTTTGAATAGTAGCGTCAAGTCCATGCTGGAA-3′, UBA-OTU 5′-GGTATTCTCCTTCTGCTCCATCTACTAGAGTGCAGAAAAGTGGGCCTG-3′ and 5′-CAGGCCCACTTTTCTGCACTCTAGTAGATGGAGCAGAAGGAGAATACC-3′, ΔZF 5′-CAAACAACCGAACTGCAGCTTCTATTGATAGCCTGAGACAAACAAC-3′ and 5′-GT TGTTTGTCTCAGGCTATCAATAGAAGCTGCAGTTCGGTTGTTTG-3′, ΔUBA5′-CCTGGACATGGATGCTCAAGTCCATGCTGGAA-3′ and 5′-TTCCAGCATGGACTTGAGCATCCATGTCCAGG-3′. The primers for generating the Cezanne sgRNA-resistant construct are: 5′-GCTCTGCTCCTGTGCTCCGGACGAAATCTGACAGAACAGCATCCATGTC-3′ and 5′-GACATGGATGCTGTTCTGTCAGATTTCGTCCGGAGCACAGGAGCAGAGC-3′.
Immunofluorescence
Immunofluorescence was carried out as previously described (Hu et al. 2011). Briefly, cells were grown on coverslips, treated or nontreated with IR. Cells were fixed with a fresh 3% paraformaldehyde/ 2% sucrose solution for 20 min at room temperature, followed by permeabilization with 0.5% Triton X-100 solution on ice for 5 min. After wash with PBS, cells were then incubated with the indicated primary antibody diluted in 1% BSA at 37°C for 1 h, followed by incubation with the secondary antibody conjugated with Alexa-488 or Alexa-555 for 1 h at 37°C. Coverslips were then washed with PBS and mounted using DAPI containing antifade solution (Invitrogen). For K63 IF staining, cells were pre-extracted using pre-extraction buffer (10 mM PIPES PH6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCL2, 1 mM EGTA, and 0.2% Triton X-100) on ice for 5 min. Images were collected with an 80i eclipse Nikon microscope using the 40× objective.
Cell lysis and chromatin fractionation
Cells were lysed using NETN buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM DTT, and protease inhibitors including 1 mM PMSF, 5 mM NaF, 1 mM Na3VO4, 50 mM β-Glycerophosphate, and protease inhibitor cocktails) on ice for 20 min, centrifuged at 4°C using 15,000 rpm for 10 min, and the supernatant was collected as total cell lysate. Chromatin fractionation was carried out as previously described (Xu et al. 2017). Briefly, 2 × 106 cells were incubated with 300 µL cold buffer A (10 mM HEPES pH 7.9, 10 mM KCL, 1.5 mM MgCL2, 0.34 M sucrose, 10% glycerol, 5 mM NaF, 1 mM Na3VO4, 1 mM DTT, protease inhibitor cocktails, and 0.1% Triton X-100) for 5 min on ice. After centrifugation at 4000 rpm at 4°C for 5 min, cell pellets were collected and washed with cold buffer A. Cell pellets were then incubated with cold buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, and protease inhibitor cocktails) for 30 min on ice. After centrifugation at 4500 rpm at 4°C for 5 min, pellets were collected and washed with buffer B, and then suspended in NETN buffer and sonicated, followed by centrifugation. The supernatant was collected as the chromatin fraction. For co-immunoprecipitation (co-IP), 293T cells transfected with the indicated plasmids were lysed using NETN buffer with 3U DNase I (NEB, M0303S). After sonication and centrifugation, total cell lysate was collected for IP with anti-HA-agarose beads.
Colony survival assay
For colony survival, the experiment was carried out as previously described (Wu et al. 2016). In brief, cells were plated at low density and treated with different doses of IR or left untreated. Cells were then incubated in a 37°C, 5% CO2 incubator for about 10 d. Colonies were fixed and stained with 0.5% crystal violet and a 20% methanol solution. Colonies having more than 50 cells were counted as a positive. Colony formation efficiency was normalized to untreated samples for the calculation of percentage of survival. Error bars represent standard deviation (SD) across triplicates.
Laser microirradiation
U2OS stably expressing GFP-Cezanne or GFP-ΔUBA Cezanne cells were seeded in cell culture plates with glass bottoms and incubated with 10 µM BrdU for 24 h. Laser ablation was performed with an ABLATE 355 nm from an Intelligent Imaging Innovations (3i) system integrated in a Nikon Inverted Ti Microscope using an Apo 60× λS oil 1.4 N.A. objective. Ablation was applied on a line using a raster block of 1 and 40% of total attenuation with a repetition of 5. Cells were pre-extracted with pre-extraction buffer before fixation and staining. Images were collected with an inverted Zeiss Observer Z1 using a Plan Neofluor 40× oil 1.3 N.A. objective and analyzed with SlideBook 6.0.
HR and NHEJ assay
A DR-GFP reporter assay and EJ2GFP reporter assay were carried out as previously described (Moynahan et al. 1999; Bennardo et al. 2008). EJ2GFP-puro was a gift from Jeremy Stark (Addgene plasmid #44024; http://n2t.net/addgene:44025;RRID:Addgene_44025). Forty-eight hours after siRNA transfection, 2 µg of I-SceI expression plasmid were transfected into cells. The percentage of GFP positive cells was analyzed by a Gallios561 flow cytometer.
qPCR
Total RNA was isolated from cells using the RNeasy RNA extraction kit (Qiagen), and cDNA was synthesized from 1 µg of total RNA using a iScript cDNA synthesis kit (Bio-Rad). Fluorescence real-time PCR analysis was conducted using iTaq Universal SYBR Green Supermix (Bio-Rad) and performed on the Bio-Rad C1000 thermal cycler (CFX-96 real-time PCR detection systems; Bio-Rad). The ribosomal protein large subunit P0 (RPLP0) mRNA level was used for normalization. The real-time PCR primers are: Cezanne 2, 5′-CACGAGCTGTAAACGGCTTCT-3′ and 5′-GCTTTCCGTAACACCAGGTCC-3′ and RPLP0, 5′-TTCATTGTGGGAGCAGAC-3′ and 5′-CAGCAGTTTCTCCAGAGC-3′.
Statistics
All data were plotted as mean values with variances as SD using GraphPad Prism 7 software. Student's t-test, nonparametric Kruskal–Wallis ANOVA, or two-way ANOVA were used for statistical analysis.
Competing interest statement
M.T.B. is a cofounder of EpiCypher.
Supplementary Material
Acknowledgments
We thank Dr. Clayton B. Woodcock for help with ITC experiments, Dr. Adriana Paulucci from the BSRB Microscopy Facility at the Department of Genetics MD Anderson Cancer Center for assistance with ABLATE microscope acquisition, training with imaging collection and analysis, and Dr. Shao-cong Sun from the Department of Immunology for critical reagents. This work was supported by the Cancer Prevention Research Institute of Texas (CPRIT) grant (RP180244 to B.W.). X.W. and B.W. are supported by National Institutes of Health (NIH) grant CA155025 and CPRIT grant RP180244. X.W. is an MD Anderson Cancer Center/Center for Cancer Epigenetics (CCE) postdoctoral fellow. D.F. is supported by NIH grant GM065334. X.C. and J.R.H. are supported by NIH grant GM114306 and CPRIT grant RR160029. C.S., J.C., and M.T.B. are supported by CPRIT grant RP180804.
Author contributions: X.W., X.C., M.T.B., and B.W. designed the study and experiments. X.W. carried out the majority of the experiments, S.L. carried out some of the experiments analyzing Cezanne2 function, C.S. and J.C. performed the protein array screen, R.S. and A.C. synthesized the probe, J.R.H. performed the ITC, D.F. and T.R.K. synthesized tri-ubiquitin material. X.W. and B.W. wrote the manuscript with input from all authors.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.332395.119.
References
- Baboshina OV, Haas AL. 1996. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem 271: 2823–2831. 10.1074/jbc.271.5.2823 [DOI] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. 2008. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4: e1000110 10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci T, Suzuki A, Grant GD, Stanley N, Cook JG, Brown NG, Emanuele MJ. 2018. Cezanne/otud7B is a cell cycle-regulated deubiquitinase that antagonizes the degradation of apc/C substrates. EMBO J 37: e98701 10.15252/embj.201798701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremm A, Freund SM, Komander D. 2010. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol 17: 939–947. 10.1038/nsmb.1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremm A, Moniz S, Mader J, Rocha S, Komander D. 2014. Cezanne (OTUD7B) regulates HIF-1α homeostasis in a proteasome-independent manner. EMBO Rep 15: 1268–1277. 10.15252/embr.201438850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JR, Soll JM, Lombardi PM, Vågbø CB, Mudge MC, Oyeniran C, Rabe R, Jackson J, Sullender ME, Blazosky E, et al. 2017. A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair. Nature 551: 389–393. 10.1038/nature24484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda CA, Kashyap TR, Nakasone MA, Krueger S, Fushman D. 2013. Unique structural, dynamical, and functional properties of k11-linked polyubiquitin chains. Structure 21: 1168–1181. 10.1016/j.str.2013.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446. 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Elia AE, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge SJ. 2015. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol Cell 59: 867–881. 10.1016/j.molcel.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enesa K, Zakkar M, Chaudhury H, Luong le A, Rawlinson L, Mason JC, Haskard DO, Dean JL, Evans PC. 2008. NF-κB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem 283: 7036–7045. 10.1074/jbc.M708690200 [DOI] [PubMed] [Google Scholar]
- Feng L, Chen J. 2012. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 19: 201–206. 10.1038/nsmb.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Díaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499: 50–54. 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. 2009. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol 11: 1363–1369. 10.1038/ncb1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, Penengo L. 2015. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep 10: 226–238. 10.1016/j.celrep.2014.12.021 [DOI] [PubMed] [Google Scholar]
- Hu X, Kim JA, Castillo A, Huang M, Liu J, Wang B. 2011. NBA1/MERIT40 and BRE interaction is required for the integrity of two distinct deubiquitinating enzyme BRCC36-containing complexes. J Biol Chem 286: 11734–11745. 10.1074/jbc.M110.200857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Brittain GC, Chang JH, Puebla-Osorio N, Jin J, Zal A, Xiao Y, Cheng X, Chang M, Fu YX, et al. 2013. OTUD7B controls non-canonical NF-κB activation through deubiquitination of TRAF3. Nature 494: 371–374. 10.1038/nature11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Wang H, Xiao Y, Jin J, Chang JH, Zou Q, Xie X, Cheng X, Sun SC. 2016. Otud7b facilitates T cell activation and inflammatory responses by regulating Zap70 ubiquitination. J Exp Med 213: 399–414. 10.1084/jem.20151426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603. 10.1038/ncb1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914. 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. 2013. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 49: 795–807. 10.1016/j.molcel.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205. 10.1126/science.1139621 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640. 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81: 203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbé S. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10: 550–563. 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900. 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150: 1182–1195. 10.1016/j.cell.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. 2013. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154: 169–184. 10.1016/j.cell.2013.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen TE, Kulathu Y, Mulder MP, Geurink PP, Maslen SL, Gersch M, Elliott PR, Burke JE, van Tol BD, Akutsu M, et al. 2016. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature 538: 402–405. 10.1038/nature19836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Solomon E. 2004. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet 13: 807–817. 10.1093/hmg/ddh095 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. 1999. Brca1 controls homology-directed DNA repair. Mol Cell 4: 511–518. 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. 2013. Mixed-linkage ubiquitin chains send mixed messages. Structure 21: 727–740. 10.1016/j.str.2013.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Durocher D. 2013. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol 14: 661–672. 10.1038/nrm3659 [DOI] [PubMed] [Google Scholar]
- Panier S, Ichijima Y, Fradet-Turcotte A, Leung CC, Kaustov L, Arrowsmith CH, Durocher D. 2012. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol Cell 47: 383–395. 10.1016/j.molcel.2012.05.045 [DOI] [PubMed] [Google Scholar]
- Paul A, Wang B. 2017. RNF8- and Ube2S-dependent ubiquitin Lysine 11-linkage modification in response to DNA damage. Mol Cell 66: 458–472.e5. 10.1016/j.molcel.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. 2001. Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533. 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G, Penengo L. 2009. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol Biol 10: 55 10.1186/1471-2199-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L. 2011. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol Cell Biol 31: 118–126. 10.1128/MCB.00818-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202. 10.1126/science.1139516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434. 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Wang B. 2012. BRCA1 tumor suppressor network: focusing on its tail. Cell Biosci 2: 6 10.1186/2045-3701-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Elledge SJ. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci 104: 20759–20763. 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198. 10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hurov K, Hofmann K, Elledge SJ. 2009. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev 23: 729–739. 10.1101/gad.1770309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Jie Z, Joo D, Ordureau A, Liu P, Gan W, Guo J, Zhang J, North BJ, Dai X, et al. 2017. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature 545: 365–369. 10.1038/nature22344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. 2009. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci 106: 18213–18218. 10.1073/pnas.0907887106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, Durocher D. 2016. The structural basis of modified nucleosome recognition by 53BP1. Nature 536: 100–103. 10.1038/nature18951 [DOI] [PubMed] [Google Scholar]
- Wu Q, Paul A, Su D, Mehmood S, Foo TK, Ochi T, Bunting EL, Xia B, Robinson CV, Wang B, et al. 2016. Structure of BRCA1-BRCT/Abraxas complex reveals phosphorylation-dependent BRCT dimerization at DNA damage sites. Mol Cell 61: 434–448. 10.1016/j.molcel.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Wu X, Wu L, Castillo A, Liu J, Atkinson E, Paul A, Su D, Schlacher K, Komatsu Y, et al. 2017. Abro1 maintains genome stability and limits replication stress by protecting replication fork stability. Genes Dev 31: 1469–1482. 10.1101/gad.299172.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.