Abstract
The above-ground plant body in different plant species can have very distinct forms or architectures that arise by recurrent redeployment of a finite set of building blocks—leaves with axillary meristems, stems or branches, and flowers. The unique architectures of plant inflorescences in different plant families and species, on which this review focuses, determine the reproductive success and yield of wild and cultivated plants. Major contributors to the inflorescence architecture are the activity and developmental trajectories adopted by axillary meristems, which determine the degree of branching and the number of flowers formed. Recent advances in genetic and molecular analyses in diverse flowering plants have uncovered both common regulatory principles and unique players and/or regulatory interactions that underlie inflorescence architecture. Modulating activity of these regulators has already led to yield increases in the field. Additional insight into the underlying regulatory interactions and principles will not only uncover how their rewiring resulted in altered plant form, but will also enhance efforts at optimizing plant architecture in desirable ways in crop species.
The colonization and radiation of plants on land was a major event in plant evolution, shaping the terrestrial biosphere and the geosphere and transforming global climate. The embryophytes, informally named land plants, originated around 430 million years ago from the last shared unicellular ancestor classified among the charophyta, a group of freshwater green algae (Wickett et al. 2014; Jill Harrison 2017). Evolution from the aquatic to the terrestrial habitat of plants required key morphological innovations (Pires and Dolan 2012). Based on recent molecular phylogenetic advances, comparative analysis pinpoints fundamental body plan features in plant evolution, such as multicellularity, plasmodesmatal cell–cell connections, specialized apical cell fates, three-dimensional apical growth, asymmetric cell division, bifurcation, diversification of indeterminate branched forms with leaves and roots, as well as axillary branching (Wang and Li 2008). Those innovations in development and the underlying genetic changes drive the radiation of the three-dimensional organization of the plant body and, thus, plant architecture (Reinhardt and Kuhlemeier 2002).
Plant architecture is a product of development and keeps changing during the life cycle of plants from young seedlings to adult plants. Plant architecture furthermore is species specific and it has long been the best means of identifying plants (Yang and Hwa 2008). The architecture of a plant is also influenced by environmental conditions such as light, temperature, humidity, and nutrient status (Harder and Prusinkiewicz 2012; Morris et al. 2017; Scheres and van der Putten 2017). Based on plant molecular and genetic studies, the equilibrium between endogenous and exogenous cues define the relative arrangement of above-ground parts of the plant, including the branching pattern (the degree of branching and internodal elongation) and shoot determinacy (the size, shape, and position of leaves, branches, and flowers). Above-ground plant architecture is thus a collection of important agronomic traits and a major determinant of ability of plants to thrive during cultivation, their yield and quality, and harvesting efficiency (Wang et al. 2018). During reproductive development, the inflorescence architecture can be determinate (i.e., terminate in a flower) or indeterminate; simple (flowers are borne on the main stem) or compound (flowers born on branches). For example, Arabidopsis and snapdragon form indeterminate simple inflorescences, whereas pea and Medicago have determinate compound inflorescences. The inflorescences of tomato are determinate and compound; while maize and rice have determinate or indeterminate compound inflorescences, respectively (Benlloch et al. 2015; Bommert and Whipple 2018). Recent advances in our understanding of the molecular-genetic regulation of plant architecture are mostly from these and other model and crop plants (Benlloch et al. 2007; Zhang and Yuan 2014; Teichmann and Muhr 2015; Bommert and Whipple 2018; Chongloi et al. 2019) and have begun to shed light on how individual aspects of plant architecture initiate, develop, and are maintained. This review will focus on these advances and the underlying genetic determinants.
LATERAL ORGAN INITIATION
Lateral organs initiate from the flanks of the stem cell pool containing shoot apical meristem (SAM) at sites of local auxin maxima (Weijers and Wagner 2016). Stem cell descendants in the SAM periphery can respond to auxin maxima by initiating primordia and so-called axillary meristems; the latter in turn can be dormant, or grow out and become branches or flowers (Prusinkiewicz et al. 2007; Martín-Fontecha et al. 2018). The phyllotaxis—the species-specific arrangement of the lateral organs along the stem and inflorescence—relies on the positioning of auxin maxima together with additional chemical (cytokinin) and physical (mechanical) cues (Kuhlemeier 2017; Truskina and Vernoux 2018; Shi and Vernoux 2019).
Recent advances have shed light on the transcriptional responses triggered by auxin sensing that enable formation of lateral organ primordia. In reproductive meristems of Arabidopsis, the auxin signal is primarily transduced by the auxin response factor (ARF) MONOPTEROS (MP) (Vernoux et al. 2010; Weijers and Wagner 2016). MP directs initiation of reproductive primordia in part by up-regulating expression of diverse transcription factors, including the YABBY transcription factor FILAMENTOUS FLOWERS (FIL), the LEAFY (LFY) transcription factor, important for floral fate, and two PLETHORA transcription factors that promote organ growth (Yamaguchi et al. 2013; Wu et al. 2015). Auxin-dependent up-regulation of FIL and LFY depends on recruitment of chromatin remodelers that open up chromatin near MP-bound sites at target loci (Wu et al. 2015). More recently, two additional ARFs [ETTIN (ETT) and ARF4] were shown to promote lateral organ initiation in parallel with MP and sever mp ett arf4 triple mutants fail to initiate any lateral organs. MP, ETT, and ARF4 jointly promote organogenesis by down-regulating the expression of class I KNOX pluripotency genes (SHOOT MERISTEMLESS [STM] and BREVIPEDICELLUS [BP]). MP represses class I KNOX genes indirectly, via its direct target FIL, whereas ETT/ARF4 repress these genes directly. STM/BP repression by the YABBY/ARF complex is mediated by chromatin compaction through histone deacetylation (Chung et al. 2019). Subsequent to flower initiation, STM is up-regulated when the flower meristem is established in stage 2 flowers by an as-yet unknown mechanism. In addition, STM is up-regulated in the boundary between the initiating flower and the meristem by mechanical signals, which enables separation of the primordium from the meristem (Landrein et al. 2015).
In inflorescences, on the basis of live imaging and reporter constructs, the auxin-positive (and STM-depleted) region comprises both the cryptic bract and the cells that give rise to the flower (Heisler et al. 2005). Burian et al. (2016) recently provided evidence that axillary meristems that give rise to branches form at sites where auxin response is high concomitant with leaf initiation, as do flowers. These studies focused on secondary branches that arise in the axils of bracts (also called cauline leaves) early during reproductive development (40-day-old short-day grown plants). The combined studies from Arabidopsis and tomato suggest similarities between initiation of bracts with associated secondary branches and flowers plus (cryptic) bracts during reproductive development.
Axillary meristem initiation during vegetative development is delayed relative to leaf initiation and requires an auxin minimum that supports low STM expression (Wang et al. 2014; Zhou et al. 2014; Shi et al. 2016). Thus, during vegetative development, lateral organ initiation and branch meristem formation are distinct events with opposite auxin requirements, which will be discussed further below.
The role of auxin and cytokinin in lateral organ initiation during reproductive development is evolutionarily conserved in the grasses (for reviews, see Barazesh and McSteen 2008; Matthes et al. 2018). In both maize and rice inflorescences, auxin and PIN markers reveal auxin accumulation in primordia that will give rise to branches, spikelets, spikelet pairs (maize), and flowers with associated (cryptic) bracts or leaves (Gallavotti et al. 2008; Galli et al. 2015; Yang et al. 2017). Likely because of genetic redundancy, specific ARFs have thus far not been linked to auxin-mediated lateral organ initiation (Galli et al. 2015). However, mutations in two Aux/IAA proteins—negative regulators of ARFs like MP—caused defects in lateral organ initiation in male and female inflorescences in maize (Galli et al. 2015). The identified Aux/IAA auxin response inhibitors act directly upstream of a classical regulator of lateral organ initiation, the bHLH transcription factor BARREN STALK1 (BA1) (Gallavotti et al. 2004). The BA1 homolog in rice, LAX PANICLE 1 (LAX1) also promotes initiation of lateral organs (Komatsu et al. 2003). In maize and rice, similar BA1/LAX1 interacting nuclear proteins with putative RAWUL/PDZ/SH3 domains called BA2 (Yao et al. 2019) or LAX2 (Tabuchi et al. 2011) were identified. LAX2 links to auxin response in other processes by blocking the interaction between Aux/IAA proteins and ARFs (Li et al. 2018). Mutants in the Arabidopsis BA1/LAX1 homolog ROX display defects in initiation of meristems in the axils of leaves during vegetative development (Yang et al. 2012). A role for YABBY family members in lateral organ initiation, as described above for Arabidopsis, is conserved in rice; simultaneous loss- or reduction-of-function in three FIL YABBY proteins leads in the most extreme cases to formation of naked inflorescence branches lacking spikelets (Tanaka et al. 2017).
In the remainder of this review, we will consider the different developmental trajectories of the axillary meristems, which we define here as populations of cells born in the axils of leaves initiated either concomitantly with leaves or much later, which have the potential to give rise to leaves, branches, and flowers. Taking recent research into account, this review will focus on mechanisms underlying axillary meristem formation, activity, and identity as major contributors to plant architecture and to agronomically desirable plant traits.
AXILLARY MERISTEM INITIATION
As outlined above, axillary meristem initiation and development during vegetative development is delayed relative to leaf initiation and furthermore regulated by transcriptional regulators largely conserved between eudicots and monocots (Teichmann and Muhr 2015; Wang and Jiao 2018).
The combined available data suggest that in eudicots (Arabidopsis/tomato) and monocots (rice) a cascade of transcription factors coordinately up-regulate the pluripotency gene STM to enable formation of the axillary meristem (e.g., see Tian et al. 2014; Tanaka et al. 2015). The Lateral suppressor (Ls) gene of tomato encodes a member of the VHIID subfamily of the plant-specific GRAS (GA INSENSITIVE, REPRESSOR OF GA1-3, SCARECROW) transcription factor that is required for axillary meristem initiation in many plant species besides tomato, such as Arabidopsis LATERAL SUPPRESSOR (LAS) and rice MONOCLUM1 (MOC1) (Greb et al. 2003; Li et al. 2003). The las mutant is unable to form lateral shoots and the moc1 mutant has a single primary stem (culm) lacking tillers during vegetative development (Greb et al. 2003; Li et al. 2003). MOC1 in rice and TaMOC1 in wheat also play a role in inflorescence branching (Li et al. 2003; Zhang et al. 2015).
The tomato Blind (Bl) gene encodes an R2R3 class MYB transcription factor and is required for axillary meristem initiation. During vegetative development, bl mutants lack axillary meristems in most leaves (Schmitz et al. 2002). Likewise, Arabidopsis REGULATORS OF AXILLARY MERISTEM (RAX) genes, homologs of Bl, control axillary meristem initiation. RAX1/MYB37, RAX2/MYB38, and RAX3/MYB84 act redundantly in this process (Müller et al. 2006). RAX1 acts in parallel with LAS in axillary meristem initiation and both genes promote establishment/maintenance of STM expression in axillary meristems (Greb et al. 2003; Keller et al. 2006; Müller et al. 2006). RAX proteins have additional roles during reproductive development (Keller et al. 2006; Müller et al. 2006). Recently, gain-of-function of five closely related WRKY transcription factors each promoted axillary meristem initiation; one of these, EXCESSIVE BRANCHES1 (EXB1/WRKY71), positively regulates RAX1, 2, and 3 transcription (Guo et al. 2015).
The class III HD-ZIP transcription factor REVOLUTA (REV) and the AP2 transcription factor DORNRÖSCHEN (DRN) also contribute to axillary meristem initiation during vegetative development (Shi et al. 2016; Zhang et al. 2018). The drn drnl double mutant fails to initiate lateral meristems, whereas REV and DRN together promote STM up-regulation. Some genes affecting axillary meristem initiation, such as RAX1 and Bl, up-regulate CUP-SHAPED COTYLEDON 1 (CUC1), CUC2, and CUC3, and these NAC transcription factors redundantly promote axillary meristem initiation (Hibara et al. 2006; Raman et al. 2008). A recent study of mobile and nonmobile STM function revealed that STM mobility is critically important to suppress axillary meristem formation by precisely regulating CUC genes (Balkunde et al. 2017). Up-regulation of STM during vegetative axillary meristem initiation is also important in other plant species. In rice, TILLERS ABSENT1 (TAB1), a WUSCHEL ortholog promotes formation of the axillary meristem via the activation of OSH1, the meristematic cells marker and the ortholog of STM (Lu et al. 2015; Tanaka et al. 2015).
AXILLARY MERISTEM OUTGROWTH
Transcriptional Control of Axillary Meristem Outgrowth
Vegetative axillary meristems, once initiated, either grow out as branches or form dormant axillary buds. Thus, the final number of branches is determined by the number of axillary meristems formed, the activity of the axillary buds, and the timing of flower formation. Endogenous cues related to developmental stage and meristem activity at the main shoot are integrated with environmental signals related to nutrient status or plant density to optimize bud outgrowth for plant survival and fitness (Domagalska and Leyser 2011). For example, to survive under adverse conditions, axillary meristems can switch to a dormant state. Once environmental or endogenous conditions change, dormant organs can resume growing (Martín-Fontecha et al. 2018).
In many plant species, TCP transcription factors promote bud dormancy. Maize TEOSINTE BRANCHED 1 (TB1) encodes a class II TCP family transcriptional regulator expressed in axillary buds and axillary branches and prevents bud outgrowth (Doebley et al. 1997; Hubbard et al. 2002). The TB1 homologs Arabidopsis BRANCHED1 (BRC1) (Aguilar-Martínez et al. 2007; Seale et al. 2017), rice FINE CULM1 (FC1/OsTB1) (Minakuchi et al. 2010), sorghum (Sorghum bicolor) SbTB1 (Kebrom et al. 2006) and Tulipa gesneriana (TgTB1) (Moreno-Pachon et al. 2018) are also expressed in axillary meristems and buds and prevent their outgrowth. Both TB1 in maize and BRC1 in Arabidopsis also affect onset of flower formation in branches (Hubbard et al. 2002; Niwa et al. 2013). A recent study in Arabidopsis shows that buds in which BRC1 has accumulated can be activated, indicating that BRC1 is not sufficient for bud growth inhibition (Seale et al. 2017).
Transcriptomes of active and dormant axillary buds in herbaceous and woody plant species have identified additional regulators of bud activity (Stafstrom et al. 1998; Tatematsu et al. 2005; Wood et al. 2013; Kebrom and Mullet 2016). These studies point to a small disordered protein of unknown function called dormancy marker gene DORMANCY ASSOCIATED PROTEIN 1 (DRM1) expressed specifically in dormant axillary buds of pea (Stafstrom et al. 1998), Arabidopsis (Tatematsu et al. 2005), wheat (Kebrom et al. 2012), sorghum (Kebrom et al. 2006), kiwifruit (Wood et al. 2013), and poplar and grapevine (Tarancón et al. 2017).
Systemic Control of Axillary Meristem Outgrowth
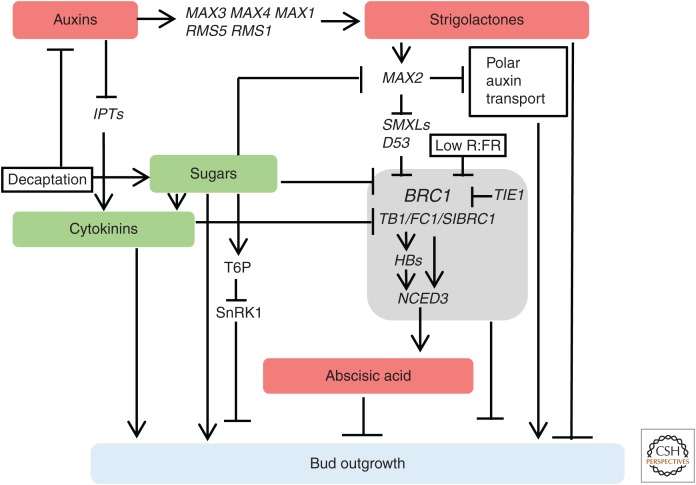
Multiple signals modulate outgrowth of dormant axillary buds, such as phytohormones (auxin, cytokinin, strigolactones, and abscisic acid [ABA]), nutrients (sugars, nitrogen), light quality (far red), and damage to the shoot apex (decapitation) (Fig. 1; Domagalska and Leyser 2011).
Figure 1.
Phytohormones, sugars, and key regulatory genes control bud outgrowth. Different types of phytohormones (auxin, cytokinin, strigolactone, and abscisic acid) as well as sugars are key endogenous determinants of bud outgrowth. The complex interplay among those regulatory pathways are summarized (see text for details).
Auxin produced in the shoot enters the rootward polar auxin transport (PAT) stream and prevents bud outgrowth in at least two ways: by modulating production of cytokinin and strigolactone, which act in the axillary buds (Dun et al. 2012; Teichmann and Muhr 2015; Dierck et al. 2016), and by regulating auxin flow out of buds (Sachs 1981; Prusinkiewicz et al. 2009; Balla et al. 2011). The latter response is less pronounced in pea (Chabikwa et al. 2019). Auxin represses the biosynthesis of cytokinins, which promote bud outgrowth, via down-regulation of ADENYLATE ISOPENTENYLTRANSFERASE (IPT) family members in the nodal stem (Tanaka et al. 2006). Auxin promotes synthesis of strigolactone, which inhibits bud outgrowth via up-regulating MORE AXILLARY GROWTH 3 (MAX3) and MAX4 expression (Hayward et al. 2009). Recently, the highly branched rms2 mutant of pea, which has lower levels of strigolacton, was shown to be caused by a defect in an ABF4/5 like auxin receptor (Ligerot et al. 2017).
Cytokinins can directly promote bud outgrowth regardless of auxin accumulation (Sachs and Thimann 1967; Müller et al. 2015). Cytokinins are mostly synthesized in the root but also locally in the shoot and are transported shootward in the xylem (Chen et al. 1985). How cytokinin promotes bud outgrowth is still not fully understood. Cytokinin inhibits accumulation of BRC1 in pea and Arabidopsis (Braun et al. 2012; Dun et al. 2012; Seale et al. 2017). In addition, cytokinin enhances polar auxin transport in the main stem by promoting accumulation of the polar auxin efflux regulators, PIN3, 4, and 7 (Waldie and Leyser 2018). Cytokinin response regulators also contribute to dormancy; with “repressive” type-A cytokinin response regulators (ARRs) promoting and “activating” type-B ARRs inhibiting bud outgrowth in Arabidopsis (Müller et al. 2015; Waldie and Leyser 2018). The molecular mechanism for this is not yet understood, but it has been proposed that the phenotypes of the cytokinin response regulator mutants may be attributable to their effects on cytokinin homeostasis (Waldie and Leyser 2018).
The hormone strigolactone inhibits branch outgrowth in Arabidopsis, pea, rice and petunia (for reviews, see Domagalska and Leyser 2011; Barbier et al. 2019). The available evidence suggests that strigolactones prevent bud outgrowth at least in part by up-regulating expression BRC1 (Dun et al. 2012) and by impacting auxin transport (see below). Strigolactone, received by Arabidopsis and rice DWARF14 (D14) (Chevalier et al. 2014), leads to degradation of strigolactone response regulators mediated by the F-box protein MAX2 in Arabidopsis and the homolog D3 in rice (Jiang et al. 2013; Zhou et al. 2013). Strigolactone response regulators important for branch outgrowth include the SUPPRESSOR OF MAX2 1-LIKE (SMXL) 6, 7, and 8 genes in Arabidopsis and co-ortholog DWARF53 (D53) in rice (Jiang et al. 2013; Zhou et al. 2013; Soundappan et al. 2015; Wang et al. 2015b). smxl 6, 7, and 8 can restore branch suppression in strigolactone biosynthesis and signaling mutants (Soundappan et al. 2015; Wang et al. 2015b). D53-like SMXLs promote branching by down-regulating BRC1 (Seale et al. 2017). However, this does not explain all of strigolactorne action as buds in brc1 brc2 double mutant are strigolactone responsive (Seale et al. 2017). FC1/OsTB1 functions downstream of strigolactones to inhibit the outgrowth of buds (Minakuchi et al. 2010). The pea Psbrc1 mutant displays an increased shoot-branching phenotype, is able to synthesize strigolactone, and does not respond to strigolactone application (Braun et al. 2012). Tomato SIBRC1a and SIBRC1b are expressed in arrested axillary buds and both are down-regulated on bud activation (Martín-Trillo et al. 2011).
Strigolactone also negatively regulates auxin flux and PIN1 polarization via the max pathway (Bennett and Leyser 2006; Bennett et al. 2006, 2016) and promotes rapid PIN1 endocytosis (Crawford et al. 2010; Shinohara et al. 2013; Xu et al. 2015). Indeed, branching defects of strigolactone biosynthesis mutants can be rescued by inhibition of PAT (Bennett and Leyser 2006; Bennett et al. 2006). Strigolactone can also inhibit branching independent of auxin (Brewer et al. 2009, 2015; Chabikwa et al. 2019).
Sugar promotes bud outgrowth and it has been suggested that the growing shoot tip may suppress branch outgrowth because it acts as a sugar sink (Barbier et al. 2015b; Kebrom 2017). Addition of sucrose to inhibited buds can rapidly release inhibition by repressing BRC1 expression (Mason et al. 2014). Studies on shoot branching in barley (Felippe and Dale 1973), sorghum (Kebrom and Mullet 2015), wheat (Kebrom et al. 2012), rose (Barbier et al. 2015a), and Arabidopsis (Otori et al. 2017) have also indicated a strong association between sucrose supply and axillary bud outgrowth. The rice monoculm mutant moc2, which is defective in branch outgrowth and has significantly reduced tiller numbers, displays reduced sucrose supply to the buds and is caused by disruption of cytosolic fructose-1,6-bisphosphatase 1 (FBP1) gene (Koumoto et al. 2013). Recent studies suggest that sucrose may act as a signal rather than an energy substrate in promoting bud outgrowth (Barbier et al. 2015b). Overexpression of Arabidopsis HEXOKINASE1 (HXK1) leads to the increased number of primary rosette branches, increased expression of ABA-related genes, and reduced expression of auxin-related genes (Kelly et al. 2012). Trehalose-6-P (T6P) accumulates to low concentrations in wild-type plants (Grennan 2007). Loss of activity in the Arabidopsis trehalose-6-P synthase 6 (TPS6), which encodes an active trehalose synthase (Chary et al. 2008), leads to reduced bud outgrowth (Schluepmann et al. 2003; Chary et al. 2008). In addition, T6P levels in the axillary buds of pea increase rapidly after decapitation, triggering onset of bud outgrowth (Fichtner et al. 2017). More importantly, signaling of carbon and nitrogen metabolic pathways might be integrated at the level of T6P to control axillary meristem outgrowth (Fichtner et al. 2017; Barbier et al. 2019). It has been proposed that T6P may link to carbon starvation syndrome, and genes related to this pathway are enriched in dormant buds of poplar and grapevine (Tarancón et al. 2017).
Little is known about hormonal control inside axillary buds during the growth-to-dormancy transition, although ABA accumulation may promote bud dormancy (Reddy et al. 2013; Yao and Finlayson 2015). ABA was proposed to be a general inhibitor of axillary bud outgrowth (Shimizu-Sato and Mori 2001; Yao and Finlayson 2015). BRC1 stimulates the ABA biosynthesis pathway by the up-regulation of HOMEOBOX PROTEIN 21 (HB21), HB40, and HB53, which in turn activate the ABA biosynthesis gene NINE-CIS-EPOXICAROTENOID DIOXIGENASE 3 (NCED3) (González-Grandío et al. 2017). TCP interactor containing EAR motif protein 1 (TIE1) interacts with BRC1 and prevents it from activating HB21, HB40, and HB53 expression (Yang et al. 2018). ABA levels increase in buds under low R/FR light and in phytochrome B mutants, which also display reduced branch outgrowth in Sorghum and Arabidopsis (Kebrom et al. 2010; Reddy et al. 2013; Yao and Finlayson 2015; Kebrom and Mullet 2016). This may be a response to plant density/shade avoidance.
AXILLARY MERISTEM IDENTITY IN THE INFLORESCENCE
Axillary Meristems that Remain Vegetative
Axillary meristems in the inflorescence can adopt a variety of different fates: They can remain vegetative, give rise to branches, or become flowers. During the vegetative-to-reproductive transition in axillary meristems, plants integrate environmental and developmental stimuli and either “decide” to irreversibly initiate reproductive development—give rise to flowering branches or flowers (most Arabidopsis cultivars)—or remain vegetative, as do a subset of axillary meristems in perennial species.
In Arabidopsis, vegetative meristems can form aerial rosettes in lieu of flowering branches. This is in all cases linked with a severe delay in the onset of reproductive development, triggered by gain-of-function of flowering repressors or by loss-of-function of flowering activators. For example, in annual Arabidopsis thaliana accession (Columbia-0), overexpressing the mobile “antiflorigen” TERMINAL FLOWER 1 (TFL1) in non-inductive photoperiod (short-day) not only severely delays onset of reproduction but produces large aerial rosettes (Ratcliffe et al. 1998). Consistent with its opposite role to TFL1, failure to fully induce FLOWERING LOCUS T (FT) also leads to formation of aerial rosettes (Liu et al. 2014). Mutations in two flowering activator MADS box transcription factors, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) and FRUITFULL (FUL) soc1 ful double mutants, form many aerial rosettes even in inductive photoperiod and resemble perennial plants (Melzer et al. 2008). Aerial rosette formation in the late flowering Sy-0 accession of Arabidopsis has been attributed to elevated levels of the flowering repressor FLOWERING LOCUS C (FLC), a known negative regulator of the mobile florigen FT (Wang et al. 2007).
These phenotypes are reminiscent of vegetative meristems in the perennial species Arabis alpina. The FLC ortholog PERPETUAL FLOWERING 1 (PEP1), the AP2 ortholog PEP2, TFL1, and miR156 promote the perennial growth habit, in which some axillary meristems remain vegetative (Wang et al. 2009, 2011; Bergonzi et al. 2013; Lazaro et al. 2018, 2019; Hyun et al. 2019). After vernalization, PEP1 represses flowering branch or flower formation in these axillary meristems (Wang et al. 2009; Lazaro et al. 2018) in a mechanism that is partially dependent on PEP2 (Lazaro et al. 2019). Plant mutants for these flowering repressors convert vegetative meristems to flowering branches or flowers. Likewise, in Arabis alpina expressing a miR156-resistant version of SPL15, most axillary meristems gave rise to flowering branches (Hyun et al. 2019). A similar phenotype was observed when TFL1 activity was reduced (Wang et al. 2011). AP2 also acts as a floral repressor in Arabidopsis (Yant 2012).
Axillary Meristems that Give Rise to Branches or Flowers
In most angiosperms, axillary meristems on the inflorescence either give rise to branches or flowers. Branches, unlike flowers, usually display internode elongation, produce leaves and secondary branches in addition to flowers and can be indeterminate (Arabidopsis) or determinate (pea and rice) (Benlloch et al. 2007). Flowers are determinate, give rise to a finite number of floral organs, and do not generally display internode elongation. In the racemes of Arabidopsis, branch fate and indeterminacy of the axillary meristems as well as the primary inflorescence meristem are promoted by TFL1 (Conti and Bradley 2007), which acts in opposition to factors that induce floral fate, most notably LEAFY (LFY) and the MADS box transcription factor APETALA1 (AP1) (for reviews, see Denay et al. 2017; Wagner 2017). LFY is expressed before formation of the first flower in bracts that subtend branches, whereas AP1 up-regulation occurs later and is restricted to young flower primordia that have committed to floral fate (Alejandra Mandel et al. 1992; Blazquez et al. 1997; Hempel et al. 1997; Yamaguchi et al. 2009). LFY directly up-regulates AP1 (Wagner et al. 1999).
As AP1 promotes determinacy, branches are formed until AP1 is up-regulated. The delay in AP1 relative to LFY up-regulation is in part attributable to a requirement for transcriptional cofactors that contribute to AP1 induction with LFY in coherent feedforward loops, such as the MYB transcription factor LATE MERISTEM IDENTITY2 (LMI2) (Pastore et al. 2011). In addition, gibberellin, which increases during cessation of vegetative development (Galvão et al. 2012; Porri et al. 2012), blocks formation of flowers (Yamaguchi et al. 2014). LFY directly up-regulates the GA-catabolic enzyme EUI-LIKE P450 A1 (ELA1) as well as GA2 oxidases to enable accumulation of GA-sensitive DELLA proteins, which complex with the SBP transcription factor SPL9 to activate AP1 in parallel with LFY (Yamaguchi et al. 2009, 2014). Gibberellin also opposes determinacy/flower formation in woody perennial trees such as orange, mango, grapevine, and diploid strawberry (for reviews, see Boss and Thomas 2002; Bangerth 2009; Tenreira et al. 2017). Finally, ubiquitination of LFY by BLADE ON PETIOLE gene BOP2 and CULLIN3 (CUL3) in seedlings is important for AP1 induction (Chahtane et al. 2018). This fits with prior findings that LFY is posttranslationally activated via ubiquitination and the F-box protein UNUSUAL FLORAL ORGANS (UFO)/CUL1 for flower patterning (Lee et al. 1997; Parcy et al. 1998; Chae et al. 2008). Likewise, in tomato and rice, ubiquitination of LFY by UFO is required broadly for LFY activity (see below).
AP1 is both a direct target of LFY and acts in parallel with LFY to specify floral fate; this is because of LFY independent inputs into AP1 activation from FT and SPL transcription factors (for review, see Wagner 2017). Other related MADS box proteins, such as CAULIFLOWER (CAL) and FRUITFULL (FUL) also promote determinacy (Ferrandiz et al. 2000). In addition, consistent with its role in floral commitment, AP1 directly silences TFL1 with assistance from the SEPALLATA family transcription factor SEP4 (Ferrandiz et al. 2000; Kaufmann et al. 2010; Liu et al. 2013; Serrano-Mislata et al. 2016; Goslin et al. 2017). Because of its role in the switch to reproductive development (inflorescence formation), the role of FT in promoting floral fate is difficult to assess (Yamaguchi et al. 2014). However, FT has been implicated in the maintenance of floral fate during reproductive development (Liu et al. 2014; Müller-Xing et al. 2014).
The above-described factors have largely similar roles in the legumes. In these compound racemes, flowers are not born on the primary inflorescence, which is indeterminate, but on branches (secondary inflorescences), which are determinate. Similar to the scenario in Arabidopsis, the pea TFL1 homolog DETERMINATE (DET) and the MtTFL1 of Medicago promote indeterminant growth of the primary inflorescence and branch fate in the axillary meristems. Accordingly, the primary inflorescences of det or mttfl1 mutants resemble a flower-bearing branch (secondary inflorescence) and misexpress VEGETATIVE1 (VEG1) and MtFUL, respectively (Foucher et al. 2003; Berbel et al. 2012; Cheng et al. 2018). In addition, paradoxically, the uppermost axillary branch meristem of mttfl1 became indeterminate, phenocopying sympodial growth (Cheng et al. 2018). It would be interesting to explore whether the latter phenotype is caused by competition between MtTFL1 and one of the other three MtTFL paralogs.
Secondary branch fate is promoted by the AGL79/FUL clade MADS box transcription factors VEG1 and MtFUL in pea and Medicago, respectively. VEG1 is expressed in young secondary inflorescences, which are replaced by nonflowering, indeterminate primary inflorescences that express DET in veg1 mutants (Berbel et al. 2012). Similar morphological and molecular phenotypes are observed in mutants of the VEG1 homolog MtFUL in Medicago (Cheng et al. 2018). Secondary inflorescence fate is also promoted by pea VEG2, a homolog of the bZIP transcription factor FD, which can complex with florigen family proteins (Périlleux et al. 2019). Together with GIGAS/FTa1 (an FT homolog), VEG2 contributes to secondary inflorescence fate in pea (Berbel et al. 2012; Sussmilch et al. 2015). In addition, veg2-2 mutants display reduced VEG1 expression (Sussmilch et al. 2015). Interestingly, in mttfl1 and det veg1 double mutants, all secondary inflorescence branches are replaced by a single flower and the primary inflorescence terminates in a flower (Berbel et al. 2012; Cheng et al. 2018). Thus, DET/MtTFL1 and VEG1/MtFUL not only repress each other (to enable distinct primary and secondary inflorescence development trajectories, respectively), but also jointly prevent floral fate by repressing PROLIFERATING INFLORESCENCE MERISTEM (PIM)/MtAP1.
PIM and MtAP1 promote floral fate together with the LFY homologs UNIFOLIATA (UNI)/SINGLE LEAFLET1 (SGL1) in pea and Medicago, as well as UFO (Hofer et al. 1997; Taylor et al. 2001, 2002; Cheng et al. 2018). However, compared with Arabidopsis, PIM/MtAP1 play a more central role (for review, see Benlloch et al. 2015). pim and mtap1 mutants form VEG1/MtFUL expressing I2 inflorescences in lieu of flowers, suggesting that PIM/MtAP1 represses VEG1/MtFUL (Berbel et al. 2012; Cheng et al. 2018). As in Arabidopsis, only plants mutant for both MtSGL and MtAP1 entirely fail to establish flowers in Medicago (Cheng et al. 2018).
Two largely independent regulatory networks control sympodial growth and inflorescence development of tomato. The former centers on TFL1, the second on LFY/UFO as well as additional factors whose Arabidopsis homologs have no apparent role in axillary meristem identity. After the transition to the reproductive phase, tomato produces a repeating sympodial unit consisting of three leaves with associated axillary meristem and a terminal cluster of flowers (the inflorescence) (Lifschitz et al. 2014). A specialized axillary meristem associated with the uppermost leaf, the sympodial meristem, repeats this entire unit of development. Mutations in the TFL1 homolog SELF PRUNING (SP) reduces the length of the sympodial unit (numbers of leaves produced) and leads to eventual termination in a cluster of flowers. Termination is more extreme in overexpressors of the tomato FT-homolog SINGLE FLOWER TRUSS (SFT) and near complete when SFT is overexpressed in sp mutants, consistent with the idea that the balance between TFL1 and FT homologs determines the switch between indeterminacy/branching and determinacy/flower formation (Lifschitz et al. 2014; Park et al. 2014; Shalit-Kaneh et al. 2018). An FT homolog that evolved to act as a flowering repressor, SP5G, also contributes to sympodial growth by repressing onset of reproductive development. sp5g sp double mutants flower earlier, have shorter sympodial units, and terminate more rapidly in a cluster of flowers (Soyk et al. 2016).
The flower-bearing tomato inflorescence is formed by a sympodial inflorescence meristem, which gives rise to an additional sympodial inflorescence meristem before terminating in a flower; this pattern continues until seven to nine flowers have been formed (Lippman et al. 2008). Branching (formation of sympodial inflorescence meristems) is reduced in the early-flowering tmf mutant, which develops a single-flowered primary inflorescence (MacAlister et al. 2012). TERMINATING FLOWER (TMF), which encodes an ALOG transcription factor (MacAlister et al. 2012), likely is not a negative regulator of branching but promotes vegetative identity. The reduced sympodial activity of the tmf tomato inflorescence is attributable to precocious activation of FALSIFLORA (FA) (LFY ortholog) and ANANTHA (AN) (UFO ortholog). Intriguingly, BOP family members physically interact with TMF and closely related ALOG factors in tomato; the slbop2 mutant in particular reduces the number of flowers formed per inflorescence (Xu et al. 2016). Complete loss of TMF/BOP activity converts all inflorescences to single flowers (Xu et al. 2016). As outlined above, BOP2 in Arabidopsis is required for the ability of LFY to specify floral fate via AP1 up-regulation (Chahtane et al. 2018). Thus, different interaction partners with opposite roles in floral fate rely on BOP activity in the inflorescence of Arabidopsis and tomato. Whether SlBOP2 posttranslationally modifies TMF is not known.
Consistent with the idea that delayed adoption of floral fate within the inflorescence promotes branch formation, inflorescences mutant for AN or UFO, FA or LFY, COMPOUND INFLORESCENCE (S) or WOX9, are more branched (Lippman et al. 2008; MacAlister et al. 2012). In an or fa mutants, the primary meristems of the sympodial inflorescence do not form flowers but remain indeterminate, and repeatedly initiate secondary sympodial inflorescence meristem, which in turn produce more sympodial inflorescence meristems (Lippman et al. 2008). Of note, the tomato and petunia homologs of LFY (FA and ABERRANT LEAF AND FLOWER [ALF]) are more broadly expressed, but their activity is restricted by UFO (AN and DOUBLE TOP [DOT]), whose accumulation is tightly spatiotemporally controlled (for review, see Moyroud et al. 2010). Inflorescences mutant for S fail to properly activate AN and initiate additional sympodial inflorescence (Lippman et al. 2008; MacAlister et al. 2012). More recently, elegant genetic analysis implicated three SEP4 MADS box transcription factors (J2, EJ2, and LIN) in regulating the tomato inflorescence architecture (Soyk et al. 2017). Combination of a weak allele of j2 with heterozygous ej2 as well as heterozygous S mutants enhanced branching without affecting flower development or fruit set and was thus able to enhance yield (Soyk et al. 2017). The roles and genetic interactions between WOX9/EVERGREEN (EVG), LFY/ALF, and UFO/DOT are conserved in petunia, which also has sympodial meristems, but forms single flowers (for review, see Castel et al. 2010). As observed in other plant species, the tomato AP1-like gene MACROCALYX (MC) promotes tomato inflorescence determinacy (Vrebalov et al. 2002; Yuste-Lisbona et al. 2016).
In the grasses, a plethora of distinct inflorescence architectures are found and recent investigations in rice, maize, wheat, barley, and brachypodium are unraveling the genetic basis of each (Bommert and Whipple 2018; Chongloi et al. 2019; Gauley and Boden 2019; Koppolu and Schnurbusch 2019). In the compound inflorescence of rice, flowers are borne on branches, with early arising branches forming secondary branches. Both primary and secondary branches then form spikelet meristems and also terminate in a spikelet meristem. Each spikelet meristem gives rise to a single-flower meristem and flower (Bommert and Whipple 2018; Chongloi et al. 2019). Maize has both male (tassel) and female (ear) inflorescences. The tassel is indeterminate, forming long branches and short branches (spikelet pairs), which in turn gives rise to two spikelet meristems. The ear does not form long branches (Bortiri and Hake 2007; Bommert and Whipple 2018). The spikelet pair and spikelet meristems are considered determinate because of their limited branching capacity (Bortiri and Hake 2007; Bommert and Whipple 2018).
In the rice inflorescence, double mutants in the AP1 and FUL clade MADS box transcription factors OsMADS14 and OsMADS15 form an inflorescence and a primary branch meristem but no secondary branches or spikelets (Wu et al. 2017). The phenotypes suggest redundant roles in promoting determinacy, as vegetative structures form in the place of spikelets. The phenotypes and expression patterns of ABERRANT PANICLE ORGANIZATION 1 (APO1/UFO homolog) and APO2/LFY homolog implicate APO1 and APO2 in promotion of indeterminacy; they prevent termination of the primary inflorescence in a spikelet as well as conversion of secondary branches into spikelets (Kyozuka et al. 1998; Rao et al. 2008; Ikeda-Kawakatsu et al. 2012), an apparently opposite function to that of LFY and UFO homologs in inflorescence development of the eudicots discussed above. Similar to rice, maize zfl2 (LFY homolog) mutants display reduced tassel branching and kernel row number (Bomblies et al. 2003; Bomblies and Doebley 2006). In addition, primary inflorescence meristem phyllotaxis of apo1 and apo2 resembled that of the vegetative meristem (Ikeda-Kawakatsu et al. 2009, 2012). Additional studies are needed to understand the role of LFY homologs in monocots and to dissect communalities and differences with eudicot LFY homologs.
Interestingly, although the role of TFL1 family members has thus far mainly been studied by gain-of-function approaches in rice and maize (Nakagawa et al. 2002; Danilevskaya et al. 2010), these studies as well as RNAi knockdown of all rice CENTRORADIALIS (RCN) homologs point to a role of TFL1 in primary inflorescence and branch meristem maintenance, as in the eudicots (Liu et al. 2013). A similar—albeit weaker—phenotype was observed in rice for mutant in mft1, one of two MOHTER OF FT AND TFL1 (MFT) proteins of the FT/TFL1/MFT family (Song et al. 2018). MFT1 overexpression resulted in increased secondary branch formation and reduced expression of spikelet meristem identity genes, including FRIZZY PANICLE (FZP) (Song et al. 2018), which is discussed in more detail below. Finally, the ALOG transcription factor TAWAWA1 (TAW1), the rice homolog of tomato TMF, prevents premature loss of primary inflorescence activity and conversion of branch meristems into spikelet meristems, at least in part by up-regulating the SHORT VEGETATIVE PHASE (SVP) homologs OsMADS22 and OsMADS55 (Yoshida et al. 2013). However, unlike tmf mutants, taw1 mutants are not early flowering. The miR156 targeted SPL family of transcription factors, especially OsSPL14, the closest homolog of Arabidopsis SPL15 and SPL9, promotes panicle branching, perhaps together with OsSPL17 (Jiao et al. 2010; Miura et al. 2010; Wang et al. 2015a). Posttranslational modifications also modulate OsSPL14 accumulation and panicle branching (Wang et al. 2017a,b). Similar roles in inflorescence branching were described for the maize orthologs TASSELSHEATH 4 (TSH4), UNBRANCHED 2 (UB2), and UB3 (Chuck et al. 2014).
The AP2/ERF transcription factors BRANCHED SILKLESS1 (BD1) in maize and FZP in rice, as well as their orthologs in barley, wheat, and brachypodium, are markers for and promote spikelet meristem formation (Chuck et al. 2002; Komatsu et al. 2003; Derbyshire and Byrne 2013; Poursarebani et al. 2015). Recently, rice grain number QTLs were mapped to fzp, which led to formation of more secondary branches (Bai et al. 2017; Fujishiro et al. 2018). Plants mutant for Arabidopsis PUCHI, the BD1 and FZP homolog, have only subtle defects in inflorescence architecture (Karim et al. 2009). Recently, it was shown that FZP down-regulates APO2/LFY in spikelet meristems (Bai et al. 2016). PANICLE PHYTOMER2 (PAP2/OsMADS34), a member of a grass-specific clade of SEP proteins, promotes spikelet meristem identity (for review, see Chongloi et al. 2019). Spikelet meristem determinacy, prevention of formation of multiple florets, is under control of microRNA miR172 and its AP2 transcription factor targets INDETERMINATE SPIKELET1 (IDS1) and SISTER OF IDS1 (SID) (Chuck et al. 2007, 2008). IDS1 and SID were first identified in maize, but have conserved roles in rice, wheat, and barley (Brown and Bregitzer 2011; Lee and An 2012; Lee et al. 2014; Debernardi et al. 2017).
The spikelet pair meristem found in the Andropogonae, which includes maize, is considered a determinate branch meristem (Whipple 2017) and gives rise to two spikelet meristems. The genetically defined RAMOSA (RA) genes promote determinacy in spikelet pair meristems (Bommert and Whipple 2018). RA1 and RA2 encode C2H2 Zn-finger proteins and LATERAL ORGAN BOUNDARY (LOB) transcription factors, respectively (Vollbrecht et al. 2005; Bortiri et al. 2006). RA3 acts downstream of RA1 and RA2 and encodes a trehalose 6 phosphate phosphatase (Satoh-Nagasawa et al. 2006; for review, see Bommert and Whipple 2018). RA1 is only found in the Andropogonae (Bommert and Whipple 2018; Chongloi et al. 2019). RA1 physically and genetically interacts with a transcriptional corepressor RAMOSA1 ENHANCER LOCUS2 (REL) of the TOPLESS family, which—as its Arabidopsis counterparts—interacts with many transcription factors (Liu et al. 2019b). rel single mutants, although pleiotropic, have shorter tassels with reduced branching (Liu et al. 2019b).
CONCLUDING REMARKS
Summary Points
Despite their divergent above-ground architectures, common themes underlie organogenesis, axillary meristem formation, activity, and fate in diverse plant species.
Organogenesis is auxin-mediated and results in initiation of (cryptic) bracts and associated axillary meristems during reproductive development. During vegetative development, axillary meristem initiation is delayed relative to later organ initiation and requires an auxin minimum as well as transcriptional cascades that eventually results in up-regulation of the STM pluripotency factor. Axillary meristems arising during vegetative development also frequently remain dormant, and their outgrowth is dependent on interactions between diverse hormonal and nutritional cues and a TCP transcription factor (Fig. 1).
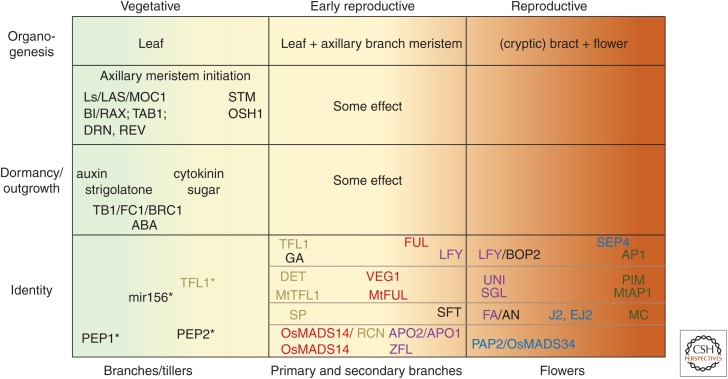
During reproductive development, meristem activity is strongly influenced by stimuli that promote flowering, with less floral promoting cues leading to vegetative fate in axillary meristems, intermediate floral cues to branching, and strong floral cues to determinacy (see also Prusinkiewicz et al. 2007). With a few exceptions, the functions of key regulators along this spectrum are conserved (Fig. 2). In tomato and rice, ALOG family transcription factors control vegetative fate and branching, respectively, whereas additional factors repress excessive branching of the spikelet pair meristem and the spikelet meristems found in maize and rice.
Figure 2.
Key regulators of plant inflorescence architecture and their function are conserved in diverse flowering plants. Regulators are grouped based on the developmental phase they act in (boxes) and from less (left) to more (right) determinate in each phase. Homologs in different species are color coded. *Vegetative axillary meristem identity was only discussed for Arabidopsis and Arabis alpina (see text for additional details).
Future Challenges and Link to Crop Yield
Several of the factors described here and their regulatory elements (critical cis regulatory elements for transcriptional regulation and microRNA binding sites) were identified as major QTLs for traits linked to yield, highlighting their agronomical importance (Simons et al. 2007; Jiao et al. 2010; Bai et al. 2017; Fujishiro et al. 2018). Genome-wide association studies (GWAS) have also implicated several of the regulators discussed above as important contributors to yield (Olsen and Wendel 2013; Guo et al. 2018). A recent study used GWAS to show that the inflorescence architecture largely determines the efficiency of grain production and yield in wheat (Guo et al. 2018). In addition, genes that contribute to the architecture of the inflorescence, such as AGL79/VEG1, CUC3, FZP, and TAW1, are potentially linked to yield enhancement (Crowell et al. 2016; Li et al. 2016; He et al. 2017; Zheng et al. 2017; Fujishiro et al. 2018; Gao et al. 2018; Ta et al. 2018). Studies in other crops, such as maize (Wu et al. 2016), chickpea (Bajaj et al. 2016), and sorghum (Kong et al. 2014), have also dissected the genetic link between inflorescence architecture and yield performance.
Targeted changes in cis regulatory elements, by CRISPR/Cas9-mediated cis element editing, for example, are emerging as a promising approach. cis regulatory mutations that reduce expression of flowering repressor SP5G in tomato led to loss of day length sensitivity and—combined with mutations in sp—to a higher harvest index in field conditions (Rodríguez-Leal et al. 2017; Soyk et al. 2017). Regulatory changes may also avoid negative epistasis by lowering rather than blocking gene function of multiple loci to “improve” inflorescence architecture. This was shown by stacking of weak alleles that minimize detrimental effects on other traits like fruit development and enhanced yield of a cherry tomato variety in greenhouse conditions (Soyk et al. 2017). That CRISPR/Cas9 approaches that target domestication genes can lead to rapid increase in fruit size and number in wild relatives was also recently shown in tomato (Zsögön et al. 2018). In addition, microRNA-resistant alleles of the OsSPL14/IPA1 transcription factor in rice enhance panicle branching and increase yield in field conditions (Jiao et al. 2010; Miura et al. 2010) as well as disease resistance (Liu et al. 2019a).
All of these approaches will profit from increased understanding of the key players and their regulatory interactions. For example, identification of direct targets of transcriptional regulators by chromatin immunoprecipitation and defining the underlying cis motifs will allow precise “rewiring” of specific regulatory interactions by CRISPR/Cas9 mutagenesis of noncoding DNA. As many regulators have multiple roles in diverse aspects of inflorescence development, more precise insight into their role in a given process/trait of interest, for example by cell-type-specific loss-of-function mutants, is important. These types of analyses are facilitated by development of new technologies, such as targeting of dCas9 fusions with strong transcriptional repressor domains (Lowder et al. 2015; Piatek et al. 2015).
Similarly, because several of the regulators discussed here act cell nonautonomously (for review, see Bommert and Whipple 2018) or are mobile (TFL1, FT, LFY, STM) (Lucas et al. 1995; Sessions et al. 2000; Conti and Bradley 2007; Jaeger and Wigge 2007), it will be critical to identify their precise sites (cell type) of activity, for example by confocal imaging of the localization of the relevant proteins. Recent advances in gene targeting in plants might be especially helpful to achieve this goal (Miki et al. 2018; Huang and Puchta 2019). The real payoff, for mechanistic insight into plant architecture and for crop improvement alike, is that these approaches—when applied to plant species with diverse inflorescence architectures—will reveal commonalities and differences in the regulatory components and their interactions and hence the “wiring plan” that gives rise to distinct inflorescence architectures.
ACKNOWLEDGMENTS
We apologize to those whose work was not cited because of space constraints. Work in the Wagner laboratory related to this topic is funded by NSF-IOS 1557529.
Footnotes
Editor: Pamela C. Ronald
Additional Perspectives on Engineering Plants for Agriculture available at www.cshperspectives.org
REFERENCES
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472. 10.1105/tpc.106.048934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejandra Mandel M, Gustafson-Brown C, Savidge B, Yanofsky MF. 1992. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273 10.1038/360273a0 [DOI] [PubMed] [Google Scholar]
- Bai X, Huang Y, Mao D, Wen M, Zhang L, Xing Y. 2016. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci Rep 6: 19022 10.1038/srep19022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Huang Y, Hu Y, Liu H, Zhang B, Smaczniak C, Hu G, Han Z, Xing Y. 2017. Duplication of an upstream silencer of FZP increases grain yield in rice. Nat Plants 3: 885–893. 10.1038/s41477-017-0042-4 [DOI] [PubMed] [Google Scholar]
- Bajaj D, Upadhyaya HD, Das S, Kumar V, Gowda CLL, Sharma S, Tyagi AK, Parida SK. 2016. Identification of candidate genes for dissecting complex branch number trait in chickpea. Plant Sci 245: 61–70. 10.1016/j.plantsci.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Balkunde R, Kitagawa M, Xu XM, Wang J, Jackson D. 2017. SHOOT MERISTEMLESS trafficking controls axillary meristem formation, meristem size and organ boundaries in Arabidopsis. Plant J 90: 435–446. 10.1111/tpj.13504 [DOI] [PubMed] [Google Scholar]
- Balla J, Kalousek P, Reinöhl V, Friml J, Procházka S. 2011. Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J 65: 571–577. 10.1111/j.1365-313X.2010.04443.x [DOI] [PubMed] [Google Scholar]
- Bangerth KF. 2009. Floral induction in mature, perennial angiosperm fruit trees: Similarities and discrepancies with annual/biennial plants and the involvement of plant hormones. Sci Hortic (Amsterdam) 122: 153–163. 10.1016/j.scienta.2009.06.014 [DOI] [Google Scholar]
- Barazesh S, McSteen P. 2008. Hormonal control of grass inflorescence development. Trends Plant Sci 13: 656–662. 10.1016/j.tplants.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Barbier F, Péron T, Lecerf M, Perez-Garcia MD, Barrière Q, Rolčík J, Boutet-Mercey S, Citerne S, Lemoine R, Porcheron B, et al. 2015a. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J Exp Bot 66: 2569–2582. 10.1093/jxb/erv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier FF, Lunn JE, Beveridge CA. 2015b. Ready, steady, go! A sugar hit starts the race to shoot branching. Curr Opin Plant Biol 25: 39–45. 10.1016/j.pbi.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Barbier FF, Dun EA, Kerr SC, Chabikwa TG, Beveridge CA. 2019. An update on the signals controlling shoot branching. Trends Plant Sci 24: 220–236. 10.1016/j.tplants.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueño F. 2007. Floral initiation and inflorescence architecture: A comparative view. Ann Bot 100: 659–676. 10.1093/aob/mcm146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Ali L, Gohari G, Millán T, Madueño F. 2015. Genetic control of inflorescence architecture in legumes. Front Plant Sci 6: 543 10.3389/fpls.2015.00543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, Leyser O. 2006. Something on the side: Axillary meristems and plant development. Plant Mol Biol 60: 843–854. 10.1007/s11103-005-2763-4 [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. 2006. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563. 10.1016/j.cub.2006.01.058 [DOI] [PubMed] [Google Scholar]
- Bennett T, Hines G, van Rongen M, Waldie T, Sawchuk MG, Scarpella E, Ljung K, Leyser O. 2016. Connective auxin transport in the shoot facilitates communication between shoot apices. PLOS Biol 14: e1002446 10.1371/journal.pbio.1002446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A, Ferrándiz C, Hecht V, Dalmais M, Lund OS, Sussmilch FC, Taylor SA, Bendahmane A, Ellis THN, Beltrán JP, et al. 2012. VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat Commun 3: 797 10.1038/ncomms1801 [DOI] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC, Loren van Themaat EV, Nordström KJV, Wang R, Schneeberger K, Moerland PD, Coupland G. 2013. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340: 1094–1097. 10.1126/science.1234116 [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Soowal LN, Lee I, Weigel D. 1997. LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844. [DOI] [PubMed] [Google Scholar]
- Bomblies K, Doebley JF. 2006. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics 172: 519–531. 10.1534/genetics.105.048595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Wang RL, Ambrose BA, Schmidt RJ, Meeley RB, Doebley J. 2003. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395. 10.1242/dev.00457 [DOI] [PubMed] [Google Scholar]
- Bommert P, Whipple C. 2018. Grass inflorescence architecture and meristem determinacy. Semin Cell Dev Biol 79: 37–47. 10.1016/j.semcdb.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Hake S. 2007. Flowering and determinacy in maize. J Exp Bot 58: 909–916. 10.1093/jxb/erm015 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S. 2006. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585. 10.1105/tpc.105.039032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Thomas MR. 2002. Association of dwarfism and floral induction with a grape “green revolution” mutation. Nature 416: 847 10.1038/416847a [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot JP, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. 2012. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol 158: 225–238. 10.1104/pp.111.182725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. 2009. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol 150: 482–493. 10.1104/pp.108.134783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Gui R, Mason MG, Beveridge CA. 2015. Strigolactone inhibition of branching independent of polar auxin transport. Plant Physiol 168: 1820–1829. 10.1104/pp.15.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RH, Bregitzer P. 2011. A Ds insertional mutant of a barley miR172 gene results in indeterminate spikelet development. Crop Sci 51: 1664–1672. 10.2135/cropsci2010.09.0532 [DOI] [Google Scholar]
- Burian A, Barbier de Reuille P, Kuhlemeier C. 2016. Patterns of stem cell divisions contribute to plant longevity. Curr Biol 26: 1385–1394. 10.1016/j.cub.2016.03.067 [DOI] [PubMed] [Google Scholar]
- Castel R, Kusters E, Koes R. 2010. Inflorescence development in petunia: Through the maze of botanical terminology. J Exp Bot 61: 2235–2246. 10.1093/jxb/erq061 [DOI] [PubMed] [Google Scholar]
- Chabikwa TG, Brewer PB, Beveridge CA. 2019. Initial bud outgrowth occurs independent of auxin flow from out of buds. Plant Physiol 179: 55–65. 10.1104/pp.18.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Tan QKG, Hill TA, Irish VF. 2008. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135: 1235–1245. 10.1242/dev.015842 [DOI] [PubMed] [Google Scholar]
- Chahtane H, Zhang B, Norberg M, LeMasson M, Thévenon E, Bakó L, Benlloch R, Holmlund M, Parcy F, Nilsson O, et al. 2018. LEAFY activity is post-transcriptionally regulated by BLADE ON PETIOLE2 and CULLIN3 in Arabidopsis. New Phytol 220: 579–592. 10.1111/nph.15329 [DOI] [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV. 2008. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol 146: 97–107. 10.1104/pp.107.107441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-M, Ertl JR, Leisner SM, Chang CC. 1985. Localization of cytokinin biosynthetic sites in pea plants and carrot roots. Plant Physiol 78: 510–513. 10.1104/pp.78.3.510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Li G, Tang Y, Wen J. 2018. Dissection of genetic regulation of compound inflorescence development in Medicago truncatula. Development 145: dev158766 10.1242/dev.158766 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P. 2014. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26: 1134–1150. 10.1105/tpc.114.122903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chongloi GL, Prakash S, Vijayraghavan U. 2019. Rice shoot and floral meristem development: An overview of developmental regulators of meristem maintenance and organ identity. J Exp Bot 70: 1719–1736. 10.1093/jxb/erz046 [DOI] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. 2002. The control of spikelet meristem identity by the branched Silkless1 gene in maize. Science 298: 1238–1241. 10.1126/science.1076920 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S. 2007. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet 39: 1517 10.1038/ng.2007.20 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S. 2008. Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019. 10.1242/dev.024273 [DOI] [PubMed] [Google Scholar]
- Chuck GS, Brown PJ, Meeley R, Hake S. 2014. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc Natl Acad Sci 111: 18775–18780. 10.1073/pnas.1407401112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Zhu Y, Wu MF, Simonini S, Kuhn A, Armenta-Medina A, Jin R, Østergaard L, Gillmor CS, Wagner D. 2019. Auxin response factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat Commun 10: 886 10.1038/s41467-019-08861-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Bradley D. 2007. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19: 767–778. 10.1105/tpc.106.049767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O. 2010. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913. 10.1242/dev.051987 [DOI] [PubMed] [Google Scholar]
- Crowell S, Korniliev P, Falcão A, Ismail A, Gregorio G, Mezey J, McCouch S. 2016. Genome-wide association and high-resolution phenotyping link Oryza sativa panicle traits to numerous trait-specific QTL clusters. Nat Commun 7: 10527 10.1038/ncomms10527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Meng X, Ananiev EV. 2010. Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol 153: 238–251. 10.1104/pp.110.154211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Lin H, Chuck G, Faris JD, Dubcovsky J. 2017. microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144: 1966–1975. 10.1242/dev.146399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denay G, Chahtane H, Tichtinsky G, Parcy F. 2017. A flower is born: An update on Arabidopsis floral meristem formation. Curr Opin Plant Biol 35: 15–22. 10.1016/j.pbi.2016.09.003 [DOI] [PubMed] [Google Scholar]
- Derbyshire P, Byrne ME. 2013. MORE SPIKELETS1 is required for spikelet fate in the inflorescence of brachypodium. Plant Physiol 161: 1291–1302. 10.1104/pp.112.212340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierck R, De Keyser E, De Riek J, Dhooghe E, Van Huylenbroeck J, Prinsen E, Van Der Straeten D. 2016. Change in auxin and cytokinin levels coincides with altered expression of branching genes during axillary bud outgrowth in chrysanthemum. PLoS ONE 11: e0161732 10.1371/journal.pone.0161732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. 1997. The evolution of apical dominance in maize. Nature 386: 485–488. 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. 2011. Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211 10.1038/nrm3088 [DOI] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. 2012. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol 158: 487–498. 10.1104/pp.111.186783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippe GM, Dale JE. 1973. Effects of shading the first leaf of barley plants on growth and carbon nutrition of the stem apex. Ann Bot 37: 45–56. 10.1093/oxfordjournals.aob.a084680 [DOI] [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. 2000. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734. [DOI] [PubMed] [Google Scholar]
- Fichtner F, Barbier FF, Feil R, Watanabe M, Annunziata MG, Chabikwa TG, Höfgen R, Stitt M, Beveridge CA, Lunn JE. 2017. Trehalose 6-phosphate is involved in triggering axillary bud outgrowth in garden pea (Pisum sativum L.). Plant J 92: 611–623. 10.1111/tpj.13705 [DOI] [PubMed] [Google Scholar]
- Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Rameau C. 2003. DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15: 2742–2754. 10.1105/tpc.015701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishiro Y, Agata A, Ota S, Ishihara R, Takeda Y, Kunishima T, Ikeda M, Kyozuka J, Hobo T, Kitano H. 2018. Comprehensive panicle phenotyping reveals that qSrn7/FZP influences higher-order branching. Sci Rep 8: 12511 10.1038/s41598-018-30395-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Enrico Pè M, Schmidt RJ. 2004. The role of barren stalk1 in the architecture of maize. Nature 432: 630 10.1038/nature03148 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. 2008. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci 105: 15196–15201. 10.1073/pnas.0805596105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Liu Q, Moss BL, Malcomber S, Li W, Gaines C, Federici S, Roshkovan J, Meeley R, Nemhauser JL, et al. 2015. Auxin signaling modules regulate maize inflorescence architecture. Proc Natl Acad Sci 112: 13372–13377. 10.1073/pnas.1516473112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Horrer D, Küttner F, Schmid M. 2012. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082. 10.1242/dev.080879 [DOI] [PubMed] [Google Scholar]
- Gao R, Wang Y, Gruber MY, Hannoufa A. 2018. miR156/SPL10 modulates lateral root development, branching and leaf morphology in Arabidopsis by silencing AGAMOUS-LIKE 79. Front Plant Sci 8: 2226 10.3389/fpls.2017.02226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauley A, Boden SA. 2019. Genetic pathways controlling inflorescence architecture and development in wheat and barley. J Integr Plant Biol 61: 296–309. 10.1111/jipb.12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E, Pajoro A, Franco-Zorrilla JM, Tarancón C, Immink RGH, Cubas P. 2017. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc Natl Acad Sci 114: E245–E254. 10.1073/pnas.1613199114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslin K, Zheng B, Serrano-Mislata A, Rae L, Ryan PT, Kwaśniewska K, Thomson B, Ó’Maoiléidigh DS, Madueño F, Wellmer F, et al. 2017. Transcription factor interplay between LEAFY and APETALA1/CAULIFLOWER during floral initiation. Plant Physiol 174: 1097–1109. 10.1104/pp.17.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schäfer E, Müller D, Herrero R, Schmitz G, Theres K. 2003. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187. 10.1101/gad.260703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grennan AK. 2007. The role of trehalose biosynthesis in plants. Plant Physiol 144: 3–5. 10.1104/pp.104.900223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Zhang J, Wang X, Han X, Wei B, Wang J, Li B, Yu H, Huang Q, Gu H, et al. 2015. The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 27: 3112–3127. 10.1105/tpc.15.00829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zhao Y, Röder MS, Reif JC, Ganal MW, Chen D, Schnurbusch T. 2018. Manipulation and prediction of spike morphology traits for the improvement of grain yield in wheat. Sci Rep 8: 14435 10.1038/s41598-018-31977-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder LD, Prusinkiewicz P. 2012. The interplay between inflorescence development and function as the crucible of architectural diversity. Ann Bot 112: 1477–1493. 10.1093/aob/mcs252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. 2009. Interactions between auxin and strigolactone in shoot branching control. Plant Physiol 151: 400–412. 10.1104/pp.109.137646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wu D, Wei D, Fu Y, Cui Y, Dong H, Tan C, Qian W. 2017. GWAS, QTL mapping and gene expression analyses in Brassica napus reveal genetic control of branching morphogenesis. Sci Rep 7: 15971 10.1038/s41598-017-15976-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz E. 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911. 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. 1997. Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124: 3845–3853. [DOI] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Ki T, Furutani M, Aida M, Tasaka M. 2006. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957. 10.1105/tpc.106.045716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N. 1997. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol 7: 581–587. 10.1016/S0960-9822(06)00257-0 [DOI] [PubMed] [Google Scholar]
- Huang TK, Puchta H. 2019. CRISPR/Cas-mediated gene targeting in plants: Finally a turn for the better for homologous recombination. Plant Cell Rep 38: 443 10.1007/s00299-019-02379-0 [DOI] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S. 2002. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162: 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Vincent C, Tilmes V, Bergonzi S, Kiefer C, Richter R, Martinez-Gallegos R, Severing E, Coupland G. 2019. A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363: 409–412. 10.1126/science.aau8197 [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Yasuno N, Oikawa T, Iida S, Nagato Y, Maekawa M, Kyozuka J. 2009. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol 150: 736–747. 10.1104/pp.109.136739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh JI, Nagato Y. 2012. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J 69: 168–180. 10.1111/j.1365-313X.2011.04781.x [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. 2007. FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054. 10.1016/j.cub.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. 2013. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401 10.1038/nature12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42: 541 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- Jill Harrison C. 2017. Development and genetics in the evolution of land plant body plans. Philos Trans R Soc B Lond Biol Sci 372: 20150490 10.1098/rstb.2015.0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MR, Hirota A, Kwiatkowska D, Tasaka M, Aida M. 2009. A role for Arabidopsis PUCHI in floral meristem identity and bract suppression. Plant Cell 21: 1360–1372. 10.1105/tpc.109.067025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, et al. 2010. Orchestration of floral initiation by APETALA1. Science 328: 85–89. 10.1126/science.1185244 [DOI] [PubMed] [Google Scholar]
- Kebrom TH. 2017. A growing stem inhibits bud outgrowth—The overlooked theory of apical dominance. Front Plant Sci 8: 1874 10.3389/fpls.2017.01874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Mullet JE. 2015. Photosynthetic leaf area modulates tiller bud outgrowth in sorghum. Plant Cell Environ 38: 1471–1478. 10.1111/pce.12500 [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Mullet JE. 2016. Transcriptome profiling of tiller buds provides new insights into PhyB regulation of tillering and indeterminate growth in sorghum. Plant Physiol 170: 2232–2250. 10.1104/pp.16.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA. 2006. Phytochrome B represses teosinte branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol 140: 1109–1117. 10.1104/pp.105.074856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Brutnell TP, Finlayson SA. 2010. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant Cell Environ 33: 48–58. 10.1111/j.1365-3040.2009.02050.x [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Chandler PM, Swain SM, King RW, Richards RA, Spielmeyer W. 2012. Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol 160: 308–318. 10.1104/pp.112.197954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P. 2006. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611. 10.1105/tpc.105.038588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly G, David-Schwartz R, Sade N, Moshelion M, Levi A, Alchanatis V, Granot D. 2012. The pitfalls of transgenic selection and new roles of AtHXK1: A high level of AtHXK1 expression uncouples hexokinase1-dependent sugar signaling from exogenous sugar. Plant Physiol 159: 47–51. 10.1104/pp.112.196105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J. 2003. LAX and SPA: Major regulators of shoot branching in rice. Proc Natl Acad Sci 100: 11765–11770. 10.1073/pnas.1932414100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Guo H, Goff VH, Lee TH, Kim C, Paterson AH. 2014. Genetic analysis of vegetative branching in sorghum. Theor Appl Genet 127: 2387–2403. 10.1007/s00122-014-2384-x [DOI] [PubMed] [Google Scholar]
- Koppolu R, Schnurbusch T. 2019. Developmental pathways for shaping spike inflorescence architecture in barley and wheat. J Integr Plant Biol 61: 278–295. 10.1111/jipb.12771 [DOI] [PubMed] [Google Scholar]
- Koumoto T, Shimada H, Kusano H, She KC, Iwamoto M, Takano M. 2013. Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1,6-bisphosphatase. Plant Biotechnol 30: 47–56. 10.5511/plantbiotechnology.12.1210a [DOI] [Google Scholar]
- Kuhlemeier C. 2017. Phyllotaxis. Curr Biol 27: R882–R887. 10.1016/j.cub.2017.05.069 [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K. 1998. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc Natl Acad Sci 95: 1979–1982. 10.1073/pnas.95.5.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Kiss A, Sassi M, Chauvet A, Das P, Cortizo M, Laufs P, Takeda S, Aida M, Traas J, et al. 2015. Mechanical stress contributes to the expression of the STM homeobox gene in Arabidopsis shoot meristems. eLife 4: e07811 10.7554/eLife.07811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Obeng-Hinneh E, Albani MC. 2018. Extended vernalization regulates inflorescence fate in Arabis alpina by stably silencing PERPETUAL FLOWERING1. Plant Physiol 176: 2819–2833. 10.1104/pp.17.01754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Giesguth M, Zhou Y, Albani MC, Coupland G, Bergonzi S, Pecinka A, Nawaz K. 2019. PERPETUAL FLOWERING2 coordinates the vernalization response and perennial flowering in Arabis alpina. J Exp Bot 70: 949–961. 10.1093/jxb/ery423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, An G. 2012. Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69: 445–461. 10.1111/j.1365-313X.2011.04804.x [DOI] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D. 1997. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol 7: 95–104. 10.1016/S0960-9822(06)00053-4 [DOI] [PubMed] [Google Scholar]
- Lee YS, Lee DY, Cho LH, An G. 2014. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 7: 31 10.1186/s12284-014-0031-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F. 2003. Control of tillering in rice. Nature 422: 618 10.1038/nature01518 [DOI] [PubMed] [Google Scholar]
- Li F, Chen B, Xu K, Gao G, Yan G, Qiao J, Li J, Li H, Li L, Xiao X, et al. 2016. A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Sci 242: 169–177. 10.1016/j.plantsci.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Li G, Yao G, Guo H, Huang H, Zhang H, Yu J, Li J, Li J, Sun X, Xu Y, et al. 2018. Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3–OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J Exp Bot 69: 4723–4737. 10.1093/jxb/ery256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Ayre BG, Eshed Y. 2014. Florigen and anti-florigen—A systemic mechanism for coordinating growth and termination in flowering plants. Front Plant Sci 5: 465 10.3389/fpls.2014.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligerot Y, de Saint Germain A, Waldie T, Troadec C, Citerne S, Kadakia N, Pillot J-P, Prigge M, Aubert G, Bendahmane A, et al. 2017. The pea branching RMS2 gene encodes the PsAFB4/5 auxin receptor and is involved in an auxin-strigolactone regulation loop. PLoS Genet 13: e1007089 10.1371/journal.pgen.1007089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D. 2008. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol 6: e288 10.1371/journal.pbio.0060288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Teo ZW, Bi Y, Song S, Xi W, Yang X, Yin Z, Yu H. 2013. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev Cell 24: 612–622. 10.1016/j.devcel.2013.02.013 [DOI] [PubMed] [Google Scholar]
- Liu L, Farrona S, Klemme S, Turck FK. 2014. Post-fertilization expression of FLOWERING LOCUS T suppresses reproductive reversion. Front Plant Sci 5: 164 10.3389/fpls.2014.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Shi Z, Zhang X, Wang M, Zhang L, Zheng K, Liu J, Hu X, Di C, Qian Q, et al. 2019a. Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nat Plants 5: 389–400. 10.1038/s41477-019-0383-2 [DOI] [PubMed] [Google Scholar]
- Liu X, Galli M, Camehl I, Gallavotti A. 2019b. RAMOSA1 ENHANCER LOCUS2-mediated transcriptional repression regulates vegetative and reproductive architecture. Plant Physiol 179: 348–363. 10.1104/pp.18.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, Paul JW, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y. 2015. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169: 971–985. 10.1104/pp.15.00636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Shao G, Xiong J, Jiao Y, Wang J, Liu G, Meng X, Liang Y, Xiong G, Wang Y, et al. 2015. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J Genet Genom 42: 71–78. 10.1016/j.jgg.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. 1995. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983. 10.1126/science.270.5244.1980 [DOI] [PubMed] [Google Scholar]
- MacAlister CA, Park SJ, Jiang K, Marcel F, Bendahmane A, Izkovich Y, Eshed Y, Lippman ZB. 2012. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat Genet 44: 1393 10.1038/ng.2465 [DOI] [PubMed] [Google Scholar]
- Martín-Fontecha ES, Tarancón C, Cubas P. 2018. To grow or not to grow, a power-saving program induced in dormant buds. Curr Opin Plant Biol 41: 102–109. 10.1016/j.pbi.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Grandío EG, Serra F, Marcel F, Rodríguez-Buey ML, Schmitz G, Theres K, Bendahmane A, Dopazo H, Cubas P. 2011. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J 67: 701–714. 10.1111/j.1365-313X.2011.04629.x [DOI] [PubMed] [Google Scholar]
- Mason MG, Ross JJ, Babst BA, Wienclaw BN, Beveridge CA. 2014. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc Natl Acad Sci 111: 6092–6097. 10.1073/pnas.1322045111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes MS, Best NB, Robil JM, Malcomber S, Gallavotti A, McSteen P. 2018. Auxin EvoDevo: Conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Mol Plant 12: 298–320. 10.1016/j.molp.2018.12.012 [DOI] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. 2008. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet 40: 1489 10.1038/ng.253 [DOI] [PubMed] [Google Scholar]
- Miki D, Zhang W, Zeng W, Feng Z, Zhu JK. 2018. CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 9: 1967 10.1038/s41467-018-04416-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, Umehara M, Luo L, Kobayashi K, Hanada A, Ueno K, Asami T, Yamaguchi S. 2010. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol 51: 1127–1135. 10.1093/pcp/pcq083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. 2010. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42: 545 10.1038/ng.592 [DOI] [PubMed] [Google Scholar]
- Moreno-Pachon NM, Mutimawurugo MC, Heynen E, Sergeeva L, Benders A, Blilou I, Hilhorst HWM, Immink RGH. 2018. Role of Tulipa gesneriana TEOSINTE BRANCHED1 (TgTB1) in the control of axillary bud outgrowth in bulbs. Plant Reprod 31: 145–157. 10.1007/s00497-017-0316-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, Mairhofer S, Burr-Hersey J, Goh T, von Wangenheim D, Atkinson B, Sturrock CJ, Lynch JP, et al. 2017. Shaping 3D root system architecture. Curr Biol 27: R919–R930. 10.1016/j.cub.2017.06.043 [DOI] [PubMed] [Google Scholar]
- Moyroud E, Kusters E, Monniaux M, Koes R, Parcy F. 2010. LEAFY blossoms. Trends Plant Sci 15: 346–352. 10.1016/j.tplants.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Müller D, Schmitz G, Theres K. 2006. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597. 10.1105/tpc.105.038745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Waldie T, Miyawaki K, To JPC, Melnyk CW, Kieber JJ, Kakimoto T, Leyser O. 2015. Cytokinin is required for escape but not release from auxin mediated apical dominance. Plant J 82: 874–886. 10.1111/tpj.12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Xing R, Clarenz O, Pokorny L, Goodrich J, Schubert D. 2014. Polycomb-Group proteins and FLOWERING LOCUS T maintain commitment to flowering in Arabidopsis thaliana. Plant Cell 26: 2457–2471. 10.1105/tpc.114.123323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J. 2002. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29: 743–750. 10.1046/j.1365-313X.2002.01255.x [DOI] [PubMed] [Google Scholar]
- Niwa M, Endo M, Araki T. 2013. Florigen is involved in axillary bud development at multiple stages in Arabidopsis. Plant Signal Behav 8: e27167 10.4161/psb.27167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Wendel JF. 2013. A bountiful harvest: Genomic insights into crop domestication phenotypes. Annu Rev Plant Biol 64: 47–70. 10.1146/annurev-arplant-050312-120048 [DOI] [PubMed] [Google Scholar]
- Otori K, Tamoi M, Tanabe N, Shigeoka S. 2017. Enhancements in sucrose biosynthesis capacity affect shoot branching in Arabidopsis. Biosci Biotechnol Biochem 81: 1470–1477. 10.1080/09168451.2017.1321954 [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. 1998. A genetic framework for floral patterning. Nature 395: 561 10.1038/26903 [DOI] [PubMed] [Google Scholar]
- Park SJ, Jiang K, Tal L, Yichie Y, Gar O, Zamir D, Eshed Y, Lippman ZB. 2014. Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat Genet 46: 1337 10.1038/ng.3131 [DOI] [PubMed] [Google Scholar]
- Pastore JJ, Limpuangthip A, Yamaguchi N, Wu M-F, Sang Y, Han S-K, Malaspina L, Chavdaroff N, Yamaguchi A, Wagner D. 2011. LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development 138: 3189–3198. 10.1242/dev.063073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Périlleux C, Bouché F, Randoux M, Orman-Ligeza B. 2019. Turning meristems into fortresses. Trends Plant Sci 24: 431–442. 10.1016/j.tplants.2019.02.004 [DOI] [PubMed] [Google Scholar]
- Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM. 2015. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13: 578–589. 10.1111/pbi.12284 [DOI] [PubMed] [Google Scholar]
- Pires ND, Dolan L. 2012. Morphological evolution in land plants: New designs with old genes. Philos Trans R Soc Lond B Biol Sci 367: 508–518. 10.1098/rstb.2011.0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G. 2012. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209. 10.1242/dev.077164 [DOI] [PubMed] [Google Scholar]
- Poursarebani N, Seidensticker T, Koppolu R, Trautewig C, Gawroński P, Bini F, Govind G, Rutten T, Sakuma S, Tagiri A, et al. 2015. The genetic basis of composite spike form in barley and “miracle-wheat.” Genetics 201: 155–165. 10.1534/genetics.115.176628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. 2007. Evolution and development of inflorescence architectures. Science 316: 1452–1456. 10.1126/science.1140429 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O. 2009. Control of bud activation by an auxin transport switch. Proc Natl Acad Sci 106: 17431–17436. 10.1073/pnas.0906696106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K. 2008. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55: 65–76. 10.1111/j.1365-313X.2008.03483.x [DOI] [PubMed] [Google Scholar]
- Rao NN, Prasad K, Kumar PR, Vijayraghavan U. 2008. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci 105: 3646–3651. 10.1073/pnas.0709059105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ. 1998. A common mechanism controls the life cycle and architecture of plants. Development 125: 1609–1615. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Holalu SV, Casal JJ, Finlayson SA. 2013. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol 163: 1047–1058. 10.1104/pp.113.221895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Kuhlemeier C. 2002. Plant architecture. EMBO Rep 3: 846 10.1093/embo-reports/kvf177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. 2017. Engineering quantitative trait variation for crop improvement by genome editing. Cell 171: 470–480.e478. 10.1016/j.cell.2017.08.030 [DOI] [PubMed] [Google Scholar]
- Sachs T. 1981. The control of the patterned differentiation of vascular tissues. In Advances in botanical research (ed. Woolhouse HW), pp. 151–262. Academic, London. [Google Scholar]
- Sachs T, Thimann KV. 1967. The role of auxins and cytokinins in the release of buds from dominance. Am J Bot 54: 136–144. 10.1002/j.1537-2197.1967.tb06901.x [DOI] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. 2006. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230. 10.1038/nature04725 [DOI] [PubMed] [Google Scholar]
- Scheres B, van der Putten WH. 2017. The plant perceptron connects environment to development. Nature 543: 337 10.1038/nature22010 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. 2003. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci 100: 6849–6854. 10.1073/pnas.1132018100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K. 2002. The tomato blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci 99: 1064–1069. 10.1073/pnas.022516199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale M, Bennett T, Leyser O. 2017. BRC1 expression regulates bud activation potential but is not necessary or sufficient for bud growth inhibition in Arabidopsis. Development 144: 1661–1673. 10.1242/dev.145649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Mislata A, Fernández-Nohales P, Doménech MJ, Hanzawa Y, Bradley D, Madueño F. 2016. Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity. Development 143: 3315–3327. 10.1242/dev.135269 [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D. 2000. Cell–cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289: 779–781. 10.1126/science.289.5480.779 [DOI] [PubMed] [Google Scholar]
- Shalit-Kaneh A, Eviatar-Ribak T, Horev G, Suss N, Aloni R, Eshed Y, Lifschitz E. 2018. The flowering hormone florigen accelerates secondary cell wall biogenesis to harmonize vascular maturation with reproductive development. bioRxiv 476028 10.1101/476028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Vernoux T. 2019. Patterning at the shoot apical meristem and phyllotaxis. Curr Topic Dev Biol 131: 81–107. 10.1016/bs.ctdb.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Shi B, Zhang C, Tian C, Wang J, Wang Q, Xu T, Xu Y, Ohno C, Sablowski R, Heisler MG, et al. 2016. Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet 12: e1006168 10.1371/journal.pgen.1006168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S, Mori H. 2001. Control of outgrowth and dormancy in axillary buds. Plant Physiol 127: 1405–1413. 10.1104/pp.010841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O. 2013. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474 10.1371/journal.pbio.1001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC. 2007. Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol 143: 697–706. 10.1104/pp.106.087957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Wang G, Hu Y, Liu H, Bai X, Qin R, Xing Y. 2018. OsMFT1 increases spikelets per panicle and delays heading date in rice by suppressing Ehd1, FZP and SEPALLATA-like genes. J Exp Bot 69: 4283–4293. 10.1093/jxb/ery232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC. 2015. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27: 3143–3159. 10.1105/tpc.15.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez JM, Lippman ZB. 2016. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet 49: 162 10.1038/ng.3733 [DOI] [PubMed] [Google Scholar]
- Soyk S, Lemmon ZH, Oved M, Fisher J, Liberatore KL, Park SJ, Goren A, Jiang K, Ramos A, van der Knaap E, et al. 2017. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 169: 1142–1155.e1112. 10.1016/j.cell.2017.04.032 [DOI] [PubMed] [Google Scholar]
- Stafstrom JP, Ripley BD, Devitt ML, Drake B. 1998. Dormancy-associated gene expression in pea axillary buds. Planta 205: 547–552. 10.1007/s004250050354 [DOI] [PubMed] [Google Scholar]
- Sussmilch FC, Berbel A, Hecht V, Vander Schoor JK, Ferrándiz C, Madueño F, Weller JL. 2015. Pea VEGETATIVE2 is an FD homolog that is essential for flowering and compound inflorescence development. Plant Cell 27: 1046–1060. 10.1105/tpc.115.136150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta KN, Khong NG, Ha TL, Nguyen DT, Mai DC, Hoang TG, Phung TPN, Bourrie I, Courtois B, Tran TTH, et al. 2018. A genome-wide association study using a Vietnamese landrace panel of rice (Oryza sativa) reveals new QTLs controlling panicle morphological traits. BMC Plant Biol 18: 282 10.1186/s12870-018-1504-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H, et al. 2011. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23: 3276–3287. 10.1105/tpc.111.088765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H. 2006. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45: 1028–1036. 10.1111/j.1365-313X.2006.02656.x [DOI] [PubMed] [Google Scholar]
- Tanaka W, Ohmori Y, Ushijima T, Matsusaka H, Matsushita T, Kumamaru T, Kawano S, Hirano HY. 2015. Axillary meristem formation in rice requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 27: 1173–1184. 10.1105/tpc.15.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Hirano HY. 2017. Three TOB1-related YABBY genes are required to maintain proper function of the spikelet and branch meristems in rice. New Phytol 215: 825–839. 10.1111/nph.14617 [DOI] [PubMed] [Google Scholar]
- Tarancón C, González-Grandío E, Oliveros JC, Nicolas M, Cubas P. 2017. A conserved carbon starvation response underlies bud dormancy in woody and herbaceous species. Front Plant Sci 8: 788 10.3389/fpls.2017.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E. 2005. Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138: 757–766. 10.1104/pp.104.057984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S, Hofer J, Murfet I. 2001. Stamina pistilloida, the pea ortholog of Fim and UFO, is required for normal development of flowers, inflorescences, and leaves. Plant Cell 13: 31–46. 10.1105/tpc.13.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Hofer JMI, Murfet IC, Sollinger JD, Singer SR, Knox MR, Ellis THN. 2002. PROLIFERATING INFLORESCENCE MERISTEM, a MADS-Box gene that regulates floral meristem identity in pea. Plant Physiol 129: 1150–1159. 10.1104/pp.001677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann T, Muhr M. 2015. Shaping plant architecture. Front Plant Sci 6: 233 10.3389/fpls.2015.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenreira T, Lange MJP, Lange T, Bres C, Labadie M, Monfort A, Hernould M, Rothan C, Denoyes B. 2017. A specific gibberellin 20-oxidase dictates the flowering-runnering decision in diploid strawberry. Plant Cell 29: 2168–2182. 10.1105/tpc.16.00949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C, et al. 2014. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol Syst Biol 10: 755 10.15252/msb.20145470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truskina J, Vernoux T. 2018. The growth of a stable stationary structure: Coordinating cell behavior and patterning at the shoot apical meristem. Curr Opin Plant Biol 41: 83–88. 10.1016/j.pbi.2017.09.011 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Besnard F, Traas J. 2010. Auxin at the shoot apical meristem. Cold Spring Harb Perspect Biol 2: a001487 10.1101/cshperspect.a001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler IV ES, Martienssen R. 2005. Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126. 10.1038/nature03892 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. 2002. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 296: 343–346. 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- Wagner D. 2017. Key developmental transitions during flower morphogenesis and their regulation. Curr Opin Genet Dev 45: 44–50. 10.1016/j.gde.2017.01.018 [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RWM, Meyerowitz EM. 1999. Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584. 10.1126/science.285.5427.582 [DOI] [PubMed] [Google Scholar]
- Waldie T, Leyser O. 2018. Cytokinin targets auxin transport to promote shoot branching. Plant Physiol 177: 803–818. 10.1104/pp.17.01691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiao Y. 2018. Axillary meristem initiation—A way to branch out. Curr Opin Plant Biol 41: 61–66. 10.1016/j.pbi.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J. 2008. Molecular basis of plant architecture. Annu Rev Plant Biol 59: 253–279. 10.1146/annurev.arplant.59.032607.092902 [DOI] [PubMed] [Google Scholar]
- Wang Q, Sajja U, Rosloski S, Humphrey T, Kim MC, Bomblies K, Weigel D, Grbic V. 2007. HUA2 caused natural variation in shoot morphology of A. thaliana. Curr Biol 17: 1513–1519. 10.1016/j.cub.2007.07.059 [DOI] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC. 2009. PEP1 regulates perennial flowering in Arabis alpina. Nature 459: 423 10.1038/nature07988 [DOI] [PubMed] [Google Scholar]
- Wang R, Albani MC, Vincent C, Bergonzi S, Luan M, Bai Y, Kiefer C, Castillo R, Coupland G. 2011. Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23: 1307–1321. 10.1105/tpc.111.083451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz EM, Jiao Y. 2014. The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067. 10.1105/tpc.114.123083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sun S, Jin J, Fu D, Yang X, Weng X, Xu C, Li X, Xiao J, Zhang Q. 2015a. Coordinated regulation of vegetative and reproductive branching in rice. Proc Natl Acad Sci 112: 15504–15509. 10.1073/pnas.1521949112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J. 2015b. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27: 3128–3142. 10.1105/tpc.15.00605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu H, Xiong G, Lu Z, Jiao Y, Meng X, Liu G, Chen X, Wang Y, Li J. 2017a. Tissue-specific ubiquitination by IPA1 interacting PROTEIN1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 29: 697–707. 10.1105/tpc.16.00879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wu K, Qian Q, Liu Q, Li Q, Pan Y, Ye Y, Liu X, Wang J, Zhang J, et al. 2017b. Non-canonical regulation of SPL transcription factors by a human OTUB1-like deubiquitinase defines a new plant type rice associated with higher grain yield. Cell Res 27: 1142 10.1038/cr.2017.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Smith SM, Li J. 2018. Genetic regulation of shoot architecture. Annu Rev Plant Biol 69: 437–468. 10.1146/annurev-arplant-042817-040422 [DOI] [PubMed] [Google Scholar]
- Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67: 539–574. 10.1146/annurev-arplant-043015-112122 [DOI] [PubMed] [Google Scholar]
- Whipple CJ. 2017. Grass inflorescence architecture and evolution: The origin of novel signaling centers. New Phytol 216: 367–372. 10.1111/nph.14538 [DOI] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA, et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc Natl Acad Sci 111: E4859 10.1073/pnas.1323926111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M, Rae GM, Wu R-M, Walton EF, Xue B, Hellens RP, Uversky VN. 2013. Actinidia DRM1—An intrinsically disordered protein whose mRNA expression is inversely correlated with spring budbreak in kiwifruit. PLoS ONE 8: e57354 10.1371/journal.pone.0057354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M-F, Yamaguchi N, Xiao J, Bargmann B, Estelle M, Sang Y, Wagner D. 2015. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4: e09269 10.7554/eLife.09269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Shi Y, Song Y, Zhang D, Li C, Buckler ES, Li Y, Zhang Z, Wang T. 2016. Joint-linkage mapping and GWAS reveal extensive genetic loci that regulate male inflorescence size in maize. Plant Biotechnol J 14: 1551–1562. 10.1111/pbi.12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Shi X, Lin X, Liu Y, Chong K, Theißen G, Meng Z. 2017. The ABCs of flower development: Mutational analysis of AP1/FUL-like genes in rice provides evidence for a homeotic (A)-function in grasses. Plant J 89: 310–324. 10.1111/tpj.13386 [DOI] [PubMed] [Google Scholar]
- Xu J, Ding C, Ding Y, Tang S, Zha M, Luo B, Wang S. 2015. A proteomic approach to analyze differential regulation of proteins during bud outgrowth under apical dominance based on the auxin transport canalization model in rice (Oryza sativa L.). J Plant Growth Regul 34: 122–136. 10.1007/s00344-014-9450-0 [DOI] [Google Scholar]
- Xu C, Park SJ, Van Eck J, Lippman ZB. 2016. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev 30: 2048–2061. 10.1101/gad.288415.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. 2009. The microRNA-regulated SBP-Box transcription factor SPL3 Is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17: 268–278. 10.1016/j.devcel.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D. 2013. A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell 24: 271–282. 10.1016/j.devcel.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D. 2014. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344: 638–641. 10.1126/science.1250498 [DOI] [PubMed] [Google Scholar]
- Yang XC, Hwa CM. 2008. Genetic modification of plant architecture and variety improvement in rice. Heredity 101: 396 10.1038/hdy.2008.90 [DOI] [PubMed] [Google Scholar]
- Yang F, Wang Q, Schmitz G, Müller D, Theres K. 2012. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J 71: 61–70. 10.1111/j.1365-313X.2012.04970.x [DOI] [PubMed] [Google Scholar]
- Yang J, Yuan Z, Meng Q, Huang G, Périn C, Bureau C, Meunier AC, Ingouff M, Bennett MJ, Liang W, et al. 2017. Dynamic regulation of auxin response during rice development revealed by newly established hormone biosensor markers. Front Plant Sci 8: 256 10.3389/fpls.2017.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Nicolas M, Zhang J, Yu H, Guo D, Yuan R, Zhang T, Yang J, Cubas P, Qin G. 2018. The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet 14: e1007296 10.1371/journal.pgen.1007296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L. 2012. Genome-wide mapping of transcription factor binding reveals developmental process integration and a fresh look at evolutionary dynamics. Am J Bot 99: 277–290. 10.3732/ajb.1100333 [DOI] [PubMed] [Google Scholar]
- Yao C, Finlayson SA. 2015. Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol 169: 611–626. 10.1104/pp.15.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Skirpan A, Wardell B, Matthes MS, Best NB, McCubbin T, Durbak A, Smith T, Malcomber S, McSteen P. 2019. The barren stalk2 gene is required for axillary meristem development in maize. Mol Plant 12: 374–389. 10.1016/j.molp.2018.12.024 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen R, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, et al. 2013. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci 110: 767–772. 10.1073/pnas.1216151110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste-Lisbona FJ, Quinet M, Fernández-Lozano A, Pineda B, Moreno V, Angosto T, Lozano R. 2016. Characterization of vegetative inflorescence (mc-vin) mutant provides new insight into the role of MACROCALYX in regulating inflorescence development of tomato. Sci Rep 6: 18796 10.1038/srep18796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yuan Z. 2014. Molecular control of grass inflorescence development. Annu Rev Plant Biol 65: 553–578. 10.1146/annurev-arplant-050213-040104 [DOI] [PubMed] [Google Scholar]
- Zhang B, Liu X, Xu W, Chang J, Li A, Mao X, Zhang X, Jing R. 2015. Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat. Sci Rep 5: 12211 10.1038/srep12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang J, Wenkel S, Chandler JW, Werr W, Jiao Y. 2018. Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145: dev158352 10.1242/dev.158352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Peng C, Liu H, Tang M, Yang H, Li X, Liu J, Sun X, Wang X, Xu J, et al. 2017. Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed (Brassica napus L.). Front Plant Sci 8: 1246 10.3389/fpls.2017.01246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, et al. 2013. D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406 10.1038/nature12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Li G, Chai M, Fu C, Cheng X, Wen J, Tang Y, Wang ZY. 2014. STM/BP-like KNOXI is uncoupled from ARP in the regulation of compound leaf development in Medicago truncatula. Plant Cell 26: 1464–1479. 10.1105/tpc.114.123885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP. 2018. De novo domestication of wild tomato using genome editing. Nat Biotechnol 36: 1211 10.1038/nbt.4272 [DOI] [PubMed] [Google Scholar]