SUMMARY
RNA molecules fold into complex three-dimensional structures that sample alternate conformations ranging from minor differences in tertiary structure dynamics to major differences in secondary structure. This allows them to form entirely different substructures with each population potentially giving rise to a distinct biological outcome. The substructures can be partitioned along an existing energy landscape given a particular static cellular cue or can be shifted in response to dynamic cues such as ligand binding. We review a few key examples of RNA molecules that sample alternate conformations and how these are capitalized on for control of critical regulatory functions.
1. Introduction
RNA plays an important role in a large number of cellular processes, and, in many cases, it is the RNA that drives function. RNA can fold into complex three-dimensional structures, enabling it to perform diverse biological functions beyond the simple transfer of genetic information. For example, in the case of mRNAs, these structural elements have been shown to regulate every aspect of the mRNA’s voyage through the cell—from transcription, splicing, and nuclear export to trafficking, localization, translation, and stability. Even ribosomal RNA (rRNA), the most abundant form of RNA in cells, samples alternate structures to control translation (Zamir et al. 1969; McGinnis et al. 2015). Although we have for a long time appreciated the direct relationship between RNA structures and the functions they regulate, we are just starting to fathom the depth and extent of the regulatory information contained in RNA. Furthermore, we are just beginning to understand that, unlike proteins, an RNA with a particular sequence has the potential to form entirely different structures and can therefore be regulated to be partitioned into distinct subpopulations, with each population giving rise to a distinct biological outcome.
The potential for RNA to form energetically similar conformations comes from the chemical simplicity of RNA molecules, which are made up of combinations of nucleotides joined together by phosphodiester bonds. To begin with, there are only four nucleotides, making it possible to find alternate identical complementary sequences for a given stretch of RNA. Second, each nucleotide is capable of making hydrogen bonding pairs with each of the others (Leontis and Westhof 1998; Leontis and Westhof 2001), with some combinations rivaling the stability of Watson–Crick base pairs (Leontis et al. 2002; Sponer et al. 2005). Third, the canonical G-C and A-U Watson–Crick pairs are nearly isosteric (Westhof 2014), making base pairs in a stem interchangeable and allowing for multiple ways of forming RNA stems—the basic building blocks of RNA structures. Finally, a structure with a seemingly energetically suboptimal base-pairing pattern can still form because of stabilization by tertiary interactions, such as loops, bulges, helical junctions, and long-range interactions (Popenda et al. 2010; Petrov et al. 2013; Chojnowski et al. 2014; Parlea et al. 2016).
All of the above characteristics allow RNA molecules to sample alternate conformations that range from minor differences in tertiary structure dynamics to major differences in secondary structure. Accordingly, alternate RNA structures can be loosely grouped into categories based on the timescales at which the transitions between structures occur—for instance: structures that differ considerably in secondary structures occur at the millisecond to second timescales; structures that differ at the base pair or tertiary structure levels occur at the microsecond to second timescales; and structures that differ because of interhelical or loop dynamics occur at the picosecond to microsecond timescales (Mustoe et al. 2014). Nature has made use of all of these transitions to control regulatory networks, with new functions using alternate RNA structures being uncovered on a regular basis.
To coexist, alternate structures by necessity have to be nearly isoenergetic. Such structures can be a priori appropriately populated along an equilibrium given a particular static cellular environment, or the partitioning can be shifted in response to dynamic cues like ligand binding and temperature changes. Whereas the use of static cues allows for structural modulation within a given energetic landscape, the use of dynamic cues changes the fundamental landscape itself and allows access to previously inaccessible structures. Many examples have been found for regulation using dynamic cues, but to date only a few examples have been discovered for regulation using static cues. In this review, we will use a few key examples of interactions that occur primarily at the microsecond and higher timescales to highlight alternate RNA structures controlled by both static and dynamic cues, and how they can be used to fine tune regulatory functions.
2. Alternate RNA structures controlled by static cues
Although it is intuitive that RNA elements can be partitioned into alternate structures in response to dynamic or changing cues, as will be discussed in more detail further on in this review, the partitioning of conformations based on a static environment is also possible and can be capitalized on for function. The simplest partitioning of alternate RNA structures is through equilibria set by fixed cellular parameters such as temperature, pH, thermodynamic stability, concentration of ions, etc. Furthermore, this partitioning can occur on every level of organization, from primary structures to tertiary structures to the binding of trans protein factors.
2.1. Primary Structure
A recent study by Kharytonchyk et al. on human immunodeficiency virus-1 (HIV-1) transcription shows the potential to use sequence heterogeneity during transcription initiation to leverage alternate RNA structures (Kharytonchyk et al. 2016). In this case, the subtle difference between starting the mRNA with one guanosine as opposed to two or three guanosines defines the biological fate of the mRNA.
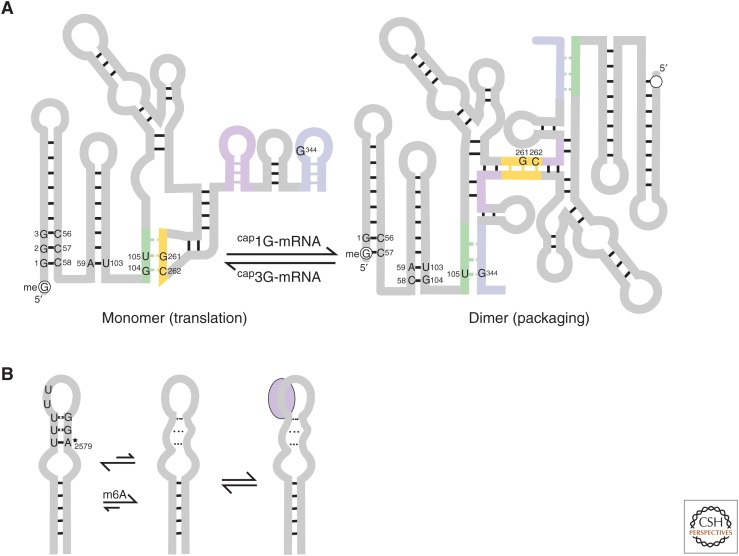
In brief, the full-length HIV mRNA plays a dual functional role: It is not only used as a transcript for the synthesis of essential Gag and Gag-Pol viral polyproteins, but also as the genetic material that gets packaged into virions during budding (D’Souza and Summers 2005). The full-length HIV mRNA also has two structural states: It not only exists in a monomeric state as per conventional mRNAs, but also in a dimeric state (Fig. 1A). It has been shown that the dual functions (translational vs. genomic) and the dual structures (monomer vs. dimer) of the viral RNA are correlated with each other. A region called the packaging signal (Ψ site) in the 5′ leader, containing nucleocapsid (NC, the genome packaging protein)-binding sites and a dimerization initiation site (DIS) (D’Souza and Summers 2005), is designed to dictate this correlation. Namely, the monomeric form has the DIS and NC-binding sites sequestered with the start codon exposed, making this mRNA conformation translation-competent. In contrast, the dimeric form exposes the NC-binding site and sequesters the start codon, making this mRNA conformation packaging-competent (Lu et al. 2011).
Figure 1.
(A) The monomeric and dimeric states of the human immunodeficiency virus 1 (HIV-1) 5′ leader. Transcripts with cap2G or cap3G favor the monomeric form, whereas transcripts with cap1G favor the dimeric form, largely because of the dissolution or formation of the C58-G104 base pair, respectively. (B) Sampling of alternate conformations by the metastasis-associated lung adenocarcinoma transcript 1 long noncoding RNA (MALAT1 lncRNA). The RNA shows increased dynamics of the upper hairpin structure on N6-methyladenosine (m6A)-modification, as indicated by the dotted lines, allowing for increased sampling of the protein-binding conformation (lavender oval). The star indicates the location for the potential m6A modification.
Kharytonchyk et al. found that the partitioning of the two conformations is defined by the variation in transcriptional start site usage (Kharytonchyk et al. 2016). Transcription of the HIV-1 genome can initiate at any of three guanosines at the transcription start site. The resulting transcripts will start with either cap1G, cap2G, or cap3G sequences immediately preceding the transactivation response (TAR) element in the 5′ leader. In the dimeric form, the terminal C58-G104 base pair of the poly(A) stem directly connects to both the TAR stem (via C58) and the DIS:U5 interaction stem (via G104) (Fig. 1A). On the other hand, in the monomeric form, G104 forms a base pair with C262. Kharytonchyk et al. showed that the equilibrium between the monomeric and dimeric forms can be manipulated simply by affecting the formation of the C58-G104 base pair. Transcripts with cap2G or cap3G at the start of the TAR stem can compete with the poly(A) stem for C58, thus freeing G104 to base-pair with C262. This extends the DIS:U5 interaction by one base pair and drives the equilibrium to the monomeric form. The extension of the DIS:U5 stem by this single additional base pair is estimated to stabilize the monomeric form by −3 to −10 kcal/mol compared with the dimeric form. Even at the lower end of this estimate, the structural equilibrium for transcripts that start with cap2G or cap3G will be shifted toward the monomeric form. Importantly, in the monomeric forms, the 5′ cap is exposed and would be available for cap recognition by the cap-binding protein eukaryotic translation initiation factor 4E (eIF4E). On the other hand, in the cap1G transcripts, the 5′ cap is sequestered as part of a base pair in the TAR stem and is not available for cap recognition. Instead, these transcripts are prone to dimerization and are packaged. It seems then that HIV-1 may have capitalized on the existence of sequence heterogeneity to partition its mRNAs into distinct pools (cap3G ⇒ monomer ⇒ cap-exposed ⇒ translation, cap1G ⇒ dimer ⇒ cap-sequestered ⇒ packaging). Although the use of sequence heterogeneity as a regulatory mechanism has been shown only in HIV-1, it is possible that other retroviruses also use this mechanism, because the existence of alternate monomeric and dimeric conformations is a common theme in retroviral biology (D’Souza and Summers 2005).
The relative populations of structures could potentially be defined by the inherent initiation properties of the RNA polymerase, which viruses may have taken advantage of during evolution. It is also possible that the relative use of each transcription start site could be altered in response to modulation by other regulatory factors present in the cell that would change the initiation properties of the polymerase. In HIV-1, Kharytonchyk et al. suggest that such temporal changes could lead to the production of cap2G /cap3G early in the viral life cycle, followed by an increase in the cap1G production for genome packaging (Kharytonchyk et al. 2016).
It is important to note that transcriptional start site heterogeneity is not limited to retroviruses. Many cellular genes also initiate transcription at heterogeneous start sites, the most pertinent example being the “twinned” TATA-driven promoters, which have start sites predominantly separated by 0–3 nucleotides (nt) (Carninci et al. 2006; Ponjavic et al. 2006). In addition, for dispersed promoters where the start sites are distributed over a larger region spanning between 50 and 100 nt in length (Juven-Gershon and Kadonaga 2010), the implications for structural heterogeneity at the 5′ end of transcripts, and the potential regulatory mechanisms at play, may be even more pronounced. The usage of specific transcriptional start sites has been shown to vary between different cell types and also between normal and disease states, suggesting that trans factors can also be involved (Ponjavic et al. 2006; Frith et al. 2008).
Although inhibition of translation by sequestration of the 5′ cap in HIV-1 may at first glance seem to be a unique system, a recent discovery of a translation inhibitory element (TIE) located in close proximity to the 5′ cap in the Hox mRNA suggests that the inhibition of cap-dependent translation may be more common than expected (Shi and Barna 2015). In this system, protein production occurs via specialized translation, with ribosomes accessing the mRNA through internal ribosome entry sites (IRES) concomitant with active inhibition of canonical translation by TIE. It has been hypothesized that differential regulation of the TIE and IRES motifs in the Hox mRNA provides the control required for ideal expression of the Hox transcripts (Shi and Barna 2015). Nevertheless, it will be interesting to see if this inhibition is also maintained by alternate RNA structures.
It is also worth noting that the concept of differences in primary sequence resulting in alternate RNA structures is not limited to transcription start sites; in fact, single point mutations can alter the entire energetic landscape. For example, riboSNitches are regulatory RNA structural elements in which single-nucleotide polymorphisms (SNPs) result in the formation of alternate RNA structures (Halvorsen et al. 2010; Ritz et al. 2012; Corley et al. 2015). RiboSNitches have been implicated in causing global RNA structural rearrangements that can be directly linked to disease states. In one example, Kutchko et al. used selective 2′-hydroxyl acylation with primer extension (SHAPE)-directed Boltzmann suboptimal sampling to show that the retinoblastoma 1 5′-untranslated region (RB1 5′ UTR) samples three major conformations (Kutchko et al. 2015). Single-nucleotide variants in RB1 that cause retinoblastoma were found to limit the amount of dynamic sampling and cause the stabilization of single conformational states, leading to an increase in protein expression (Kutchko et al. 2015). Other potential riboSNitches have been identified through combined analyses using human genome-wide association studies, RNA-folding algorithms, and genome-wide enzymatic structure probing (Halvorsen et al. 2010; Wan et al. 2014).
In the last few years, it has also become evident that posttranscriptional RNA modifications serve as another layer of primary structure-based regulation, with modifications implicated in splicing, localization, translation, and decay (Lewis et al. 2017; Roundtree et al. 2017). Some forms of RNA modification include N6-methyladenosine (m6A) (Lavi et al. 1977), N1-methyladenosine (m1A) (Dominissini et al. 2016), 5-methylcytosine (m5C) (Desrosiers et al. 1974), 5-hydroxymethylcytosine (hm5C) (Guerrero et al. 2014), pseudouridine (Ψ) (Cohn 1960), and 2′-O-methylated ribose (2′-OMe) (Schibler and Perry 1977), with m6A as the most commonly found in mRNA. Such modifications change the folding landscape of RNA by altering their base- pairing and stacking capabilities, electrostatics, and sterics (Chawla et al. 2015). For example, in m6A, Watson–Crick base-pairing leads to the methyl group being forced into an anticonformation, in which steric hindrance between the methyl group and the N7 leads to destabilization of the interaction (Roost et al. 2015). In conjunction, the m6A modification also stabilizes single strands because of its enhanced base-stacking capabilities (Roost et al. 2015). Zhou et al. describe a mechanism in which the MALAT1 lncRNA (metastasis-associated lung adenocarcinoma transcript 1 long noncoding RNA) has a hairpin motif that samples alternate conformations (Fig. 1B) (Zhou et al. 2016). In the presence of m6A modifications, the stem architecture is retained but the hairpin loop is more dynamic and samples a conformation similar to that of the protein heterogenous nuclear ribonucleoprotein C (hnRNPC)-bound form. This destabilized conformation allows for better access to the protein-binding site, thereby increasing protein affinity by eightfold. (m6A ⇒ ↑loop dynamics ⇒ ↑protein-binding site access ⇒ ↑protein affinity.) Mechanisms that leverage the sampling of alternate conformations within an altered folding landscape to drive regulation may be more general and need to be further investigated.
2.2. Secondary Structure
A few examples are beginning to emerge in which RNA domains sample conformations with significant differences in secondary structure. Additionally, structural complexity can involve more than just two equilibrating structures. One example of this involves mutually exclusive splicing, a form of alternative splicing in which only one exon in a set is spliced in at the exclusion of the others (Jin et al. 2011; Pohl et al. 2013). Splicing can dramatically increase the diversity of potential protein products that can be made from a single sequence. The most extensive example of mutually exclusive splicing is the insect Dscam gene, which contains four groups of mutually exclusive exons with 12, 48, 33, and two alternate exons, respectively, each group of which will select only one alternate exon (Schmucker et al. 2000). Mutually exclusive splicing can provide an advantage for helping proteins maintain similar overall folds while swapping out individual sequences. Examples of mutually exclusive splicing have been found throughout eukaryotes (Pohl et al. 2013), with a variety of mechanisms proposed including spliceosome incompatibility (Letunic et al. 2002), nonsense-mediated decay combined with splicing factors that enhance one site while suppressing another (Jones et al. 2001), steric hindrance on short introns (Smith and Nadal-Ginard 1989), and more recently, the use of alternative RNA structures (Graveley 2005; Jin et al. 2011; Yang et al. 2011).
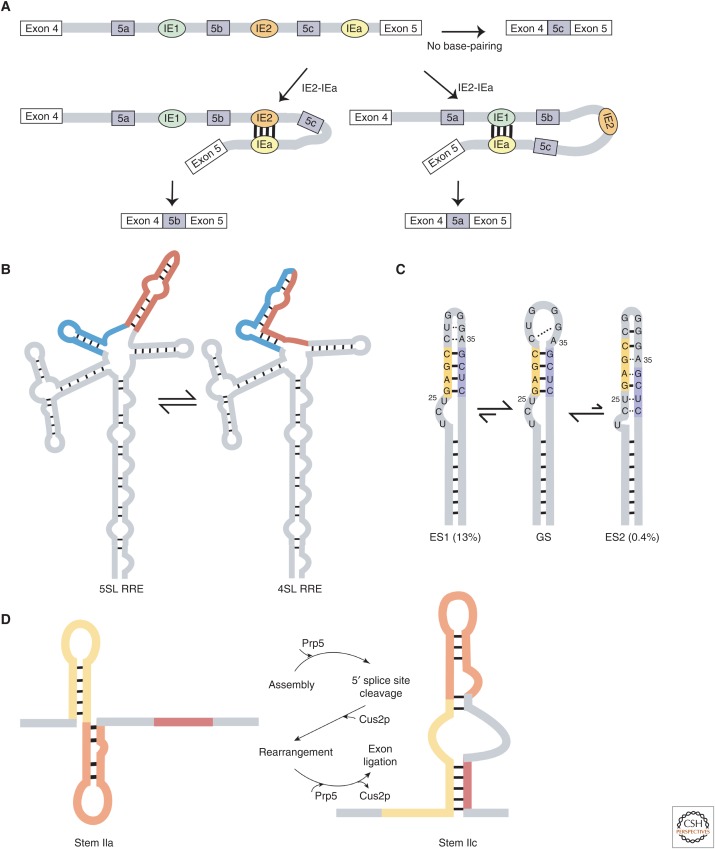
Yang et al. show that the mutually exclusive splicing of exon 5a, 5b, or 5c in the 14-3-3ξ mRNA in Drosophila melanogaster is regulated by three competing conformations (Fig. 2A) (Yang et al. 2011). Two of the conformations involve alternate base-pairing between intronic elements (IEs), in which IEa, located downstream from 5c, can pair with either IE1 or IE2, located downstream from 5a and 5b, respectively. The resulting alternate stem loops (SLs) vary in their relative locations of exons, with either 5b and 5c or only 5c in the loop. The exon located immediately proximal to the stem is activated, whereas exons sequestered in the loop are simultaneously suppressed. The interintronic base-pairings are strong enough that they can occur between partners as far as 1,000–12,000 nt away. The third conformation is formed when the base-pairings are suppressed altogether, which results in the activation of exon 5c. Other genes with mutually exclusive splicing, such as Dscam (Graveley 2005; Anastassiou et al. 2006; May et al. 2011; Yue et al. 2016), Mhc (Yang et al. 2011), and MRP (Yue et al. 2017), also use mechanisms involving the formation of structures that are mutually exclusive (Jin et al. 2018). As in the above examples, Yang et al. suggest that the particular usage of each splice site can be additionally modulated by enhancers and silencers that vary between cell types and developmental stages.
Figure 2.
(A) Mutually exclusive splicing in the 14-3-3ξ mRNA is controlled by three structures that differ in their base-pairing pattern: Intronic element a (IEa) forms base pairs with either IE1 or IE2 or neither. (B) The HIV-1 Rev response element (RRE) partitions into two equally populated structures with either four or five stem loops (SLs). (C) The HIV-1 transactivation response (TAR) element has two “excited” states that differ from the “ground” state (GS). Excited state 1 (ES1) maintains the same overall stem architecture but differs in tertiary interactions at the apical loop, whereas ES2 shows reshuffled base pairs, resulting in a large-scale repairing of the TAR secondary structure. (D) The stem II region on the spliceosome U2 small nuclear RNA (snRNA) can transition between two alternate conformations that each serve a different role in the process of splicing.
Although conformational equilibriums are being found in many systems, in some of these examples, the significance of the structural heterogeneity has yet to be defined. For example, in the case of the HIV-1 Rev Response Element (RRE), a unique motif responsible for exporting unspliced mRNA, two different structures with either four or five SLs form spontaneously in vitro and in vivo (Fig. 2B) (Sherpa et al. 2015). Both structures are present during infection in approximately equal amounts because of similar free energies of folding. However, the five SL structure has been shown to have higher infection rates, likely because of higher Rev/RRE-dependent mRNA export activity. Sherpa et al. suggest that the near neutral energetics provide a sensitive way to modulate the equilibrium, which may be used to dictate the course of infection in a particular cell by altering levels of cellular factors (Sherpa et al. 2015). Similarly, subtle differences in primary sequence during virus evolution can lead to differences in infection severity between patients (Sherpa et al. 2015).
Another example of a well characterized structural equilibrium is the HIV TAR element. In addition to the predominant “ground” state (GS) structure, TAR can also adopt two alternate “excited” states, ES1 and ES2 (Fig. 2C) (Lee et al. 2014). ES1 differs from the GS in the apical loop and is present at 13%, with a timescale of 45 µsec. ES2 differs from the GS by reshuffling its base pairs and is present at ∼0.4% with a timescale of 2 msec. Although the functional significance of the complex dynamics shown by the TAR stem have yet to be determined, Lee et al. suggest that the excited structures may be involved in mediating the ability of the TAR element to perform its various critical functions, including transcriptional elongation and latency (Lee et al. 2014).
The above structural equilibriums have the potential to be modulated by protein effectors, and the resulting protein–RNA complexes can influence the relative conformational populations. In fact, examples exist in which such protein-derived stabilization of specific conformations along an energetic landscape is required for vital functions. For example, the stem II region of the spliceosome U2 small nuclear RNA (snRNA) spontaneously transitions between two mutually exclusive alternate conformations (stems IIa and IIc) that toggle back and forth throughout the process of splicing (Fig. 2D) (Rodgers et al. 2016). Although stem IIa is necessary for spliceosome assembly and formation of the active site, stem IIc is required for the two catalytic steps of splicing (Hilliker et al. 2007; Perriman and Ares 2007; Rodgers et al. 2016). Currently, the Cus2p (cold-sensitive U2 snRNA suppressor 2) and the DEAD-box adenosine triphosphatase (ATPase) Prp5p proteins are the two candidates that have been implicated in orchestrating these structural transitions (Rodgers et al. 2016). Rodgers et al. show that Cus2p stabilizes stem IIa and suggest that Prp5p may be involved in the removal of Cus2p, a model in which Cus2p and Prp5p may work in tandem to organize the interconversion of stems IIa and IIc during splicing. It is important to note that the equilibrium itself is preexisting and can be accessed without protein factors, even if protein factors present in the cell can help facilitate the transitions. Cus2p and Prp5p have yet to be shown to be dynamically changing.
2.3. Tertiary Structure
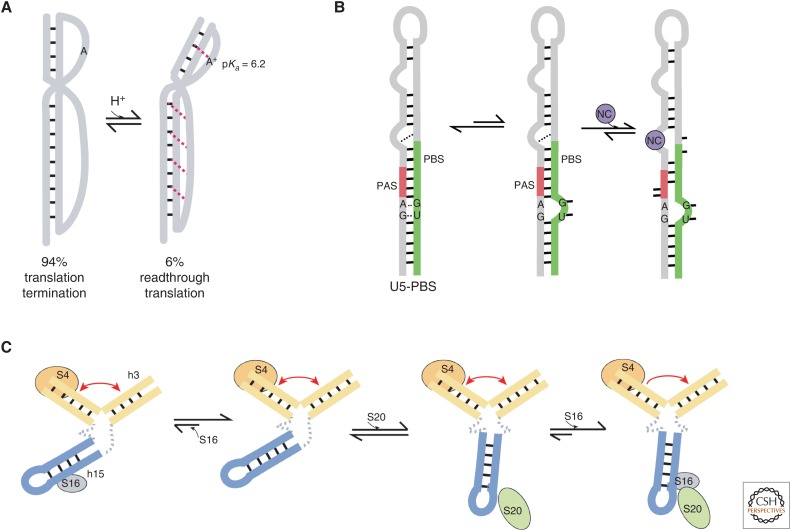
Many RNA viruses maintain appropriate viral protein ratios via translational recoding (Houck-Loomis et al. 2011; Walsh and Mohr 2011). For example, translational recoding in the murine leukemia virus (MLV) produces ∼95% Gag (structural) polyprotein by canonical termination at the stop codon, and ∼5% extended Gag-Pol (Pol, enzymatic) polyprotein by stop codon readthrough that occurs when ribosomes encounter a signal on the mRNA (Philipson et al. 1978; Houck-Loomis et al. 2011). Houck-Loomis et al. found that in MLV the frequency of stop codon readthrough is regulated by a dynamic structural equilibrium-based mechanism (Houck-Loomis et al. 2011), in which a pseudoknotted recoding signal on the mRNA is maintained in a preexisting, protonation-dependent equilibrium between two functional alternate conformations—an active conformation that allows recoding and an alternate inactive conformation that terminates translation. Importantly, the pKa of the equilibrium is such that at physiological pH, the populations of the active and inactive forms correlate with the critical levels of recoding and termination, respectively—in other words, the RNA signal uses the physiological pH of the host cell to automatically partition its conformations into substates that govern explicit biological output.
In MLV, the protonation of the loop 1 adenine residue leads to the formation of hydrogen bond interactions with a base pair in stem 2, leading to the formation of a triple base interaction (Fig. 3A). This information is then transmitted through changes in the helical junction to loop 2 and stem 1, which together form a triple helix. Therefore, in this example the alternate structures have the same secondary structures, but dramatically different tertiary structures (A17H+ ⇒ triple base ⇒ tertiary structure ⇒ stop codon read-through translation; A17 ⇒ lack of tertiary structure ⇒ translation termination at the stop codon). This type of proton-driven equilibrium may be a common mechanism for regulating recoding in diverse viruses. For example, this mechanism has been observed in the beet western yellows virus (BWYV) luteovirus (Houck-Loomis et al. 2011) and the severe acute respiratory syndrome (SARS) coronavirus (V D’Souza, unpubl.). Furthermore, both of these viruses genetically recode the mRNA message not by stop codon readthrough but by −1 programmed ribosomal frameshifting (Su et al. 1999; Thiel et al. 2003), suggesting that the control of translational recoding through structural partitioning may be a broader mechanism.
Figure 3.
(A) The murine leukemia virus (MLV) readthrough RNA pseudoknot is partitioned into two alternate structures, depending on the protonation state. The inactive structure is unprotonated, with stem 2 stacked on top of stem 1. The active structure is protonated at the loop 1 adenine, forming a triple-base interaction that causes a bend at the junction, which allows for tertiary interactions to be formed between loop 2 and stem 1. (B) The U5–primer-binding site (PBS) of MLV exchanges between two conformations, one of which is bound by the nucleocapsid (NC). NC binding frees select nucleotides in the PBS and polyadenylation signal (PAS) regions for access by the appropriate complementary regions on the transfer RNA (tRNA; not pictured), to anneal and form the reverse transcription initiation complex. (C) The 16S ribosomal RNA (rRNA) samples many conformations including native and non-native forms. Protein binding simultaneously reduces conformational sampling and enriches the native forms.
In general, adenines and cytosines are protonatable, but their pKas (∼3.7 and ∼4.5, respectively) are far removed from physiological pH. However, the electrostatic environment surrounding a nucleotide in the folded structure of an RNA molecule can significantly shift its pKa toward neutrality. The physiological pH of cells stays fairly constant, allowing the structural signal of the mRNA to be programmed with a pKa value that is in line with the desired protein ratios. In the case of MLV and SARS, the pKas of the protonation events are ∼6.23 and ∼6.65 (V D’Souza, unpubl.), respectively, which result in the desired ratios of 95:5 and 85:15 for structural and enzymatic proteins. The pKa shift toward neutrality has been shown to be true for many biologically important RNAs (Cai and Tinoco 1996; Butcher et al. 2000; Huppler et al. 2002; Cornish et al. 2005; Gong et al. 2007; Houck-Loomis et al. 2011; Cash et al. 2013; Pechlaner et al. 2015). A recent example of a guanosine triphosphate (GTP)-sensing aptamer shows that the pKa can be shifted as high as 8.7 (Wolter et al. 2017), which would in principle allow for a full range of modulation of alternate structures at physiological proton concentration.
In some cases, proteins assist in the alteration of the energy barriers between conformations along an existing landscape to enrich a particular conformation, as seen in the example of the MLV NC protein chaperone activity. In retroviruses, reverse transcription requires annealing of complementary sequences on a host transfer RNA (tRNA) primer and on the U5–primer-binding site (U5–PBS) region on the viral genome to form the reverse transcription initiation complex (Fig. 3B) (Harada et al. 1979; Wain-Hobson et al. 1985). The complementary sequences in both molecules are sequestered by intramolecular base pairs (Mougel et al. 1993; Paillart et al. 2004; Wilkinson et al. 2008) that must be broken before new intermolecular interactions can be formed, which is orchestrated by the binding of the NC protein (Levin et al. 2010). Miller et al. showed that the U5–PBS samples two conformations that differ at the tetraloop and at an internal loop. In the major conformation, both of the loops are structured, with the internal loop sequestered via cross-strand base-pairing, whereas in the minor conformation part of the tetraloop and part of the internal loop both extrude out of the structure (Miller et al. 2014). The NC tails are capable of pushing the structural equilibrium to the minor destabilized population, which allows the tRNA to access the appropriate complementary sequences on the U5–PBS for annealing. It is important to note that NC can shift only a preexisting equilibrium, without which it would not otherwise be able to access the U5–PBS for remodeling.
Another example is the U1A protein, which binds to the U1 snRNA hairpin II via conformational capture, whereby the free structure samples many conformations including a rigid state similar to the protein-bound state (Shajani et al. 2007). Similarly, the cyclic adenosine monophosphate response element-binding protein 2 (CBP2) protein binds the initially unstructured bI5 group I intronic RNA in a nonspecific manner and induces conformational fluctuations, before binding specifically and stabilizing the final native structure (Bokinsky et al. 2006). Even in this example, the free RNA is able to transiently access the structured state in the absence of protein (Bokinsky et al. 2006).
Finally, some proteins can bind multiple conformations instead of just favoring one. For example, Abeysirigunawardena et al. show that ribosomal protein bS16 can bind both a native and non-native form of the 16S rRNA during ribosome assembly, although bS16 binding nevertheless drives the equilibrium to the required native structure (Fig. 3C) (Abeysirigunawardena et al. 2017). During ribosome assembly, the sequential binding of proteins stabilizes the rRNA into conformations that allow for binding of the next protein. Proteins uS4 and bS20 both bind the 16S 5′ domain, whereas protein bS16 binds the interface between the uS4 and bS20 domains. Upon uS4 binding, the complex shows a slow exchange equilibrium in which the h3 helix converts between the native and non-native “flipped” conformations. Binding of bS20 does not affect the equilibrium of the h3 helix, but switches the conformation of helix h15, which increases the probability of stable bS16 binding. Binding of bS16 stabilizes the native conformation of the h3 helix to allow for further assembly of 30S ribosome proteins. The binding of proteins does not lock in any particular RNA conformation, but rather limits the dynamics on the energy landscape to a narrower set of productive intermediate structures while selecting against unproductive conformations.
3. Alternate RNA structures controlled by dynamic cues
Alternate RNA structures can also be triggered by dynamic cues, allowing for the accessing of states that would have otherwise been inaccessible; these types of alternate RNA structures are loosely grouped under the term riboswitches. Historically, riboswitches were defined as RNAs that bound metabolites such as purine (Batey 2012; Porter et al. 2014), lysine (Garst et al. 2008; Serganov et al. 2008; Smith-Peter et al. 2015), thiamine pyrophosphate (TPP) (Thore et al. 2006; Bocobza et al. 2007; Anthony et al. 2012; Li and Breaker 2013; Uhm et al. 2018), S-adenosyl methionine (SAM) (Price et al. 2014; Trausch et al. 2014; Wostenberg et al. 2015), preQ1 (Eichhorn et al. 2014), adenosyl cobalamin (Johnson et al. 2012; Peselis and Serganov 2012; Schaffer et al. 2014), glycine (Huang et al. 2010; Butler et al. 2011; Erion and Strobel 2011; Baird and Ferre-D’Amare 2013), fluoride (Baker et al. 2012; Ren et al. 2012; Li et al. 2013; Zhao et al. 2017), and tetrahydrofolate (Ames et al. 2010; Huang et al. 2011; Trausch et al. 2011). However, this definition has since been expanded to more broadly include RNAs that change structure in response to dynamic cues such as temperature (Kortmann and Narberhaus 2012; Krajewski and Narberhaus 2014), pH (Nechooshtan et al. 2009), tRNA binding (Zhang and Ferre-D’Amare 2013; Henkin 2014), and metal ion binding (Cromie et al. 2006; Dann et al. 2007; Furukawa et al. 2015; Price et al. 2015; Wedekind et al. 2017).
Riboswitches generally have two domains—a ligand-binding aptamer and a gene expression platform that can modify gene expression based on the structure of the ligand-binding aptamer (Schwalbe et al. 2007). In general, the most widely accepted mechanism is that riboswitches can exist as two conformational structures depending on whether they are ligand-bound or ligand-free. These two structures produce two different functions in response to changing metabolite levels in the cell, thereby enabling regulation. Usually the riboswitch functions as part of a feedback loop pathway for ligand concentration, in which the ligand sensed by the riboswitch is directly related to the function of the riboswitch-regulated gene, either to cause a decrease of expression of ligand-producing proteins or to cause an increase of expression of ligand-clearing proteins (Mandal and Breaker 2004; Mandal et al. 2004; Porter et al. 2014). Although most riboswitches loosely abide by this general model, recent studies have suggested that some riboswitches are not binary on/off switches, but function by finely tuning relative populations to act as molecular “dimmers” (Baird and Ferre-D’Amare 2010; Baird et al. 2010; Helmling et al. 2017).
Several different mechanisms for riboswitch function have been proposed, many of which were only able to be characterized using new techniques and technologies. In this review, we will not be covering the entire range of riboswitch structural and mechanistic diversity, as these have been extensively covered in other reviews (Haller et al. 2011b; Breaker 2012; Serganov and Nudler 2013; Peselis and Serganov 2014; Sherwood and Henkin 2016), but instead will focus on a few key examples in transcription, translation, and splicing to illustrate how these systems use alternate RNA structures to perform gene regulation and how new technologies have made some of these observations possible.
3.1. Transcription
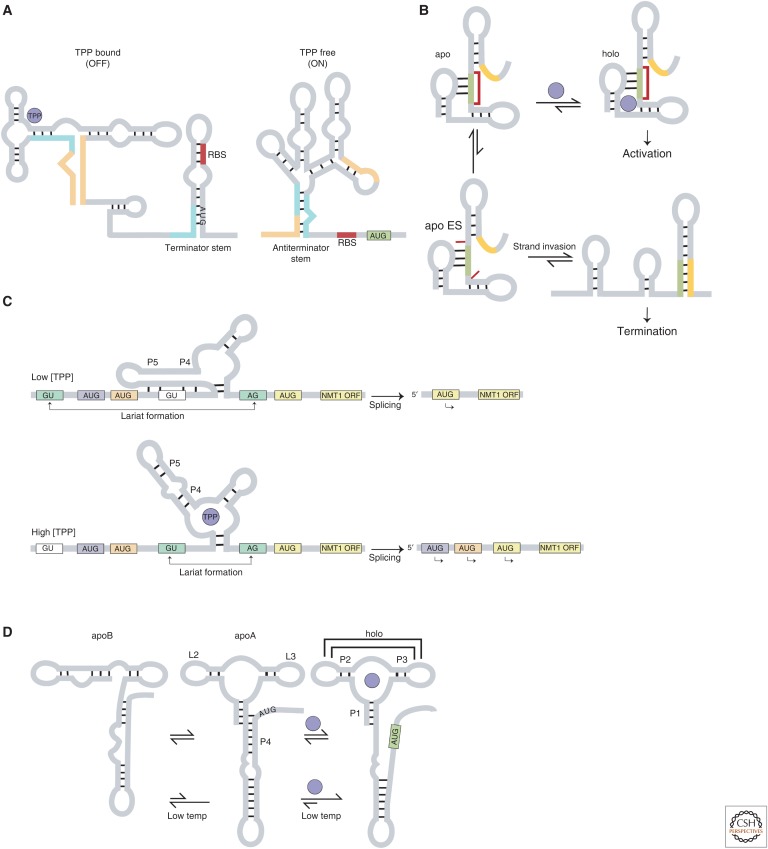
Riboswitches can control transcription termination as a cis-acting switch using two mutually exclusive base-paired stems: One conformation (terminator stem) causes transcription termination, whereas the other (antiterminator stem) allows transcription to proceed (Fig. 4A) (Mandal et al. 2004; Porter et al. 2014). Many riboswitches use a version of the conformational capture mechanism. Once folded, the thermodynamic barrier between two competing structures is high enough that they cannot equilibrate within the timescale of transcription. Therefore, by necessity, whether transcription will terminate or proceed must be regulated by structural partitioning during co-transcriptional folding (Noeske et al. 2007; Haller et al. 2011a; Frieda and Block 2012; Lai et al. 2013; Peselis and Serganov 2014; Gong et al. 2015; Watters et al. 2016; Gong et al. 2017). In most instances, this decision-making occurs during transcriptional pausing, which can be used as a checkpoint for riboswitch-driven regulation (Wickiser et al. 2005a; Wickiser et al. 2005b; Greenleaf et al. 2008). We will first describe the general co-transcriptional pausing mechanism using the Escherichia coli TPP thiC riboswitch as an example, and then describe a variation on the mechanism using the Bacillus cereus fluoride riboswitch.
Figure 4.
(A) The Escherichia coli thiamine pyrophosphate (TPP) thiC riboswitch has two conformations, ligand-bound (OFF) and ligand-free (ON). The ribosome-binding site (RBS) and the AUG start codon are sequestered in the ligand-bound state and exposed in the ligand-free state, respectively. (B) The Bacillus cereus fluoride riboswitch has apo and holo conformations with highly similar structures. The apo conformation can transiently access a short-lived excited state (apo ES) in which a highly conserved reverse Hoogsteen “linchpin” base pair is broken, enabling strand invasion and leading to formation of the terminator stem. (C) The Neurospora crassa TPP NMT1 riboswitch regulates the expression of alternate upstream open reading frames (ORFs) by using alternate base-pairing involving residues in the P4 and P5 stems to change the formation of the lariat. (D) The Vibrio vulnificus adenine add riboswitch operates using a three-state mechanism rather than canonical two-state mechanism, allowing it to maintain its ligand concentration dependence at temperatures ranging from 37°C to 10°C. The apoA conformation is similar to the holo conformation, whereas apoB differs significantly.
Chauvier et al. found that in the thiC riboswitch, the antiterminator stem conformation forms only in the presence of long pausing steps during transcription (Chauvier et al. 2017). In this riboswitch, ligand affinity is high prior to transcription reaching the transcription pause site (TPS), which is located near the translation start site. At the TPS, ligand affinity decreases by ∼100-fold, ensuring that any ligand-free structures that reach the TPS will no longer preferentially bind ligand but can now form the alternate metastable antiterminator stem. This results in completion of transcription and subsequent translation. Even with a decreased ligand affinity, the pausing step is still necessary to provide enough time to fold into the metastable state (ligand-free at TPS ⇒ pausing ⇒ antiterminator stem ⇒ transcription). On the other hand, ligand-bound complexes that reach the TPS have folded into the terminator stem conformation and will primarily result in transcription termination. In addition, any ligand-bound complexes that do not terminate are nevertheless inhibited for translation initiation because of the sequestering of the translation start site in the terminator stem conformation. This provides a fail-safe mechanism to maximally ensure that the protein product is not made (ligand-bound at TPS ⇒ pausing ⇒ terminator stem ⇒ transcription termination/translation start site sequestered).
It is worth noting that transcriptional pausing itself can be modulated by ligand concentration, such as for the pH-responsive alx riboswitch involved in maintaining homeostasis in E. coli (Nechooshtan et al. 2009). In this example, ligand binding during cotranscriptional pausing regulates translation initiation rather than transcription termination. The RNA-folding dynamics during transcription are directly driven by pH: Transcriptional pausing is prolonged in alkaline conditions, allowing for better refolding of transcribed RNA into an active structure where the ribosome-binding site (RBS) is exposed rather than sequestered. Note that although the alx riboswitch is pH-dependent, it operates in an environment with a dynamically changing pH, as opposed to the previously discussed pH-dependent MLV readthrough system which used the unchanging physiological pH in the cell as a static cue.
In the B. cereus fluoride riboswitch, Zhao et al. discovered a switching mechanism counter to the current paradigm (Fig. 4B) (Zhao et al. 2017). Whereas most riboswitches involve the formation of alternate ligand-bound (holo) and ligand-free (apo) conformations with different secondary structures, in this example, Zhao et al. found that the two states have highly similar structures. The only difference between the two conformations is that the apo conformation has an additional structural equilibrium wherein it samples a comparatively short-lived (τ ∼3 msec) ES that the holo conformation does not. Only a small population of the apo conformation (∼1%) samples the ES, which unlocks a highly conserved reverse Hoogsteen “linchpin” base pair present in the apo and holo conformations. This unlocking event exposes the antiterminator stem to strand invasion and allows for the structural transition to the terminator state. In the absence of fluoride, the apo state (τ ∼200 msec) cannot access the holo state; therefore, although only a small population samples the ES, because of the high favorability of strand invasion, the overall equilibrium proceeds toward termination (low fluoride ⇒ apo state ⇒ excited state ⇒ unlocked linchpin ⇒ terminator stem ⇒ transcription termination). On the other hand, the holo conformation is stabilized by ligand binding (τ ∼2.4 sec) and therefore does not access the ES, but instead completes transcription (fluoride ⇒ holo state with antiterminator stem ⇒ transcription). Zhao et al. suggest that this mechanism could potentially be more widespread and that it would be prudent to reexamine previously characterized systems to determine if any are regulated by the presence of as yet unobserved ESs.
3.2. Splicing
Unlike bacterial TPP riboswitches that regulate transcription and translation (Miranda-Rios et al. 2001; Mironov et al. 2002; Winkler et al. 2002), eukaryotic TPP riboswitches are often located in the introns of pre-mRNAs and regulate splicing (Kubodera et al. 2003; Cheah et al. 2007; Li and Breaker 2013; Mukherjee et al. 2018). These are currently the only known class of eukaryotic riboswitches (Serganov and Nudler 2013). All of the mechanisms characterized to date involve alternate base-pairing interactions that selectively mask or expose splice sites (Cheah et al. 2007; Croft et al. 2007; Li and Breaker 2013). For instance, the TPP riboswitch located in an intron in the NMT1 gene in the fungus Neurospora crassa regulates the expression of upstream open reading frames (ORFs) (Cheah et al. 2007). Two alternate conformations are formed with the second 5′ splice site either base-paired with residues in the P4 and P5 stems (ligand-free) or exposed (ligand-bound) (Fig. 4C). Sequestration of the second 5′ splice site allows the lariat to form between the first 5′ splice site and the 3′ splice site, giving rise to a short mRNA product and resulting in functional NMT1 translation. On the other hand, exposure of the 5′ splice site under high TPP concentrations results in a longer spliced mRNA product which contains upstream ORFs that compete with the primary ORF to produce an overall lower NMT1 expression. In another example, the TPP riboswitch found in the 3′ UTRs of the THIC gene in Arabidopsis thaliana and in a variety of other plant species, uses a similar mechanism but controls gene expression via alternate 3′ end processing (Wachter et al. 2007). This mechanism also uses the sequestration of splice sites, but operates by retaining or removing an intronic transcript-processing signal, thus leading to a longer or shorter 3′ UTR, respectively (Bocobza et al. 2007; Wachter et al. 2007).
3.3. Translation
Analogous to transcriptional riboswitches, many translational riboswitches use alternate structures that either sequester or expose the Shine–Dalgarno sequence in response to dynamic cues. However, unlike transcriptional riboswitches, translational riboswitches can recognize ligand recognition either during or after transcription and can equilibrate within the timescale of translation and the lifetime of the mRNA (Peselis and Serganov 2014). Most regulation occurs via a simple structural equilibrium with ligand-bound and ligand-free conformations. In this section, we will use the adenine add riboswitch discovered in the human pathogenic bacterium Vibrio vulnificus to illustrate an example of a noncanonical translation initiation riboswitch involving three alternate structures (Reining et al. 2013).
V. vulnificus occupies a range of habitats and can survive in temperatures ranging from ∼10°C in marine habitats to 37°C in human hosts (Reining et al. 2013). It needs to maintain functional riboswitch activity through the entire range of temperatures. Reining et al. show that the add riboswitch uses a three-state mechanism, as opposed to the canonical two-state mechanism, to couple ligand and temperature sensing. In the three-state mechanism, there is one ligand-bound (holo) structure and two ligand-free (apoA and apoB) structures (Fig. 4D).
The holo conformation has three helices (P1, P2, P3) organized by a three-way junction and stabilized by long-range interactions between loops L2 and L3. Upon adenine binding, P1 is extended by three additional base pairs, which causes the dismantling of the P4 helix and exposes the Shine–Dalgarno sequence for ribosome binding. The apoA and apoB conformations differ significantly from each other although both are ligand-free structures. The apoA conformation is similar to the holo conformation and is binding-competent. However, it lacks L2–L3 interactions and contacts across the junction but has a partially formed P4 stem that sequesters the Shine–Dalgarno sequence. In contrast, the apoB conformation has key motifs rearranged to form a completely different secondary structure, which makes it insensitive to ligand binding.
Reining et al. measured and simulated how efficiently a riboswitch can switch between active and inactive conformations, and defined “switching efficiency” as the difference in the percentage of holo population at two different ligand concentrations. In a two-state mechanism, the riboswitch shows an increased ligand affinity at lower temperatures, leading to a corresponding increase in the holo population. In contrast, the three-state mechanism is able to use the equilibrium between apoB and apoA to counteract the increased ligand affinity of apoA as temperature decreases. Namely, at lower temperatures, the equilibrium favors apoB, thereby sequestering part of the population into a binding-incompetent state (↓temperature ⇒ ↑ligand affinity ⇒ ↑binding-incompetent apoB; ↑temperature ⇒ ↓ligand affinity ⇒ ↑binding-competent). In their simulation, the three-state mechanism is able to produce a 67% switching efficiency at 5°C compared with 83% at 30°C, whereas in the two-state mechanism, the riboswitch is only able to produce a 14% switching efficiency at 5°C. Therefore, the three-state mechanism is able to maintain a more consistent ligand concentration dependence over a wider range in temperatures.
4. New technologies for investigating alternate RNA structures
In recent years, advances in new technology have enabled the characterization of previously unobserved alternate RNA conformations. Nuclear magnetic resonance (NMR) remains the most powerful technique for probing RNA structural dynamics and continues to be developed for visualizing alternate conformations even in large RNAs.
Typically, NMR experiments are challenging for oligonucleotides larger than approximately 50 residues, predominantly because of spectral overlap. Lu et al. developed a new NMR approach called long-range probing by adenosine interaction detection (lr-AID) and used it to characterize the monomeric (356 nt) and dimeric (712 nt) conformations of the HIV-1 5′ leader (described above) (Lu et al. 2011). Specifically, to confirm the DIS:AUG interaction in the dimer, they replaced three adjacent base pairs within the expected stem with AU base pairs (5′-AUU; 3′-AAU). This had two advantages: First, as expected, the adenine H2 from the central AU base pair experienced an upfield chemical shift away from the spectral overlap of the other nucleotides; second, this adenine H2 gave nuclear Overhauser effects (NOEs) to the H1s of the other two adenines, providing direct proof of stem formation. Use of these signature chemical shift patterns is widely applicable to the study of long-range interactions within large RNA structures; in theory, any number of base pairs within a stem not involved in tertiary interactions can be interchanged without altering the overall fold, because of the isosteric nature of Watson–Crick base pairs.
NMR is also a powerful technique for characterizing the timescales and populations of alternate RNA structures (μsec–msec). Whereas relaxation dispersion experiments are routinely used to study structures exchanging at fast timescales (μsec) (Hoogstraten et al. 2000; Blad et al. 2005), and their use in conjunction with a low spin lock field has been used to obtain information on medium timescales (μsec–msec) (Massi et al. 2004; Hansen et al. 2009; Dethoff et al. 2012), the challenge has been in observing transitions in slow timescales (msec). Slow timescales have recently become accessible through the use of chemical exchange saturation transfer (CEST) NMR combined with a low spin lock field (Zhao et al. 2014). In fact, as described above, Zhao et al. used CEST NMR to visualize a lowly populated ES in the B. cereus fluoride riboswitch and to describe the existence of a new kinetic switching mechanism (Zhao et al. 2017).
Beyond NMR, other techniques such as chemical modification-based experiments are being advanced to prove the existence of in vitro characterized equilibria in cells. One example is dimethyl sulfate mutational profiling with sequencing (DMS-MaPseq), a form of secondary structure determination distinct from previous structure probing techniques in that it encodes modifications as mutations instead of truncations (Zubradt et al. 2017). The recording of multiple mutation sites per RNA molecule via deep sequencing allows for the analysis of each individual RNA molecule rather than a population average. This enables clustering of distinct RNA substructures within a population, as was shown using the two human mitochondrial ribosomal protein small 21 (MRPS21) riboSNitch alleles (Zubradt et al. 2017).
Recently, an RNA duplex-mapping technique called PARIS (psoralen analysis of RNA interactions and structures) was used to identify new potential alternate base-pairing interactions in cells (Lu et al. 2016). PARIS uses chemical cross-linking combined with deep sequencing to determine which RNA sequences are paired to each other with near base pair resolution. PARIS can be used to obtain transcriptome-wide information, and was used to discover several alternate base-pairing partners in the 3′ UTR of tubulin β class I (TUBB) mRNA, and in the MALAT1 and X-inactive specific transcript (XIST) lncRNAs (Lu et al. 2016). Although there are no known functions associated with these alternate RNA structures as of yet, these observations provide new investigative directions.
The development of new computational techniques is also facilitating the identification of potential new alternate RNA structures, particularly within riboswitches. Many classes of riboswitches share common structural motifs (Breaker 2012) and can therefore be used for computational screening of transcriptomic data to identify new riboswitches (Drory Retwitzer et al. 2015). Also, evolutionary studies are being used to support the functional significance of alternate RNA structures by computational mapping of sequence co-variation across species (Ritz et al. 2013). In instances in which two conformations exist, covariation of base pairs for both conformations in homologous sequences are expected only if they are functionally important.
5. Concluding remarks
Although we have long known about the richness of regulatory information contained in RNA, we have just begun to scrape the surface of the depth and extent of complexity that can be contained within individual RNA domains. The handful of examples described above gives a glimpse into the potential contained in alternate RNA structures to drive key biological functions. The development of new technologies on two major fronts has been key to opening up this field: Screening techniques have enabled us to realize how widespread the prevalence of alternate RNA structures is, whereas structural techniques have allowed for the identification and detailed investigation of such substructures. The combined power of these new technologies and their continued further development is promising for the future discovery of novel alternate RNA structures and the mechanisms by which they function.
ACKNOWLEDGMENTS
We would like to thank Aaron Fernandes for his assistance in the creation of the figures.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
References
- Abeysirigunawardena SC, Kim H, Lai J, Ragunathan K, Rappe MC, Luthey-Schulten Z, Ha T, Woodson SA. 2017. Evolution of protein-coupled RNA dynamics during hierarchical assembly of ribosomal complexes. Nat Commun 8: 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames TD, Rodionov DA, Weinberg Z, Breaker RR. 2010. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem Biol 17: 681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiou D, Liu H, Varadan V. 2006. Variable window binding for mutually exclusive alternative splicing. Genome Biol 7: R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony PC, Perez CF, Garcia-Garcia C, Block SM. 2012. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci 109: 1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Ferre-D’Amare AR. 2010. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA 16: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Ferre-D’Amare AR. 2013. Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches. RNA 19: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Kulshina N, Ferre-D’Amare AR. 2010. Riboswitch function: Flipping the switch or tuning the dimmer? RNA Biol 7: 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. 2012. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335: 233–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT. 2012. Structure and mechanism of purine-binding riboswitches. Q Rev Biophys 45: 345–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blad H, Reiter NJ, Abildgaard F, Markley JL, Butcher SE. 2005. Dynamics and metal ion binding in the U6 RNA intramolecular stem-loop as analyzed by NMR. J Mol Biol 353: 540–555. [DOI] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. 2007. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev 21: 2874–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky G, Nivon LG, Liu S, Chai G, Hong M, Weeks KM, Zhuang X. 2006. Two distinct binding modes of a protein cofactor with its target RNA. J Mol Biol 361: 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2012. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 4: a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SE, Allain FH, Feigon J. 2000. Determination of metal ion binding sites within the hairpin ribozyme domains by NMR. Biochemistry 39: 2174–2182. [DOI] [PubMed] [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA. 2011. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol 18: 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Tinoco I Jr. 1996. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35: 6026–6036. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635. [DOI] [PubMed] [Google Scholar]
- Cash DD, Cohen-Zontag O, Kim NK, Shefer K, Brown Y, Ulyanov NB, Tzfati Y, Feigon J. 2013. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc Natl Acad Sci 110: 10970–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvier A, Picard-Jean F, Berger-Dancause JC, Bastet L, Naghdi MR, Dube A, Turcotte P, Perreault J, Lafontaine DA. 2017. Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat Commun 8: 13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla M, Oliva R, Bujnicki JM, Cavallo L. 2015. An atlas of RNA base pairs involving modified nucleobases with optimal geometries and accurate energies. Nucleic Acids Res 43: 6714–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. 2007. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447: 497–500. [DOI] [PubMed] [Google Scholar]
- Chojnowski G, Walen T, Bujnicki JM. 2014. RNA Bricks—A database of RNA 3D motifs and their interactions. Nucleic Acids Res 42: D123–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn WE. 1960. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: Isolation, structure, and chemical characteristics. J Biol Chem 235: 1488–1498. [PubMed] [Google Scholar]
- Corley M, Solem A, Qu K, Chang HY, Laederach A. 2015. Detecting riboSNitches with RNA folding algorithms: A genome-wide benchmark. Nucleic Acids Res 43: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Hennig M, Giedroc DP. 2005. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated–1 ribosomal frameshifting. Proc Natl Acad Sci 102: 12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. 2007. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci 104: 20770–20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. 2006. An RNA sensor for intracellular Mg2+. Cell 125: 71–84. [DOI] [PubMed] [Google Scholar]
- D’Souza V, Summers MF. 2005. How retroviruses select their genomes. Nat Rev Microbiol 3: 643–655. [DOI] [PubMed] [Google Scholar]
- Dann CE 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. 2007. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130: 878–892. [DOI] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci 71: 3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM. 2012. Visualizing transient low-populated structures of RNA. Nature 491: 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. 2016. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drory Retwitzer M, Kifer I, Sengupta S, Yakhini Z, Barash D. 2015. An efficient minimum free energy structure-based search method for riboswitch identification based on inverse RNA folding. PLoS One 10: e0134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn CD, Kang M, Feigon J. 2014. Structure and function of preQ1 riboswitches. Biochim Biophys Acta 1839: 939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion TV, Strobel SA. 2011. Identification of a tertiary interaction important for cooperative ligand binding by the glycine riboswitch. RNA 17: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda KL, Block SM. 2012. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338: 397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A. 2008. A code for transcription initiation in mammalian genomes. Genome Res 18: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Ramesh A, Zhou Z, Weinberg Z, Vallery T, Winkler WC, Breaker RR. 2015. Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol Cell 57: 1088–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT. 2008. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem 283: 22347–22351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B, Chen JH, Chase E, Chadalavada DM, Yajima R, Golden BL, Bevilacqua PC, Carey PR. 2007. Direct measurement of a pKa near neutrality for the catalytic cytosine in the genomic HDV ribozyme using Raman crystallography. J Am Chem Soc 129: 13335–13342. [DOI] [PubMed] [Google Scholar]
- Gong S, Wang Y, Zhang W. 2015. Kinetic regulation mechanism of pbuE riboswitch. J Chem Phys 142: 015103. [DOI] [PubMed] [Google Scholar]
- Gong S, Wang Y, Wang Z, Zhang W. 2017. Co-transcriptional folding and regulation mechanisms of riboswitches. Molecules 22: 1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. 2005. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. 2008. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319: 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero A, Dallas DC, Contreras S, Chee S, Parker EA, Sun X, Dimapasoc L, Barile D, German JB, Lebrilla CB. 2014. Mechanistic peptidomics: Factors that dictate specificity in the formation of endogenous peptides in human milk. Mol Cell Proteomics 13: 3343–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. 2011a. Conformational capture of the SAM-II riboswitch. Nat Chem Biol 7: 393–400. [DOI] [PubMed] [Google Scholar]
- Haller A, Souliere MF, Micura R. 2011b. The dynamic nature of RNA as key to understanding riboswitch mechanisms. Acc Chem Res 44: 1339–1348. [DOI] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A. 2010. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet 6: e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. 2009. Extending the range of microsecond-to-millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R1ρ NMR spectroscopy. J Am Chem Soc 131: 3818–3819. [DOI] [PubMed] [Google Scholar]
- Harada F, Peters GG, Dahlberg JE. 1979. The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J Biol Chem 254: 10979–10985. [PubMed] [Google Scholar]
- Helmling C, Wacker A, Wolfinger MT, Hofacker IL, Hengesbach M, Furtig B, Schwalbe H. 2017. NMR structural profiling of transcriptional intermediates reveals riboswitch regulation by metastable RNA conformations. J Am Chem Soc 139: 2647–2656. [DOI] [PubMed] [Google Scholar]
- Henkin TM. 2014. The T box riboswitch: A novel regulatory RNA that utilizes tRNA as its ligand. Biochim Biophys Acta 1839: 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. 2007. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraten CG, Wank JR, Pardi A. 2000. Active site dynamics in the lead-dependent ribozyme. Biochemistry 39: 9951–9958. [DOI] [PubMed] [Google Scholar]
- Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D’Souza VM. 2011. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ. 2010. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell 40: 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Ishibe-Murakami S, Patel DJ, Serganov A. 2011. Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch. Proc Natl Acad Sci 108: 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE. 2002. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat Struct Biol 9: 431–435. [DOI] [PubMed] [Google Scholar]
- Jin Y, Yang Y, Zhang P. 2011. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol 8: 450–457. [DOI] [PubMed] [Google Scholar]
- Jin Y, Dong H, Shi Y, Bian L. 2018. Mutually exclusive alternative splicing of pre-mRNAs. Wiley Interdiscip Rev RNA 9: e1468. [DOI] [PubMed] [Google Scholar]
- Johnson JE Jr., Reyes FE, Polaski JT, Batey RT. 2012. B12 cofactors directly stabilize an mRNA regulatory switch. Nature 492: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RB, Wang F, Luo Y, Yu C, Jin C, Suzuki T, Kan M, McKeehan WL. 2001. The nonsense-mediated decay pathway and mutually exclusive expression of alternatively spliced FGFR2IIIb and -IIIc mRNAs. J Biol Chem 276: 4158–4167. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT. 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharytonchyk S, Monti S, Smaldino PJ, Van V, Bolden NC, Brown JD, Russo E, Swanson C, Shuey A, Telesnitsky A, et al. 2016. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci 113: 13378–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J, Narberhaus F. 2012. Bacterial RNA thermometers: Molecular zippers and switches. Nat Rev Microbiol 10: 255–265. [DOI] [PubMed] [Google Scholar]
- Krajewski SS, Narberhaus F. 2014. Temperature-driven differential gene expression by RNA thermosensors. Biochim Biophys Acta 1839: 978–988. [DOI] [PubMed] [Google Scholar]
- Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. 2003. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett 555: 516–520. [DOI] [PubMed] [Google Scholar]
- Kutchko KM, Sanders W, Ziehr B, Phillips G, Solem A, Halvorsen M, Weeks KM, Moorman N, Laederach A. 2015. Multiple conformations are a conserved and regulatory feature of the RB1 5′ UTR. RNA 21: 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D, Proctor JR, Meyer IM. 2013. On the importance of cotranscriptional RNA structure formation. RNA 19: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi U, Fernandez-Munoz R, Darnell JE Jr. 1977. Content of N-6 methyl adenylic acid in heterogeneous nuclear and messenger RNA of HeLa cells. Nucleic Acids Res 4: 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dethoff EA, Al-Hashimi HM. 2014. Invisible RNA state dynamically couples distant motifs. Proc Natl Acad Sci 111: 9485–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 1998. Conserved geometrical base-pairing patterns in RNA. Q Rev Biophys 31: 399–455. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Bork P. 2002. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet 11: 1561–1567. [DOI] [PubMed] [Google Scholar]
- Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. 2010. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol 7: 754–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CJ, Pan T, Kalsotra A. 2017. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat Rev Mol Cell Biol 18: 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Breaker RR. 2013. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Res 41: 3022–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Smith KD, Davis JH, Gordon PB, Breaker RR, Strobel SA. 2013. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc Natl Acad Sci 110: 19018–19023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, et al. 2011. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. 2004. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 11: 29–35. [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. 2004. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306: 275–279. [DOI] [PubMed] [Google Scholar]
- Massi F, Johnson E, Wang C, Rance M, Palmer AG 3rd. 2004. NMR R1 ρ rotating-frame relaxation with weak radio frequency fields. J Am Chem Soc 126: 2247–2256. [DOI] [PubMed] [Google Scholar]
- May GE, Olson S, McManus CJ, Graveley BR. 2011. Competing RNA secondary structures are required for mutually exclusive splicing of the Dscam exon 6 cluster. RNA 17: 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis JL, Liu Q, Lavender CA, Devaraj A, McClory SP, Fredrick K, Weeks KM. 2015. In-cell SHAPE reveals that free 30S ribosome subunits are in the inactive state. Proc Natl Acad Sci 112: 2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SB, Yildiz FZ, Lo JA, Wang B, D’Souza VM. 2014. A structure-based mechanism for tRNA and retroviral RNA remodelling during primer annealing. Nature 515: 591–595. [DOI] [PubMed] [Google Scholar]
- Miranda-Rios J, Navarro M, Soberón M. 2001. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci 98: 9736–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747–756. [DOI] [PubMed] [Google Scholar]
- Mougel M, Tounekti N, Darlix JL, Paoletti J, Ehresmann B, Ehresmann C. 1993. Conformational analysis of the 5′ leader and the gag initiation site of Mo-MuLV RNA and allosteric transitions induced by dimerization. Nucleic Acids Res 21: 4677–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Retwitzer MD, Barash D, Sengupta S. 2018. Phylogenomic and comparative analysis of the distribution and regulatory patterns of TPP riboswitches in fungi. Sci Rep 8: 5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe AM, Brooks CL, Al-Hashimi HM. 2014. Hierarchy of RNA functional dynamics. Annu Rev Biochem 83: 441–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. 2009. A pH-responsive riboregulator. Genes Dev 23: 2650–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. 2007. Interplay of ’induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res 35: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. 2004. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J Biol Chem 279: 48397–48403. [DOI] [PubMed] [Google Scholar]
- Parlea LG, Sweeney BA, Hosseini-Asanjan M, Zirbel CL, Leontis NB. 2016. The RNA 3D motif atlas: Computational methods for extraction, organization and evaluation of RNA motifs. Methods 103: 99–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechlaner M, Donghi D, Zelenay V, Sigel RK. 2015. Protonation-dependent base flipping at neutral pH in the catalytic triad of a self-splicing bacterial group II intron. Angew Chem Int Ed Engl 54: 9687–9690. [DOI] [PubMed] [Google Scholar]
- Perriman RJ, Ares M Jr. 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev 21: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peselis A, Serganov A. 2012. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat Struct Mol Biol 19: 1182–1184. [DOI] [PubMed] [Google Scholar]
- Peselis A, Serganov A. 2014. Themes and variations in riboswitch structure and function. Biochim Biophys Acta 1839: 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AI, Zirbel CL, Leontis NB. 2013. Automated classification of RNA 3D motifs and the RNA 3D motif atlas. RNA 19: 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson L, Andersson P, Olshevsky U, Weinberg R, Baltimore D, Gesteland R. 1978. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: Enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell 13: 189–199. [DOI] [PubMed] [Google Scholar]
- Pohl M, Bortfeldt RH, Grutzmann K, Schuster S. 2013. Alternative splicing of mutually exclusive exons—A review. Biosystems 114: 31–38. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. 2006. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol 7: R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M, Szachniuk M, Blazewicz M, Wasik S, Burke EK, Blazewicz J, Adamiak RW. 2010. RNA FRABASE 2.0: An advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures. BMC Bioinformatics 11: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter EB, Marcano-Velazquez JG, Batey RT. 2014. The purine riboswitch as a model system for exploring RNA biology and chemistry. Biochim Biophys Acta 1839: 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price IR, Grigg JC, Ke A. 2014. Common themes and differences in SAM recognition among SAM riboswitches. Biochim Biophys Acta 1839: 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price IR, Gaballa A, Ding F, Helmann JD, Ke A. 2015. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol Cell 57: 1110–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reining A, Nozinovic S, Schlepckow K, Buhr F, Furtig B, Schwalbe H. 2013. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature 499: 355–359. [DOI] [PubMed] [Google Scholar]
- Ren A, Rajashankar KR, Patel DJ. 2012. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature 486: 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz J, Martin JS, Laederach A. 2012. Evaluating our ability to predict the structural disruption of RNA by SNPs. BMC Genomics 13: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz J, Martin JS, Laederach A. 2013. Evolutionary evidence for alternative structure in RNA sequence co-variation. PLoS Comput Biol 9: e1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers ML, Tretbar US, Dehaven A, Alwan AA, Luo G, Mast HM, Hoskins AA. 2016. Conformational dynamics of stem II of the U2 snRNA. RNA 22: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. 2015. Structure and thermodynamics of N6-methyladenosine in RNA: A spring-loaded base modification. J Am Chem Soc 137: 2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer MF, Choudhary PK, Sigel RK. 2014. The AdoCbl-riboswitch interaction investigated by in-line probing and surface plasmon resonance spectroscopy (SPR). Methods Enzymol 549: 467–488. [DOI] [PubMed] [Google Scholar]
- Schibler U, Perry RP. 1977. The 5′-termini of heterogeneous nuclear RNA: A comparison among molecules of different sizes and ages. Nucleic Acids Res 4: 4133–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671–684. [DOI] [PubMed] [Google Scholar]
- Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. 2007. Structures of RNA switches: Insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl 46: 1212–1219. [DOI] [PubMed] [Google Scholar]
- Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. 2008. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455: 1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Drobny G, Varani G. 2007. Binding of U1A protein changes RNA dynamics as observed by 13C NMR relaxation studies. Biochemistry 46: 5875–5883. [DOI] [PubMed] [Google Scholar]
- Sherpa C, Rausch JW, Le Grice SF, Hammarskjold ML, Rekosh D. 2015. The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res 43: 4676–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood AV, Henkin TM. 2016. Riboswitch-mediated gene regulation: Novel RNA architectures dictate gene expression responses. Annu Rev Microbiol 70: 361–374. [DOI] [PubMed] [Google Scholar]
- Shi Z, Barna M. 2015. Translating the genome in time and space: Specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu Rev Cell Dev Biol 31: 31–54. [DOI] [PubMed] [Google Scholar]
- Smith CW, Nadal-Ginard B. 1989. Mutually exclusive splicing of α-tropomyosin exons enforced by an unusual lariat branch point location: Implications for constitutive splicing. Cell 56: 749–758. [DOI] [PubMed] [Google Scholar]
- Smith-Peter E, Lamontagne AM, Lafontaine DA. 2015. Role of lysine binding residues in the global folding of the lysC riboswitch. RNA Biol 12: 1372–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponer JE, Spackova N, Leszczynski J, Sponer J. 2005. Principles of RNA base pairing: Structures and energies of the trans Watson–Crick/sugar edge base pairs. J Phys Chem B 109: 11399–11410. [DOI] [PubMed] [Google Scholar]
- Su L, Chen L, Egli M, Berger JM, Rich A. 1999. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat Struct Biol 6: 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, et al. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84: 2305–2315. [DOI] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. 2006. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science 312: 1208–1211. [DOI] [PubMed] [Google Scholar]
- Trausch JJ, Ceres P, Reyes FE, Batey RT. 2011. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure 19: 1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Xu Z, Edwards AL, Reyes FE, Ross PE, Knight R, Batey RT. 2014. Structural basis for diversity in the SAM clan of riboswitches. Proc Natl Acad Sci 111: 6624–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhm H, Kang W, Ha KS, Kang C, Hohng S. 2018. Single-molecule FRET studies on the cotranscriptional folding of a thiamine pyrophosphate riboswitch. Proc Natl Acad Sci 115: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. 2007. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19: 3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. 1985. Nucleotide sequence of the AIDS virus, LAV. Cell 40: 9–17. [DOI] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2011. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9: 860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. 2014. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505: 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Strobel EJ, Yu AM, Lis JT, Lucks JB. 2016. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol 23: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind JE, Dutta D, Belashov IA, Jenkins JL. 2017. Metalloriboswitches: RNA-based inorganic ion sensors that regulate genes. J Biol Chem 292: 9441–9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E. 2014. Isostericity and tautomerism of base pairs in nucleic acids. FEBS Lett 588: 2464–2469. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM. 2005a. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44: 13404–13414. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. 2005b. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18: 49–60. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956. [DOI] [PubMed] [Google Scholar]
- Wolter AC, Weickhmann AK, Nasiri AH, Hantke K, Ohlenschlager O, Wunderlich CH, Kreutz C, Duchardt-Ferner E, Wohnert J. 2017. A stably protonated adenine nucleotide with a highly shifted pKa value stabilizes the tertiary structure of a GTP-binding RNA aptamer. Angew Chem Int Ed Engl 56: 401–404. [DOI] [PubMed] [Google Scholar]
- Wostenberg C, Ceres P, Polaski JT, Batey RT. 2015. A highly coupled network of tertiary interactions in the SAM-I riboswitch and their role in regulatory tuning. J Mol Biol 427: 3473–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhan L, Zhang W, Sun F, Wang W, Tian N, Bi J, Wang H, Shi D, Jiang Y, et al. 2011. RNA secondary structure in mutually exclusive splicing. Nat Struct Mol Biol 18: 159–168. [DOI] [PubMed] [Google Scholar]
- Yue Y, Yang Y, Dai L, Cao G, Chen R, Hong W, Liu B, Shi Y, Meng Y, Shi F, et al. 2016. Long-range RNA pairings contribute to mutually exclusive splicing. RNA 22: 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Hou S, Wang X, Zhan L, Cao G, Li G, Shi Y, Zhang P, Hong W, Lin H, et al. 2017. Role and convergent evolution of competing RNA secondary structures in mutually exclusive splicing. RNA Biol 14: 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir A, Miskin R, Elson D. 1969. Interconversions between inactive and active forms of ribosomal subunits. FEBS Lett 3: 85–88. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ferre-D’Amare AR. 2013. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500: 363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Hansen AL, Zhang Q. 2014. Characterizing slow chemical exchange in nucleic acids by carbon CEST and low spin-lock field R1ρ NMR spectroscopy. J Am Chem Soc 136: 20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Guffy SL, Williams B, Zhang Q. 2017. An excited state underlies gene regulation of a transcriptional riboswitch. Nat Chem Biol 13: 968–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, Pan T. 2016. N6-methyladenosine modification in a long noncoding RNA hairpin predisposes is conformation to protein binding. J Mol Biol 428: 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. 2017. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods 14: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]