Abstract
Background and objectives
Diabetes is a chronic disease characterized by high blood sugar. It may cause many complicated disease like stroke, kidney failure, heart attack, etc. About 422 million people were affected by diabetes disease in worldwide in 2014. The figure will be reached 642 million in 2040. The main objective of this study is to develop a machine learning (ML)-based system for predicting diabetic patients.
Materials and methods
Logistic regression (LR) is used to identify the risk factors for diabetes disease based on p value and odds ratio (OR). We have adopted four classifiers like naïve Bayes (NB), decision tree (DT), Adaboost (AB), and random forest (RF) to predict the diabetic patients. Three types of partition protocols (K2, K5, and K10) have also adopted and repeated these protocols into 20 trails. Performances of these classifiers are evaluated using accuracy (ACC) and area under the curve (AUC).
Results
We have used diabetes dataset, conducted in 2009–2012, derived from the National Health and Nutrition Examination Survey. The dataset consists of 6561 respondents with 657 diabetic and 5904 controls. LR model demonstrates that 7 factors out of 14 as age, education, BMI, systolic BP, diastolic BP, direct cholesterol, and total cholesterol are the risk factors for diabetes. The overall ACC of ML-based system is 90.62%. The combination of LR-based feature selection and RF-based classifier gives 94.25% ACC and 0.95 AUC for K10 protocol.
Conclusion
The combination of LR and RF-based classifier performs better. This combination will be very helpful for predicting diabetic patients.
Keywords: Diabetes, Classification, Machine learning, Naïve Bayes, Decision tree, Random forest, Adaboost
Introduction
Diabetes mellitus (DM) is commonly known as diabetes. It is a group of metabolic disorders which are characterized by the high blood sugar [1–3]. Diabetes can lead to many serious long-term complicated disease like cardiovascular disease, stroke, kidney failure, heart attack, peripheral arterial disease, blood vessels, and nerves [4, 5]. About 122 million people were affected by diabetes in worldwide in 1980 and this figure was reached about 422 million in 2014 [6]. The figure will be reached about 642 million in 2040 [7]. Moreover, there were directly about 1.6 million deaths due to diabetes [8]. Therefore; it is an alarming figure to us. The number of diabetic patients is increased day by day as a result deaths are also increased day by day. Diabetes can be divided into three types as (i) type I diabetes (T1D), (ii) type II diabetes (T2D), and (iii) gestational diabetes (GD) [9]. T1D are normally in young adults whose age is less than 30 years. The symptoms of T1D are polyuria, thirst, constant hunger, weight loss, vision changes and fatigue [10]. T2D occurs in adults over 45 years which are often associated with obesity, hypertension, dyslipidemia, arteriosclerosis, and other diseases [11]. The third type of diabetes is gestational diabetes. Actually pregnant women are affected by gestational diabetes.
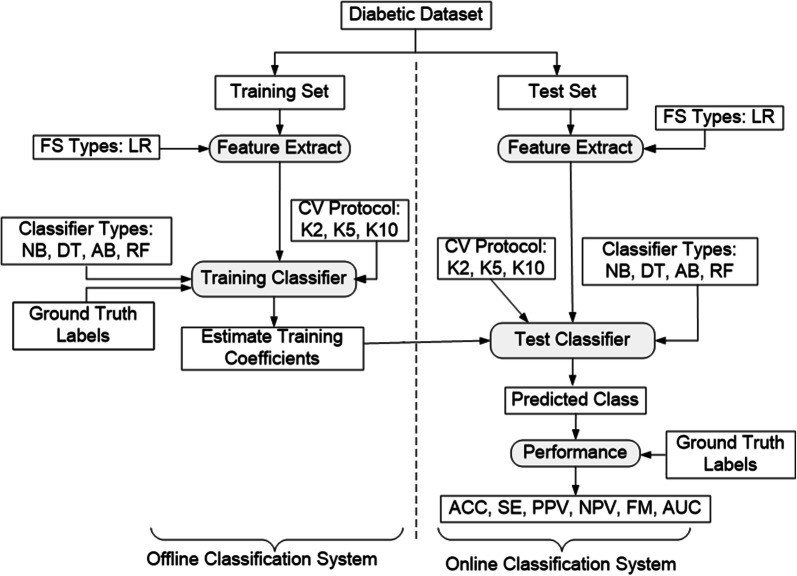
The analysis of diabetes data is a challenging issue because most of the medical data are nonlinear, non-normal, correlation structured, and complex in nature [12]. The ML-based systems have dominated in the field of medical healthcare [12–21] and medical imaging such as stroke, coronary artery disease, and cancer [22–26]. Moreover, ML-based systems can be used as both feature selection techniques (FST) and classifiers. It also helps the people to accurately diagnosis of diabetes and the best classifier is the most important problems for accurate diabetes risk stratification. There were various ML-based systems used to classify and predict of diabetic disease like linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), naïve Bayes (NB), support vector machine (SVM), artificial neural network (ANN), feed-forward neural network (FFNN), Adaboost (AB), decision tree (DT), J48, random forest (RF), Gaussian process classification (GPC), logistic regression (LR), and k-nearest neighborhood (KNN) and so on [12, 13, 17–30]. The overview of the proposed ML-based system is shown in Fig. 1. We hypothesize that the combination of logistic regression (LR) based FST along with the training-based four classifiers can be accurately diabetes risk stratification.
Fig. 1.
Overview of the proposed ML-based system
Thus, in this study we have adopted LR model to identify the risk factors of diabetes disease based on p-value and odds ratio (OR). We also adopted four applicable and important ML-based classifiers as: NB, DT, AB, and RF. The main objective of this study is to identify the most significant factors of diabetes disease based on LR model and develop a ML-based system for the accurate risk stratification of diabetes disease. Finally, we thus summarize the main contributions in this study as follows:
To identify the risk factors of diabetes disease using LR model based p-values and OR.
To choose the best ML-based system by selecting the best protocol (three partition protocols: K2, K5, and K10) and classifier (four classifiers: NB, DT, AB, and RF) combination based on accuracy (ACC), sensitivity (SE), positive predictive value (PPV), negative predictive value (NPV), F-measure (FM), and area under the curve (AUC).
To validate our proposed ML-based system, we demonstrate the same performance using Indian liver patient dataset.
The paper is organized as follows: “Materials and methods” section represents materials and methods, including description of the dataset, statistical analysis, machine learning system, feature selection techniques, data partitioning, prediction model, and performance evaluations of the classifiers. The results are discussed in “Results” section. “Discussion” section represents the discussions in detail along with key difference between our proposed ML-based system and previous work, strength and extension of the study, and summary of the current study. Finally conclusion is presented in “Conclusion” section.
Materials and methods
Data
The diabetes 2009–2012 dataset have been used in this study, derived from the National Health and Nutrition Examination Survey (NHANES). NHANES is an ongoing and cross-sectional study and population sample survey of United States (US) population. The dataset consisted of 9858 respondents. Respondents were identified as a diabetic patient if they met with at least one of the following criteria: plasma fasting glucose ≥ 126 mg/dL, serum glucose ≥ 200 mg/dL, glycohemoglobin ≥ 6.5%. There were about 760 diabetic respondents and 9098 control respondents. There were some missing values and unusual observations in the dataset. Excluding the missing values and unusual observations from the dataset, there were a total of 6561 respondents with 657 diabetic and 5904 controls. Detailed description of the dataset is shown in Table 9 in Appendix 1. The dataset is public domain survey and freely available in online.
Table 9.
Description of the diabetes database
| SN | Factors | Description | Categorical |
|---|---|---|---|
| 1 | Age | Age in years | Continuous |
| 2 | Gender | Gender of the patients | (i) Male; (ii) female |
| 3 | Race | Race | (i) Black; (ii) Hispanic; (iii) Mexican; (iv) other; (v) White |
| 4 | Education | Education level of the patients | (i) 8th grade; (ii) 9–11th grade; (iii) college grad; (iv) high school; (v) some college |
| 5 | Marital status | Marital status of the patients | (i) Divorced; (ii) live partner; (iii) married; (iv) never married; (v) separated; (vi) widowed. |
| 6 | Occupation | Occupation of the patients | (i) looking work; (ii) not working; (iii) working |
| 7 | Weight | Weight in kilogram (kg) | Continuous |
| 8 | Height | Height in meter (m) | Continuous |
| 9 | BMI | Body mass index (kg/m2) | Continuous |
| 10 | Systolic BP | Systolic blood pressure (mm Hg) | Continuous |
| 11 | Diastolic BP | Diastolic blood pressure (mm Hg) | Continuous |
| 12 | Direct cholesterol | Direct cholesterol (mg/dL) | Continuous |
| 13 | Total cholesterol | Total cholesterol (mg/dL) | Continuous |
| 14 | Physical activity | Physical activity | (i) No; (ii) yes. |
| 15 | Outcome | Class label | (i) Diabetic; (ii) control |
Statistical analysis
The baseline characteristics of the study population are presented as mean ± SD (standard deviation) for continuous variables and number (percentage) for the categorical variables, respectively. Differences in variables between diabetic patients and control are analyzed by independent t-test for continuous variables and Chi square test for categorical variables. All of the tests are two-tailed and considered as significant factors whose p-values are less than 0.05. The demographic and clinical characteristics of the diabetic patients are described in Table 1. Data are analyzed using Stata-version 14.10 and R-i386 3.6.1.
Table 1.
Demographic and clinical characteristics of diabetic patients
| Factors | Overall (6561) | Diabetic (657) | Control (5904) | p valuea |
|---|---|---|---|---|
| Age (years) | 47.18 ± 16.79 | 59.85 ± 13.22 | 45.77 ± 16.56 | < 0.001 |
| Gender, male n (%) | 3257 (49.64) | 361 (54.95) | 2896 (49.05) | < 0.001 |
| Race, white n (%) | 4474 (68.19) | 400 (60.88) | 4074 (69.00) | < 0.001 |
| Education, college n (%) | 3994 (60.87) | 337 (51.29) | 3657 (61. 94) | < 0.001 |
| Marital Status, married n (%) | 4132 (62.98) | 409 (62.25) | 3723 (63.06) | < 0.001 |
| Occupation, working n (%) | 4084 (62.25) | 272 (41.40) | 3812 (64.57) | < 0.001 |
| Weight (kg) | 82.48 ± 21.24 | 92.40 ± 25.17 | 81.38 ± 20.47 | < 0.001 |
| Height (m) | 1.69 ± 0.10 | 1.68 ± 0.11 | 1.69 ± 0.10 | < 0.001 |
| BMI (kg/m2) | 28.78 ± 6.65 | 32.70 ± 8.10 | 28.34 ± 6.32 | < 0.001 |
| Systolic BP (mm Hg) | 120.86 ± 16.96 | 128.30 ± 19.42 | 120.04 ± 16.46 | < 0.001 |
| Diastolic BP (mm Hg) | 70.24 ± 12.32 | 68.37 ± 14.05 | 70.46 ± 12.10 | < 0.001 |
| Direct cholesterol (mg/dL) | 1.37 ± 0.42 | 1.24 ± 0.35 | 1.38 ± 0.42 | < 0.001 |
| Total cholesterol (mg/dL) | 5.07 ± 1.05 | 4.80 ± 1.15 | 5.10 ± 1.04 | < 0.001 |
| Physical activity, yes n (%) | 3497 (53.30) | 249 (37.90) | 3248 (55.01) | < 0.001 |
The continuous variables are expressed as mean ± SD and the categorical variables expressed as n (%)
BMI body mass index, BP blood pressure
ap value is obtained from an independent t-test for continuous variable and a Chi- square test for a categorical variable
Machine learning system
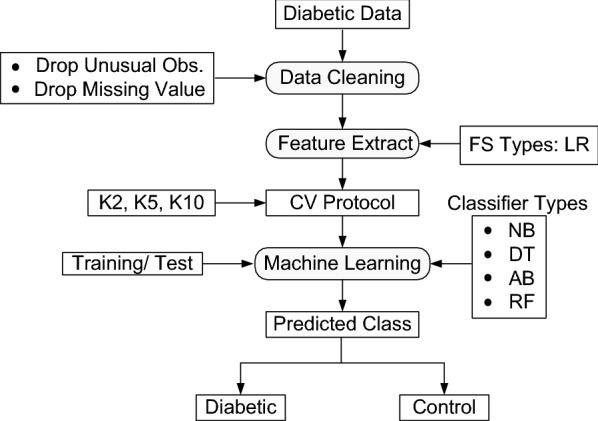
The main objective of the ML-based system is to classify and predict of the diabetes disease. The overview of the proposed ML-based systems has been shown in Fig. 1. The training/test set paradigm of the entire ML-based systems has been shown in Fig. 2. The first step is to divide the dataset into two sets such as training set and test set. The training and test sets are separated by dotted line as online and offline classification system. In the second step, the most significant risk factors of diabetes disease using LR model based on p-value and OR. The next stage is to adapt three partition protocols (K2, K5, and K10). And each protocol is repeated into 20 trials (T). We have also adopted four classifiers as: NB, DT, AB, and RF, respectively. The fourth step is to estimate the training classifier coefficients, and then the test classifiers have been applied to classify the patients into two categories as diabetic vs. control. Finally the performances of the classifiers are evaluated using six performance parameters, namely: ACC, SE, PPV, NPV, FM, and AUC.
Fig. 2.
The training/test set paradigm of the ML-based system
Feature selection
In machine learning and statistics, feature selection is known as variable selection. It is mainly exercised for choosing the relevant factors to use in ML-based system. There are five reasons for selecting the best subset of predictors as: (i) run the ML-based system easily and interpret the results, (ii) avoid curse of dimensionality, (iii) save computational cost as well as time, (iv) reduce the over-fitting, and (v) improve the classification accuracy. There were some commonly used FST in ML/statistics namely: RF [13, 31, 32], LR [33, 34], mutual information (MI) [13, 35], principal component analysis (PCA) [13, 35, 36], analysis of variance (ANOVA) [13, 37, 38], and Fisher’s discriminant ratio (FDR) [13, 35, 39]. In this study, we have used LR model to identify the risk factor for diabetic disease based on p-value (p < 0.05) and OR.
Logistic regression
Logistic regression (LR) is a supervised learning while the predictors are continuous/discrete and response variable is dichotomous (diabetic vs. control). LR model is used to estimate the probability of a binary response based on one or more predictors. LR also measures the relationship between response and one or more predictors by estimating probability logit function. The logit of response variable (Y) is the linear combination of the predictors (X) which can be written as follows:
| 1 |
where is defined as the probability for Y = 1 (diabetic) and 1 − Pj (control) is defined when Y = 0. βi’s are the unknown regression coefficient, K is the total number of predictors (14 factors) and Xi’s are the predictors and . We estimate the regression coefficients by maximum likelihood estimator (MLE) and get easily OR by the exponent of the regression coefficients. We can easily test the regression coefficients/ORs by applying z-test and select the features corresponding to the regression coefficients/ORs whose p-values are less than 0.05.
Data partitioning
Data partitioning is known as cross-validation (CV) protocol. It is mainly used for dividing the given dataset into two subsets as: (i) training set and (ii) validation/test set. There are lots of CV protocols, used for partitioning the dataset to reduce the variability. The tenfold CV protocol is commonly used in both ML and statistics, whereas the dataset is divided into ten equal parts while the nine parts are used as a training set for ML-based system and remaining part is used as a validation/test set. This protocol is repeated into 20 times while run the ML-based system for test set and calculate the classification accuracy at each protocol and then take the average. This is termed in the manuscript as K10 where 10 indicates the number of total partition during ML-based system. Similarly, the well-known data partitioning protocols are K2, K4, and K5, respectively, depending on the percentage (%) of the training set as 50%, 75%, and 80%, while rest of the parts are treated as a test set. In this study, we have used three partition protocols as K2, K5, and K10, respectively.
Prediction model
In this study, we have used four sets of ML-based classifiers as NB, DT, AB, and RF. The brief discussions of these classifiers are discussed as follows.
Naïve Bayes
Naïve Bayes (NB) is a simple probabilistic classifier based on Bayes theorem. The main assumption of NB is features are mutually independent [40]. In the recent study, it is used to diagnosis of different types of disease especially diabetes disease [12, 13]. The NB classifies the data using Bayes theorem as follows:
| 2 |
where, z is the response variable, w1, w2,…, wn are input variables; P(z|w1, …, wn) is the conditional probability distribution of z given w1, w2,…, wn; P(z) is the marginal probability distribution of z; is the conditional probability distribution of given z; p (wi) is the marginal probability distribution of wi; π is the product symbol. We found the probability of z given the set of inputs and picked up the output with maximum probability. The corresponding classifier, a Bayes classifier, is the function that assigns a class label as follows:
| 3 |
Decision tree
Decision tree (DT) is a supervised learning that can be used as regression tree while the response variable is continuous and as classification tree while the response variable is categorical. Whereas the input variables are any types as like graph, text, discrete, continuous, and so on in the case of both regression and classification. A decision tree is a tree structure based model which describes the classification process based on input features [41]. The steps of DT as follows: (i) construct a tree with its nodes as input features; (ii) select the feature to predict the output from the input features whose gives the highest information gain; (iii) repeat the above steps to form sub trees based on features which was not used in the above nodes [30].
Adaboost
Adaboost (AB) means adaptive boosting, a ML-based technique. Freund and Schapire introduced AB algorithm in 1996 [42] and won Gödel prize in 2003. It is used in conjunction with different types of algorithm to improve classifier’s performance. AB is very sensitive to noisy data and outliers. In some problems, it is less susceptible to the over-fitting problem than other learning algorithms. Every learning algorithm tends to suit some problem types better than others, and typically has many different parameters and configurations to adjust before it achieves optimal performance on a dataset. AB is known as the best out-of-the-box classifier [34].
Random forest
Random forest (RF) is a ML-based classifier by constructing decision trees. This algorithm was first proposed by Breiman [43]. RF can be also used as both regression and classification. RF can be used in several biomedicine research [20, 44], especially diagnosis of diabetes [12, 13]. The steps of RF as follows:
Step 1: divide the dataset into two parts as training set as well as test set. From the given training set, create a new dataset using the bootstrapping method.
Step 2: construct a DT based on the results of step 1.
Step 3: repeat step1 and step2 and produce many trees which consist of a forest.
Step 4: use every tree in the forest to vote for given input variables.
Step 5: compute the mean votes for each class. The class which gives the highest votes that are belongs to the classification label for the given input variables.
Finally compute the classification accuracy of RF-based classifier.
Performance evaluations
Many statistical parameters may be used to compare the performance of the classifiers. In this study, we have used ACC, SE, PPV, NPV, and FM. ACC is the proportion of the sum of the true positive and true negative against total number of population. SE is the proportion of the positive condition against the predicted condition is positive. PPV is the proportion of the predicted positive condition against the true condition is positive. NPV is the proportion of the predicted negative condition against the true condition is negative. FM is defined as the harmonic mean of the precision and recall.
Results
Demographics and clinical characteristics of diabetic patients
The patient’s demographics and clinical characteristics are shown in Table 1. A total of 657 (11%) from the pool of 6561 subjects are diabetic patients. In this study, we have taken 50% male and the overall age of the respondents are 47.18 ± 16.79 years. There are 55% male diabetic patients with average age 59.85 ± 13.22 years. It is observed that all attributes are highly statistically (p < 0.001) associated with diabetes.
Feature extraction using logistic regression
Table 2 shows that the effect of selected factors on diabetes using logistic regression. Its shows that age, education, BMI, systolic BP, diastolic BP, direct cholesterol, and total cholesterol are statistically significant factors for diabetes disease at 5% level of significance and the rest of the factors are insignificant. These seven factors are used for ML-based system to classify and predict of diabetes disease.
Table 2.
Effect of selected factors on the diabetes using logistic regression
| Factors | OR | p value | 95% CI for OR | |
|---|---|---|---|---|
| Lower | Upper | |||
| Age (years) | 1.055 | < 0.001 | 1.047 | 1.064 |
| Gender | ||||
| Female (ref) | 1.000 | |||
| Male | 1.270 | 0.086 | 0.967 | 1.670 |
| Race | ||||
| Black (ref) | 1.000 | |||
| Hispanic | 0.746 | 0.206 | 0.470 | 1.167 |
| Mexican | 0.858 | 0.465 | 0.567 | 1.290 |
| Other | 1.109 | 0.623 | 0.732 | 1.670 |
| White | 0.469 | 0.263 | 0.358 | 0.619 |
| Education | ||||
| 8th grade (ref) | 1.000 | |||
| 9–11th grade | 0.577 | 0.005 | 0.394 | 0.844 |
| College grad | 0.641 | 0.019 | 0.441 | 0.932 |
| High school | 0.746 | 0.010 | 0.526 | 1.060 |
| Some college | 0.786 | 0.030 | 0.559 | 1.112 |
| Marital status | ||||
| Divorced (ref) | 1.000 | |||
| Live partner | 0.562 | 0.240 | 0.335 | 0.915 |
| Married | 0.780 | 0.083 | 0.591 | 1.038 |
| Never married | 0.600 | 0.112 | 0.404 | 0.887 |
| Separated | 0.985 | 0.957 | 0.552 | 1.709 |
| Widowed | 0.541 | 0.162 | 0.368 | 0.796 |
| Occupation | ||||
| Looking working (ref) | 1.000 | |||
| Not working | 1.061 | 0.838 | 0.619 | 1.932 |
| Working | 0.776 | 0.371 | 0.457 | 1.402 |
| Weight (kg) | 0.974 | 0.135 | 0.940 | 1.008 |
| Height (m) | 1.029 | 0.155 | 0.989 | 1.072 |
| BMI (kg/m2) | 1.170 | 0.002 | 1.061 | 1.292 |
| Systolic BP (mm Hg) | 1.007 | 0.007 | 1.002 | 1.013 |
| Diastolic BP (mm Hg) | 0.994 | 0.008 | 0.986 | 1.001 |
| Direct cholesterol (mg/dL) | 0.510 | < 0.001 | 0.381 | 0.678 |
| Total cholesterol (mg/dL) | 0.809 | < 0.001 | 0.738 | 0.885 |
| Physical activity | ||||
| No (ref) | 1.000 | |||
| Yes | 0.906 | 0.319 | 0.747 | 1.099 |
ref reference
Performance analysis of machine learning system
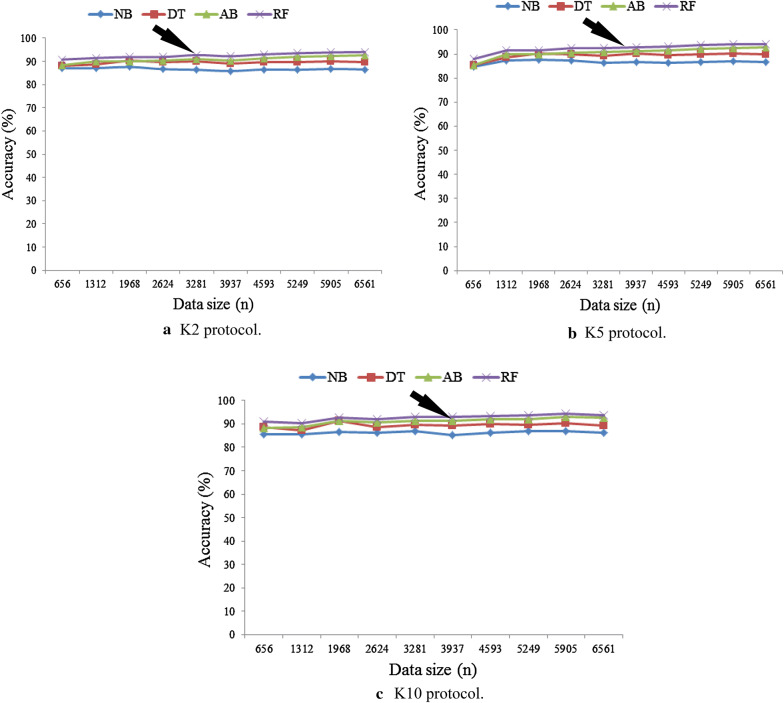
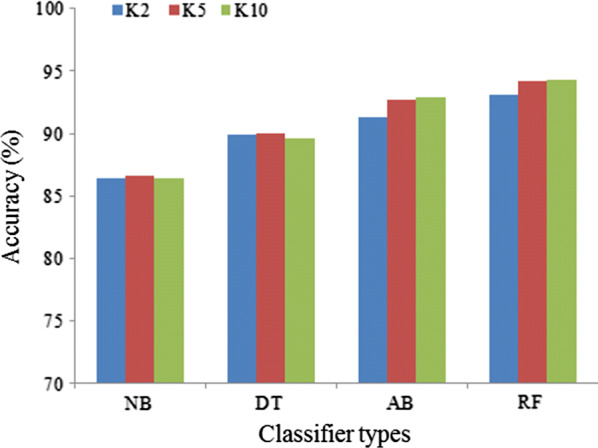
The comparison of the classification accuracy of four classifiers are shown in Fig. 3 for three partition protocols (K2, K5, and K10) while keeping the data size fixed (n = 6561). It also shows that the accuracy of all classifiers is increased as increasing the number of partition protocol from K2 to K5 to K10. It is observed that RF-based classifier performs better for all protocols compared to other classifiers. It is also observed that RF-based classifier gives the highest classification accuracy of 94.25% for K10 protocol whereas NB classifier gives the lowest classification accuracy of 86.70% for K10 protocol. The corresponding results are presented in Table 3. Moreover, the four performance evaluation parameters as SE, PPV, NPV, and FM of four classifiers for three partition protocols are shown in Table 4. Also the best performers of RF-based classifiers among four classifiers are validated using FM (see the last column of Table 4). We observe that RF-based classifier gives the highest FM of 96.88% for K10 protocol.
Fig. 3.
Comparison of accuracy of four classifiers for three partition protocols keeping data size fixed
Table 3.
Comparison of accuracy (%) of four classifiers for three partition protocols
| Classifier types | Protocol types | ||
|---|---|---|---|
| K2 | K5 | K10 | |
| NB | 86.42 | 86.61 | 86.70 |
| DT | 89.90 | 89.97 | 89.65 |
| AB | 91.32 | 92.72 | 92.93 |
| RF | 93.12 | 94.15 | 94.25 |
Bold values indicate the proposed method results
NB naïve Bayes, DT decision tree, AB adaboost, RF random forest, ACC accuracy, K2 twofold cross-validation protocol, K5 fivefold cross-validation protocol, K10 tenfold cross-validation protocol
Table 4.
Four performance evaluation parameters for 4 classifiers
| Protocol types | Classifier types | Performance evaluation parameters | |||
|---|---|---|---|---|---|
| SE (%) | PPV (%) | NPV (%) | FM (%) | ||
| K2 | NB | 92.11 | 92.75 | 33.16 | 92.43 |
| DT | 99.12 | 90.56 | 37.28 | 94.64 | |
| AB | 96.04 | 94.42 | 57.91 | 95.22 | |
| RF | 99.56 | 93.25 | 89.98 | 96.30 | |
| K5 | NB | 92.05 | 93.02 | 33.71 | 92.53 |
| DT | 99.18 | 90.59 | 38.00 | 94.68 | |
| AB | 96.60 | 95.39 | 64.93 | 95.99 | |
| RF | 99.54 | 94.29 | 91.53 | 96.84 | |
| K10 | NB | 92.13 | 92.74 | 34.08 | 92.43 |
| DT | 99.48 | 90.06 | 40.25 | 94.52 | |
| AB | 96.81 | 95.40 | 67.49 | 96.09 | |
| RF | 99.57 | 94.34 | 92.59 | 96.88 | |
Bold values indicate the proposed method results
SE sensitivity, PPV positive predictive value, NPV negative predictive value, FM F-measure
Figure 4 shows the effect of varying data size on the classification accuracy of four classifiers for three partition protocols. We divide the actual data size (n) into ten parts as follows: 656, 1312, 1968, 2624, 3281, 3937, 4593, 52,49, 5905, and 6561. Now, we take these training sets and calculate the classification accuracy of four classifiers for three partition protocols. It is observed that the net generalization yields the generalization cutoff of 70% of the cohort (patient pool of 4593 patients). That means we need at least 70% patients to achieve the generalization. It is observed that the classification accuracy is increased by increasing the data size (n). Figure 4 also shows the classification accuracy of all classifiers increases with increases in data size and RF-based classifier is performed better than others. In Appendix 2 shows the system accuracy of four classifiers varying of data sizes for K2 protocol (Table 10 in Appendix 2), K5 protocol (Table 11 in Appendix 2), and K10 protocol (Table 12 in Appendix 2). Then the system mean accuracy is calculated by averaging the classification accuracy of all classifiers over varying data sizes for three partition protocols (K2, K5, and K10). Table 5 shows the system mean accuracy of four classifiers for three protocols. It is also showed that RF-based classifier is performed better compared to others.
Fig. 4.
Effect of varying data size: Accuracy vs data size (n) for three partition protocols. a K2 protocol; b K5 protocol; and c K10 protocol. Medical arrows indicate the proposed method’s result
Table 10.
System accuracy of 4 classifiers varying data sizes for K2 protocol
| Data size | Classifier types | |||
|---|---|---|---|---|
| NB | DT | AB | RF | |
| 656 | 84.75 | 85.34 | 85.25 | 87.82 |
| 1312 | 87.20 | 88.66 | 89.82 | 91.46 |
| 1968 | 87.48 | 90.30 | 90.00 | 91.57 |
| 2624 | 87.16 | 89.80 | 90.37 | 92.26 |
| 3281 | 86.35 | 89.14 | 90.72 | 92.45 |
| 3937 | 86.70 | 90.09 | 91.17 | 92.88 |
| 4593 | 86.23 | 89.65 | 91.59 | 93.19 |
| 5249 | 86.77 | 89.93 | 91.94 | 93.62 |
| 5905 | 86.85 | 90.07 | 92.37 | 93.96 |
| 6561 | 86.58 | 89.78 | 92.61 | 94.10 |
Bold values indicate the accuracy stability of all classifiers at 70% and above of the dataset for K2, K5 and K10 protocols
Table 11.
System accuracy of 4 classifiers varying data sizes for K5 protocol
| Data size | Classifier types | |||
|---|---|---|---|---|
| NB | DT | AB | RF | |
| 656 | 84.75 | 85.34 | 85.25 | 87.82 |
| 1312 | 87.20 | 88.66 | 89.82 | 91.46 |
| 1968 | 87.48 | 90.30 | 90.00 | 91.57 |
| 2624 | 87.16 | 89.80 | 90.37 | 92.26 |
| 3281 | 86.35 | 89.14 | 90.72 | 92.45 |
| 3937 | 86.70 | 90.09 | 91.17 | 92.88 |
| 4593 | 86.23 | 89.65 | 91.59 | 93.19 |
| 5249 | 86.77 | 89.93 | 91.94 | 93.62 |
| 5905 | 86.85 | 90.07 | 92.37 | 93.96 |
| 6561 | 86.58 | 89.78 | 92.61 | 94.10 |
Bold values indicate the accuracy stability of all classifiers at 70% and above of the dataset for K2, K5 and K10 protocols
Table 12.
System accuracy of 4 classifiers varying data sizes for K10 protocol
| Data size | Classifier types | |||
|---|---|---|---|---|
| NB | DT | AB | RF | |
| 656 | 85.61 | 88.64 | 88.18 | 90.91 |
| 1312 | 85.50 | 87.33 | 88.55 | 90.46 |
| 1968 | 86.55 | 91.22 | 91.42 | 92.69 |
| 2624 | 86.37 | 88.47 | 90.53 | 92.06 |
| 3281 | 86.80 | 89.70 | 91.22 | 93.11 |
| 3937 | 85.30 | 89.19 | 91.24 | 92.89 |
| 4593 | 86.25 | 90.07 | 91.98 | 93.51 |
| 5249 | 86.76 | 89.73 | 92.10 | 93.66 |
| 5905 | 87.07 | 90.20 | 92.94 | 94.31 |
| 6561 | 86.22 | 89.21 | 92.59 | 93.86 |
Bold values indicate the accuracy stability of all classifiers at 70% and above of the dataset for K2, K5 and K10 protocols
Table 5.
System mean accuracy (%) of four classifiers for three partition protocols
| Protocol types | Classifier types | |||
|---|---|---|---|---|
| NB | DT | AB | RF | |
| K2 | 86.67 | 89.52 | 90.79 | 92.54 |
| K5 | 86.61 | 89.28 | 90.58 | 92.33 |
| K10 | 86.24 | 89.38 | 91.08 | 92.75 |
Bold values indicate the proposed method results
Receiver operating characteristics (ROC) analysis
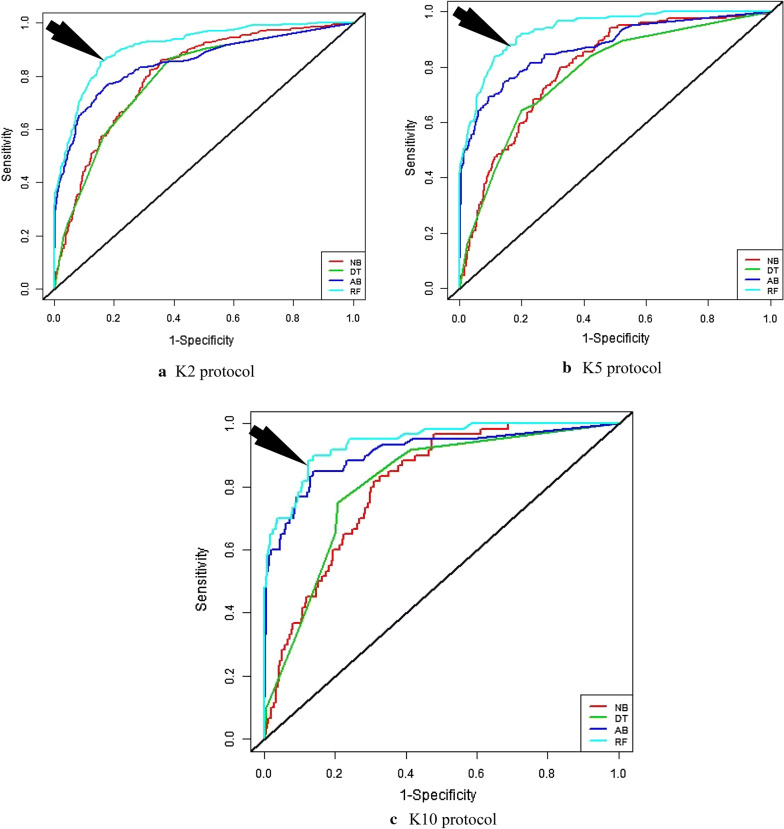
Receiver operating characteristics (ROC) is a graphical plot that is created by plotting sensitivity versus ‘1-specificity’. The area under the curve (AUC) which is computed from ROC curve is the indicator to evaluate the performance of the classifiers [45]. The value of the AUC lies between ‘0’ to ‘1’. R-i386 3.6.1 statistical software was used to compute the value of AUC from ROC curves. Figure 5 shows ROC curves of four classifiers for three partition protocols (K2, K5, and K10). It is observed that RF-based classifier is better for all partition protocols than NB, DT, and AB, respectively. The corresponding AUC values are presented in Table 6. It is also observed that the AUC of RF-based classifier for K10 protocol is 0.95 while NB, DT and AB are 0.82, 0.78, and 0.90, respectively.
Fig. 5.
ROC curves of four classifiers for three partition protocols: a K2 protocol; b K5 protocol; and c K10 protocol. Medical arrows indicate the proposed method results
Table 6.
Comparison of AUC of four classifiers for three partition protocols
| Classifier types | Protocol types | ||
|---|---|---|---|
| K2 | K5 | K10 | |
| NB | 0.80 | 0.81 | 0.82 |
| DT | 0.78 | 0.78 | 0.78 |
| AB | 0.86 | 0.89 | 0.90 |
| RF | 0.91 | 0.94 | 0.95 |
Bold values indicate the proposed method results
AUC area under the curve
Validations of the proposed method
To validate our proposed method, we have used Indian liver patient’s dataset. The dataset has been taken from the University of California, Irvine (UCI) machine learning data repository [46]. The dataset consists of ten attributes and 583 patients. There are 416 liver patients and 167 non-liver patients. Our result denominates that RF based classifier gives the highest accuracy compared to others. Therefore, we may conclude that our proposed method is the best classifier for both diabetes dataset and Indian liver patient’s dataset. It indicates that our proposed method is validated (see Table 7).
Table 7.
Validation of the proposed method using liver patient’s dataset
| Classifier types | Protocol types | ||
|---|---|---|---|
| K2 | K5 | K10 | |
| NB | 55.85 | 55.38 | 55.21 |
| DT | 66.83 | 67.29 | 67.62 |
| AB | 69.24 | 69.60 | 70.45 |
| RF | 70.42 | 70.44 | 70.59 |
Bold values indicate the proposed method results
Discussion
In this study, we have presented the ML-based system risk stratification to classify the patients into two classes as diabetic and control. Moreover, LR-based model has been adopted to determine the high risk factors of diabetes disease. The high risk factors have been selected based on p-values and odds ratio (OR). Moreover, four classifiers have been also adapted and compared their performance based on ACC, SE, PPV, NPV, FM, and AUC, respectively. Further, three partition protocols have been applied for generalization of classification and this process has been repeated for T = 20 times to reduce the variability. Also we have validated the ML-based system using Indian liver patient’s dataset. The next section represents the key comparison between proposed ML-based system and previous work.
Key difference between proposed ML-based system and previous work
Several papers in the literature are focused on the identification of high risk factors as well as sophisticated classification the diabetes disease. Zou et al. [47] studied on the diagnosis of diabetes dataset. The dataset was taken from the hospital physical examination data in Luzhou, China. The dataset contained 14 attributes and consisted of 220,680 patients. Among them, 151,598 (69%) patients were diabetic and 69,082 (31%) were control. They applied PCA and minimum redundancy maximum relevance (mRMR) to reduce the dimensionality and also K5 cross-validation protocol adopted to examine the data. They also applied three classifiers as: DT, NN, and RF to classify the diabetes patients and demonstrated that RF based classifier gave the highest classification accuracy of 80.84%. Maniruzzaman et al. [12] applied LDA, QDA, NB, and Gaussian process classification (GPC) on the Pima Indian diabetes (PID) dataset to classify the diabetic patients. They adopted two partition protocols as K5, and K10. They showed that GP-based classifier with radial basis kernel (RBF) gave the highest classification accuracy approximately 82.00%. The same authors firstly identified the outliers in the PID dataset using inter-quartile range (IQR). If the outliers were detected, then they were replaced the outliers by the median. They also replaced the missing value by median. They adopted six feature selection techniques as PCA, LR, MI, ANOVA, FDR, and RF to extract the features. They also adopted ten classifiers as: LDA, QDA, NB, NN, GPC, SVM, AB, LR, DT, and RF to classify the diabetes patients. They found that the combination of RF based feature selection technique and RF based classifier gave the highest classification accuracy of 92.26% compared to others [13].
Ahuja et al. [30] used PID dataset in his study. The dataset consisted of 768 patients and 10 attributes. The dataset had some missing values and they were replaced the missing values by median. LDA was used to extract the feature selection. They applied five classification algorithms as: SVM, multi-layer perceptron (MLP), LR, RF, and DT. They showed that LDA with MLP based classifier gave the highest classification accuracy of 78.70%. Sisodia et al. [29] did not apply any feature selection techniques to extract the feature. They performed K10 protocol and applied SVM, NB, DT classifiers and got the highest accuracy 76.30% of NB classifier compared to others. Yu et al. [48] took diabetes dataset from the 1999–2004 US NHANES to develop a SVM model to classify diabetes patients. The dataset consisted of 6214 patients (1461 diabetic patients and 4853 patients were control). Firstly, they optimized the kernel and chose the best kernel for SVM based on the classification accuracy. They adopted K10 cross-validation protocol and four types of kernel as: linear, polynomial, sigmoid, and RBF respectively. They got that SVM with RBF kernel gave the highest classification accuracy of 83.50%. Semerdjian et al. [49] used 5515 total samples were available from the NHANES 1999 to 2004 dataset. They identified the highest risk factors based on RF. Five set of classifiers (LR, KNN, RF, Gradient boosting (GB), and RF) adopted to predict the diabetes status based on the 16 attributes and the performance of GB based classifier was higher (AUC: 0.84) compared to others. Mohapatra et al. [50] applied MLP and found that MLP gave the classification accuracy of 77.50%. Pei et al. [51] also applied DT and gave 94.20% classification accuracy. However, in this study; we have selected LR based feature selection technique to identify the risk factors of diabetes disease. The results show that the combination of LR and RF-based classifier gives 94.25% ACC and 0.95 AUC are the highest as compared to conventional techniques (see Table 8).
Table 8.
Key difference between proposed study and previous studies in literature
| SN | C1 | C2 | C3 | C4 | C5 | C7 | C8 | C9 | C10 | C11 | C12 | C13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Authors | Year | Dataset | DS | FST | Classifiers | PT | ACC | AUC | FM | Validations | G Vs. M | |
| 1 | Zou et al. [47] | 2018 | Diabetes | 220,680 | PCA | DT, NN, | K5 | 80.84 | No | No | Yes | No |
| mRMR | RF | |||||||||||
| 2 | Maniruzzaman et al. [12] | 2017 | PID | 768 | No | LDA, QDA | K5 | 81.97 | Yes | No | No | No |
| NB, GPC | K10 | |||||||||||
| 3 | Maniruzzaman et al. [13] | 2018 | PID | 768 | RF | LDA, QDA | K2 | 92.26 | Yes | No | No | Yes |
| LR | NB, NN | K4 | ||||||||||
| MI,PCA | GPC, SVM | K5 | ||||||||||
| ANOVA | AB, LR | K10 | ||||||||||
| FDR | DT, RF | JK | ||||||||||
| 4 | Ahuja et al. [30] | 2019 | PID | 768 | LDA | SVM, MLP | K2, K4 | 78.70 | No | Yes | No | No |
| LR, DT, RF | K5, K10 | |||||||||||
| 5 | Sisodia et al. [28] | 2018 | PID | 768 | No | SVM, NB, DT | K10 | 76.30 | Yes | Yes | No | No |
| 6 | Yu et al. [48] | 2010 | NHANES | 6314 | No | SVM | K10 | 83.50 | Yes | No | No | No |
| Diabetic | ||||||||||||
| 7 | Semerdjian et al. [49] | 2017 | NHANES | 5515 | RF | LR, KNN, RF GB, SVM | K10 | NA | Yes | No | No | |
| Diabetic | ||||||||||||
| 8 | Mohapatra et al. [50] | 2018 | PID | 768 | NA | MLP | No | 77.50 | No | No | No | No |
| 9 | Pei et al. [51] | 2018 | Diabetes | 10,436 | CS | DT | Tr: 70% | 94.20 | Yes | No | No | No |
| Test: 30% | ||||||||||||
| 10 | Proposed | 2019 | NHANES | 6561 | LR | NB, DT, AB, RF | K2, K5, K10 | 94.25 | Yes | Yes | Yes | Yes |
| Diabetic |
Bold value indicates the proposed method results
FST feature selection techniques, PT protocol types, DS data size, ACC accuracy in %, AUC area under the curve, FM F-measure, PCA principal component analysis, mRMR minimum redundancy maximum relevance, LDA linear discriminant analysis, QDA quadratic discriminant analysis, NB Naïve Bayes, GPC Gaussian process classification, SVM support vector machine, AB Adaboost, LR logistic regression, MI mutual information, ANOVA analysis of variance, FDR Fisher discriminant analysis, DT decision tree, NN neural network, RF random forest, KNN K-nearest neighborhood, GB gradient boosting, MLP multilayer perceptron, K2 twofold cross-validation protocol, K4 fourfold cross validation protocol, K5 fivefold cross-validation protocol, K10 tenfold cross-validation protocol, JK Jackknife protocol, G generalization, M memorization, PID Pima Indian diabetes, NHANES National Health and Nutrition Examination Survey, Tr training set
Strength and extension of the study
This paper shows the risk stratification and classification of diabetes patients while there are 6561 respondents with 657 diabetics and 5904 controls. Our result demonstrates that the overall accuracy of ML-based system is 90.62%. Moreover, the combination of LR based feature selection technique and RF-based classifier gives the highest classification accuracy of 94.25% and 0.95 AUC for K10 protocol. Nevertheless, the presented system can still be improved. Further, preprocessing techniques may be used to replace missing values by various missing value imputation techniques like: mean or median, expectation maximization (EM) algorithm, K-nearest neighbors (KNN), fuzzy K-means (FKM), and singular value decomposition (SVD). Moreover, there are various techniques of feature extraction, feature selection (PCA, different statistical tests, FDR, RF, etc.), and classifiers, namely: NN, GPC, SVM, deep learning (DL) and so on.
Summary of the current study
In this section, we summarize the current study at a glance as follows:
- Data:
- Extraction: Extract the dataset from the National Health and Nutrition Examination Survey (NHANES) into a dta (Stata) format.
- Data cleaning: Drop the missing values and outliers from the dataset.
- Feature extraction: Identify the high risk factors using LR for prediction.
- Modeling:
-
Model selection: The popular predictive models were selected for this project:(i) Naïve Bayes; (ii) Decision tree; (iii) Adaboost; and (iv) Random forest
- Data split: Discussed in the “data partitioning” section.
-
- Evaluation:
- Metrics: Reporting of accuracy, sensitivity, positive predictive value, negative predictive value, F-measure, and area under the curve to evaluate the classifiers.
- Interpretation: examining results of metrics to compare the classifiers and finally conclude the experiments.
Conclusion
Diabetes mellitus (DM) is commonly known as diabetes. It is a group of metabolic orders which are characterized by the high blood sugar. Our hypothesis was used in ML based system using LR-RF combination for feature selection technique and classifier gave the highest classification accuracy. Our results demonstrated that our proposed combination reached an accuracy of 94.25% for K10 protocol. Moreover, a comparative analysis was conducted using four classifiers, one feature selection technique, and three partition protocol (total 12) experiments. It would be interesting in future to see classification of other kinds of medical data to be adapted in such a framework creating a cost-effective and time-saving option for both diabetic patients and doctors.
Acknowledgements
The authors would like to acknowledge the contribution of Statistics Discipline, Science, Engineering and Technology School, Khulna University, Khulna-9208, Bangladesh. The authors also thank to the editor and reviewers for their comments and positive critique.
Appendix 1
See Table 9.
Appendix 2
Appendix 3
See Table 13.
Table 13.
List of abbreviations
| SN | Acronyms | Full form | SN | Acronyms | Full form |
|---|---|---|---|---|---|
| 1 | LR | Logistic regression | 25 | FST | Feature selection technique |
| 2 | DM | Diabetes mellitus | 26 | PT | Protocol types |
| 3 | OR | Odds ratio | 27 | mRMR | Minimum redundancy maximum relevance |
| 4 | NB | Naïve Bayes | 28 | LDA | Linear discriminant analysis |
| 5 | AB | Adaboost | 29 | QDA | Quadratic discriminant analysis |
| 6 | RF | Random forest | 30 | GPC | Gaussian process classification |
| 7 | DT | Decision tree | 31 | SVM | Support vector machine |
| 8 | ACC | Accuracy | 32 | NN | Neural network |
| 9 | SE | Sensitivity | 33 | KNN | K-nearest neighborhood |
| 10 | PPV | Positive predictive value. | 34 | GB | Gradient boosting |
| 11 | NPV | Negative predictive value | 35 | G | Generalization |
| 12 | FM | F-Measure | 36 | M | Memorization |
| 13 | AUC | Area under the curve | 37 | JK | Jackknife protocol |
| 14 | CI | Confidence interval | 38 | K2 | Two-fold cross-validation protocol |
| 15 | ML | Machine learning | 39 | K4 | Fourfold cross-validation protocol |
| 16 | SD | Standard deviation | 40 | K5 | Fivefold cross-validation protocol |
| 17 | MI | Mutual information | 41 | K10 | Tenfold cross-validation protocol |
| 18 | PCA | Principal component analysis | 42 | PID | Pima Indian diabetes |
| 19 | ANOVA | Analysis of variance | 43 | MLP | Multilayer perceptron |
| 20 | FDR | Fisher discriminant ratio | 44 | p-value | Probability value |
| 21 | Tr. | Training | 45 | DS | Data size |
| 22 | EM | Expectation maximization | 46 | DL | Deep learning |
| 23 | FKM | Fuzzy K-means | 47 | SVD | Singular value decomposition |
| 24 | NHANES | National Health and Nutrition Examination Survey |
Author contributions
Md. Maniruzzaman: Statistical analysis, draft the original manuscript, and principal investigator and management of the project. Md. Jahanur Rahman: Acquisition of data, interpretation of the results and methodology; Benojir Ahammed: Machine learning concepts and design. Md. Menhazul Abedin: Data preprocessing, English writing, strategy, and interpretation.
Funding
No fund received for this project.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
No ethics approval is required for this dataset.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Supplement 1):S62–S69. doi: 10.2337/dc10-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarwar N, Gao P, Seshasai SR. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease. Lancet. 2010;375(9733):2215–2222. doi: 10.1016/S0140-6736(10)60484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lonappan A, Bindu G, Thomas V, Jacob J, Rajasekaran C, Mathew KT. Diagnosis of diabetes mellitus using microwaves. J Electromagn Waves Appl. 2007;21(10):1393–1401. [Google Scholar]
- 4.Krasteva A, Panov V, Krasteva A, Kisselova A, Krastev Z. Oral cavity and systemic diseases—diabetes mellitus. Biotechnol Biotechnol Equip. 2011;25(1):2183–2186. [Google Scholar]
- 5.Nathan DM. Long-term complications of diabetes mellitus. N Engl J Med. 1993;328(23):1676–1685. doi: 10.1056/NEJM199306103282306. [DOI] [PubMed] [Google Scholar]
- 6.NCD Risk Factor Collaboration (NCD-RisC) Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 192 million participants. Lancet. 2016;387(10026):1377–1396. doi: 10.1016/S0140-6736(16)30054-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmet P, Alberti KG, Magliano DJ, Bennett PH. Diabetes mellitus statistics on prevalence and mortality: facts and fallacies. Nat Rev Endocrinol. 2016;12(10):616. doi: 10.1038/nrendo.2016.105. [DOI] [PubMed] [Google Scholar]
- 8.Bharath C, Saravanan N, Venkatalakshmi S. Assessment of knowledge related to diabetes mellitus among patients attending a dental college in Salem city—a cross sectional study. Braz Dental Sci. 2017;20(3):93–100. [Google Scholar]
- 9.Danaei G, Finucane MM, Lu Y, Singh GM, Cowan MJ, Paciorek CJ, Rao M. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet. 2011;378(9785):31–40. doi: 10.1016/S0140-6736(11)60679-X. [DOI] [PubMed] [Google Scholar]
- 10.Iancu, I., Mota, M., & Iancu, E. Method for the analysing of blood glucose dynamics in diabetes mellitus patients. In 2008 IEEE international conference on automation, quality and testing, robotics, vol. 3; 2008. pp. 60–65.
- 11.Robertson G, Lehmann ED, Sandham W, Hamilton D. Blood glucose prediction using artificial neural networks trained with the AIDA diabetes simulator: a proof-of-concept pilot study. J Electr Comput Eng. 2012;2011:2–13. [Google Scholar]
- 12.Maniruzzaman M, Kumar N, Abedin MM, Islam MS, Suri HS, El-Baz AS, Suri JS. Comparative approaches for classification of diabetes mellitus data: machine learning paradigm. Comput Methods Programs Biomed. 2017;152:23–34. doi: 10.1016/j.cmpb.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Maniruzzaman M, Rahman MJ, Al-MehediHasan M, Suri HS, Abedin MM, El-Baz A, Suri JS. Accurate diabetes risk stratification using machine learning: role of missing value and outliers. J Med Syst. 2018;42(5):92. doi: 10.1007/s10916-018-0940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava SK, Singh SK, Suri JS. Healthcare text classification system and its performance evaluation: a source of better intelligence by characterizing healthcare text. J Med Syst. 2018;42(5):97. doi: 10.1007/s10916-018-0941-6. [DOI] [PubMed] [Google Scholar]
- 15.Luo G. Automatically explaining machine learning prediction results: a demonstration on type 2 diabetes risk prediction. Health Inf Sci Syst. 2016;4(1):2. doi: 10.1186/s13755-016-0015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shakeel PM, Baskar S, Dhulipala VS, Jaber MM. Cloud based framework for diagnosis of diabetes mellitus using K-means clustering. Health Inf Sci Syst. 2018;6(1):16. doi: 10.1007/s13755-018-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo G. MLBCD: a machine learning tool for big clinical data. Health Inf Sci Syst. 2015;3(1):3. doi: 10.1186/s13755-015-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo G. PredicT-ML: a tool for automating machine learning model building with big clinical data. Health Inf Sci Syst. 2016;4(1):5. doi: 10.1186/s13755-016-0018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahle G. Ethiopic maternal care data mining: discovering the factors that affect postnatal care visit in Ethiopia. Health Inf Sci Syst. 2016;4(1):4. doi: 10.1186/s13755-016-0017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah S, Luo X, Kanakasabai S, Tuason R, Klopper G. Neural networks for mining the associations between diseases and symptoms in clinical notes. Health Inf Sci Syst. 2019;7(1):1. doi: 10.1007/s13755-018-0062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauder RA, Khoshgoftaar TM. The effects of varying class distribution on learner behavior for medicare fraud detection with imbalanced big data. Health Inf Sci Syst. 2018;6(1):9. doi: 10.1007/s13755-018-0051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deniz E, Şengür A, Kadiroğlu Z, Guo Y, Bajaj V, Budak Ü. Transfer learning based histopathologic image classification for breast cancer detection. Health Inf Sci Syst. 2018;6(1):18. doi: 10.1007/s13755-018-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashour AS, Hawas AR, Guo Y. Comparative study of multiclass classification methods on light microscopic images for hepatic schistosomiasis fibrosis diagnosis. Health Inf Sci Syst. 2018;6(1):7. doi: 10.1007/s13755-018-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banchhor SK, Londhe ND, Araki T, Saba L, Radeva P, Laird JR, Suri JS. Wall-based measurement features provides an improved IVUS coronary artery risk assessment when fused with plaque texture-based features during machine learning paradigm. Comput Biol Med. 2017;91:198–212. doi: 10.1016/j.compbiomed.2017.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Kuppili V, Biswas M, Sreekumar A, Suri HS, Saba L, Edla DR, Suri JS. Extreme learning machine framework for risk stratification of fatty liver disease using ultrasound tissue characterization. J Med Syst. 2017;41(10):152. doi: 10.1007/s10916-017-0797-1. [DOI] [PubMed] [Google Scholar]
- 26.Banchhor SK, Londhe ND, Araki T, Saba L, Radeva P, Khanna N, Suri JS. Calcium detection, its quantification, and grayscale morphology-based risk stratification using machine learning in multimodality big data coronary and carotid scans: a review. Comput Biol Med. 2018;101:184–198. doi: 10.1016/j.compbiomed.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Bashir S, Qamar U, Khan FH. IntelliHealth: a medical decision support application using a novel weighted multi-layer classifier ensemble framework. J Biomed Inform. 2016;59:185–200. doi: 10.1016/j.jbi.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Zou Q, Liu B, Liu X. Exploratory predicting protein folding model with random forest and hybrid features. Curr Proteomics. 2014;11:289–299. [Google Scholar]
- 29.Sisodia D, Sisodia DS. Prediction of diabetes using classification algorithms. Procedia Comput Sci. 2018;132:1578–1585. [Google Scholar]
- 30.Ahuja R, Vivek V, Chandna M, Virmani S, Banga A. Comparative study of various machine learning algorithms for prediction of Insomnia. In: Advanced classification techniques for healthcare analysis; 2019. p. 234–257.
- 31.Genuer R, Poggi JM, Tuleau-Malot C. Variable selection using random forests. Pattern Recogn Lett. 2010;31(14):2225–2236. [Google Scholar]
- 32.Degenhardt F, Seifert S, Szymczak S. Evaluation of variable selection methods for random forests and omics data sets. Brief Bioinform. 2017;20(2):492–503. doi: 10.1093/bib/bbx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Austin PC, Tu JV. Automated variable selection methods for logistic regression produced unstable models for predicting acute myocardial infarction mortality. J Clin Epidemiol. 2004;57(11):1138–1146. doi: 10.1016/j.jclinepi.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Maniruzzaman M, Suri HS, Kumar N, Abedin MM, Rahman MJ, El-Baz A, Suri JS. Risk factors of neonatal mortality and child mortality in Bangladesh. J Glob Health. 2018;8(1):1–16. doi: 10.7189/jogh.08.010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput Methods Programs Biomed. 2017;150:9–22. doi: 10.1016/j.cmpb.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: a first comparative study of its kind. Comput Methods Programs Biomed. 2016;126:98–109. doi: 10.1016/j.cmpb.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 37.Elssied NOF, Ibrahim O, Osman AH. A Novel feature selection based on one-way ANOVA F-Test for e-mail spam classification. Res J Appl Sci Eng Technol. 2014;7(3):625–638. [Google Scholar]
- 38.Shaharum SM, Sundaraj K, Helmy K. Performance analysis of feature selection method using ANOVA for automatic wheeze detection. Jurnal Teknologi. 2015;77(7):2015. [Google Scholar]
- 39.Wang S, Li D, Song X, Wei Y, Li H. A feature selection method based on improved fisher’s discriminant ratio for text sentiment classification. Expert Syst Appl. 2011;38(7):8696–8702. [Google Scholar]
- 40.Cover TM. Geometrical and statistical properties of systems of linear inequalities with applications in pattern recognition. IEEE Trans Electron Comput. 1965;14(3):326–334. [Google Scholar]
- 41.Quinlan JR. Induction of decision trees. Mach Learn. 1986;1(1):81–106. [Google Scholar]
- 42.Hu W, Hu W, Maybank S. Adaboost-based algorithm for network intrusion detection. IEEE Trans Syst Man Cybern B. 2008;38(2):577–583. doi: 10.1109/TSMCB.2007.914695. [DOI] [PubMed] [Google Scholar]
- 43.Breiman L. Random forest. Mach Learn. 2001;45:5–32. [Google Scholar]
- 44.Liao Z, Ju Y, Zou Q. Prediction of G protein-coupled receptors with SVM-prot features and random forest. Scientifica. 2016;2016:1–10. doi: 10.1155/2016/8309253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acharya UR, Chua CK, Lim TC, Dorithy, Suri JS. Automatic identification of epileptic EEG signals using nonlinear parameters. J Mech Med Biol. 2009;9(4):539–553. [Google Scholar]
- 46.Ramana BV, Babu MSP, Venkateswarlu NB. A critical comparative study of liver patients from USA and INDIA: an exploratory analysis. Int J Comput Sci Issues. 2012;9(3):506. [Google Scholar]
- 47.Zou Q, Qu K, Luo Y, Yin D, Ju Y, Tang H. Predicting diabetes mellitus with machine learning techniques. Front Genet. 2018;9(515):1–10. doi: 10.3389/fgene.2018.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu W, Liu T, Valdez R, Gwinn M, Khoury MJ. Application of support vector machine modeling for prediction of common diseases: the case of diabetes and pre-diabetes. BMC Med Inform Decis Mak. 2010;10(1):16–23. doi: 10.1186/1472-6947-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semerdjian J, Frank S. An ensemble classifier for predicting the onset of type II diabetes. arXiv:1708.07480 (2017).
- 50.Mohapatra SK, Swain JK, Mohanty MN. Detection of diabetes using multilayer perceptron. In: International conference on intelligent computing and applications, 2019, pp. 109–116.
- 51.Pei D, Zhang C, Quan Y, Guo Q. Identification of potential type II diabetes in a chinese population with a sensitive decision tree approach. J Diabetes Res. 2019;2019:1–7. doi: 10.1155/2019/4248218. [DOI] [PMC free article] [PubMed] [Google Scholar]