Abstract
Understanding the reaction mechanism of Cas9 is crucial for the application of programmable gene editing. Despite the availability of the structures of Cas9 in apo- and substrate-bound forms, the catalytically active structure is still unclear. Our first attempt to explore the catalytic mechanism of Cas9 HNH domain has been based on the reasonable assumption that we are dealing with the same mechanism as endonuclease VII, including the assumption that the catalytic water is in the first shell of the Mg2+. Trying this mechanism with the cryo-EM structure forced us to induce significant structural change driven by the movement of K848 (or other positively charged residue) close to the active site to facilitate the proton transfer step. In the present study, we explore a second reaction mechanism where the catalytic water is in the second shell of the Mg2+ and assume that the cryo-EM structure by itself is a suitable representation of a catalytic-ready structure. The alternative mechanism indicates that if the active water is from the second shell then the calculate reaction barrier is lower compared with the when the corresponding barrier when the water comes from the first shell.
Introduction
The Cas9 protein (CRISPR-associated protein 9), derived from type II CRISPR (clustered regularly interspaced short palindromic repeats) bacterial immune systems, can cleave DNA with distinct specificity once provided appropriately paired sgRNAs.1 It has gained a tremendous attention as a powerful tool in genome editing.1–4 Despite its successful usage as genome editing tool, a catalytically inactive Cas9 can help to target effector domains and modulate endogenous transcription guided by RNA. With flexible RNA-guided, Cas9 can also be used in genome positioning system.4–6
Cas9 has two nuclease domains: HNH domain and RuvC domain that are designed to cleave the target DNA (tDNA) strand and the non-target DNA (ntDNA) strand respectively. The HNH Domain has a ββα-metal fold while the RuvC domain has an RNase H fold and shares structural similarity with the members of retroviral integrase superfamily.1
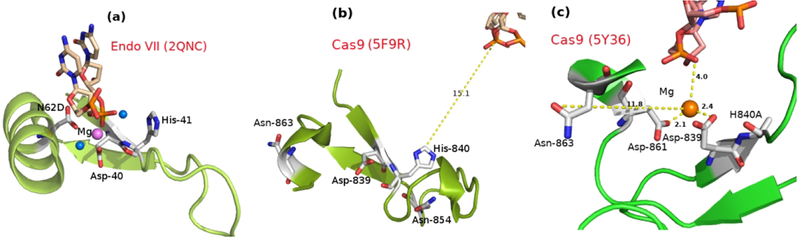
The structural changes that are involved in the activation of CRISPR have been discussed extensively. But from a mechanistic point of view, what is mostly missing is the structure of the catalytically competent HNH domain of Cas9 and the information about structural changes that lead to this structure. The plasticity of the HNH domain has been evident in experiments in which the HNH domain adopts different conformation and position7,8. The catalytic site of the tDNA is more than 10Å away from the active site (see Figure 1(b)) in the latest X-ray structure8. The difference in the conformation and position of the HNH domain in two available Cas9 structures (with an incomplete nontarget DNA strand7 and with both unwound DNA strands8) demonstrated the plasticity of the HNH. In the recent cryo-EM (5Y36.pdb),9 solved at 5.2 Å resolution, the gap between catalytic phosphodiester bond to the active site is smaller (see Figure 1(c)). This structure has provided the most promising information, and the MD simulation used in ref 9, tried to determine the closest position of the HNH domain to the cleavage site.
Figure 1:
(a) The active site of the Endo VII (2QNC.pdb). Catalytic residues His-41, Asp-40 and Asp-62 are shown as stick and colored by element (C: white; N: blue; O: red). The water molecules and Mg2+ are shown as marine and violet sphere respectively. One nucleic acid (as a part of DNA stand) is shown as stick and colored by element (C: wheat; N: blue; O: red; P: orange). The protein backbone (ββα) is shown as cartoon representation. (b) The active site of the Cas9 HNH domain (5F9R.pdb). Residues Asp-839, His-840 and Asn-863, which can be corresponded to residues Asp-40, His-41 and Asn-62 in Endo VII. The residues are shown as stick and colored by element (C: white; N: blue; O: red). The distance between the phosphodiester center from His840 is shown in yellow dashed line and the distance is reported in Å unit. (c) The active site of the Cas9 HNH domain from cryo-EM (5Y36.pdb). Residues Asp-839, H840A and Asn-863, which are the representative to the residues Asp-40, His-41 and Asn-62 in Endo VII, are shown as stick and colored by element (C: white; N: blue; O: red). The distance between the Mg2+ and other catalytic site residues are shown in yellow dashed lines and the distances are reported in Å unit. This figure is generated by PyMol.
However, the reaction mechanism cannot be established without investigating the free energy profile of the chemical reaction. The relevance of those structures cannot be established without calculating the free energy of the chemical reaction that would occur in the corresponding structures. Furthermore, the detailed available structural information is far too limited and not enough to take the cryo-EM as a basis for modeling the active form of Cas9. The missing information includes the positions of the divalent metal ion, active water molecules and the exact position of key residues. Additionally, Cas9-sgRNA-dsDNA is very large, complex and dynamics, which brings much more challenges for computational studies.
In our earlier study,10 we have examined a mechanism where the catalytic water is in the first shell of the Mg2+. This mechanism was found to reproduce the observed kinetics of Endonuclease VII. However, in the active site model that corresponds to the cryo-EM structure we could not obtain any reasonable barrier without bringing the positively charged residue to the active site region. Moving K848 to the active site facilitates the proton transfer step and led to a reasonable activation barrier. In this work we explored a second reaction mechanism where the catalytic water is in the second shell of the Mg2+ and assume that the cryo-EM structure by itself is a suitable representation of a catalytic-ready structure.
By fine-tuning the active site of the cryo-EM structure (5Y36.pdb) and using that in our EVB calculations (by assuming that the nucleophilic water is coming from the second solvent sphere of Mg2+), we obtained results that has shown good agreement to the experimentally found rate enhancement by Cas9.
Methods
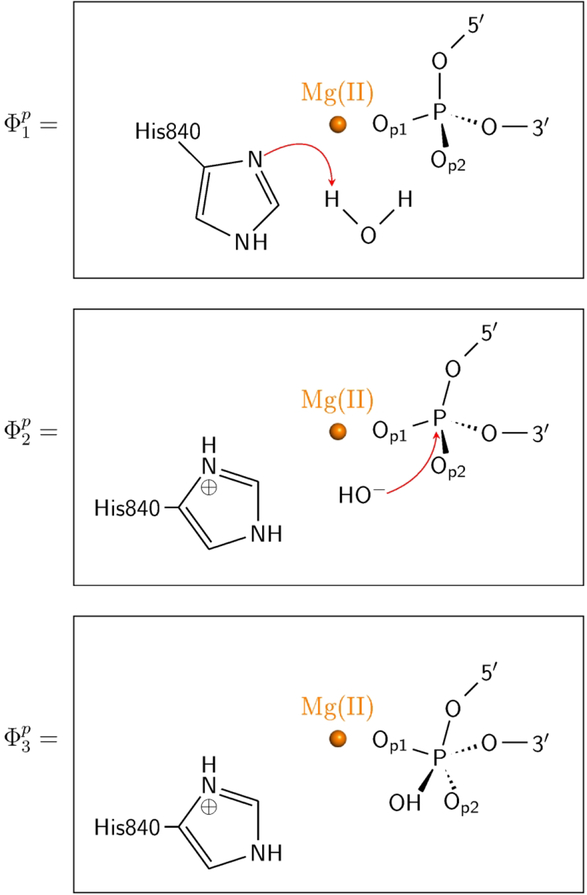
In order to understand the possible mechanism of Cas9, it is crucial to look for well understood related reactions. In the previous work10 we made the reasonable assumption that the closest reaction mechanism is that of Endo VII. A similar assumption is also made in our current study. The corresponding calibration of the reference reaction in water considered a direct PT from a water molecule to a His residue. The empirical valence bond approach11–12 was used to simulate the reaction mechanisms as shown in Scheme 1 (see Figure 2). The calibration on the water reference reaction was based on the previous study see ref13, and the key energetics is given in Table 1.
Figure 2:
Three EVB resonance states (ϕ1P, ϕ2P, ϕ3P) used for describing the first two steps of phosphodiester bond hydrolysis by HNH domain of Cas9.
Table 1.
Calculated free energy (kcal/mol) for the catalytic reaction of the Cas9 HNH domain. The ∆G‡obs is the relevant experimental value.
| ΔG‡,PT | ΔG‡,NA | ΔG‡ | ΔG‡obs | |||
|---|---|---|---|---|---|---|
| water in 1st shell | endo VII | wild type | 2.75 | 20.4 | 23.02 | 21.48 |
| H43N | 17.47 | 26.05 | 41.12 | |||
| K848 far | 27.49 | 20.73 | 48.22 | |||
| Cas9 | K848 near/ionized | 3.64 | 17.37 | 21.01 | 17.8 | |
| K848 near/unionized | 4.38 | 21.97 | 26.35 | |||
| water in 2ed shell | water | 11.2 | 33 | 44.2 | ||
| Cas9 | 3 | 13.8 | 16.8 | 17.8 |
Data for the active water in the first shell is from our previous study (see Ref. 10).
In building the starting structure in the catalytic active form, we took the cryo-EM structure (5Y36.pdb) and reconstituted the active center first by mutating Ala840 back to His840, and then superimposed the active center with the Endo VII14 to check the differences (See Figure 1(a) and 1(c)).
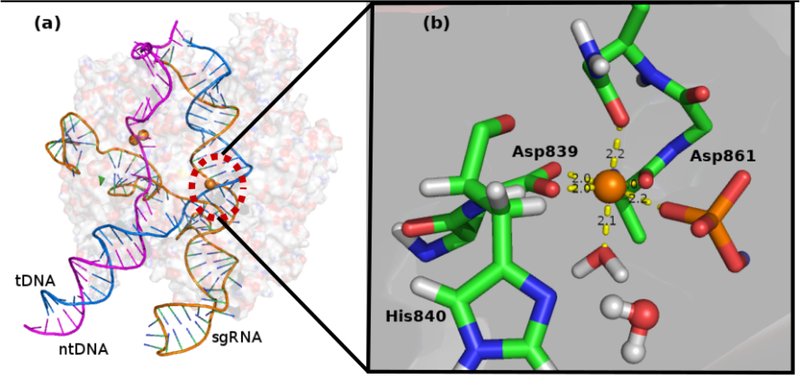
A constrained force of 10.0 kcal/mol was applied to keep the distance between atom OD1 of Asn863 and catalytic Mg2+ atom around 2.1Å and was relaxed for 40ps. In the following rounds of relaxation, the constrained force was gradually removed. After this step the system has reached the equilibrium with a constraint force removed gradually. Further a total of 200 ps relaxation was done with 4 representative structures (after every 50 ps) were selected for the EVB calculations. One final model and fine-tuned active center is shown in Figure 3. The atoms from amino acids shown in Scheme 1 (see Figure 2) are considered as the EVB atoms.
Figure 3:
(a) Modeled structural overview of Cas9-sgRNA-dsDNA system. (b) Fine-tuned HNH active site overview. Residues Asp839, His840, Asp861 and Asn863 are shown as stick and colored by element (C: green; N: blue; O: red). The first shell water molecule is shown as stick, and the nucleophilic water is shown as ball and stick. Mg2+ is shown as orange sphere. The nucleic acid is shown as stick and colored by element (C: blue; N: blue; O: red; P: orange). This figure is generated by PyMol.21
The geometric center of the EVB reacting atoms was set as the center of the simulation sphere. The active site region along with the Cas9-RNA-DNA complex were immersed in a 32 Å sphere of water molecules using the surface-constraint all-atom solvent (SCAAS) type boundary condition.15 Outside of this 32 Å region there was a 2 Å surface of Langevin dipoles and then a bulk continuum. The local reaction field (LRF) method16 was used to treat the long-range electrostatics. Atoms beyond this sphere were fixed at their initial positions as in the Cryo-EM structure, and no electrostatic interaction from outside of the sphere was considered. Our system consists of 32,125 atoms including 1,289 molaris-generated water molecules. The protonation states for the residue within 12 Å from the center of the simulation were determined by calculating the pKa’s using our coarse grained (CG) model17 with a macroscopic charge dielectric for the effect of ionizable residues beyond this range. For the EVB free energy calculations, the free energy perturbation/umbrella sampling approach (FEP/US)11 was used with 11 frames of 200 ps each where the time-step is 1 fs. All the calculations in this study used the Enzymix module of the MOLARIS-XG package.18 All EVB parameters are taken from our previous study13. The nonbonded parameters AMg=72 KJ/mol·Å12 and BMg=15 KJ/mol·Å6 are used for Mg2+ with the formula Vab =Ai Aj/r12−Bi Bj/r6. The atoms outside EVB region are represented by the Enzymix force field.19
Results and discussion
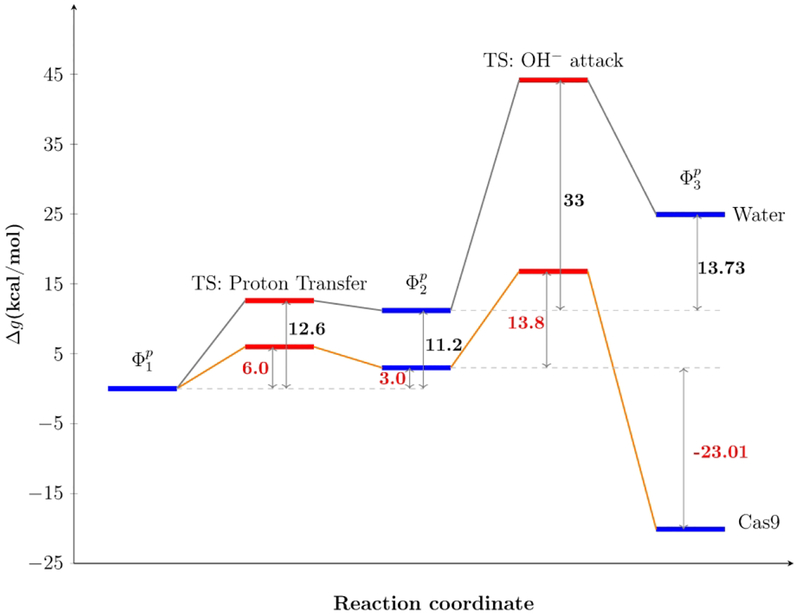
Our calculated results are in good agreement with the experiment (see Table 1). After calibrating the EVB parameters with respect to the reactions in water (both PT and NA steps), we have used the same parameters for studying the reaction in protein and obtained 3.0 and 13.8 kcal/mol as reaction free energy for the proton transfer step and activation free energy of the nucleophilic step, respectively. Thus, our calculation shows that the reaction in protein is stabilized by almost 27 kcal/mol with respect to the reaction in water.
Comparing with our previous study10, in which the active water molecule is in the first solvation shell of Mg2+, in this work by taking the water from the second solvation shell, brings down the activation energy of the rate determining step. The activation free energy for the nucleophilic attack by OH− decreases due to the weaker electrostatic attraction from Mg2+ for the water in second solvation shell compared with the first shell. Interestingly, it is found that in this proposed mechanism the movement of the K848 to the active site is not required, which present an open problem.
As discussed in our previous work,10 it is not straightforward to explore the effect of mutations on the catalysis of the phosphodiester bond hydrolysis by Cas9, for example, the engineered Cas912 (eCas9; K848A/K1003A/R1060A), shows a reduced off target activity and also slows down the intrinsic cleavage rate. Apparently, there is a strong correlation between the DNA unwinding and the cleavage rate. However, the DNA unwinding is affected by (1) the interaction between the negatively-charged DNA backbone and the positively charged residues from Cas9 DNA binding groove and (2) the mismatches between the PAM-distal DNA-RNA base mismatches. The mutation of the positively charged residues in eCas9 to Ala hindered the binding affinity and thus the unwinding process. The PAM-distal mismatches have been reported to be greater for eCas9 than Cas9. Our previous study has explored the effect of the absence of K848 in the binding groove by moving the positive charged residues to the active site. Our results10 has captured the slower reaction rate in the absence of K848 in the binding groove. Figure 4d in Ref 12 indicates nearly constant differences in the reaction rate between the WT and eCas9 above an unwound fraction of 50%. Our study has shed light on the interplay between the specificity, cleavages as well as DNA unwound process. The DNA unwinding is hindered by the mutation of positive residues in eCas9 PAM-distal DNA-RNA base mismatches. The impact of PAM-distal mismatches is greater for eCas9. Thus, they suggested that the enhancement in the cleavage specificity of eCas9 is largely due to destabilization of the unwound states by mutating the positive residues. Our previous results correspond to the same slower reaction rate in the absence of K848, suggesting the possible presence of a positive residue such as K848 near the cleavage site. In fact, Figure 4d of ref12 shows a nearly constant difference in the cleavage rate between the WT and eCas9 above an unwound fraction of 50%. While different dynamics and DNA unwound fractions between the WT and eCas9 seem to be correlated with the specificity, it also seems that there is a constant factor that makes a difference at the cleavage stage regardless of the mismatches.
Finally, we have avoided here the question of off-target, on-target control, since it most probably occurs at the open state; and being expressed by the barrier for moving to the closed form. This crucial effect cannot be explored with a structure that represents the closed form, which in the present case was assumed to be the cryo-EM structure.
Concluding remarks
The nature of the catalytic reaction of the HNH domain of Cas9 is far from obvious, in part due to the unavailability of a relevant high-resolution structure. In our earlier study,10 we calibrate the mechanism on the strongly related mechanism of endo VII. It was found that bringing a positive charged K848 residues to the active site facilitates structural change that activates the reaction. The results were consistent with the main experimental observations, but it remained tentative due to the absence of high-resolution structure. Here we consider the alternative, that the cryo-EM structure is closed to the active structure. In this case we had to modify the mechanism of Endo VII and to have a proton transfer from a water molecule in the second solvation shell of the Mg2+ ion. The considered mechanism appeared to reproduce a rate constant in the range of the observed rate constant.
As mentioned earlier, the X-ray structure failed to capture the catalytic-ready structure of Cas9 HNH domain, even in the latest advancement8 the catalytic phosphodiester bond is still more than 10Å away from the active site. The new cryo-EM structure9 has bridged the gap and the cleavage site of the phophodiester is close to the active site. However, when the catalytic essential residues His840 was mutated to Ala840 the Asn-863 (which corresponds to the catalytically essential residue Asn-62 in Endo VII) is more than 10Å from the Mg2+ atom. In our study, we have reconstituted the active site based on the Endo VII and applied a constraint to bring the OD1 atom of Asn-863 to Mg2+ ion during a long relaxation. With the resulting active site the EVB simulations, where the nucleophilic water has been assumed to be coming from the second solvation shell of Mg2+, has accounted for the observed activation barrier of the Cas9 HNH domain.
The fact that two mechanisms can account for the rate of acceleration of Cas9 means that more structural information is clearly needed. In Particular, it seems that an issue that was not addressed in this work is of major importance. Namely elucidating the nature of the DNA unwinding may provide a direct information on the control of the off-target activity.
Figure 4:
Calculated reaction free energy profile in solution and in Cas9 for proton transferand nucleophilic attack.
Acknowledgment
We thank the University of Southern California High Performance Computing and Communication Center for computational resources. This work was supported by National Science Foundation Grant MCB 1707167, National Institute of Health R01-AI055926, and Agency for Science, Technology and Research (A*STAR) International Fellowship (AIF) provided for L. N. Z. Additionally, L. N. Z. and D. M. would love to thank Dr. Mikolaj Feliks, Dr. Han W. Yoon, Dr. Zhen Tao Chu for guidance in EVB simulation as well as helpful discussion. L. N. Z. would love to thank Dr. Philipp Kaldis for his support and discussions.
References
- 1.Nishimasu H; Ran FA; Hsu PD; Konermann S; Shehata SI; Dohmae N; Ishitani R; Zhang F; Nureki O, Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156 (5), 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang HF; La Russa M; Qi LS, CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 2016, 85, 227–264. [DOI] [PubMed] [Google Scholar]
- 3.Jiang FG; Doudna JA, CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys 2017, 46, 505–529. [DOI] [PubMed] [Google Scholar]
- 4.Kearns NA; Genga RMJ; Enuameh MS; Garber M; Wolfe SA; Maehr R, Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development 2014, 141 (1), 219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi LS; Larson MH; Gilbert LA; Doudna JA; Weissman JS; Arkin AP; Lim WA, Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152 (5), 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maji B; Moore CL; Zetsche B; Volz SE; Zhang F; Shoulders MD; Choudhary A, Multidimensional chemical control of CRISPR-Cas9. Nat Chem Biol 2017, 13 (1), 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders C; Niewoehner O; Duerst A; Jinek M, Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513 (7519), 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang FG; Taylor DW; Chen JS; Kornfeld JE; Zhou KH; Thompson AJ; Nogales E; Doudna JA, Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 2016, 351 (6275), 867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huai C; Li G; Yao RJ; Zhang Y; Cao M; Kong LL; Jia CQ; Yuan H; Chen HY; Lu DR; Huang Q, Structural insights into DNA cleavage activation of CRISPR-Cas9 system. Nat Commun 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon H; Zhao LN; Warshel A, Exploring the Catalytic Mechanism of Cas9 Using Information Inferred from Endonuclease VII. Acs Catal 2019, 9 (2), 1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warshel A; Weiss RM, An Empirical Valence Bond Approach for Comparing Reactions in Solutions and in Enzymes. J Am Chem Soc 1980, 102 (20), 6218–6226. [Google Scholar]
- 12.Kamerlin SCL; Warshel A, The EVB as a quantitative tool for formulating simulations and analyzing biological and chemical reactions. Faraday Discuss 2010, 145, 71–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aqvist J; Warshel A, Calculations of Free-Energy Profiles for the Staphylococcal Nuclease Catalyzed Reaction. Biochemistry-Us 1989, 28 (11), 4680–4689. [DOI] [PubMed] [Google Scholar]
- 14.Biertumpfel C; Yang W; Suck D, Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 2007, 449 (7162), 616–U14. [DOI] [PubMed] [Google Scholar]
- 15.Warshel A; King G, Polarization Constraints in Molecular-Dynamics Simulation of Aqueous-Solutions - the Surface Constraint All Atom Solvent (Scaas) Model. Chem Phys Lett 1985, 121 (1–2), 124–129. [Google Scholar]
- 16.Lee FS; Warshel A, A Local Reaction Field Method for Fast Evaluation of Long-Range Electrostatic Interactions in Molecular Simulations. J Chem Phys 1992, 97 (5), 3100–3107. [Google Scholar]
- 17.Vicatos S; Rychkova A; Mukherjee S; Warshel A, An effective Coarse-grained model for biological simulations: Recent refinements and validations. Proteins 2014, 82 (7), 1168–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warshel A; Chu Z; Villa J; Strajbl M; Schutz C; Shurki A; Vicatos S; Chakrabarty S; Plotnikov N; Schopf P, Molaris-Xg, v 9.11; University of Southern California, Los Angeles, CA, 2012. [Google Scholar]
- 19.Lee FS; Chu ZT; Warshel A Microscopic and semimicroscopic calculations of electrostatic energies in proteins by the POLARIS and ENZYMIX programs. Journal of Computational Chemistry 1993, 14, 161–185. [Google Scholar]
- 20.Singh D; Wang YB; Mallon J; Yang O; Fei JY; Poddar A; Ceylan D; Bailey S; Ha T, Mechanisms of improved specificity of engineered Cas9s revealed by single-molecule FRET analysis. Nat Struct Mol Biol 2018, 25 (4), 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLano WL, The PyMOL Molecular Graphics System, Version 1.4. 2011, Schrodinger, LLC. [Google Scholar]