Abstract
Background
Radiogenomics is the extension of radiomics through the combination of genetic and radiomic data. Because genetic testing remains expensive, invasive, and time-consuming, and thus unavailable for all patients, radiogenomics may play an important role in providing accurate imaging surrogates which are correlated with genetic expression, thereby serving as a substitute for genetic testing.
Main body
In this article, we define the meaning of radiogenomics and the difference between radiomics and radiogenomics. We provide an up-to-date review of the radiomics and radiogenomics literature in oncology, focusing on breast, brain, gynecological, liver, kidney, prostate and lung malignancies. We also discuss the current challenges to radiogenomics analysis.
Conclusion
Radiomics and radiogenomics are promising to increase precision in diagnosis, assessment of prognosis, and prediction of treatment response, providing valuable information for patient care throughout the course of the disease, given that this information is easily obtainable with imaging. Larger prospective studies and standardization will be needed to define relevant imaging biomarkers before they can be implemented into the clinical workflow.
Keywords: Radiomics, Radiogenomics, Molecular profiling, Precision medicine
Keypoints
Current radiomic and radiogenomic studies are limited to few common cancers.
Radiogenomics may provide accurate imaging biomarkers, substituting for genetic testing.
Radiomics/radiogenomics biomarkers may predict risk and outcomes.
Radiomics/radiogenomics biomarkers may be used to personalize treatment options.
Larger prospective studies and standardization are needed to validate radiomics/radiogenomics biomarkers.
Background
Personalized medicine is yielding increasingly precise disease treatments and prevention strategies for groups of individuals based on their genetic makeup, environment, and lifestyle. To enable personalized medicine, more precise and personalized genetic-based approaches (genomics, transcriptomics, proteomics, metabolomics, etc.) are used. In oncology, the goal of using such approaches is to allow more individual-level information rather than population-level or aspecific clinical information (tumor stage, age, gender, etc.) to select the most successful cancer treatment regimen for each patient [1].
Molecular tumor characterization can be performed using genomic and proteomic approaches, but this requires using tissue sampling from invasive surgery or biopsy [2]. However, even when molecular characterization is performed using tissue sampling, samples may not be accurate for the entire lesion as they are often obtained from a small portion of a heterogeneous lesion with inherent selection bias during biopsy [3]. Currently, large-scale genome cancer characterization that would allow genetic testing for every individual is not feasible due to high costs, the considerable time burden, and technically complex data analysis and interpretation [3].
Imaging can provide a more comprehensive view of the tumor in its entirety via radiomics and radiogenomics. Whereas radiomics analysis extracts large volumes of quantitative data from medical images and amalgamates these together with clinical and patient data into mineable shared databases [4–6], radiogenomics is the extension of radiomics through the combination of genetic and radiomic data. Detailed reviews of the process of radiomics analysis (image acquisition, volume of interest selection, segmentation, feature extraction and quantification, database building, classifier modeling, and data sharing) are described in detail by Pinker et al. [7], Gillies et al. [8], Sala et al. [5], and Lambin et al. [4].
Briefly, in radiomics, firstly, a region of interest (ROI) containing either the whole tumor or sub-regions within the tumor is identified from multimodality imaging data. ROIs are then segmented with operator edits and are eventually rendered in three dimensions (3D). High dimension features are extracted from these ROIs that include semantic and agnostic features [8]. Semantic features are morphological features that are commonly used in radiology reports to describe lesions such as size, location, vascularity, spiculation, and necrosis. Agnostic features are more complex mathematically extracted quantitative features which can be divided into first order statistical outputs (which describe distribution of value inside a single voxel), second order statistical outputs (describe interrelations between voxels), and higher order statistical outputs (extract repetitive and non-repetitive patterns within an image trough filter grids).
These features extracted from these rendered volumes generate a report, which is placed in a database along with other data, such as clinical and genomic data such as genes, mutations, and expression patterns.
Radiogenomics entails the correlation between quantitative or qualitative imaging features and the genomic data obtained from analysis of tissue as well as other clinical data, thus allowing the discovery of imaging surrogates that can serve as a substitute for genetic testing. Radiogenomics studies can be either exploratory or hypothesis-driven. Imaging features that are associated with single oncogenic defects of a tumor can be used to support treatment selection and monitoring as well as predict treatment outcomes [3, 5, 9, 10]. Hence, radiogenomics represents a promising novel approach to enable more personalized patient care [3, 4, 8, 9, 11–13].
Main structure
Because genetic testing remains expensive, invasive, and time-consuming, and thus unavailable for all patients, radiogenomics may play an important role in providing accurate imaging surrogates which are correlated with genetic expression, thereby serving as a substitute for genetic testing [9, 13, 14]. These imaging surrogates can be used to predict response to therapy and the potential for early metastasis as well as to personalize treatment options [15–17].
So far, there are numerous genomic and clinical biomarkers identified from various adult cancers that have been collected in The Cancer Genome Atlas (TCGA). These biomarkers have been linked to the corresponding imaging data present in The Cancer Imaging Archive (TCIA) [17–19]. However, due to the lack of image sample registration (i.e., genetic test results cannot be matched to a specific location on imaging), the imaging data in the TCIA is so far limited for clinical use [3]. Given that imaging acquisition protocols are becoming more homogeneous and outcome data more robust, growing publicly available databases including the TCGA and TCIA will become more useful and will allow further radiogenomic studies [3].
Current radiomic and radiogenomic studies are limited to few types of common cancers. In this review, we will present the current data as pertains to radiomics and radiogenomics in glioblastoma multiforme (GBM), non-small cell lung cancer (NSCLC), hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma, breast cancer (BC), prostate cancer, renal cell carcinoma, cervical cancer, and ovarian cancer and discuss their role and possible future applications in oncology.
Brain
GBM remains the most common and the most fatal primary brain tumor in adults [20]. It is characterized by tremendous molecular and genomic heterogeneity, which leads to treatment resistance. Radiomics and radiogenomics research in the brain have thus far focused on GBM.
Through the TCGA, the genomic profile of GBM has been thoroughly assessed, resulting in its division into four distinct molecular subtypes: classical, mesenchymal, proneural, and neural. These subtypes are associated with different outcomes and tumor progression patterns [21, 22]. Another more recent stratification divided GBM into three core pathways according to RTK/RAS/PI [3] K, p53, and RB signaling alterations [23], showing a better correlation with outcomes. GBM magnetic resonance imaging (MRI) data from the TCIA has been matched with genetic data contained in the TCGA, enabling radiogenomics studies.
Using MRI, survival, and genetic data on 92 patients with GBM from the TCGA, Rao et al. [24] showed that a combination of three distinct features (volume-class, hemorrhage, and T1/fluid-attenuated inversion recovery (FLAIR)-envelope ratio) was able to stratify patient survival in a statistically significant manner. Between patients with poorer survival and patients with a better prognosis, there were significant differences in genes and microRNAs regulating invasion and proliferation (median difference of survival of 8 months).
Zinn et al. [25] used microRNA and genetic data of 78 GBMs from the TCGA to identify associations with quantitative MRI FLAIR features from the TCIA. The results suggested that MRI FLAIR can serve as an imaging surrogate for GBMs highly enriched in genes and microRNAs involved in cellular migration/invasion. In both discovery and validation sets, POSTN was the top upregulated gene and was associated with high FLAIR volumes, shorter progression-free survival, and shorter overall survival. POSTN upregulation is thought to induce tumor invasion through epithelial to mesenchymal transformation.
Colen et al. [26] reported in a study of 104 TCGA GBMs with MRI data from the TCIA that three MRI features could predict a worse prognosis: ependymal enhancement (10.6 versus 18.6 months, p = 0.0018), deep white matter tract involvement (10.9 months versus 19.9 months, p < 0.0008), and enhancement across midline (9 months versus 14.3 months (p < 0.0003). Ependymal enhancement and deep white matter tract involvement demonstrated significant association with mitochondrial dysfunction (p < 0.0001), MYC oncogene activation, and NFKBIA inhibition. A year later, using gene expression profiles from the TCGA and MRI data from the TCIA, Colen et al. [27] reported that patients with GBM with little necrotic component on MRI had a high prevalence of X-linked genes, while patients with high volumes of necrosis on MRI had a high prevalence of Y-linked genes. The authors also demonstrated that female patients with GBM characterized by low volumes of necrosis on MRI had a significant survival advantage.
To date, using TCGA/TCIA data or institutional data, tumor volume remains the most common MRI feature extracted from GBM and has been shown to be correlated with genomic data [25, 27–31]. Gutman et al. [31] carried out a volumetric analysis on the MRI of 76 GBMs profiled in the TCGA and found that tumors with TP53 mutation had smaller enhancing and necrotic volumes (p ≤ 0.017) while tumors with RB1 mutation were associated with less edema (p = 0.015). Using institutional data, Diehn et al. [32] found that the ratio of enhancing to non-enhancing volume was significantly correlated with EGFR overexpression (evaluated with immunohistochemistry) (p = 0.019). The enhancing phenotype was correlated with overexpression of angiogenesis and tumor hypoxia-related genes such as VEGF, ADM, and PLAUR (p = 0.012).
Barajas et al. [33] correlated MRI parameters with genetic features (investigated with RNA microarrays) in enhancing vs peritumoral non-enhancing GBM biopsy samples. Results showed that T2 dynamic susceptibility-weighted, perfusion-weighted, and diffusion-weighted measurements were different between biopsy regions and were correlated with histopathologic features of aggressiveness.
A study by Jamshidi et al. [34] in 23 patients with GBM identified associations between genes and MRI features such as contrast-to-necrosis ratio with KLK3 and RUNX3, subventricular zone involvement with Ras oncogenes RAP2 and TYMS, and vasogenic edema with oncogenes FOXP1 and PIK3IP1.
Pope et al. [35] found that incomplete enhancing GBM on MRI was associated with increased levels of the oligodendroglioma markers OLIG2 and ASCL1 compared with completely enhancing imaging GBM. Histopathology confirmed this finding, showing a higher percentage of the oligodendroglioma histologic component in the incomplete enhancing group.
Pope et al. [36] also used magnetic resonance spectroscopy (MRS) in patients with glioma to measure the level of the oncometabolite 2-HG which is associated with mutations of IDH1 and IDH2. Patients with IDH1 mutation are more likely to progress to malignant gliomas. MRS detected elevated 2-HG levels in gliomas with IDH1 mutations compared with those without the mutation (p = 0.003). Tumors with IDH1 mutations showed elevated levels choline (p = 0.01) and decreased levels of glutathione (p = 0.03).
Hu et al. [37] demonstrated correlations between diffusion tensor imaging and dynamic susceptibility contrast perfusion metrics and mutations in EGFR, PDGFRA, PTEN, CDKN2A, RB1, and TP53 (p < 0.03). Accuracies of the predictive models ranged from 37.5% for TP53 to 87.5% for RB1. A similar study [38] involving MRI texture features was able to characterize local EGFR mutation status as well as predict patient survival in 65 GBMs.
In Jain et al. [39], CT perfusion parameters were correlated with angiogenesis-related genes. The authors found that 19 of 92 angiogenesis-related genes were significantly correlated with permeability surface area product and nine genes were significantly correlated with cerebral blood volume.
Breast
Multiple studies have been conducted in the breast with promising results for BC analysis of genomic signatures, molecular subtype characterization, and clinically used recurrence scores. Most have relied mainly on tumor features in dynamic contrast-enhanced (DCE)-MRI, although diffusion-weighted imaging (DWI) has been used, for example, for characterization of molecular subtypes [40–42].
The first radiogenomic MRI study of the breast was published in 2012 by Yamamoto et al. [43] who investigated the potential of radiogenomics to correlate gene expression patterns with DCE-MRI characteristics. They found that 21 of 26 imaging characteristics were significantly associated with 71% of the approximately 52,000 variably expressed genes in BC. The same investigators also conducted another study [44] to examine the relationship between quantitative DCE-MRI imaging phenotypes, early metastasis, and long non-coding RNA expression using RNA sequencing. They found eight long non-coding RNAs that correlated with the enhancing rim fraction score which was in turn associated with early metastasis and poor metastasis-free survival in patients with BC.
Using a radiogenomics approach, Zhu et al. [45] investigated DCE-MRI characteristics (tumor size, shape, and morphology) of 91 BCs and their correlation with genomic features such as protein expression and mutations. Tumor size revealed that larger cancers have upregulated pathways, while blurred tumor margins and irregular shape were associated with more aggressive tumors.
Associations between MRI characteristics and molecular BC subtypes have been investigated in several studies. In a systematic review and meta-analysis published in 2014, Elias et al. [46] reported that higher enhancement within the tumor is associated with the luminal B subtype, while HER2-enriched cancers are more likely to show fast initial enhancement or wash-out kinetics with circumscribed margins. Elsewhere, triple-negative cancers have been associated with high T2 signal intensity and the presence of rim enhancement [47, 48]. In DWI, HER2-enriched tumors showed the highest ADC values, while luminal B/HER2-negative cancers showed the lowest [40–42].
Computer-extracted radiomic features have themselves been associated with molecular BC subtypes. Mazurowski et al. [49] showed that in DCE-MRI, extracted MRI features that relate to an increased ratio of tumor-to-background parenchymal enhancement were associated with HER2-positive cancers. This difference might be due to the increased vascularization found in HER2-positive subtypes mediated by VEGF which leads to increased vessel diameter, vascular permeability, and extracellular fluid. Grimm et al. [50] found correlations between extracted imaging features and luminal A and B breast cancer subtypes. In a more recent study by Grimm et al. [51], non-mass enhancement and qualitative BI-RADS descriptors were evaluated on DCE-MRI in 278 patients with breast cancer; results showed significant correlations between mass shape and basal cancers, mass margin and HER2 cancers, and internal enhancement and luminal B cancers. In another study, Yamaguchi et al. [52] assessed the relationship between the delayed phase of enhancement of DCE-MRI and molecular subtypes, finding that ER-positive and/or PgR-positive and HER2-negative cancers demonstrated less washout. A recent study by Leithner et al. [53] showed that radiomic features extracted from DWI were able to separate breast cancers based on molecular subtype and receptor status with high accuracy (> 90%). Accuracy was superior for radiomics features extracted directly from the apparent diffusion coefficient (ADC) map.
Classifier models using tumor phenotypes to differentiate between molecular subtypes have been evaluated by Li et al. [54] with promising results (Fig. 1). However, in a similar study, Waugh et al. [55] found an accuracy of only 57.2%. More studies need to be conducted in this regard to validate these preliminary findings.
Fig. 1.
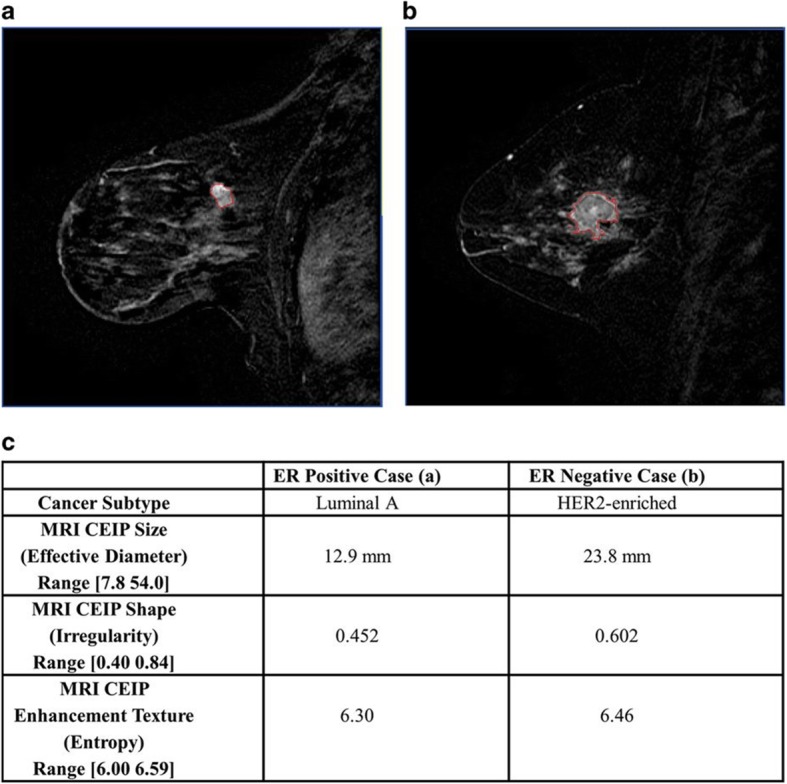
The computer segmentation method in example cases of one estrogen receptor positive tumor and one estrogen receptor negative tumor. The tumor segmentation outlines are shown along with computer-extracted image phenotype (CEIP) values (and ranges) for size, irregularity, and contrast enhancement heterogeneity. Reprinted under a Creative Commons license from: NPJ Breast Cancer. 2016;2. pii: 16012. Epub 2016 May 11. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA, Fan C, Conzen SD, Zuley M, Net JM, Sutton E, Whitman GJ, Morris E, Perou CM, Ji Y, Giger ML
Previous studies have also correlated imaging characteristics with clinically available prognostic genomic assays which provide a clinical score for the risk of recurrence. As Oncotype Dx gene-expression score, MammaPrint, and PAM50 have been shown to predict recurrence in early-stage ER-positive/HER2-negative invasive cancers, these correlations between imaging characteristics and assay recurrence scores could have important implications in patient management. Woodard et al. [56] evaluated BC recurrence in ER-positive patients using OncotypeDx through the association of BI-RADS mammography and MRI features. Indistinct mass margins and fine linear branching calcifications on mammography were associated with a higher recurrence score, while breast density on mammography was inversely associated with the recurrence score. Spiculated mass margins and non-mass enhancement on MRI were associated with a lower recurrence score. Several radiomic imaging models assessing risk of recurrence using several assays have been developed [43–45, 57–65]. Combining imaging and pathology information, Sutton et al. [59] developed a model that correlated with the OncotypeDx Recurrence Score which was predictive of recurrence and therapeutic outcome (Fig. 2). A study by Li et al. [62] evaluated whether computer-extracted imaging phenotypes could predict cancer recurrence using MammaPrint, OncotypeDx, and PAM50/Prosigna. They found significant correlations between tumor size, heterogeneity, and higher risk for recurrence (Figs. 3 and 4). In this and other studies using radiomics computer-extraction methods [57, 62, 63], enhancement heterogeneity has been related to a high risk of recurrence in tumor assays. In two studies [57, 59], rapid contrast uptake predicted high-risk Oncotype Dx.
Fig. 2.
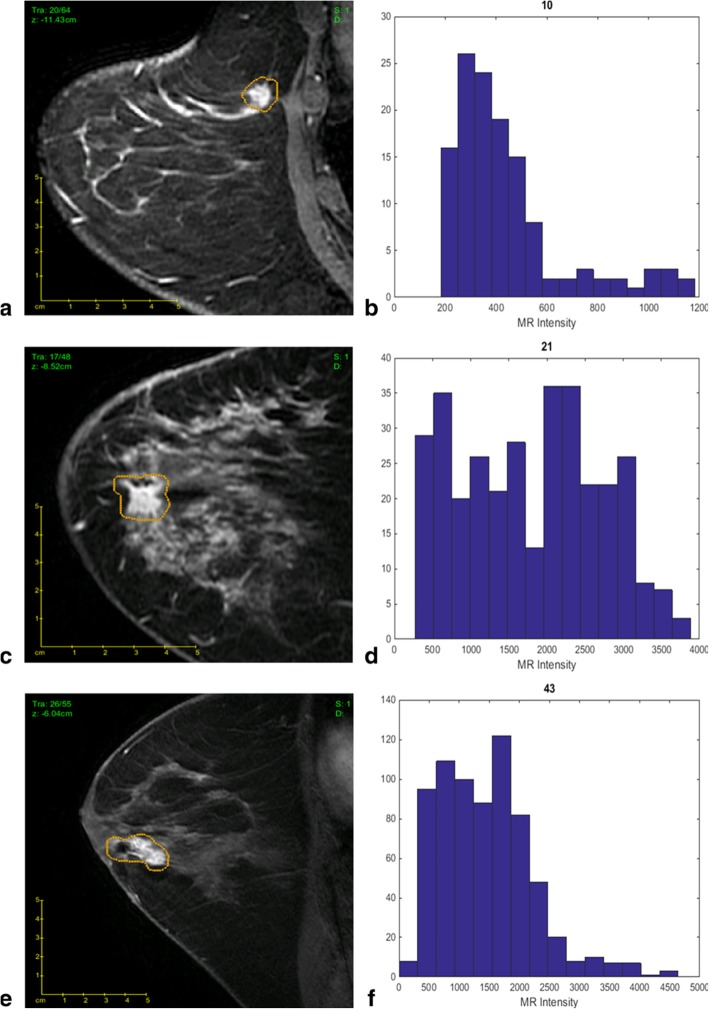
The best-fit linear regression model allows imaging features to differentiate tumors with different Oncotype Dx Recurrence Score (ODxRS). a Sagittal T1-weighted fat-suppressed post-contrast MRI of an invasive ductal nuclear grade 1 carcinoma with an ODxRS of 10 and (b) corresponding kurtosis histogram, which demonstrates the frequency of MR intensity. c Sagittal T1-weighted fat-suppressed postcontrast MRI of an invasive ductal nuclear grade 2 carcinoma with an ODxRS of 21 and (d) corresponding kurtosis histogram. e Sagittal T1-weighted fat-suppressed postcontrast MRI of an invasive ductal nuclear grade 3 carcinoma with an ODxRS of 43 and (f) corresponding kurtosis histogram. Reprinted with permission from: J Magn Reson Imaging. 2015 Nov;42 [5]:1398–406. doi: 10.1002/jmri.24890 Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. Sutton EJ, Oh JH, Dashevsky BZ, Veeraraghavan H, Apte AP, Thakur SB, Deasy JO, Morris EA.
Fig. 3.
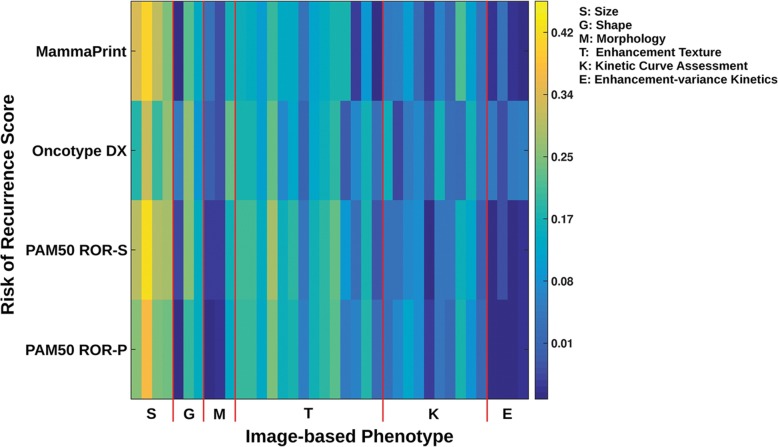
Correlation heat map based on univariate linear regression analysis between each individual MR imaging phenotype and the recurrence predictor models of MammoPrint, Oncotype DX, PAM50 ROR-S, and PAM50 ROR-P. In this color scale, yellow indicates higher correlation as compared with blue and the different gene assays served as the “reference standard” in this study. Some phenotypes correlate similarly (i.e., similar color on the color scale) across the risk estimate models, while others do not. Reprinted with permission from: Radiology. 2016 Nov;281 [2]:382–391. Epub 2016 May 5. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, Conzen SD, Whitman GJ, Sutton EJ, Net JM, Ganott M, Huang E, Morris EA, Perou CM, Ji Y, Giger ML
Fig. 4.
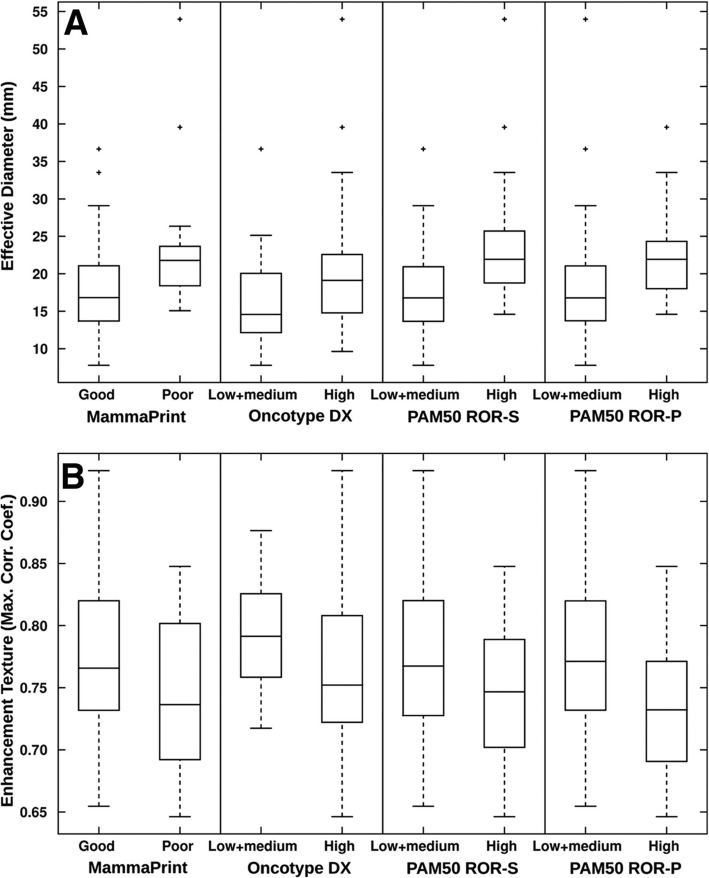
Box and whisker plots show the relationship of the MRI based phenotypes of (a) size (effective diameter) and (b) enhancement texture (maximum correlation coefficient) with the recurrence predictor models of MammaPrint, Oncotype DX, PAM50 ROR-S, and PAM50 ROR-P. A positive correlation between the selected MR imaging phenotypes of size (effective diameter) and negative correlation with enhancement texture (maximum correlation coefficient) and increasing levels of risk of recurrence for MammaPrint, Oncotype DX, PAM50 ROR-S, and PAM50 ROR-P were observed. A low value of this enhancement texture feature indicates a more heterogeneous enhancement pattern. Reprinted with permission from: Radiology. 2016 Nov;281 [2]:382–391. Epub 2016 May 5. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, Conzen SD, Whitman GJ, Sutton EJ, Net JM, Ganott M, Huang E, Morris EA, Perou CM, Ji Y, Giger ML
There is potential for radiogenomics studies regarding MR perfusion characteristics and genetic expression. One previous study [65] using genomics analysis has associated MR perfusion parameters with early metastasis or with differential gene expression when monitoring anti-VEGF treatment.
Yamamoto et al. [43] evaluated a qualitative imaging model including tumor heterogeneity and enhancement for prediction of expression of immune-response genes, and high-level analysis revealed 21 imaging traits that were globally significantly correlated with 71% of the total genes (3717/5231 genes) measured in BC patients.
Gynecological tumors
To date, radiomics and radiogenomics have been applied in cervical cancer and in high grade serous ovarian cancer, providing valuable prognostic information.
DCE-MRI pharmacokinetic parameters have been associated with both genetic and outcome data in patients with cervical cancer. In Norway, Halle et al. [66] conducted an assessment of pre-treatment DCE-MRI pharmacokinetic parameters with global gene expression data in 78 patients who underwent chemotherapy. Through gene set analysis of 42/78 tumors, they found that the ABrix parameter correlated with hypoxia gene sets. Through immunohistochemistry analysis of the remaining 32/78 tumors, they found that a low ABrix was associated with upregulated HIF1α protein expression. A DCE-MRI hypoxia gene signature consisting of 31 hypoxia genes upregulated in tumors with low ABrix was constructed, showing prognostic value in an independent cohort of 109 patients. As reported by same group of authors [67] a year later, several pharmacokinetic parameters, also derived from pre-treatment DCE-MRI in 78 patients with cervical cancer who underwent chemotherapy, may be used to identify patients at risk of treatment failure: ABrix and Ktrans were associated with poor clinical outcome, while patients with high Kel had longer survival.
The TCGA Research Network previously introduced the Classification of Ovarian Cancer (CLOVAR) describing four prognostic genomic subtypes: differentiated, immunoreactive, mesenchymal, and proliferative [68, 69]. In a hypothesis-generating single-institution study, Vargas et al. [70] conducted a retrospective radiogenomics study of 46 patients with stage IIIC or IV high grade serous ovarian cancer. They reported that preoperative CT features evaluated by two radiologists were associated with the CLOVAR genomic subtypes of high grade serous ovarian cancer and were predictive of survival. Specifically, the presence of mesenteric infiltration and diffuse peritoneal involvement at baseline on CT were associated with CLOVAR mesenchymal subtype. The presence of mesenteric infiltration was also shown to provide important prognostic information as it correlated with shorter progression-free survival and overall survival. To validate the preliminary findings from this single-institution study, a multi-institutional study was then conducted by Vargas et al. [71] using TCIA CT images of 92 patients with high-grade serous ovarian cancer. The relationship between CT features and time-to-disease progression and CLOVAR profiles were assessed. The presence of peritoneal disease in the right upper quadrant, supradiaphragmatic lymphadenopathy, multiple peritoneal disease sites, and non-visualization of a discrete ovarian mass were associated with a shorter time-to-disease progression. The presence of multiple peritoneal disease sites and the presence of disease within the pouch of Douglas were associated with the mesenchymal subtype which has the worst prognosis among CLOVAR subtypes.
In another study by Vargas et al. [72], the authors used radiomics to derive inter-site spatial heterogeneity metrics across multiple metastatic lesions from preoperative CT of 38 patients with stage IIIC–IV high grade serous ovarian cancer. Several of these metrics were associated with the amplification of CCNE1, as well as a shorter overall survival and incomplete surgical resection. Previously, CCNE1 amplification itself has been associated with higher chemoresistance [73] and treatment failure [74].
Liver
HCC, the most common primary liver cancer, can be managed with a variety of treatment approaches such as embolization, radiation, surgery, and pharmacological therapy [7]. Currently, the approach is selected based on only the number and size of liver lesions [7]. In this context, radiomics and radiogenomics can serve an important role in helping to select the best clinical management for each patient [7]. The studies below indicate that they are particularly promising to allow for a better prediction of prognosis or treatment response.
The presence of microscopic venous invasion (MVI) is a well-established parameter associated with poor prognosis in HCC but its diagnosis is difficult with only conventional imaging [75, 76]. Pathologic examination of the explanted tissue after surgery is currently the only way to diagnose MVI. Since 2002, several groups have addressed this issue. Chen et al. [77] found a correlation between the presence of MVI and a 91-gene expression signature assessed through microarray analysis. Later, Segal et al. [78] identified two imaging features on CT—the presence of “internal arteries” and absence of “hypodense halos”— that were associated with these same 91 genes. In another study by Banerjee et al. [79] in 157 patients with HCC who eventually underwent surgical resection, these two imaging features along with “tumor–liver difference” were shown to be both highly predictive of the presence of histological MVI as well as associated with early disease recurrence and poor overall survival, thus showing that these features may be helpful to select candidates who will benefit less from surgical treatment or liver transplant. On MRI, rather than CT, Renzulli et al. [80] found that features including peritumoral enhancement and non-smooth margins were shown to be promising for the prediction of pathologic MVI in 125 patients with 140 nodules diagnosed with HCC.
Taouli et al. [81] found significant correlations between certain imaging traits in contrast-enhanced CT (26 patients) or MRI (12 patients) and gene signatures of aggressive HCC phenotype, with an infiltrative pattern on imaging having the highest number of positive associations. Although no correlations were found between gene expression signatures and enhancement ratios in this study, they proposed that their findings should be validated with DCE-MRI in the future to overcome the inherent limitations to measurements of tumoral enhancement on arterial and portal venous phases alone as used in their study.
When using multiparametric MRI, Hectors et al. [82] did not find differences among genetic subclasses in 14 patients, and none of the parameters could distinguish between HCC grades. However, they found significant correlations between several MRI parameters and individual gene expression levels, e.g., poor tumor perfusion on DCE-MRI correlated with high expression of VEGF-A. Multiparametric MRI with quantitative parameters is potentially a powerful tool for radiomics and radiogenomics study of HCC but the challenges to the reproducibility of advanced MRI techniques including DCE-MRI, DWI, and BOLD imaging in the liver presents a handicap.
Intrahepatic cholangiocarcinoma (ICC) is a less common tumor in the liver; therefore, only a few studies have been published to evaluate the relationship of ICC imaging features with genetic data. Sadot et al. [83] used texture analysis on the pre-treatment CT of 25 ICC patients, comparing the extracted quantitative and qualitative imaging phenotypes with hypoxia biomarkers such as HIF-1α, VEGF, EGFR, and CD24 measured in pre-treatment biopsies. Qualitative variables, including “tumor liver difference” and “attenuation heterogeneity,” were found to be correlated with VEGF expression, while CD24 expression was correlated with biliary dilatation.
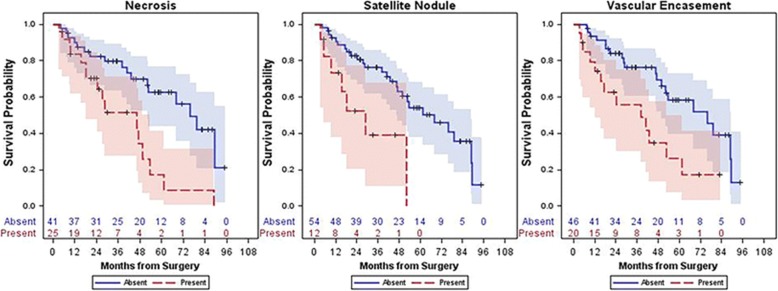
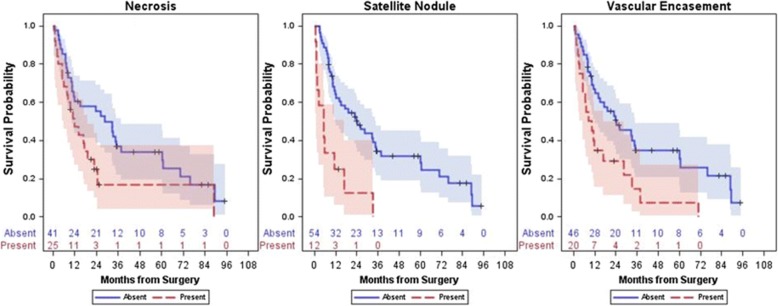
In 2011, Kim et al. [84] demonstrated that ICC enhancement in the arterial phase on CT was associated with fewer necrotic areas and longer disease-free survival after surgical resection. In 2017, Fujita et al. reached a similar in their study involving 47 ICC patients who underwent arterial phase CT. The 47 patients were divided into three groups: hypovascular, rim-enhancing, and hypervascular. Patients with hypovascular tumors had more instances of lymphatic, biliary, and perineural invasion and poorer disease-free survival than patients with hypervascular or rim-enhancing tumors [85]. More recently, Aherne et al. [86] identified a strong correlation between necrosis or vascular encasement on CT and decreased overall survival. A link was also identified between larger tumor sizes (or the presence of satellite nodules) and a reduction in progression-free survival (Figs. 5 and 6). This study also assessed associations between CT imaging features and genetic pathways (IDH1, chromatin, and RAS-MAPK) but did not find any significant associations.
Fig. 5.
Kaplan–Meier survival curves which illustrate that the presence of necrosis, satellite nodules, and vascular encasement were all associated with decreased survival. Reprinted with permission from: Aherne EA, Pak LM, Goldman DA, Gonen M, Jarnagin WR, Simpson AL, and Do RK. Intrahepatic cholangiocarcinoma: can imaging phenotypes predict survival and tumor genetics? Abdom Radiol, 2018, 43 [10]:2665
Fig. 6.
Kaplan–Meier survival curves which illustrate that the presence of satellite nodules and vascular encasement were associated with decreased disease-free survival. Reprinted with permission from: Aherne EA, Pak LM, Goldman DA, Gonen M, Jarnagin WR, Simpson AL, and Do RK. Intrahepatic cholangiocarcinoma: can imaging phenotypes predict survival and tumor genetics? Abdom Radiol, 2018, 43 [10]:2665
Kidney
The recent advances in genetics have led to the discovery of multiple mutations or genetic alterations in clear cell renal cell carcinomas (RCCs), including mutations or alterations of the genes encoding polybromo-1 protein (PBRM1), BRCA1-associated protein 1 (BAP1), SET domain containing 2 enzyme (SETD2), and lysine-specific demethylase 5C (KDM5C) [87–91], resulting in increased interest in the use of radiogenomics for assessing clear cell RCCs [92–95].
Even though the most common and well-known mutation identified in clear cell RCCs is the VHL tumor suppressor gene, it has no prognostic or predictive value in patients with clear cell RCC [96]. The second most commonly identified mutation in clear cell RCC is the PBRM1 tumor suppressor gene [92, 97–99]. A recent meta-analysis [100] of 2942 RCC patients from seven studies reported that a mutation in or decreased expression of PBRM1 is associated with poor survival, advanced TNM categories, advanced tumor stage, and a higher Fuhrman nuclear grade. A study by Kocak et al. [101] suggested that high-dimensional CT texture analysis is promising to distinguish clear cell RCCs with PBRM1 mutation and those without PBRM1 mutation.
Karlo et al. [93] conducted a preliminary radiogenomics study in 233 patients with clear cell RCC which showed associations between CT features and underlying mutations in several genes (VHL, PBRM1, BAP1, SETD2, and KDM5C). Well-defined tumor margins, nodular tumor enhancement, and intratumoral vascularization were associated with VHL mutations, while renal vein invasion was significantly associated with KDM5C and BAP1 mutations. Mutations of VHL and PBRM1 were more common in solid than in cystic tumors.
Using TCGA and TCIA data, Shinagare et al. [94] conducted a study of 103 clear cell RCCs (81 cases were evaluated with CT, 19 cases with MRI, and 3 cases with CT and MRI), showing that ill-defined tumor margins and calcifications were associated with BAP1 mutation.
Several studies have shown that KDM5C mutation is associated with decreased survival in patients with clear cell RCC, though paradoxically it is associated with prolonged survival in patients with metastatic disease [102–105].
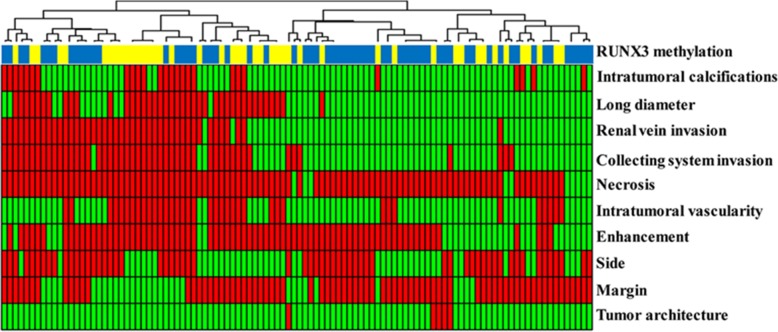
Apart from associating imaging features with genetic mutations, they can also be associated with biomarkers such as DNA methylation, which is known to play a key role in cancer development and has potential prognostic and diagnostic value [106–109]. DNA methylation in tumor suppressor genes including RUNX3 negatively impacts survival in some cancers [110–115]. An exploratory study by Cen et al. [116] divided RCC patients into high RUNX3 methylation and low RUNX3 methylation groups. Higher levels of RUNX3 methylation were linked to decreased survival time (Fig. 7). Independent imaging predictors of high methylation of RUNX3 such as ill-defined margins, high intratumoral vascularity, and left laterality of the lesions are shown in Fig. 8.
Fig. 7.
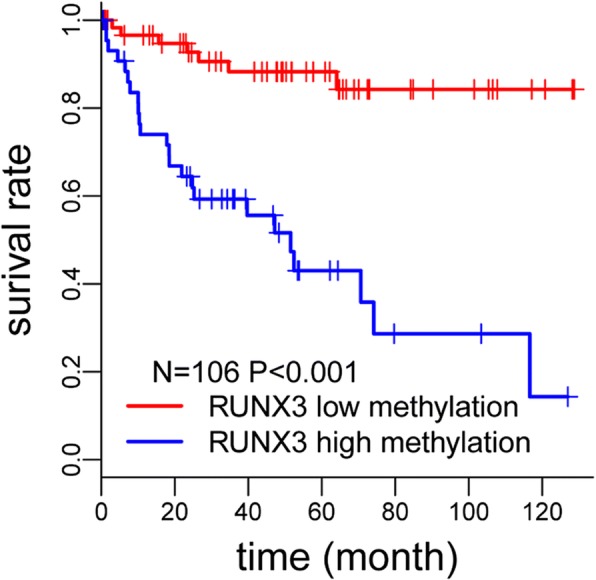
Univariate Cox regression analysis of prognostic factors for overall survival is summarized. Reprinted with permission from: Cen et al. [116] PubMed PMID: 30877466
Fig. 8.
Hierarchical clustering yielded distinct groups of RUNX3 promoter methylation status and CT features. Red positive, green negative. Reprinted with permission from: Cen et al. [116] PubMed PMID: 30877466
To build a radiogenomic risk score (RRS), a study by Jamshidi et al. [117] combined clinical data, genetic data extracted from genomic analysis, and preoperative CT data of clear cell RCC. Survival analysis confirmed that the high-RRS group had significantly lower disease-specific survival rates than the low RRS, independent of disease stage and grade.
Lung
In the past decades, several subtypes of lung carcinoma harboring specific mutations (most notably EGFR, KRAS, and ALK mutations) have been identified, allowing for the development of therapies specifically targeting the mutated pathway [118].
The NSCLC lung cancer subtype is the leading cause of cancer death, accounting for more than 85% of all lung cancer cases. Genetic expression data of NSCLC is abundantly available in public databases. Approximately 15% of all NSCLCs in patients from European ethnicities and 50% of NSCLCs in never-smokers are EGFR-positive. The most frequent EGFR mutations (sensitizing activating mutations) are associated with tumor sensitivity to EGFR tyrosine kinase inhibitors (gefitinib, erlotinib, and afatinib) [119]. ALK mutations are less common (< 7% of all NSCLCs) and are more frequent in never/former smokers. Crizotinib was the first drug approved for NSCLC harboring ALK rearrangements, while ceritinib and alectinib have been approved only in the USA and Japan [118, 120, 121]. Approximately 20–25% of NSCLC harbor KRAS mutations and are associated with smoking and adenocarcinoma histology [122]. KRAS has been associated with poor response to standard treatments. Therapeutic strategies involving KRAS are currently under research [123].
Contradictory results have been found when considering the imaging features of different lung cancer subtypes. Glynn et al. did not find an association between CT imaging and EGFR- or KRAS-positive tumors [124]. Others, however, found associations. Lee et al. [125], using CT and 18-FDG PET/CT, demonstrated that tumors > 2.4 cm in diameter, an uptake of > 5.0, or ground glass opacity proportion of ≤ 50% within the lesion were associated with EGFR overexpression. The same group of authors [126] also showed that tumor morphology on CT could differentiate between the two most common subtypes of EGFR mutation, exon 19 deletions, and exon 21 mis-sense mutations; specifically, the ground glass portion of lesions were higher in lesions with exon 21 missense mutations compared with both wild type lesions and tumors with exon 19 deletions. Yano et al. [127] suggested an association between the size of a lesions ground glass component and the likelihood of an EGFR mutation [127].
In a study by Rizzo et al. [128], EGFR mutation was shown to be associated with CT features such as the presence of air bronchogram, pleural retraction, small lesion size, and absence of fibrosis, whereas ALK mutation was associated with pleural effusion. Round shape, nodules in non-tumor lobes, and smoking were variables linked to KRAS mutation. For this study, ALK gene rearrangement was detected by fluorescence in situ hybridization while EGFR and KRAS mutations were evaluated with DNA amplification of exons 18 through 21 and exon 2 and 3 respectively.
A study by Halpenny [129] showed that ALK-positive tumors tended to be bigger with a more solid consistency and involved more thoracic lymphadenopathy. This study showed no correlation between ALK mutation and presence of pleural effusion on CT.
Other, less frequent mutations such as RET and ROS1, which comprise 1–2% of all lung adenocarcinomas, have also been assessed for associations with imaging features. A study by Plodkowski et al. [130] included a group of patients with pathologically confirmed lung adenocarcinomas of any stage with a RET or ROS1 rearrangement tested via fluorescence in-situ hybridization or next-generation sequencing and a control group of EGFR-positive lung cancers. CT features such as the presence of an effusion, lung metastases, adenopathy, and extra-thoracic disease were recorded. Peripheral tumors were seen most likely in patients with ROS1 rearrangements (65% vs 32% in EGFR-positive cancers, p = 0.04). Solid tumors with spiculation were most likely RET- and ROS1-rearranged and EGFR-mutant, and these rarely presented with cavitations or calcifications.
Nair et al. [131] have examined differential genome wide expression across varying FDG uptake levels in patients with NSCLC to identify individual genes and gene expression signatures associated with prognostically relevant FDG uptake features. Their analysis suggested an existing correlation between NFΚB signaling and FDG uptake. NFΚB signaling seems to be increased in inflammatory and malignant conditions since it is enhanced by lactate as a result of glycolysis [132].
Prostate
Prostate cancer is the most common cancer in men in the western world [133, 134]. However, only some prostate cancers exhibit a particularly more aggressive behavior, where they tend to metastasize and resist treatment [135]. Early detection of the more aggressive prostate cancers is important to optimally manage this subgroup [7]. To date, prostate risk stratification relies on clinical examination, biopsy data (e.g., Gleason grade), and serological markers (e.g., prostate-specific antigen) [136]. Genomic [137] and quantitative MRI-derived imaging biomarkers [138] have been shown to be helpful, not only in risk stratification, but also in detection [139, 140], staging [141], characterization [142–145], and treatment planning and follow-up [146, 147], thus setting the stage for radiogenomic studies.
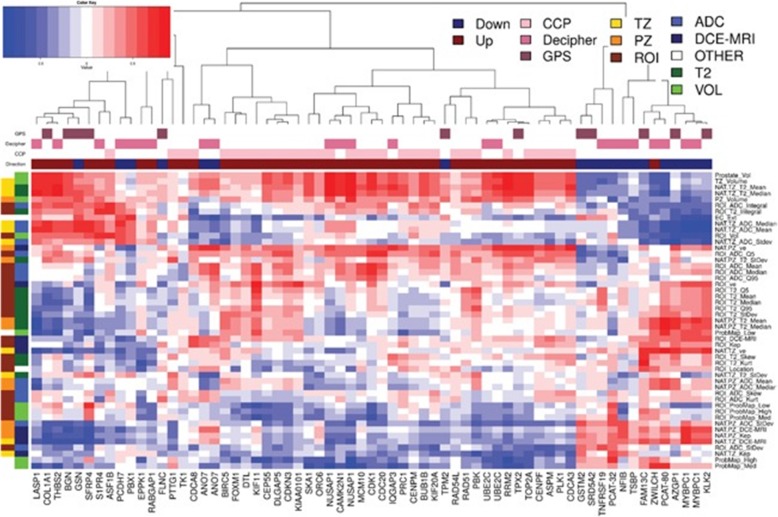
Stoyanova et al. [15] hypothesized that radiomic features may be used to characterize imaging phenotypes or habitats in the prostate, thereby improving risk stratification. Distinct habitats on multiparametric MRI analysis were identified based on reduced diffusion and increased perfusion. From radiogenomic analysis, they reported that several radiomic imaging features were significantly associated with genes related to aggressive behavior (Fig. 9), and ADC values in particular were the most strongly associated with distinct biological processes.
Fig. 9.
Pearson’s correlation analysis of imaging features and 65 genes from commercially available prostate cancer classifiers. Hierarchical clustering on Pearson’s correlation distance between radiomic features and genes from commercially available prostate cancer classifiers: CCP (Cell Cycle Progression), Decipher and GPS (Genomic Prostate Score). Genes in these signatures that are up-expressed in aggressive cancers are indicated by a dark red box over the gene’s column while those that are down-expressed are indicated with a blue box. Groups of radiomic features are indicated along the dendrogram on the left. Group1 (left) connects the radiomic feature with location (TZ, PZ and ROI); group 2 is related to the image modality/function: T2w, ADC, and DCE-MRI. Reprinted under a creative commons license from: Buerki C, Castillo R, Jorda M, Ashab HA, Kryvenko ON, Punnen S, Parekh D, Abramowitz MC, Gillies RJ, Davicioni E, Erho N, Ishkanian A. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostatebiopsies. Oncotarget. 2016 Aug 16;7 [33]:53362–53,376. doi: 10.18632/oncotarget.10523
Renard-Penna et al. [16] correlated MRI features (Prostate Imaging Reporting and Data System (PI-RADS) scores, lesion diameter, and mean ADC values) in 106 suspicious lesions with the Prolaris® Cell Cycle Progression score, finding a significant correlation between PI-RADS and CCP scores (ρ = 0.26, p = 0.007).
McCann et al. [148] correlated MRI features from 45 peripheral zone prostate cancer lesions in 30 patients with PTEN expression on prostatectomy specimens. They found only a weak correlation between kep and PTEN expression (r = − 0.35, p = 0.02).
Jamshidi et al. [149] conducted a study whereby spatial mapping between pre-operative multiparametric MRI and the resected prostate gland was performed. Whole-exome DNA sequencing data was performed on multiple regions of interest representing abnormal vs. abnormal regions. MRI features identified high-grade lesions correctly in all patients. However, there were no significant differences in mutation profiles between histopathologically normal tissue, high-grade prostate cancer, MRI-normal, and MRI-suspicious regions (p = 0.3). The authors suggested that the baseline mutation spectrum among non-cancerous tissue within prostate may be wider than hypothesized.
Challenges
Radiogenomics is an emerging field that correlates tumor genotypes with imaging phenotypes. Over the past 10 years, numerous studies have been published on radiogenomics of various cancers, yet the implementation of radiogenomic in clinical practice is still not routinely done. This is due to several limitations associated with radiogenomic analysis.
Gene expression and signaling pathways are extremely complex, and it is difficult to match the large amount of data from whole-genome sequencing with imaging data (in literature there are only few studies that use the whole genome data). The dimensionality of genomic data should be reduced to match that obtained from imaging studies.
Differences in quantitative imaging features are not only related to gene expression but can also be related to other factors such as patient characteristics or imaging technique. Inter- and intra-institutional heterogeneity of datasets due to different hardware and scan protocols limits the generalizability of results. The limitations associated with interobserver variation make qualitative imaging features even less preferable.
Another factor limiting generalizability and reproducibility of the results stems from the often-small patient cohort and the retrospective nature of radiogenomic studies.
Larger prospective studies and standardization will be necessary to validate the potential of radiogenomics, define relevant imaging biomarkers, and define which radiogenomics associations can be meaningfully implemented in the clinical routine.
Conclusion
Radiomics and radiogenomics are promising to increase precision in diagnosis, assessment of prognosis, and prediction of treatment response, providing valuable information for patient care throughout the course of the disease, given that this information is easily obtainable with imaging. With personal medicine playing an increasing role in clinical practice, radiogenomics in particular can allow fast and non-invasive genotype identification and be applied to all cancer types. Further research in radiogenomics should be conducted to obtain larger datasets with more accurate information for the standardization to provide meaningful and clinically applicable results, and standardization will be necessary to validate the potential of radiogenomics and to define relevant imaging biomarkers before they can be implemented into the clinical workflow.
Acknowledgments
The authors gratefully acknowledge the support in manuscript writing and editing from Joanne Chin.
Abbreviations
- ADC
Apparent diffusion coefficient
- BC
Breast cancer
- CCP
Cell Cycle Progression
- CLOVAR
Classification of Ovarian Cancer
- CT
Computed tomography
- DCE
Dynamic contrast-enhanced
- DSC
Dynamic susceptibility contrast
- DTI
Diffusion tensor imaging
- DWI
Diffusion-weighted imaging
- FLAIR
Fluid-attenuated inversion recovery
- GBM
Glioblastoma multiforme
- HCC
Hepatocellular carcinoma
- ICC
Intrahepatic cholangiocarcinoma
- MRI
Magnetic resonance imaging
- MVI
Microvascular invasion
- PI-RADS
Prostate Imaging Reporting and Data System
- RCC
Renal cell carcinoma
- ROI
Region of interest
- RRS
Radiogenomic risk score
- TCGA
The Cancer Genome Atlas
- TCIA
The Cancer Imaging Archive
- TTP
Time-to-disease progression
Authors’ contributions
RLG performed the literature review. RLG and DL drafted the manuscript. EAM and KP read and edited the manuscript. All authors approved the final manuscript.
Funding
This study received funding from the NIH/NCI Cancer Center Support Grant (P30 CA008748), the Breast Cancer Research Foundation, Susan G. Komen, Spanish Foundation Alfonso Martin Escudero, the European School of Radiology, the Jubilaumsfonds of the Austrian National Bank, the 2020 - Research and Innovation Framework Programme PHC-11-2015 # 667211-2, and the H2020-FETOPEN-2018-2019-2020-01 # 828978. The funding sources had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript should be declared.
Availability of data and materials
Not applicable
Ethics approval and consent to participate
Not applicable
Competing interests
Katja Pinker received payment for activities not related to the present article including lectures including service on speakers bureaus and for travel/accommodations/meeting expenses unrelated to activities listed from the European Society of Breast Imaging (MRI educational course, annual scientific meeting) and the IDKD 2019 (educational course). Elizabeth A Morris has received a grant from GRAIL Inc. The rest of the authors declare no potential competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.El Naqa I, Napel S, Zaidi H (2018) Radiogenomics is the future of treatment response assessment in clinical oncology Med Phys 45(10):4325–4328 [DOI] [PMC free article] [PubMed]
- 2.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70(2):232–241. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 3.Bai HX, Lee AM, Yang L et al (2016) Imaging genomics in cancer research: limitations and promises. Br J Radiol 89(1061):20151030 [DOI] [PMC free article] [PubMed]
- 4.Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446 [DOI] [PMC free article] [PubMed]
- 5.Sala E, Mema E, Himoto Y et al (2017) Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol 72(1):3–10 [DOI] [PMC free article] [PubMed]
- 6.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, et al. Background, current role, and potential applications of radiogenomics. J Magn Reson Imaging JMRI. 2018;47(3):604–620. doi: 10.1002/jmri.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278(2):563–577 [DOI] [PMC free article] [PubMed]
- 9.Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol: JACR. 2015;12(8):862–866. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 10.European Society of Radiology (ESR) Medical imaging in personalised medicine: a white paper of the research committee of the European Society of Radiology (ESR) Insights imaging. 2015;6(2):141–155. doi: 10.1007/s13244-015-0394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold CJ, Lewin JS, Wibmer AG, Thrall JH, Krestin GP, Dixon AK, et al. Imaging in the age of precision medicine: summary of the Proceedings of the 10th Biannual Symposium of the International Society for Strategic Studies in radiology. Radiology. 2016;279(1):226–238. doi: 10.1148/radiol.2015150709. [DOI] [PubMed] [Google Scholar]
- 12.Thrall JH. Moreton lecture: imaging in the age of precision medicine. J Am Coll Radiol. 2015;12(10):1106–1111. doi: 10.1016/j.jacr.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Kuo MD, Jamshidi N. Behind the numbers: decoding molecular phenotypes with radiogenomics--guiding principles and technical considerations. Radiology. 2014;270(2):320–325. doi: 10.1148/radiol.13132195. [DOI] [PubMed] [Google Scholar]
- 14.Bigos KL, Weinberger DR. Imaging genetics--days of future past. Neuroimage. 2010;53(3):804–809. doi: 10.1016/j.neuroimage.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 15.Stoyanova R, Pollack A, Takhar M, Lynne C, Parra N, Lam LL, et al. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies. Oncotarget. 2016;7(33):53362–53376. doi: 10.18632/oncotarget.10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renard-Penna R, Cancel-Tassin G, Comperat E, Varinot J, Leon P, Roupret M, et al. Multiparametric magnetic resonance imaging predicts postoperative pathology but misses aggressive prostate cancers as assessed by cell cycle progression score. J Urol. 2015;194(6):1617–1623. doi: 10.1016/j.juro.2015.06.107. [DOI] [PubMed] [Google Scholar]
- 17.Mehta S, Shelling A, Muthukaruppan A, Lasham A, Blenkiron C, Laking G, et al. Predictive and prognostic molecular markers for cancer medicine. Ther Adv Med Oncol. 2010;2(2):125–148. doi: 10.1177/1758834009360519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Cancer Institute, National Human Genome Research Institute (2014) The Cancer Genome Atlas (TCGA)
- 19.National Cancer Institute Cancer Imaging Program Frederick National Laboratory for Cancer Research (2013) The Cancer Imaging Archive (TCIA) Available from: http://cancerimagingarchive.net/
- 20.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao A, Rao G, Gutman DA, Flanders AE, Hwang SN, Rubin DL, et al. A combinatorial radiographic phenotype may stratify patient survival and be associated with invasion and proliferation characteristics in glioblastoma. J Neurosurg. 2016;124(4):1008–1017. doi: 10.3171/2015.4.JNS142732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zinn PO, Mahajan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One. 2011;6(10):e25451. doi: 10.1371/journal.pone.0025451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colen RR, Vangel M, Wang J, Gutman DA, Hwang SN, Wintermark M, et al. Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project. BMC Med Genomics. 2014;7:30. doi: 10.1186/1755-8794-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colen RR, Wang J, Singh SK, Gutman DA, Zinn PO. Glioblastoma: imaging genomic mapping reveals sex-specific oncogenic associations of cell death. Radiology. 2015;275(1):215–227. doi: 10.1148/radiol.14141800. [DOI] [PubMed] [Google Scholar]
- 28.Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273(1):168–174. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zinn PO, Sathyan P, Mahajan B, Bruyere J, Hegi M, Majumder S, et al. A novel volume-age-KPS (VAK) glioblastoma classification identifies a prognostic cognate microRNA-gene signature. PLoS One. 2012;7(8):e41522. doi: 10.1371/journal.pone.0041522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naeini KM, Pope WB, Cloughesy TF, Harris RJ, Lai A, Eskin A, et al. Identifying the mesenchymal molecular subtype of glioblastoma using quantitative volumetric analysis of anatomic magnetic resonance images. Neuro Oncol. 2013;15(5):626–634. doi: 10.1093/neuonc/not008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutman DA, Dunn WD, Jr, Grossmann P, Cooper LA, Holder CA, Ligon KL, et al. Somatic mutations associated with MRI-derived volumetric features in glioblastoma. Neuroradiology. 2015;57(12):1227–1237. doi: 10.1007/s00234-015-1576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105(13):5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barajas RF, Jr, Hodgson JG, Chang JS, Vandenberg SR, Yeh RF, Parsa AT, et al. Glioblastoma multiforme regional genetic and cellular expression patterns: influence on anatomic and physiologic MR imaging. Radiology. 2010;254(2):564–576. doi: 10.1148/radiol.09090663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamshidi N, Diehn M, Bredel M, Kuo MD. Illuminating radiogenomic characteristics of glioblastoma multiforme through integration of MR imaging, messenger RNA expression, and DNA copy number variation. Radiology. 2014;270(1):1–2. doi: 10.1148/radiol.13130078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pope WB, Chen JH, Dong J, Carlson MR, Perlina A, Cloughesy TF, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology. 2008;249(1):268–277. doi: 10.1148/radiol.2491072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu LS, Ning S, Eschbacher JM, Baxter LC, Gaw N, Ranjbar S, et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol. 2017;19(1):128–137. doi: 10.1093/neuonc/now135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Narang S, Martinez JJ, Rao G, Rao A. Associating spatial diversity features of radiologically defined tumor habitats with epidermal growth factor receptor driver status and 12-month survival in glioblastoma: methods and preliminary investigation. J Med Imaging (Bellingham) 2015;2(4):041006. doi: 10.1117/1.JMI.2.4.041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain R, Poisson L, Narang J, Scarpace L, Rosenblum ML, Rempel S, et al. Correlation of perfusion parameters with genes related to angiogenesis regulation in glioblastoma: a feasibility study. AJNR Am J Neuroradiol. 2012;33(7):1343–1348. doi: 10.3174/ajnr.A2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim EJ, Kim SH, Park GE, Kang BJ, Song BJ, Kim YJ, et al. Histogram analysis of apparent diffusion coefficient at 3.0t: correlation with prognostic factors and subtypes of invasive ductal carcinoma. J Magn Reson Imaging. 2015;42(6):1666–1678. doi: 10.1002/jmri.24934. [DOI] [PubMed] [Google Scholar]
- 41.Martincich L, Deantoni V, Bertotto I, Redana S, Kubatzki F, Sarotto I, et al. Correlations between diffusion-weighted imaging and breast cancer biomarkers. Eur Radiol. 2012;22(7):1519–1528. doi: 10.1007/s00330-012-2403-8. [DOI] [PubMed] [Google Scholar]
- 42.Park SH, Choi HY, Hahn SY. Correlations between apparent diffusion coefficient values of invasive ductal carcinoma and pathologic factors on diffusion-weighted MRI at 3.0 tesla. J Magn Reson Imaging. 2015;41(1):175–182. doi: 10.1002/jmri.24519. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR Am J Roentgenol. 2012;199(3):654–663. doi: 10.2214/AJR.11.7824. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto S, Han W, Kim Y, Du L, Jamshidi N, Huang D, et al. Breast cancer: radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology. 2015;275(2):384–392. doi: 10.1148/radiol.15142698. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y, Li H, Guo W, Drukker K, Lan L, Giger ML, et al. Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Sci Rep. 2015;5:17787. doi: 10.1038/srep17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elias SG, Adams A, Wisner DJ, Esserman LJ, van't Veer LJ, Mali WP, et al. Imaging features of HER2 overexpression in breast cancer: a systematic review and meta-analysis. Cancer Epidemiol Biomark Prev. 2014;23(8):1464–1483. doi: 10.1158/1055-9965.EPI-13-1170. [DOI] [PubMed] [Google Scholar]
- 47.Grimm LJ, Johnson KS, Marcom PK, Baker JA, Soo MS. Can breast cancer molecular subtype help to select patients for preoperative MR imaging? Radiology. 2015;274(2):352–358. doi: 10.1148/radiol.14140594. [DOI] [PubMed] [Google Scholar]
- 48.Uematsu T. MR imaging of triple-negative breast cancer. Breast cancer. 2011;18(3):161–164. doi: 10.1007/s12282-010-0236-3. [DOI] [PubMed] [Google Scholar]
- 49.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273(2):365–372. doi: 10.1148/radiol.14132641. [DOI] [PubMed] [Google Scholar]
- 50.Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging. 2015;42(4):902–907. doi: 10.1002/jmri.24879. [DOI] [PubMed] [Google Scholar]
- 51.Grimm LJ, Zhang J, Baker JA, Soo MS, Johnson KS, Mazurowski MA. Relationships between MRI breast imaging-reporting and data system (BI-RADS) lexicon descriptors and breast cancer molecular subtypes: internal enhancement is associated with luminal B subtype. Breast J. 2017;23(5):579–582. doi: 10.1111/tbj.12799. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi K, Abe H, Newstead GM, Egashira R, Nakazono T, Imaizumi T, et al. Intratumoral heterogeneity of the distribution of kinetic parameters in breast cancer: comparison based on the molecular subtypes of invasive breast cancer. Breast cancer. 2015;22(5):496–502. doi: 10.1007/s12282-013-0512-0. [DOI] [PubMed] [Google Scholar]
- 53.Leithner D, Bernard-Davila B, Martinez DF, Horvat JV, Jochelson MS, Marino MA et al (2019) Radiomic signatures derived from diffusion-weighted imaging for the assessment of breast cancer receptor status and molecular subtypes. Mol Imaging Biol. 10.1007/s11307-019-01383-w [DOI] [PMC free article] [PubMed]
- 54.Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA et al (2016) Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2:1-10 10.1038/npjbcancer.2016.12 [DOI] [PMC free article] [PubMed]
- 55.Waugh SA, Purdie CA, Jordan LB, Vinnicombe S, Lerski RA, Martin P, et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol. 2016;26(2):322–330. doi: 10.1007/s00330-015-3845-6. [DOI] [PubMed] [Google Scholar]
- 56.Woodard GA, Ray KM, Joe BN, Price ER. Qualitative radiogenomics: association between oncotype DX test recurrence score and BI-RADS mammographic and breast MR imaging features. Radiology. 2018;286(1):60–70. doi: 10.1148/radiol.2017162333. [DOI] [PubMed] [Google Scholar]
- 57.Ashraf AB, Daye D, Gavenonis S, Mies C, Feldman M, Rosen M, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology. 2014;272(2):374–384. doi: 10.1148/radiol.14131375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siamakpour-Reihani S, Owzar K, Jiang C, Scarbrough PM, Craciunescu OI, Horton JK, et al. Genomic profiling in locally advanced and inflammatory breast cancer and its link to DCE-MRI and overall survival. Int J Hyperth. 2015;31(4):386–395. doi: 10.3109/02656736.2015.1016557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sutton EJ, Oh JH, Dashevsky BZ, Veeraraghavan H, Apte AP, Thakur SB, et al. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging. 2015;42(5):1398–1406. doi: 10.1002/jmri.24890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Navarro P, Gonzalez-Neira A, Pita G, Diaz-Uriarte R, Tais Moreno L, Ederra M, et al. Genome wide association study identifies a novel putative mammographic density locus at 1q12-q21. Int J Cancer. 2015;136(10):2427–2436. doi: 10.1002/ijc.29299. [DOI] [PubMed] [Google Scholar]
- 61.Li H, Giger ML, Sun C, Ponsukcharoen U, Huo D, Lan L, et al. Pilot study demonstrating potential association between breast cancer image-based risk phenotypes and genomic biomarkers. Med Phys. 2014;41(3):031917. doi: 10.1118/1.4865811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology. 2016;281(2):382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wan T, Bloch BN, Plecha D, Thompson CL, Gilmore H, Jaffe C, et al. A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting OncotypeDX risk scores. Sci Rep. 2016;6:21394. doi: 10.1038/srep21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dialani V, Gaur S, Mehta TS, Venkataraman S, Fein-Zachary V, Phillips J, et al. Prediction of low versus high recurrence scores in estrogen receptor-positive, lymph node-negative invasive breast cancer on the basis of radiologic-pathologic features: comparison with Oncotype DX test recurrence scores. Radiology. 2016;280(2):370–378. doi: 10.1148/radiol.2016151149. [DOI] [PubMed] [Google Scholar]
- 65.Mehta S, Hughes NP, Li S, Jubb A, Adams R, Lord S, et al. Radiogenomics monitoring in breast cancer identifies metabolism and immune checkpoints as early actionable mechanisms of resistance to anti-angiogenic treatment. EBioMedicine. 2016;10:109–116. doi: 10.1016/j.ebiom.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halle C, Andersen E, Lando M, Aarnes EK, Hasvold G, Holden M, et al. Hypoxia-induced gene expression in chemoradioresistant cervical cancer revealed by dynamic contrast-enhanced MRI. Cancer Res. 2012;72(20):5285–5295. doi: 10.1158/0008-5472.CAN-12-1085. [DOI] [PubMed] [Google Scholar]
- 67.Andersen EK, Hole KH, Lund KV, Sundfor K, Kristensen GB, Lyng H, et al. Pharmacokinetic parameters derived from dynamic contrast enhanced MRI of cervical cancers predict chemoradiotherapy outcome. Radiother Oncol. 2013;107(1):117–122. doi: 10.1016/j.radonc.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 68.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14(16):5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 69.Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123(1):517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vargas HA, Micco M, Hong SI, Goldman DA, Dao F, Weigelt B, et al. Association between morphologic CT imaging traits and prognostically relevant gene signatures in women with high-grade serous ovarian cancer: a hypothesis-generating study. Radiology. 2015;274(3):742–751. doi: 10.1148/radiol.14141477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vargas HA, Huang EP, Lakhman Y, Ippolito JE, Bhosale P, Mellnick V, et al. Radiogenomics of high-grade serous ovarian cancer: multireader multi-institutional study from the Cancer Genome Atlas Ovarian Cancer Imaging Research Group. Radiology. 2017;285(2):482–492. doi: 10.1148/radiol.2017161870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vargas HA, Veeraraghavan H, Micco M, Nougaret S, Lakhman Y, Meier AA, et al. A novel representation of inter-site tumour heterogeneity from pre-treatment computed tomography textures classifies ovarian cancers by clinical outcome. Eur Radiol. 2017;27(9):3991–4001. doi: 10.1007/s00330-017-4779-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521(7553):489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 74.Etemadmoghadam D, deFazio a, Beroukhim R, Mermel C, George J, Getz G, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15(4):1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chandarana H, Robinson E, Hajdu CH, Drozhinin L, Babb JS, Taouli B. Microvascular invasion in hepatocellular carcinoma: is it predictable with pretransplant MRI? AJR Am J Roentgenol. 2011;196(5):1083–1089. doi: 10.2214/AJR.10.4720. [DOI] [PubMed] [Google Scholar]
- 76.Griffin N, Addley H, Sala E, Shaw AS, Grant LA, Eldaly H, et al. Vascular invasion in hepatocellular carcinoma: is there a correlation with MRI? Br J Radiol. 2012;85(1014):736–744. doi: 10.1259/bjr/94924398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13(6):1929–1939. doi: 10.1091/mbc.02-02-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25(6):675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 79.Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62(3):792–800. doi: 10.1002/hep.27877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Renzulli M, Brocchi S, Cucchetti A, Mazzotti F, Mosconi C, Sportoletti C, et al. Can current preoperative imaging be used to detect microvascular invasion of hepatocellular carcinoma? Radiology. 2016;279(2):432–442. doi: 10.1148/radiol.2015150998. [DOI] [PubMed] [Google Scholar]
- 81.Taouli B, Hoshida Y, Kakite S, Chen X, Tan PS, Sun X, et al. Imaging-based surrogate markers of transcriptome subclasses and signatures in hepatocellular carcinoma: preliminary results. Eur Radiol. 2017;27(11):4472–4481. doi: 10.1007/s00330-017-4844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hectors SJ, Wagner M, Bane O, Besa C, Lewis S, Remark R, et al. Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci Rep. 2017;7(1):2452. doi: 10.1038/s41598-017-02706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sadot E, Simpson AL, Do RK, Gonen M, Shia J, Allen PJ, et al. Cholangiocarcinoma: correlation between molecular profiling and imaging phenotypes. PLoS One. 2015;10(7):e0132953. doi: 10.1371/journal.pone.0132953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim SA, Lee JM, Lee KB, Kim SH, Yoon SH, Han JK, et al. Intrahepatic mass-forming cholangiocarcinomas: enhancement patterns at multiphasic CT, with special emphasis on arterial enhancement pattern--correlation with clinicopathologic findings. Radiology. 2011;260(1):148–157. doi: 10.1148/radiol.11101777. [DOI] [PubMed] [Google Scholar]
- 85.Fujita N, Asayama Y, Nishie A, Ishigami K, Ushijima Y, Takayama Y, et al. Mass-forming intrahepatic cholangiocarcinoma: enhancement patterns in the arterial phase of dynamic hepatic CT - correlation with clinicopathological findings. Eur Radiol. 2017;27(2):498–506. doi: 10.1007/s00330-016-4386-3. [DOI] [PubMed] [Google Scholar]
- 86.Aherne EA, Pak LM, Goldman DA, Gonen M, Jarnagin WR, Simpson AL, et al. Intrahepatic cholangiocarcinoma: can imaging phenotypes predict survival and tumor genetics? Abdom Radiol (NY) 2018;43(10):2665–2672. doi: 10.1007/s00261-018-1505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70(11):4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 89.Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2011;44(1):17–19. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 90.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karlo CA, Di Paolo PL, Chaim J, Hakimi AA, Ostrovnaya I, Russo P, et al. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology. 2014;270(2):464–471. doi: 10.1148/radiol.13130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shinagare AB, Vikram R, Jaffe C, Akin O, Kirby J, Huang E, et al. Radiogenomics of clear cell renal cell carcinoma: preliminary findings of the Cancer Genome Atlas-Renal Cell Carcinoma (TCGA-RCC) Imaging Research Group. Abdom Imaging. 2015;40(6):1684–1692. doi: 10.1007/s00261-015-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alessandrino F, Shinagare AB, Bosse D, Choueiri TK, Krajewski KM. Radiogenomics in renal cell carcinoma. Abdom Radiol (NY) 2019;44(6):1990–1998. doi: 10.1007/s00261-018-1624-y. [DOI] [PubMed] [Google Scholar]
- 96.Kim BJ, Kim JH, Kim HS, Zang DY. Prognostic and predictive value of VHL gene alteration in renal cell carcinoma: a meta-analysis and review. Oncotarget. 2017;8(8):13979–13985. doi: 10.18632/oncotarget.14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45(8):860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 98.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–Dd83. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. J Clin Oncol. 2014;32(18):1968–1976. doi: 10.1200/JCO.2012.45.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Z, Peng S, Guo L, Xie H, Wang A, Shang Z, et al. Prognostic and clinicopathological value of PBRM1 expression in renal cell carcinoma. Clin Chim Acta. 2018;486:9–17. doi: 10.1016/j.cca.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 101.Kocak B, Durmaz ES, Ates E, Ulusan MB. Radiogenomics in clear cell renal cell carcinoma: machine learning-based high-dimensional quantitative CT texture analysis in predicting PBRM1 mutation status. AJR Am J Roentgenol. 2019;212(3):W55–w63. doi: 10.2214/AJR.18.20443. [DOI] [PubMed] [Google Scholar]
- 102.Tennenbaum DM, Manley BJ, Zabor E, Becerra MF, Carlo MI, Casuscelli J, et al. Genomic alterations as predictors of survival among patients within a combined cohort with clear cell renal cell carcinoma undergoing cytoreductive nephrectomy. Urol Oncol. 2017;35(8):532.e7–532e13. doi: 10.1016/j.urolonc.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu W, Fu Q, An H, Chang Y, Zhang W, Zhu Y et al (2015) Decreased expression of SETD2 predicts unfavorable prognosis in patients with nonmetastatic clear-cell renal cell carcinoma. Medicine (Baltimore) 94(45):e2004 [DOI] [PMC free article] [PubMed]
- 104.Hoffmann I, Roatsch M, Schmitt ML, Carlino L, Pippel M, Sippl W, et al. The role of histone demethylases in cancer therapy. Mol Oncol. 2012;6(6):683–703. doi: 10.1016/j.molonc.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Manley BJ, Reznik E, Ghanaat M, Kashan M, Becerra MF, Casuscelli J, et al. Characterizing recurrent and lethal small renal masses in clear cell renal cell carcinoma using recurrent somatic mutations. Urol Oncol. 2019;37(1):12–17. doi: 10.1016/j.urolonc.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wei JH, Haddad A, Wu KJ, Zhao HW, Kapur P, Zhang ZL, et al. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat Commun. 2015;6:8699. doi: 10.1038/ncomms9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Evelonn EA, Degerman S, Kohn L, Landfors M, Ljungberg B, Roos G. DNA methylation status defines clinicopathological parameters including survival for patients with clear cell renal cell carcinoma (ccRCC) Tumour Biol. 2016;37(8):10219–10228. doi: 10.1007/s13277-016-4893-5. [DOI] [PubMed] [Google Scholar]
- 108.Fisel P, Kruck S, Winter S, Bedke J, Hennenlotter J, Nies AT, et al. DNA methylation of the SLC16A3 promoter regulates expression of the human lactate transporter MCT4 in renal cancer with consequences for clinical outcome. Clin Cancer Res. 2013;19(18):5170–5181. doi: 10.1158/1078-0432.CCR-13-1180. [DOI] [PubMed] [Google Scholar]
- 109.Joosten SC, Deckers IA, Aarts MJ, Hoeben A, van Roermund JG, Smits KM, et al. Prognostic DNA methylation markers for renal cell carcinoma: a systematic review. Epigenomics. 2017;9(9):1243–1257. doi: 10.2217/epi-2017-0040. [DOI] [PubMed] [Google Scholar]
- 110.Zheng J, Mei Y, Xiang P, Zhai G, Zhao N, Xu C, et al. DNA methylation affects metastasis of renal cancer and is associated with TGF-beta/RUNX3 inhibition. Cancer Cell Int. 2018;18:56. doi: 10.1186/s12935-018-0554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pan C, Xiang L, Pan Z, Wang X, Li J, Zhuge L, et al. MiR-544 promotes immune escape through downregulation of NCR1/NKp46 via targeting RUNX3 in liver cancer. Cancer Cell Int. 2018;18:52. doi: 10.1186/s12935-018-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen F, Liu X, Cheng Q, Zhu S, Bai J, Zheng J. RUNX3 regulates renal cell carcinoma metastasis via targeting miR-6780a-5p/E-cadherin/EMT signaling axis. Oncotarget. 2017;8(60):101042–101056. doi: 10.18632/oncotarget.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y, Qin X, Wu J, Qi B, Tao Y, Wang W, et al. Association of promoter methylation of RUNX3 gene with the development of esophageal cancer: a meta analysis. PLoS One. 2014;9(9):e107598. doi: 10.1371/journal.pone.0107598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yan C, Kim YW, Ha YS, Kim IY, Kim YJ, Yun SJ, et al. RUNX3 methylation as a predictor for disease progression in patients with non-muscle-invasive bladder cancer. J Surg Oncol. 2012;105(4):425–430. doi: 10.1002/jso.22087. [DOI] [PubMed] [Google Scholar]
- 115.Richiardi L, Fiano V, Vizzini L, De Marco L, Delsedime L, Akre O, et al. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J Clin Oncol. 2009;27(19):3161–3168. doi: 10.1200/JCO.2008.18.2485. [DOI] [PubMed] [Google Scholar]
- 116.Cen D, Xu L, Zhang S, Chen Z, Huang Y, Li Z et al (2019) Renal cell carcinoma: predicting RUNX3 methylation level and its consequences on survival with CT features. Eur Radiol 29(10):5415–5422 [DOI] [PubMed]
- 117.Jamshidi N, Jonasch E, Zapala M, Korn RL, Aganovic L, Zhao H, et al. The radiogenomic risk score: construction of a prognostic quantitative, noninvasive image-based molecular assay for renal cell carcinoma. Radiology. 2015;277(1):114–123. doi: 10.1148/radiol.2015150800. [DOI] [PubMed] [Google Scholar]
- 118.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jorge SE, Kobayashi SS, Costa DB. Epidermal growth factor receptor (EGFR) mutations in lung cancer: preclinical and clinical data. Braz J Med Biol Res. 2014;47(11):929–939. doi: 10.1590/1414-431X20144099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370(13):1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Seto T, Kiura K, Nishio M, Nakagawa K, Maemondo M, Inoue A, et al. CH5424802 (RO5424802) for patients with ALK-rearranged advanced non-small-cell lung cancer (AF-001JP study): a single-arm, open-label, phase 1-2 study. The Lancet Oncology. 2013;14(7):590–598. doi: 10.1016/S1470-2045(13)70142-6. [DOI] [PubMed] [Google Scholar]
- 122.Rodenhuis S, Slebos RJ. Clinical significance of ras oncogene activation in human lung cancer. Cancer Res. 1992;52(9 Suppl):2665s–2669s. [PubMed] [Google Scholar]
- 123.Gerber DE, Gandhi L, Costa DB (2014) Management and future directions in non-small cell lung cancer with known activating mutations. Am Soc Clin Oncol Educ Book 34:e353–e365 [DOI] [PubMed]
- 124.Glynn C, Zakowski MF, Ginsberg MS. Are there imaging characteristics associated with epidermal growth factor receptor and KRAS mutations in patients with adenocarcinoma of the lung with bronchioloalveolar features? J Thorac Oncol. 2010;5(3):344–348. doi: 10.1097/JTO.0b013e3181ce9a7a. [DOI] [PubMed] [Google Scholar]
- 125.Lee Y, Lee HJ, Kim YT, Kang CH, Goo JM, Park CM, et al. Imaging characteristics of stage I non-small cell lung cancer on CT and FDG-PET: relationship with epidermal growth factor receptor protein expression status and survival. Korean J Radiol. 2013;14(2):375–383. doi: 10.3348/kjr.2013.14.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee HJ, Kim YT, Kang CH, Zhao B, Tan Y, Schwartz LH, et al. Epidermal growth factor receptor mutation in lung adenocarcinomas: relationship with CT characteristics and histologic subtypes. Radiology. 2013;268(1):254–264. doi: 10.1148/radiol.13112553. [DOI] [PubMed] [Google Scholar]
- 127.Yano M, Sasaki H, Kobayashi Y, Yukiue H, Haneda H, Suzuki E, et al. Epidermal growth factor receptor gene mutation and computed tomographic findings in peripheral pulmonary adenocarcinoma. J Thorac Oncol. 2006;1(5):413–416. doi: 10.1097/01243894-200606000-00006. [DOI] [PubMed] [Google Scholar]
- 128.Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M, et al. CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer. Eur Radiol. 2016;26(1):32–42. doi: 10.1007/s00330-015-3814-0. [DOI] [PubMed] [Google Scholar]
- 129.Halpenny DF, Riely GJ, Hayes S, Yu H, Zheng J, Moskowitz CS, et al. Are there imaging characteristics associated with lung adenocarcinomas harboring ALK rearrangements? Lung Cancer. 2014;86(2):190–194. doi: 10.1016/j.lungcan.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Plodkowski AJ, Drilon A, Halpenny DF, O'Driscoll D, Blair D, Litvak AM, et al. From genotype to phenotype: are there imaging characteristics associated with lung adenocarcinomas harboring RET and ROS1 rearrangements? Lung Cancer. 2015;90(2):321–325. doi: 10.1016/j.lungcan.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nair VS, Gevaert O, Davidzon G, Napel S, Graves EE, Hoang CD, et al. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res. 2012;72(15):3725–3734. doi: 10.1158/0008-5472.CAN-11-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71(22):6921–6925. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- 133.Lewis DR, Chen HS, Cockburn MG, Wu XC, Stroup AM, Midthune DN, et al. Early estimates of SEER cancer incidence, 2014. Cancer. 2017;123(13):2524–2534. doi: 10.1002/cncr.30630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Leal J, Hamdy F, Wolstenholme J. Estimating age and ethnic variation in the histological prevalence of prostate cancer to inform the impact of screening policies. Int J Urol. 2014;21(8):786–792. doi: 10.1111/iju.12458. [DOI] [PubMed] [Google Scholar]
- 135.Galletti G, Leach BI, Lam L, Tagawa ST. Mechanisms of resistance to systemic therapy in metastatic castration-resistant prostate cancer. Cancer Treat Rev. 2017;57:16–27. doi: 10.1016/j.ctrv.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 136.D'Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280(11):969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 137.Moschini M, Spahn M, Mattei A, Cheville J, Karnes RJ. Incorporation of tissue-based genomic biomarkers into localized prostate cancer clinics. BMC Med. 2016;14:67. doi: 10.1186/s12916-016-0613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wibmer A, Verma S, Vargas HA. Role of MRI in the risk assessment of primary prostate cancer. Top Magn Reson Imaging. 2016;25(3):133–138. doi: 10.1097/RMR.0000000000000081. [DOI] [PubMed] [Google Scholar]
- 139.Sidhu HS, Benigno S, Ganeshan B, Dikaios N, Johnston EW, Allen C, et al. Textural analysis of multiparametric MRI detects transition zone prostate cancer. Eur Radiol. 2017;27(6):2348–2358. doi: 10.1007/s00330-016-4579-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bates A, Miles K. Prostate-specific membrane antigen PET/MRI validation of MR textural analysis for detection of transition zone prostate cancer. Eur Radiol. 2017;27(12):5290–5298. doi: 10.1007/s00330-017-4877-x. [DOI] [PubMed] [Google Scholar]
- 141.Krishna S, Lim CS, McInnes MDF, Flood TA, Shabana WM, Lim RS, et al. Evaluation of MRI for diagnosis of extraprostatic extension in prostate cancer. J Magn Reson Imaging. 2018;47(1):176–185. doi: 10.1002/jmri.25729. [DOI] [PubMed] [Google Scholar]
- 142.Rosenkrantz AB, Triolo MJ, Melamed J, Rusinek H, Taneja SS, Deng FM. Whole-lesion apparent diffusion coefficient metrics as a marker of percentage Gleason 4 component within Gleason 7 prostate cancer at radical prostatectomy. J Magn Reson Imaging. 2015;41(3):708–714. doi: 10.1002/jmri.24598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rozenberg R, Thornhill RE, Flood TA, Hakim SW, Lim C, Schieda N. Whole-tumor quantitative apparent diffusion coefficient histogram and texture analysis to predict Gleason score upgrading in intermediate-risk 3 + 4 = 7 prostate cancer. AJR Am J Roentgenol. 2016;206(4):775–782. doi: 10.2214/AJR.15.15462. [DOI] [PubMed] [Google Scholar]
- 144.Nketiah G, Elschot M, Kim E, Teruel JR, Scheenen TW, Bathen TF, et al. T2-weighted MRI-derived textural features reflect prostate cancer aggressiveness: preliminary results. Eur Radiol. 2017;27(7):3050–3059. doi: 10.1007/s00330-016-4663-1. [DOI] [PubMed] [Google Scholar]
- 145.Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol. 2015;25(10):2840–2850. doi: 10.1007/s00330-015-3701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rosenkrantz AB, Ream JM, Nolan P, Rusinek H, Deng FM, Taneja SS. Prostate cancer: utility of whole-lesion apparent diffusion coefficient metrics for prediction of biochemical recurrence after radical prostatectomy. AJR Am J Roentgenol. 2015;205(6):1208–1214. doi: 10.2214/AJR.15.14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gnep K, Fargeas A, Gutierrez-Carvajal RE, Commandeur F, Mathieu R, Ospina JD, et al. Haralick textural features on T2 -weighted MRI are associated with biochemical recurrence following radiotherapy for peripheral zone prostate cancer. J Magn Reson Imaging. 2017;45(1):103–117. doi: 10.1002/jmri.25335. [DOI] [PubMed] [Google Scholar]
- 148.McCann SM, Jiang Y, Fan X, Wang J, Antic T, Prior F, et al. Quantitative multiparametric MRI features and PTEN expression of peripheral zone prostate cancer: a pilot study. AJR Am J Roentgenol. 2016;206(3):559–565. doi: 10.2214/AJR.15.14967. [DOI] [PubMed] [Google Scholar]
- 149.Jamshidi N, Margolis DJ, Raman S, Huang J, Reiter RE, Kuo MD. Multiregional radiogenomic assessment of prostate microenvironments with multiparametric MR imaging and DNA whole-exome sequencing of prostate glands with adenocarcinoma. Radiology. 2017;284(1):109–119. doi: 10.1148/radiol.2017162827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable