Abstract
Cells invest in an extensive network of factors to maintain protein homeostasis (proteostasis) and prevent the accumulation of potentially toxic protein aggregates. This proteostasis network (PN) comprises the machineries for the biogenesis, folding, conformational maintenance, and degradation of proteins with molecular chaperones as central coordinators. Here, we review recent progress in understanding the modular architecture of the PN in mammalian cells and how it is modified during cell differentiation. We discuss the capacity and limitations of the PN in maintaining proteome integrity in the face of proteotoxic stresses, such as aggregate formation in neurodegenerative diseases. Finally, we outline various pharmacological interventions to ameliorate proteostasis imbalance.
Proteins are the most versatile macromolecules and responsible for almost all cellular functions. An average human cell expresses ∼10,000–13,000 different protein species (Bekker-Jensen et al. 2017; Kulak et al. 2017) with copy numbers varying over several orders of magnitude, from a few molecules to tens of thousands. To maintain protein homeostasis, or proteostasis (Balchin et al. 2016), cells must ensure that these proteins fold and assemble correctly and exist in the right cellular locale at appropriate abundance.
Most proteins must adopt a unique, thermodynamically stable three-dimensional structure, the functionally active state, which is determined by the amino acid sequence (Anfinsen 1973). Proteins must fold to this native state at biologically relevant timescales, a process that is increasingly well understood, at least for small model proteins that fold rapidly without detectable intermediate states (Dobson and Karplus 1999; Hartl and Hayer-Hartl 2009). However, proteins with complex domain folds and multidomain proteins, which make up the major part of the proteome, frequently populate folding intermediates that expose hydrophobic amino acid residues. These proteins are at risk of misfolding and aggregation within the highly crowded environment of the cell (Ellis and Minton 2006). Once folded, proteins must maintain their correct three-dimensional shape, which is complicated by the fact that functionality often requires considerable conformational flexibility. As a result, the folded states of many proteins are only marginally stable (or metastable) in the cellular milieu. Moreover, a considerable fraction of proteins (∼30% in human cells) are either completely devoid of ordered structure (intrinsically disordered proteins) or contain substantial unstructured regions (Dunker et al. 2008). These proteins typically only adopt folded structures dependent on binding to partner proteins, which emphasizes the importance of correct concentration and localization. Even natively structured proteins are constantly conformationally challenged in the face of a variety of endogenous and exogenous stresses. Proteins that fail to adopt or maintain their folded structure, as well as proteins that have served their function, must be removed by degradation to prevent deleterious consequences.
To maintain protein homeostasis and proper proteome balance, cells use interconnected modules of factors collectively referred to as the proteostasis network (PN). This network is comprised of the cellular machineries for the biogenesis and degradation of proteins with molecular chaperones as central coordinators mediating protein folding and conformational maintenance. Although early studies in yeast have enabled a basic understanding of the eukaryotic PN (Albanèse et al. 2006; Finley et al. 2012), the PN in human cells is predicted to be far more complex (Klaips et al. 2018), owing to the diverse requirements of cell- and tissue-specific proteomes (Wilhelm et al. 2014; Uhlen et al. 2015).
Loss of proteostasis is linked to aging and several medical conditions associated with the formation of toxic protein aggregates, prominently including numerous neurodegenerative diseases like Alzheimer's and Huntington's disease and other pathologies affecting many different cell types. The familial forms of these diseases have an underlying genetic cause in which mutations increase the likelihood of specific disease proteins to misfold and aggregate. Classic examples are forms of amyotrophic lateral sclerosis associated with mutations in superoxide dismutase and a group of disorders, including Huntington's disease, that are caused by expanded polyglutamine tracts in otherwise unrelated proteins. Importantly, most of these pathologies manifest in an age-dependent manner (age-of-onset diseases), consistent with a decline in proteostasis capacity during aging being a critical risk factor (Taylor and Dillin 2011; Labbadia and Morimoto 2015; Higuchi-Sanabria et al. 2018). Moreover, despite the presence of the respective mutation in all somatic tissues in most cases, specific cell types and tissues are exclusively or preferentially affected (Fu et al. 2018), suggesting an underlying heterogeneity between cell types in proteostasis capacity and the ability to respond to proteotoxic stress (Sala et al. 2017).
In this review, we discuss recent progress in our understanding of the modular organization of the PN and its capacity to maintain proteome integrity in the face of stress conditions. Using the example of stem cells, we examine PN plasticity and rewiring during cell differentiation. We further discuss how the burden of toxic protein aggregates in disease may cause proteostasis collapse. Finally, we outline various pharmacological interventions to ameliorate proteostasis imbalance as an attractive avenue toward therapeutic strategies for some proteinopathies.
ORGANIZATION OF THE PROTEOSTASIS NETWORK
Protein-folding accuracy is fundamental to all cells. However, maintaining fidelity of protein synthesis and folding is confounded by the fact that the underlying genetic and biochemical systems are inherently error prone, which leads to the constant production of a certain amount of misfolded proteins. This problem is further aggravated by genetic variation (single-nucleotide polymorphisms) and the effects of environmental stress. The resulting aberrant proteins must be detected and degraded by the PN to prevent them from engaging in unwanted interactions and toxic protein aggregation. Chronic aggregate production can overwhelm the cellular capacity to maintain proteostasis and respond properly to proteotoxic stress (Olzscha et al. 2011; Roth et al. 2014). To maintain a balanced and healthy proteome, cells have evolved an integrated network of protein quality-control factors (the PN) tending to the synthesis, folding, maintenance, and timely turnover of proteins. For any given protein, the PN comprises all cellular factors required for folding of the newly synthesized protein and conformational maintenance at the correct location and appropriate concentration, and finally for controlled degradation once the protein has fulfilled its function or has undergone irreversible misfolding.
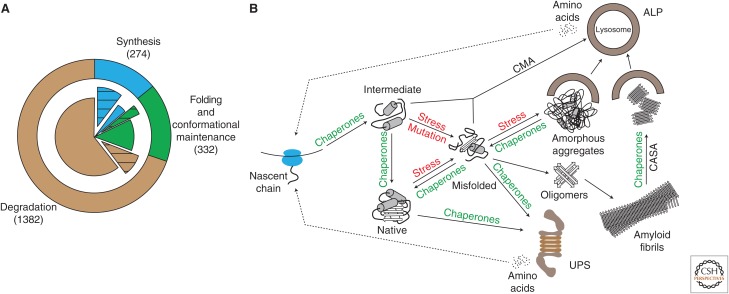
In its simplest form, the PN can be described by three major modules that govern each of these processes (Fig. 1A,B): (1) protein synthesis, (2) folding and conformational maintenance by molecular chaperones and cochaperones, and (3) protein degradation by the ubiquitin–proteasome system (UPS) and the autophagy–lysosomal pathway (ALP). These three modular branches coordinately tune the health of the proteome with molecular chaperones being central to their interconnectivity. Besides PN components that are required generally, such as ribosomes, proteasomes, and lysosomes, there is likely a requirement for “specific” factors tailored to certain proteins (or sets of proteins) with distinct properties. Additionally, proteostasis within organelles such as the endoplasmic reticulum (ER) and mitochondria requires specialized factors and chaperones for protein trafficking and folding (Hetz et al. 2015; Moehle et al. 2018). Because of this inherent complexity, the composition of the PN remains poorly defined. However, advances in understanding the functional annotation of components belonging to each individual module allows an estimation of a total size of ∼2000 components in human cells (Fig. 1A; Klaips et al. 2018).
Figure 1.
Architecture of the proteostasis network (PN). (A) The three modules of the PN (outer ring): synthesis (blue, 274 factors), folding and conformational maintenance (green, 332 factors), and degradation (brown, 1382 factors). The inner pie chart shows the partitioning of each module into essential (hatched sectors, synthesis [203/274], folding [78/332], and degradation [178/1382]) and nonessential components (filled sectors). (B) Central role of molecular chaperones in the PN. The nascent polypeptide chain emerging from the ribosomal tunnel is prevented from misfolding and aggregation by chaperones, including components that bind to the ribosome (trigger factor in bacteria, nascent chain–associated complex [NAC] and ribosome-associated complex [RAC] in eukarya). A second tier of chaperones (Hsp70, Hsp90, chaperonins) does not interact with the ribosome directly and mediates co- or posttranslational folding. Proteins that misfold because of mutations or under conditions of stress are selectively degraded either by the ubiquitin–proteasome system (UPS) or chaperone-mediated autophagy (CMA) and chaperone-assisted selective autophagy (CASA). Misfolded proteins may aggregate to soluble oligomers, amorphous aggregates, or amyloid fibrils when basal chaperone and degradation capacity is exceeded. Aggregates may be sequestered into insoluble inclusions, which may be dissociated into fragments by specialized chaperones (Hsp104 in yeast, Hsp70/Hsp40/Hsp110 in metazoans) for subsequent clearance by the autophagy–lysosomal pathway (ALP).
PROTEIN SYNTHESIS
Cytosolic ribosomes and associated factors produce the bulk of the cellular proteome at an average rate of five to six amino acids per second (Ingolia et al. 2011), generating more than a billion protein molecules per human cell (Milo 2013) with an average size of 560 amino acids (Wolff et al. 2014; Balchin et al. 2016). However, this process is error-prone with a misincorporation rate of 1 in 104 amino acids (Zaher and Green 2009). As a result, roughly 1 in 20 protein molecules contain a sequence error, potentially causing misfolding or reduced stability. Defective protein products can also arise as a result of posttranscriptional splicing errors or production of defective messenger RNA (mRNA). For example, erroneous mRNA molecules that lack stop codons cause ribosome stalling during translation. In these cases, the defective mRNAs are removed by cellular RNA surveillance pathways (Doma and Parker 2007; Isken and Maquat 2007), and the translated nascent protein chain is degraded by the ribosome-associated quality-control (RQC) pathway (Brandman and Hegde 2016; Joazeiro 2017), which senses ribosome-stalled polypeptides and engages UPS components to degrade them. Failure of the RQC machinery leads to protein aggregation and proteotoxic stress (Choe et al. 2016; Yonashiro et al. 2016; Izawa et al. 2017) and age-dependent neurodegeneration in a mouse model (Chu et al. 2009). Transfer RNA (tRNA) availability may limit translation rates and result in protein aggregation, perhaps by increasing the likelihood of misreading or frameshifts, or increasing the risk that nascent chains occupy nonproductive folding intermediates (Nedialkova and Leidel 2015). Notably, the rate of protein synthesis is also dynamically regulated in response to a variety of stimuli, both globally and in a protein-specific manner. During conditions of proteotoxic stress, a programmed decrease in protein synthesis (Spriggs et al. 2010) is initiated (Figs. 1B and 2) to alleviate the load of newly synthesized proteins that must be folded by the PN. During conformational stress in the ER or activation of the integrated stress response, this is accomplished by phosphorylation of the translation initiation factor eIF2α (Pakos-Zebrucka et al. 2016; Karagöz et al. 2019).
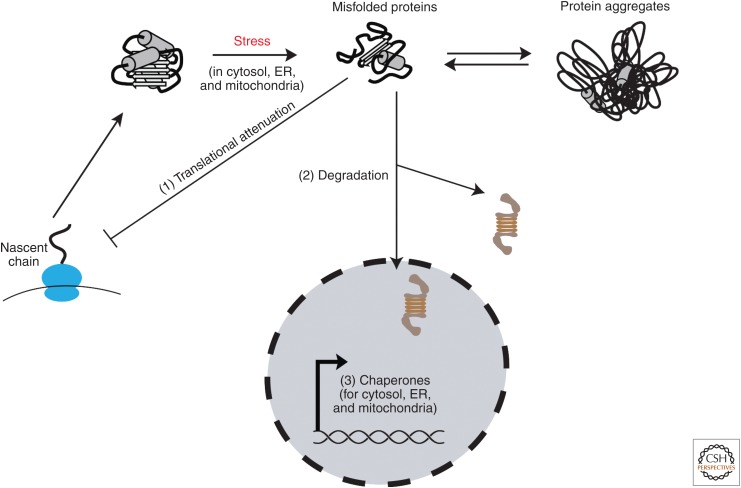
Figure 2.
Proteostasis capacity and stress responses. During proteotoxic stress in different cellular compartments, proteostasis capacity is adjusted to meet increased cellular requirements. Global translational attenuation (1) reduces the burden on the folding machineries to free chaperone capacities for assistance in clearance of misfolded and aggregated species (2). Concomitantly, transcriptional stress response pathways are activated (3) to increase chaperone pools available for folding and degradation. ER, Endoplasmic reticulum.
FOLDING AND CONFORMATIONAL MAINTENANCE
As shown recently, some small proteins can undergo partial or complete folding within the exit tunnel of the ribosome (Holtkamp et al. 2015; Nilsson et al. 2015). However, for the majority of larger proteins, folding is thought to begin once a chain segment sufficient to specify a cooperatively folding domain has emerged from the ribosome, followed by completion of folding after chain release (Balchin et al. 2016). Multidomain proteins may fold their domains cotranslationally in a sequential manner, thereby reducing the problem of folding a large protein to the folding of smaller modules (Kim et al. 2013).
Both co- and posttranslational folding is assisted by molecular chaperones (Fig. 1B), which prevent or reverse misfolding events and favor productive folding pathways. Molecular chaperones are defined as factors that assist in the folding (and often assembly) of another protein without being part of its final structure (Hartl 1996).
The human genome encodes ∼332 chaperones and cochaperones (referred to as the “chaperome”) (Fig. 1A; Brehme et al. 2014). Chaperones are broadly grouped by molecular weight and many are classified as heat shock proteins (Hsps) because their induction depends on exposure of cells to elevated temperature and other forms of proteome stress. The main classes of chaperones (88 proteins) include the ATP-dependent Hsp70s, Hsp90s, Hsp60s (also called chaperonins), and Hsp100s, and the ATP-independent small HSPs (sHsps). They are assisted by regulatory cochaperones (244 proteins) that further impart substrate specificity and selectivity. The major groups of cochaperones are the Hsp40s (49 proteins) as regulators of the Hsp70s, and the tetratricopeptide repeat proteins (TPR) (114) as regulators of the Hsp90 system (Table 1).
Table 1.
Chaperone classification in human cells
| Chaperone class | Number of genes |
|---|---|
| HSP40 | 49 |
| HSP60 | 15 |
| HSP70 | 27 |
| HSP90 | 46 |
| HSP100 | 14 |
| sHSP | 10 |
| TPR | 114 |
| Others (prefoldin, calnexin, oxidoreductases, prohibitin) | 57 |
Data adapted from Brehme et al. (2014).
HSP, Heat shock protein; sHSP, small heat shock protein; TPR, tetratricopeptide repeat protein.
Chaperones serve as the main conductors of proteostasis by directly influencing the “foldedness” of the proteome. They typically do so by binding to hydrophobic polypeptide segments that are exposed by nonnative proteins but are buried in the native state, thus, preventing aggregation and maintaining the polypeptide in a folding-competent state. The ATP-dependent chaperones (Hsp70, Hsp90, chaperonins) promote folding through ATP-regulated cycles of binding and release during which the client protein reaches its final structure, sometimes through an accelerated folding pathway (Hartl et al. 2011; Balchin et al. 2016). In some cases, proteins are altogether unable to fold without the aid of chaperones, defining the limit of the Anfinsen dogma. One such case is the cytoskeletal protein actin (Balchin et al. 2018).
Chaperones act in the folding of newly synthesized polypeptides either in association with the ribosome (e.g., the mammalian ribosome-associated complex [RAC] and specialized Hsp70s [HSP70L1]; Preissler and Deuerling 2012; Kramer et al. 2018) or downstream from the ribosome after polypeptide release, such as the classical Hsp70s, Hsp90s, and the TRiC/CCT chaperonin (Fig. 1B). TRiC (and the chaperonins in mitochondria, Hsp60, and bacteria, GroEL) are large ∼1 MDa double-ring complexes that transiently enclose a single molecule of unfolded substrate protein in a cage-like structure, allowing folding to occur unimpaired by aggregation (Balchin et al. 2016; Hayer-Hartl et al. 2016). The ATP-independent sHsps function as “holdases” in stabilizing proteins against aggregation or in shielding the surfaces of aggregates (Carra et al. 2017).
Molecular chaperones cooperate in cellular folding pathways as shown for the bacterial and eukaryotic cytosol and in other systems (Langer et al. 1992; Frydman et al. 1994; Frydman 2001). Indeed, different chaperones interact to form modules and functional supercomplexes (Rizzolo et al. 2017). Specific chaperone pathways for the diverse array of protein clients can be perceived as a sequential percolator. Proteins that fold rapidly after interaction with upstream chaperones, like the Hsp70s, would not require more specialized downstream chaperones, whereas proteins that fail to fold with the upstream chaperones will filter down to Hsp90 or the chaperonin to attain their folded functional state (Hartl 1996).
Quantitative analyses of client proteins for specific chaperones in simpler model systems like Escherichia coli (Kerner et al. 2005; Oh et al. 2011; Calloni et al. 2012) have revealed features of client flux through the bacterial chaperone network. Similar studies in Saccharomyces cerevisiae have yielded a systems view of the eukaryotic chaperone network (Zhao et al. 2005; Gong et al. 2009; Willmund et al. 2013). Chaperone-dependent proteins are frequently larger than average, containing multiple domains or domains with complex fold topologies that are typically characterized by many long-range interactions. Salient features of the eukaryotic chaperone network have emerged from systematic mapping of the interactions of chaperones and cochaperones with their clients. It has been shown for yeast cells that the “chaperome” can be divided into components that are mainly devoted to the folding of newly synthesized proteins and those that function in conformational maintenance (Albanèse et al. 2006). The former includes the ribosome-binding chaperones that are not stress inducible, whereas the latter include the classic stress-inducible chaperones.
In humans and other mammals, the chaperome has expanded to match the needs of a larger and more complex proteome (Powers and Balch 2013). Although the PN of human cells has not yet been comprehensively characterized, recent studies focused on the substrate interactome of Hsp90, a chaperone system thought to be specifically devoted to kinases and other signaling proteins. In an initial survey of ∼2000 candidates comprising kinases, transcription factors (TFs) and E3 ligases, ∼200 of 355 kinases and ∼120 of 426 ubiquitin E3 ligases analyzed were found to use Hsp90 for folding or conformational regulation, whereas TFs showed no such preference (50 Hsp90 substrates among 1100 TFs tested) (Taipale et al. 2012). The finding that E3 ligases are clients of the folding machinery further emphasizes the interconnectivity of the folding and degradation modules of the PN. A more comprehensive investigation (Taipale et al. 2014), including ∼70 chaperones, cochaperones, and protein quality-control factors, revealed a hierarchical organization of the chaperone network centered on the interconnected Hsp70 and Hsp90 chaperone systems. More recently, an integrated chaperone network that supports cell survival was observed in cancer cells to be essential for tumor maintenance. This complex called the “epichaperome” (Rodina et al. 2016) is formed by an enhanced physical interaction of the Hsp70 and Hsp90 machineries along with their cochaperones.
A key question regarding chaperone networks is how client flux is redirected when one chaperone system is overwhelmed. Similar to findings in bacterial cells in which mutual compensation among chaperone systems is observed (Deuerling et al. 1999; Teter et al. 1999; Calloni et al. 2012), inhibition of Hsp90 in human cells promoted client binding to Hsc70, the constitutively expressed Hsp70 (Taipale et al. 2014). Among the BAG family cochaperones, which function as nucleotide exchange factors of Hsp70, only BAG2 proved to have a client range similar to Hsc70, suggesting that it is a general cofactor involved in the folding pathway of Hsc70 substrate proteins. The interaction spectrum of Hsc70 also correlates with that of the E3 ligase CHIP (carboxy terminus of Hsp70-interacting protein), again pointing to a functional cooperation between chaperones and the UPS (Esser et al. 2004; Kundrat and Regan 2010). Although these studies have provided initial insights into how substrate proteins are distributed within the chaperome, it remains to be established how exactly client specificity and selectivity is achieved. Within the Hsp70 system, the Hsp40 cofactors (∼50 different proteins in human) play a central role in this process (Table 1; Kampinga and Craig 2010).
Although the majority of chaperone systems function to prevent protein aggregation, certain chaperones are specialized to act as “disaggregases” and actively dissolve protein aggregates (Fig. 1B; Mogk et al. 2018). A well-studied example is yeast Hsp104, which performs disaggregation in an ATP-driven manner (Parsell et al. 1994) in cooperation with Hsp70 and Hsp40 (Mogk et al. 2015). In metazoans, which lack Hsp104, disaggregation of stress-induced aggregates is enabled by Hsp70 in cooperation with Hsp110 (an Hsp70-like protein that functions as nucleotide exchange factor for Hsp70), Hsp40s, and sHsps (Nillegoda et al. 2015; Żwirowski et al. 2017).
PROTEIN DEGRADATION
The steady-state concentration of proteins is the result of a balance between protein synthesis and degradation. Cells are equipped with efficient protein degradation machineries that recycle proteins once they are no longer needed, and remove misfolded proteins to prevent the formation of potentially toxic aggregates. Eukaryotic cells use two main degradation machineries, the UPS and the ALP (Förster et al. 2013; Cohen-Kaplan et al. 2016; Dikic 2017; Varshavsky 2017). The UPS serves as the major pathway of protein degradation and is responsible for ∼80% of protein turnover (Zhao et al. 2015; Collins and Goldberg 2017). Central to the UPS is the 26S proteasome, a large multisubunit protease complex (Förster et al. 2013; Livneh et al. 2016). The 26S proteasome recognizes substrates after their covalent modification with the small protein ubiquitin. Ubiquitylation of target proteins is a highly regulated process that is catalyzed in an ATP-dependent manner by a cascade of ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3; Finley 2009; Zheng and Shabek 2017). Of the ∼2000 PN components, the UPS constitutes the largest group with ∼850 proteins (Klaips et al. 2018), ∼600 of which are E3 ligases (Fig. 1A; Li et al. 2008). The catalytic sites of the proteasome are buried within the central cavity of the 20S catalytic unit, and can only be accessed if the protein substrate is able to pass a narrow entrance into the catalytic core. To achieve this passage, most proteins need to be unfolded in an ATP-dependent reaction catalyzed by the 19S regulatory particle that caps the 20S core (Bhattacharyya et al. 2014). Recent cryoelectron microscopy analyses of the proteasome with substrate protein engaged provide mechanistic insight into this complex process (Beck et al. 2012; de la Peña et al. 2018; Dong et al. 2019). Owing to the requirement for substrate unfolding, the proteasome is unable to degrade aggregated proteins directly, and disaggregation by chaperone machinery is a prerequisite. More generally, chaperones function to stabilize misfolded proteins in a nonaggregated state competent for ubiquitin tagging and delivery to the proteasome (Esser et al. 2004). A chaperone-bound protein may be triaged for degradation by ubiquitylation when (re)folding is unsuccessful, resulting in a prolonged chaperone residence time. The E3 ligase CHIP interacts directly with Hsp70 to ubiquitylate chaperone-bound proteins. Up to a certain point, this modification may still be reversed by ubiquitin hydrolases (Mevissen and Komander 2017). How these opposing reactions are fine-tuned is central to understanding the mechanisms of protein quality control.
Despite the presence of the UPS, protein misfolding nevertheless leads to the formation of insoluble aggregates and larger inclusions, especially under stress conditions. In contrast to the UPS, autophagy coupled to lysosomal degradation has the capacity to directly remove such aggregates (Fig. 1B). In this process, aggregates or fragments thereof are engulfed by double-membrane structures (autophagosomes), which then fuse with the lysosome (Galluzzi et al. 2017). Although the core machinery of autophagy consists of only 17 proteins (Dikic 2017), the ALP system, including accessory factors, comprises a total of ∼500 components (García-Prat et al. 2016). Aggregated proteins accumulate in ubiquitin-positive deposits to which the autophagic machinery is recruited by chaperones in a process known as chaperone-assisted selective autophagy (CASA) (Carra et al. 2008; Gamerdinger et al. 2009; Arndt et al. 2010). Under basal conditions in mammalian cells, soluble proteins can also be degraded by a specific form of autophagy called chaperone-mediated autophagy (CMA), which involves substrate recognition and unfolding by Hsc70 and direct lysosomal translocation by binding to the lysosomal receptor LAMP2A, without the need for autophagosome formation (Fig. 1B; Kaushik and Cuervo 2018). Whereas both the UPS and ALP display a certain degree of specificity regarding their range and diversity of substrates, they are functionally interconnected and often compensate one another under conditions in which one pathway is saturated or malfunctional (Suraweera et al. 2012; Zhang et al. 2016; Dikic 2017)—another reflection of the network character of the PN.
STRESS RESPONSE PATHWAYS
Many natively folded proteins are metastable under normal cellular growth conditions (i.e., they reside at the limit of thermodynamic stability or contain substantial intrinsically disordered regions). Exposing cells to proteome stress, such as heat stress, then results in (partial) protein unfolding and possible aggregation. To deal with such stress situations, cells have evolved compartment-specific transcriptional stress response pathways (Fig. 2), prominently including the cytosolic heat stress response (HSR) (Morimoto 2011) and unfolded protein response pathways of the ER (UPRER) (Walter and Ron 2011; Karagöz et al. 2019) and mitochondria (UPRMito) (Melber and Haynes 2018; Naresh and Haynes 2019).
Activation of the HSR leads to the induction of molecular chaperones (Hsps) and other quality-control components, which act to prevent protein aggregation and mediate protein refolding or degradation via the UPS (Richter et al. 2010). The selective induction of molecular chaperones is accompanied by a global decrease in protein synthesis, particularly through attenuation of translational elongation (Liu et al. 2013; Shalgi et al. 2013), thus reducing the burden on the folding machinery and making proteostasis capacity available for conformational maintenance and clearance of misfolded proteins (Fig. 2). Induction of the HSR is not limited to acute abiotic stresses, such as heat or a change in pH, but is also mediated by other proteotoxic insults, such as the expression of certain misfolding mutant proteins at normal cellular growth temperatures (Geiler-Samerotte et al. 2011).
The eukaryotic HSR is mediated by heat shock factor 1 (HSF1) as the master TF (Vabulas et al. 2010; Anckar and Sistonen 2011). Under normal conditions, HSF1 exists as a monomer bound by chaperones (Zou et al. 1998; Neef et al. 2014; Zheng et al. 2016). Although HSF1 controls the expression of thousands of genes, its key function under conditions of proteotoxic stress is the tight regulation of a small set of chaperone genes (Mahat et al. 2016; Solís et al. 2016). The current model of the HSR posits that production of misfolded proteins under conditions of stress titrates away the chaperones bound to HSF1, which allows HSF1 to trimerize and become transcriptionally active. The resulting increase in chaperone levels provides a negative feedback loop that deactivates HSF1 once a sufficient amount of free chaperones is restored (Krakowiak et al. 2018). Additional mechanisms of attenuation of the HSR include HSF1 degradation and cycles of acetylation and deacetylation (Westerheide et al. 2009; Raychaudhuri et al. 2014).
Although it is unclear what fraction of the mammalian proteome denatures and misfolds during heat stress (Wallace et al. 2015), the level of ubiquitylated proteins rapidly increases in stressed cells (Fujimuro et al. 1997), suggesting that degradation of misfolded proteins via the UPS is critical in reestablishing proteome balance (Parag et al. 1987). Nascent polypeptides synthesized prior to heat stress, possibly still bound to ribosomes in unfolded states, are thought to represent the major vulnerable fraction of the proteome that is targeted for degradation (Medicherla and Goldberg 2008; Xu et al. 2016). A comprehensive genome-wide deletion screen for induction of the HSR in yeast (Brandman et al. 2012) under nonstress conditions revealed an enrichment of individual chaperones and members of the UPS, among other inducers, pointing to a strong cross talk between the two PN modules. Future genome-wide efforts to study proteostasis in mammalian cells will be facilitated by the advent of pooled CRISPR screens (Shalem et al. 2014) and haploid genetic screens (Brockmann et al. 2017). Although protein degradation via the UPS has been classically associated with the HSR, studies in the nematode Caenorhabditis elegans (Kumsta et al. 2017) and mammalian cells (Watanabe et al. 2017) showed a direct link between the HSR and autophagy, thus providing further evidence for a highly interconnected PN with modular flexibility.
THE PROTEOSTASIS NETWORK IN CELL DIFFERENTIATION
All existing tissues and cell types within the human body are a result of well-defined programs of stem cell division and differentiation. Although embryonic stem cells (ESCs) represent the reservoir of pluripotent cells and thus shape development early in life, postnatal maintenance of tissue homeostasis is conducted by specialized multipotent adult stem cells (e.g., neuronal stem cells and satellite stem cells in muscles) within their respective niches. Regardless of their origins, stem cells must continuously replenish their numbers via asymmetric cell division (segregating cell fate determinants into only one of the two daughter cells) (Knoblich 2008), while maintaining a damage-free proteome. At the same time, they have to remodel their proteomes as they differentiate into various fates, which emphasize the importance of proteostasis control and PN rewiring in development (Vilchez et al. 2014).
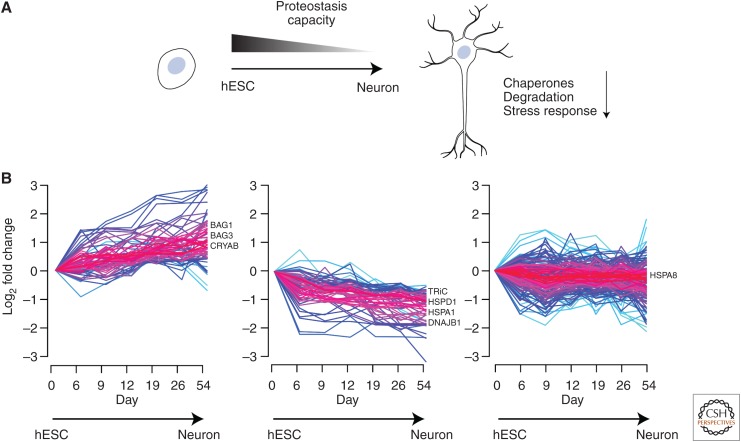
Stem cells are thought to have a higher proteostasis capacity than differentiated cells. Lower rates of protein synthesis in adult hematopoietic, neural, epidermal, and muscle stem cells (Signer et al. 2014; Llorens-Bobadilla et al. 2015; Blanco et al. 2016; Zismanov et al. 2016) presumably result in a reduced burden on the folding module of the PN. In contrast, ESCs tend to have higher translational rates (Ingolia et al. 2011; You et al. 2015). This difference can be attributed to the higher proliferative activity of ESCs (García-Prat et al. 2017). Important differences have also been noted in the composition of the stem cell chaperome (Fig. 3). ESCs have higher levels of a subset of chaperones, such as the chaperonin TRiC/CCT, which decreases as cells differentiate into neuronal progenitors and neurons (Noormohammadi et al. 2016). The high demand for TRiC/CCT may be related to the folding requirements of actin (Yam et al. 2008; Balchin et al. 2018), which is increasingly needed in proliferating ESCs for cytoskeletal synthesis and integrity. Mouse ESCs have also been found to have increased levels of stress-inducible cytosolic Hsp70 (HSPA1), a strong target of HSF1 (Saretzki et al. 2004, 2008). Hsp70 levels decrease as the cells differentiate, which may point to a dampened stress response. Indeed, neural differentiation from progenitor cells results in a marked reduction of HSR inducibility (Fig. 3A; Yang et al. 2008), suggesting that neurons may inherently be more vulnerable to proteome stress. It remains to be seen whether this is generally true for other differentiated tissues.
Figure 3.
Proteostasis network (PN) rewiring and decline in proteostasis capacity during cell differentiation. (A) Human embryonic stem cells (hESCs) have a higher proteostasis capacity as compared to differentiated cells, such as neurons. As hESCs differentiate to neurons, the chaperonins (cytosolic TRiC/CCT and mitochondrial Hsp60) decrease in abundance and there is a general decline in degradation capacity. Additionally, differentiated neurons are less efficient in mounting a stress response in comparison to their progenitors and stem cells. (B) Chaperome trajectories during neuronal differentiation of hESCs (332 proteins analyzed). The chaperome landscape changes in abundance (indicated as log2 fold change) as hESCs are differentiated into neurons. Chaperones can increase (left), decrease (middle), or remain unchanged in abundance (right) during this process. Prominent examples of each cluster are shown (BAG1/BAG3, regulators of Hsp70/Hsc70; CRYAB, α-crystallin B chain; TRiC, cytosolic chaperonin; HSPD1, mitochondrial Hsp60; HSPA1, Hsp70; DNAJB1, Hsp40 regulator of Hsp70/Hsc70; HSPA8, Hsc70). Data for time-dependent changes in chaperone abundance values were derived from Sequence Read Archive (SRA) accession No. GSE86985 (Yao et al. 2017). TPM (transcripts per million) values were directly used to calculate clustering of chaperone trajectories using the fuzzy c-means algorithm Mfuzz (Kumar and Futschik 2007) with “m” values set at 2 and a minimum membership cutoff of 0.8. The gradient from red to blue indicates high to low membership scores.
Stem cells are also equipped with enhanced protein degradation capacity, presumably allowing for the effective removal of misfolded proteins. Studies in ESCs showed higher levels of proteasome activity associated with an increase of the 19S regulatory subunit PSMD11, which is limiting for 26S proteasome assembly (Vilchez et al. 2012). Proteasome activity decreased during differentiation into several lineages.
An extreme case of PN rewiring and proteome remodeling is the recently described mechanism of terminal differentiation of reticulocytes into erythrocytes (red blood cells) within the hematopoietic lineage (Nguyen et al. 2017). Erythrocytes are unique cells whose proteome is composed to almost 98% of a single protein—hemoglobin. As reticulocytes differentiate into erythrocytes, most of their proteome is degraded and replaced by hemoglobin. Surprisingly, the determinant of such a global process is a single E2-E3 hybrid ligase, UBE2O, which encodes activities for multiple components of a quality-control pathway—substrate recognition (in lieu of a chaperone), ubiquitin transfer (E2), and ubiquitin conjugation (E3). UBE2O was also found to be responsible for the removal of ribosomal proteins via the proteasome, a hallmark of erythrocyte development. UBE2O also clears unassembled globin chains, thus, serving as an essential quality-control factor during this transition (Yanagitani et al. 2017). This unique process exemplifies extensive rewiring of PN modules.
AMELIORATING PROTEINOPATHIES
PN deregulation and decline during aging has been recognized as a major driver of many proteinopathies, especially affecting the more vulnerable populations of postmitotic neuronal cells (Dillin and Cohen 2011; Gidalevitz et al. 2011; Morimoto 2019). These age-onset pathologies include Alzheimer's, Parkinson's, and Huntington's diseases, as well as amyotrophic lateral sclerosis and other pathologies. All these disorders are characterized by the formation of soluble, oligomeric, and insoluble fibrillar aggregates that exert cytotoxic effects (Chiti and Dobson 2017; Iadanza et al. 2018). Of particular importance in understanding the progressive nature of these diseases is that the aggregates subvert the function of the PN at several levels, for example, by sequestering critical chaperone components and degradation machinery into aggregate inclusions (Rubinsztein 2006; Park et al. 2013; Yu et al. 2014; Guo et al. 2018; Thibaudeau et al. 2018), overburdening the UPS (Bence et al. 2001; Bennett et al. 2007; Hipp et al. 2012), and reducing the cellular capacity to respond to additional stresses (Labbadia et al. 2011; Olzscha et al. 2011; Roth et al. 2014). As a result, affected cells gradually lose the ability to cope with misfolding proteins, which in turn increases aggregate burden, setting in motion a vicious cycle that ends in proteostasis collapse and cell death (Hipp et al. 2014).
Because the PN is inherently modular and adaptable to stress, exploring ways to boost proteostasis could provide therapeutically viable options to decrease disease severity and slow progression (Balch et al. 2008; Powers et al. 2009; Hipp et al. 2014). Evidence has been provided that molecular chaperones, especially Hsp70 and Hsp40, can prevent formation of amyloid fibers (Muchowski et al. 2000) or disaggregate them (Gao et al. 2015), showing the potential of modulating the PN to combat proteinopathies in vivo. Accordingly, pharmacological induction or genetic overexpression of molecular chaperones has been shown to prevent aggregation in cellular (Sittler et al. 2001) and animal models (Labbadia et al. 2012; Kakkar et al. 2016; Nagy et al. 2016; Scior et al. 2018).
Although boosting chaperone activities has shown promise, tuning the other PN modules may also enhance proteostasis capacity. Small molecule-based modulation of the UPS (Lee et al. 2010; Leestemaker et al. 2017) and autophagy (Sarkar et al. 2009; Barmada et al. 2014; Kuo et al. 2015) augments clearance of protein aggregates and improves cellular health. Furthermore, stress responses to protein misfolding often culminate in translational attenuation to reduce downstream burden on the folding and degradation arm of the PN (Fig. 2). Reducing the level of newly synthesized proteins is therefore another promising approach to combat protein misfolding. Indeed, the pharmacologic prolongation of stress-induced translational attenuation has been shown to be beneficial in protein-misfolding disorders (Tsaytler et al. 2011; Das et al. 2015).
CONCLUDING REMARKS
Maintenance of proteostasis is central to cellular health, and its impairment is causally linked to neurodegenerative proteinopathies and numerous other pathologies. Advances over the past two decades have laid the foundation for our understanding of the architecture of the PN. The modular and adaptive nature of the PN enables cells to handle changing proteomic requirements during cellular development and endure adverse conditions brought about by various endogenous and environmental stresses. Although we currently have only a bird's-eye view of how the PN is organized and how many components may be involved in proteostasis maintenance, future studies will hopefully tease apart the complex and specific PN connectivities in tissues that are particularly vulnerable to proteostasis collapse. Progress resulting from such studies will be especially relevant in developing urgently needed treatments for the group of neurodegenerative proteinopathies. Protein misfolding and aggregation in these diseases result in a gain-of-function toxicity, which eventually overwhelms the capacity of the PN to maintain proteostasis. Enhancing proteostasis capacity through modular tuning of the PN offers viable strategies for therapeutic intervention.
ACKNOWLEDGMENTS
Work in the authors' laboratory is supported by the European Commission under Grant FP7 GA ERC-2012-SyG_318987-ToPAG, and the Deutsche Forschungsgemeinschaft (German Research Foundation) within the framework of the Munich Cluster for Systems Neurology.
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Albanèse V, Yam AY, Baughman J, Parnot C, Frydman J. 2006. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124: 75–88. 10.1016/j.cell.2005.11.039 [DOI] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. 2011. Regulation of HSF1 function in the heat stress response: Implications in aging and disease. Annu Rev Biochem 80: 1089–1115. 10.1146/annurev-biochem-060809-095203 [DOI] [PubMed] [Google Scholar]
- Anfinsen CB. 1973. Principles that govern the folding of protein chains. Science 181: 223–230. 10.1126/science.181.4096.223 [DOI] [PubMed] [Google Scholar]
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK, et al. 2010. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol 20: 143–148. 10.1016/j.cub.2009.11.022 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. 2008. Adapting proteostasis for disease intervention. Science 319: 916–919. 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. 2016. In vivo aspects of protein folding and quality control. Science 353: aac4354 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]
- Balchin D, Miličić G, Strauss M, Hayer-Hartl M, Hartl FU. 2018. Pathway of actin folding directed by the eukaryotic chaperonin TRiC. Cell 174: 1507–1521 e1516. 10.1016/j.cell.2018.07.006 [DOI] [PubMed] [Google Scholar]
- Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, et al. 2014. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol 10: 677–685. 10.1038/nchembio.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, et al. 2012. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci 109: 14870–14875. 10.1073/pnas.1213333109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen DB, Kelstrup CD, Batth TS, Larsen SC, Haldrup C, Bramsen JB, Sørensen KD, Høyer S, Ørntoft TF, Andersen CL, et al. 2017. An optimized shotgun strategy for the rapid generation of comprehensive human proteomes. Cell Syst 4: 587–599e584. 10.1016/j.cels.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292: 1552–1555. 10.1126/science.292.5521.1552 [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. 2007. Global changes to the ubiquitin system in Huntington's disease. Nature 448: 704–708. 10.1038/nature06022 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Yu H, Mim C, Matouschek A. 2014. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol 15: 122–133. 10.1038/nrm3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortés-Garrido R, Gkatza N, et al. 2016. Stem cell function and stress response are controlled by protein synthesis. Nature 534: 335–340. 10.1038/nature18282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Hegde RS. 2016. Ribosome-associated protein quality control. Nat Struct Mol Biol 23: 7–15. 10.1038/nsmb.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151: 1042–1054. 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehme M, Voisine C, Rolland T, Wachi S, Soper JH, Zhu Y, Orton K, Villella A, Garza D, Vidal M, et al. 2014. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep 9: 1135–1150. 10.1016/j.celrep.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann M, Blomen VA, Nieuwenhuis J, Stickel E, Raaben M, Bleijerveld OB, Altelaar AFM, Jae LT, Brummelkamp TR. 2017. Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature 546: 307–311. 10.1038/nature22376 [DOI] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang HC, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU. 2012. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep 1: 251–264. 10.1016/j.celrep.2011.12.007 [DOI] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, Lambert H, Landry J. 2008. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem 283: 1437–1444. 10.1074/jbc.M706304200 [DOI] [PubMed] [Google Scholar]
- Carra S, Alberti S, Arrigo PA, Benesch JL, Benjamin IJ, Boelens W, Bartelt-Kirbach B, Brundel B, Buchner J, Bukau B, et al. 2017. The growing world of small heat shock proteins: From structure to functions. Cell Stress Chaperones 22: 601–611. 10.1007/s12192-017-0787-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. 2017. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu Rev Biochem 86: 27–68. 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- Choe YJ, Park SH, Hassemer T, Körner R, Vincenz-Donnelly L, Hayer-Hartl M, Hartl FU. 2016. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531: 191–195. 10.1038/nature16973 [DOI] [PubMed] [Google Scholar]
- Chu J, Hong NA, Masuda CA, Jenkins BV, Nelms KA, Goodnow CC, Glynne RJ, Wu H, Masliah E, Joazeiro CA, et al. 2009. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci 106: 2097–2103. 10.1073/pnas.0812819106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Kaplan V, Livneh I, Avni N, Cohen-Rosenzweig C, Ciechanover A. 2016. The ubiquitin-proteasome system and autophagy: coordinated and independent activities. Int J Biochem Cell Biol 79: 403–418. 10.1016/j.biocel.2016.07.019 [DOI] [PubMed] [Google Scholar]
- Collins GA, Goldberg AL. 2017. The logic of the 26S proteasome. Cell 169: 792–806. 10.1016/j.cell.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das I, Krzyzosiak A, Schneider K, Wrabetz L, D'Antonio M, Barry N, Sigurdardottir A, Bertolotti A. 2015. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348: 239–242. 10.1126/science.aaa4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña AH, Goodall EA, Gates SN, Lander GC, Martin A. 2018. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362: eaav0725 10.1126/science.aav0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. 1999. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696. 10.1038/23301 [DOI] [PubMed] [Google Scholar]
- Dikic I. 2017. Proteasomal and autophagic degradation systems. Annu Rev Biochem 86: 193–224. 10.1146/annurev-biochem-061516-044908 [DOI] [PubMed] [Google Scholar]
- Dillin A, Cohen E. 2011. Ageing and protein aggregation-mediated disorders: From invertebrates to mammals. Philos Trans R Soc Lond B Biol Sci 366: 94–98. 10.1098/rstb.2010.0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM, Karplus M. 1999. The fundamentals of protein folding: Bringing together theory and experiment. Curr Opin Struct Biol 9: 92–101. 10.1016/S0959-440X(99)80012-8 [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. 2007. RNA quality control in eukaryotes. Cell 131: 660–668. 10.1016/j.cell.2007.10.041 [DOI] [PubMed] [Google Scholar]
- Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, Lu Y, Finley D, Mao Y. 2019. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565: 49–55. 10.1038/s41586-018-0736-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. 2008. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18: 756–764. 10.1016/j.sbi.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Ellis RJ, Minton AP. 2006. Protein aggregation in crowded environments. Biol Chem 387: 485–497. 10.1515/BC.2006.064 [DOI] [PubMed] [Google Scholar]
- Esser C, Alberti S, Höhfeld J. 2004. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim Biophys Acta 1695: 171–188. 10.1016/j.bbamcr.2004.09.020 [DOI] [PubMed] [Google Scholar]
- Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513. 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Ulrich HD, Sommer T, Kaiser P. 2012. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192: 319–360. 10.1534/genetics.112.140467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster F, Unverdorben P, Śledź P, Baumeister W. 2013. Unveiling the long-held secrets of the 26S proteasome. Structure 21: 1551–1562. 10.1016/j.str.2013.08.010 [DOI] [PubMed] [Google Scholar]
- Frydman J. 2001. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu Rev Biochem 70: 603–647. 10.1146/annurev.biochem.70.1.603 [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. 1994. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature 370: 111–117. 10.1038/370111a0 [DOI] [PubMed] [Google Scholar]
- Fu H, Hardy J, Duff KE. 2018. Selective vulnerability in neurodegenerative diseases. Nat Neurosci 21: 1350–1358. 10.1038/s41593-018-0221-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. 1997. Dynamics of ubiquitin conjugation during heat-shock response revealed by using a monoclonal antibody specific to multi-ubiquitin chains. Eur J Biochem 249: 427–433. 10.1111/j.1432-1033.1997.00427.x [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, et al. 2017. Molecular definitions of autophagy and related processes. EMBO J 36: 1811–1836. 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. 2009. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J 28: 889–901. 10.1038/emboj.2009.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B. 2015. Human Hsp70 disaggregase reverses Parkinson's-linked α-synuclein amyloid fibrils. Mol Cell 59: 781–793. 10.1016/j.molcel.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Prat L, Martínez-Vicente M, Perdiguero E, Ortet L, Rodríguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, et al. 2016. Autophagy maintains stemness by preventing senescence. Nature 529: 37–42. 10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- García-Prat L, Sousa-Victor P, Muñoz-Canoves P. 2017. Proteostatic and metabolic control of stemness. Cell Stem Cell 20: 593–608. 10.1016/j.stem.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci 108: 680–685. 10.1073/pnas.1017570108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Prahlad V, Morimoto RI. 2011. The stress of protein misfolding: From single cells to multicellular organisms. Cold Spring Harb Perspect Biol 3: e009704 10.1101/cshperspect.a009704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, Houry WA. 2009. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: Implications to protein folding pathways in the cell. Mol Syst Biol 5: 275 10.1038/msb.2009.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Lehmer C, Martínez-Sánchez A, Rudack T, Beck F, Hartmann H, Pérez-Berlanga M, Frottin F, Hipp MS, Hartl FU, et al. 2018. In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell 172: 696–705 e612. 10.1016/j.cell.2017.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. 1996. Molecular chaperones in cellular protein folding. Nature 381: 571–579. 10.1038/381571a0 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. 2009. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16: 574–581. 10.1038/nsmb.1591 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hayer-Hartl M, Bracher A, Hartl FU. 2016. The GroEL-GroES chaperonin machine: A nano-cage for protein folding. Trends Biochem Sci 41: 62–76. 10.1016/j.tibs.2015.07.009 [DOI] [PubMed] [Google Scholar]
- Hetz C, Chevet E, Oakes SA. 2015. Proteostasis control by the unfolded protein response. Nat Cell Biol 17: 829–838. 10.1038/ncb3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Frankino PA, Paul JW III, Tronnes SU, Dillin A. 2018. A futile battle? Protein quality control and the stress of aging. Dev Cell 44: 139–163. 10.1016/j.devcel.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, Brandeis M, Kopito RR. 2012. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J Cell Biol 196: 573–587. 10.1083/jcb.201110093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Park SH, Hartl FU. 2014. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol 24: 506–514. 10.1016/j.tcb.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Holtkamp W, Kokic G, Jager M, Mittelstaet J, Komar AA, Rodnina MV. 2015. Cotranslational protein folding on the ribosome monitored in real time. Science 350: 1104–1107. 10.1126/science.aad0344 [DOI] [PubMed] [Google Scholar]
- Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE. 2018. A new era for understanding amyloid structures and disease. Nat Rev Mol Cell Biol 19: 755–773. 10.1038/s41580-018-0060-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. 2007. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856. 10.1101/gad.1566807 [DOI] [PubMed] [Google Scholar]
- Izawa T, Park SH, Zhao L, Hartl FU, Neupert W. 2017. Cytosolic protein Vms1 links ribosome quality control to mitochondrial and cellular homeostasis. Cell 171: 890–903.e818. 10.1016/j.cell.2017.10.002 [DOI] [PubMed] [Google Scholar]
- Joazeiro CAP. 2017. Ribosomal stalling during translation: Providing substrates for ribosome-associated protein quality control. Annu Rev Cell Dev Biol 33: 343–368. 10.1146/annurev-cellbio-111315-125249 [DOI] [PubMed] [Google Scholar]
- Kakkar V, Månsson C, de Mattos EP, Bergink S, van der Zwaag M, van Waarde M, Kloosterhuis NJ, Melki R, van Cruchten RTP, Al-Karadaghi S, et al. 2016. The S/T-rich motif in the DNAJB6 chaperone delays polyglutamine aggregation and the onset of disease in a mouse model. Mol Cell 62: 272–283. 10.1016/j.molcel.2016.03.017 [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Karagöz GE, Acosta-Alvear D, Walter P. 2019. The unfolded protein response: Detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. 2018. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19: 365–381. 10.1038/s41580-018-0001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122: 209–220. 10.1016/j.cell.2005.05.028 [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82: 323–355. 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Klaips CL, Jayaraj GG, Hartl FU. 2018. Pathways of cellular proteostasis in aging and disease. J Cell Biol 217: 51–63. 10.1083/jcb.201709072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. 2008. Mechanisms of asymmetric stem cell division. Cell 132: 583–597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Krakowiak J, Zheng X, Patel N, Feder ZA, Anandhakumar J, Valerius K, Gross DS, Khalil AS, Pincus D. 2018. Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response. eLife 7: e31668 10.7554/eLife.31668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Shiber A, Bukau B. 2018. Mechanisms of cotranslational maturation of newly synthesized proteins. Annu Rev Biochem 10.1146/annurev-biochem-013118-111717. [DOI] [PubMed] [Google Scholar]
- Kulak NA, Geyer PE, Mann M. 2017. Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Mol Cell Proteomics 16: 694–705. 10.1074/mcp.O116.065136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Futschik ME. 2007. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2: 5–7. 10.6026/97320630002005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Chang JT, Schmalz J, Hansen M. 2017. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat Commun 8: 14337 10.1038/ncomms14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrat L, Regan L. 2010. Balance between folding and degradation for Hsp90-dependent client proteins: A key role for CHIP. Biochemistry 49: 7428–7438. 10.1021/bi100386w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo SY, Castoreno AB, Aldrich LN, Lassen KG, Goel G, Dančík V, Kuballa P, Latorre I, Conway KL, Sarkar S, et al. 2015. Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc Natl Acad Sci 112: E4281–E4287. 10.1073/pnas.1512289112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. 2015. The biology of proteostasis in aging and disease. Annu Rev Biochem 84: 435–464. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Cunliffe H, Weiss A, Katsyuba E, Sathasivam K, Seredenina T, Woodman B, Moussaoui S, Frentzel S, Luthi-Carter R, et al. 2011. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Invest 121: 3306–3319. 10.1172/JCI57413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Novoselov SS, Bett JS, Weiss A, Paganetti P, Bates GP, Cheetham ME. 2012. Suppression of protein aggregation by chaperone modification of high molecular weight complexes. Brain 135: 1180–1196. 10.1093/brain/aws022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. 1992. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683–689. 10.1038/356683a0 [DOI] [PubMed] [Google Scholar]
- Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al. 2010. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467: 179–184. 10.1038/nature09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leestemaker Y, de Jong A, Witting KF, Penning R, Schuurman K, Rodenko B, Zaal EA, van de Kooij B, Laufer S, Heck AJR, et al. 2017. Proteasome activation by small molecules. Cell Chem Biol 24: 725–736 e727. 10.1016/j.chembiol.2017.05.010 [DOI] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. 2008. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3: e1487 10.1371/journal.pone.0001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Han Y, Qian SB. 2013. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell 49: 453–463. 10.1016/j.molcel.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A. 2016. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res 26: 869–885. 10.1038/cr.2016.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. 2015. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain Injury. Cell Stem Cell 17: 329–340. 10.1016/j.stem.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. 2016. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell 62: 63–78. 10.1016/j.molcel.2016.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Goldberg AL. 2008. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol 182: 663–673. 10.1083/jcb.200803022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A, Haynes CM. 2018. UPRmt regulation and output: A stress response mediated by mitochondrial-nuclear communication. Cell Res 28: 281–295. 10.1038/cr.2018.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen TET, Komander D. 2017. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem 86: 159–192. 10.1146/annurev-biochem-061516-044916 [DOI] [PubMed] [Google Scholar]
- Milo R. 2013. What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 35: 1050–1055. 10.1002/bies.201300066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Shen K, Dillin A. 2018. Mitochondrial proteostasis in the context of cellular and organismal health and aging. J Biol Chem 10.1074/jbc.TM117.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Kummer E, Bukau B. 2015. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci 2: 22 10.3389/fmolb.2015.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Bukau B, Kampinga HH. 2018. Cellular handling of protein aggregates by disaggregation machines. Mol Cell 69: 214–226. 10.1016/j.molcel.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Morimoto RI. 2011. The heat shock response: Systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol 76: 91–99. 10.1101/sqb.2012.76.010637 [DOI] [PubMed] [Google Scholar]
- *.Morimoto RI. 2019. Cell non-autonomous regulation of proteostasis in aging and disease. Cold Spring Harb Perspect Biol. 10.1101/cshperspect.a034074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. 2000. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci 97: 7841–7846. 10.1073/pnas.140202897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy M, Fenton WA, Li D, Furtak K, Horwich AL. 2016. Extended survival of misfolded G85R SOD1-linked ALS mice by transgenic expression of chaperone Hsp110. Proc Natl Acad Sci 113: 5424–5428. 10.1073/pnas.1604885113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Naresh NU, Haynes CM. 2019. Signaling and regulation of the mitochondrial unfolded protein response. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA. 2015. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161: 1606–1618. 10.1016/j.cell.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Jaeger AM, Gomez-Pastor R, Willmund F, Frydman J, Thiele DJ. 2014. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep 9: 955–966. 10.1016/j.celrep.2014.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Prado MA, Schmidt PJ, Sendamarai AK, Wilson-Grady JT, Min M, Campagna DR, Tian G, Shi Y, Dederer V, et al. 2017. UBE2O remodels the proteome during terminal erythroid differentiation. Science 357: eaan0218 10.1126/science.aan0218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, et al. 2015. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524: 247–251. 10.1038/nature14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson OB, Hedman R, Marino J, Wickles S, Bischoff L, Johansson M, Müller-Lucks A, Trovato F, Puglisi JD, O'Brien EP, et al. 2015. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep 12: 1533–1540. 10.1016/j.celrep.2015.07.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noormohammadi A, Khodakarami A, Gutierrez-Garcia R, Lee HJ, Koyuncu S, König T, Schindler C, Saez I, Fatima A, Dieterich C, et al. 2016. Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat Commun 7: 13649 10.1038/ncomms13649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. 2011. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147: 1295–1308. 10.1016/j.cell.2011.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144: 67–78. 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. 2016. The integrated stress response. EMBO Rep 17: 1374–1395. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parag HA, Raboy B, Kulka RG. 1987. Effect of heat shock on protein degradation in mammalian cells: Involvement of the ubiquitin system. EMBO J 6: 55–61. 10.1002/j.1460-2075.1987.tb04718.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, Hayer-Hartl M, Hartl FU. 2013. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154: 134–145. 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372: 475–478. 10.1038/372475a0 [DOI] [PubMed] [Google Scholar]
- Powers ET, Balch WE. 2013. Diversity in the origins of proteostasis networks—A driver for protein function in evolution. Nat Rev Mol Cell Biol 14: 237–248. 10.1038/nrm3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78: 959–991. 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- Preissler S, Deuerling E. 2012. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci 37: 274–283. 10.1016/j.tibs.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Loew C, Körner R, Pinkert S, Theis M, Hayer-Hartl M, Buchholz F, Hartl FU. 2014. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell 156: 975–985. 10.1016/j.cell.2014.01.055 [DOI] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. 2010. The heat shock response: Life on the verge of death. Mol Cell 40: 253–266. 10.1016/j.molcel.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Rizzolo K, Huen J, Kumar A, Phanse S, Vlasblom J, Kakihara Y, Zeineddine HA, Minic Z, Snider J, Wang W, et al. 2017. Features of the chaperone cellular network revealed through systematic Interaction mapping. Cell Rep 20: 2735–2748. 10.1016/j.celrep.2017.08.074 [DOI] [PubMed] [Google Scholar]
- Rodina A, Wang T, Yan P, Gomes ED, Dunphy MP, Pillarsetty N, Koren J, Gerecitano JF, Taldone T, Zong H, et al. 2016. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature 538: 397–401. 10.1038/nature19807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DM, Hutt DM, Tong J, Bouchecareilh M, Wang N, Seeley T, Dekkers JF, Beekman JM, Garza D, Drew L, et al. 2014. Modulation of the maladaptive stress response to manage diseases of protein folding. PLoS Biol 12: e1001998 10.1371/journal.pbio.1001998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC. 2006. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443: 780–786. 10.1038/nature05291 [DOI] [PubMed] [Google Scholar]
- Sala AJ, Bott LC, Morimoto RI. 2017. Shaping proteostasis at the cellular, tissue, and organismal level. J Cell Biol 216: 1231–1241. 10.1083/jcb.201612111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saretzki G, Armstrong L, Leake A, Lako M, von Zglinicki T. 2004. Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells. Stem Cells 22: 962–971. 10.1634/stemcells.22-6-962 [DOI] [PubMed] [Google Scholar]
- Saretzki G, Walter T, Atkinson S, Passos JF, Bareth B, Keith WN, Stewart R, Hoare S, Stojkovic M, Armstrong L, et al. 2008. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells 26: 455–464. 10.1634/stemcells.2007-0628 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. 2009. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ 16: 46–56. 10.1038/cdd.2008.110 [DOI] [PubMed] [Google Scholar]
- Scior A, Buntru A, Arnsburg K, Ast A, Iburg M, Juenemann K, Pigazzini ML, Mlody B, Puchkov D, Priller J, et al. 2018. Complete suppression of Htt fibrilization and disaggregation of Htt fibrils by a trimeric chaperone complex. EMBO J 37: 282–299. 10.15252/embj.201797212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, Burge CB. 2013. Widespread regulation of translation by elongation pausing in heat shock. Mol Cell 49: 439–452. 10.1016/j.molcel.2012.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509: 49–54. 10.1038/nature13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittler A, Lurz R, Lueder G, Priller J, Lehrach H, Hayer-Hartl MK, Hartl FU, Wanker EE. 2001. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington's disease. Hum Mol Genet 10: 1307–1315. 10.1093/hmg/10.12.1307 [DOI] [PubMed] [Google Scholar]
- Solís EJ, Pandey JP, Zheng X, Jin DX, Gupta PB, Airoldi EM, Pincus D, Denic V. 2016. Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol Cell 63: 60–71. 10.1016/j.molcel.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. 2010. Translational regulation of gene expression during conditions of cell stress. Mol Cell 40: 228–237. 10.1016/j.molcel.2010.09.028 [DOI] [PubMed] [Google Scholar]
- Suraweera A, Münch C, Hanssum A, Bertolotti A. 2012. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 48: 242–253. 10.1016/j.molcel.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. 2012. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150: 987–1001. 10.1016/j.cell.2012.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, Larsen B, Choi H, Berger B, Gingras AC, Lindquist S. 2014. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 158: 434–448. 10.1016/j.cell.2014.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. 2011. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol 3: a004440 10.1101/cshperspect.a004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. 1999. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97: 755–765. 10.1016/S0092-8674(00)80787-4 [DOI] [PubMed] [Google Scholar]
- Thibaudeau TA, Anderson RT, Smith DM. 2018. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat Commun 9: 1097 10.1038/s41467-018-03509-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaytler P, Harding HP, Ron D, Bertolotti A. 2011. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 332: 91–94. 10.1126/science.1201396 [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. 2015. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. 2010. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol 2: a004390 10.1101/cshperspect.a004390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. 2017. The ubiquitin system, autophagy, and regulated protein degradation. Annu Rev Biochem 86: 123–128. 10.1146/annurev-biochem-061516-044859 [DOI] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, et al. 2012. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489: 304–308. 10.1038/nature11468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Simic MS, Dillin A. 2014. Proteostasis and aging of stem cells. Trends Cell Biol 24: 161–170. 10.1016/j.tcb.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. 2015. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162: 1286–1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: From stress pathway to homeostatic regulation. Science 334: 1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Tsujimura A, Taguchi K, Tanaka M. 2017. HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy 13: 133–148. 10.1080/15548627.2016.1248018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI. 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323: 1063–1066. 10.1126/science.1165946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. 2014. Mass-spectrometry-based draft of the human proteome. Nature 509: 582–587. 10.1038/nature13319 [DOI] [PubMed] [Google Scholar]
- Willmund F, del Alamo M, Pechmann S, Chen T, Albanèse V, Dammer EB, Peng J, Frydman J. 2013. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152: 196–209. 10.1016/j.cell.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Weissman JS, Dillin A. 2014. Differential scales of protein quality control. Cell 157: 52–64. 10.1016/j.cell.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Xu G, Pattamatta A, Hildago R, Pace MC, Brown H, Borchelt DR. 2016. Vulnerability of newly synthesized proteins to proteostasis stress. J Cell Sci 129: 1892–1901. 10.1242/jcs.176479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. 2008. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol 15: 1255–1262. 10.1038/nsmb.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Juszkiewicz S, Hegde RS. 2017. UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357: 472–475. 10.1126/science.aan0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Oza J, Bridges K, Chen KY, Liu AY. 2008. Neural differentiation and the attenuated heat shock response. Brain Res 1203: 39–50. 10.1016/j.brainres.2008.01.082 [DOI] [PubMed] [Google Scholar]
- Yao Z, Mich JK, Ku S, Menon V, Krostag AR, Martinez RA, Furchtgott L, Mulholland H, Bort S, Fuqua MA, et al. 2017. A single-cell roadmap of lineage bifurcation in human ESC models of embryonic brain development. Cell Stem Cell 20: 120–134. 10.1016/j.stem.2016.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R, Tahara EB, Bengtson MH, Khokhrina M, Lorenz H, Chen KC, Kigoshi-Tansho Y, Savas JN, Yates JR, Kay SA, et al. 2016. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife 5: e11794 10.7554/eLife.11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You KT, Park J, Kim VN. 2015. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev 29: 2004–2009. 10.1101/gad.267112.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Shibata Y, Shah B, Calamini B, Lo DC, Morimoto RI. 2014. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc Natl Acad Sci 111: E1481–E1490. 10.1073/pnas.1321811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. 2009. Fidelity at the molecular level: Lessons from protein synthesis. Cell 136: 746–762. 10.1016/j.cell.2009.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Shen S, Qu J, Ghaemmaghami S. 2016. Global analysis of cellular protein flux quantifies the selectivity of basal autophagy. Cell Rep 14: 2426–2439. 10.1016/j.celrep.2016.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. 2005. Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120: 715–727. 10.1016/j.cell.2004.12.024 [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhai B, Gygi SP, Goldberg AL. 2015. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci 112: 15790–15797. 10.1073/pnas.1521919112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Shabek N. 2017. Ubiquitin ligases: Structure, function, and regulation. Annu Rev Biochem 86: 129–157. 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
- Zheng X, Krakowiak J, Patel N, Beyzavi A, Ezike J, Khalil AS, Pincus D. 2016. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 5: e18638 10.7554/eLife.18638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AE, Crist C. 2016. Phosphorylation of eIF2α is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 18: 79–90. 10.1016/j.stem.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94: 471–480. 10.1016/S0092-8674(00)81588-3 [DOI] [PubMed] [Google Scholar]
- Żwirowski S, Kłosowska A, Obuchowski I, Nillegoda NB, Piróg A, Ziętkiewicz S, Bukau B, Mogk A, Liberek K. 2017. Hsp70 displaces small heat shock proteins from aggregates to initiate protein refolding. EMBO J 36: 783–796. 10.15252/embj.201593378 [DOI] [PMC free article] [PubMed] [Google Scholar]