Abstract
The AP-2γ transcription factor, encoded by the TFAP2C gene, regulates the expression of estrogen receptor-alpha (ERα) and other genes associated with hormone response in luminal breast cancer. Little is known about the role of AP-2γ in other breast cancer subtypes. A subset of HER2+ breast cancers with amplification of the TFAP2C gene locus becomes addicted to AP-2γ. Herein we sought to define AP-2γ gene targets in HER2+ breast cancer and identify genes accounting for physiologic effects of growth and invasiveness regulated by AP-2γ. Comparing HER2+ cell lines that demonstrated differential response to growth and invasiveness with knockdown of TFAP2C, we identified a set of 68 differentially expressed target genes. CDH5 and CDKN1A were among the genes differentially regulated by AP-2γ and that contributed to growth and invasiveness. Pathway analysis implicated the MAPK13/p38δ and retinoic acid regulatory nodes, which were confirmed to display divergent responses in different HER2+ cancer lines. To confirm the clinical relevance of the genes identified, the AP-2γ gene signature was found to be highly predictive of outcome in HER2+ breast cancer patients. We conclude that AP-2γ regulates a set of genes in HER2+ breast cancer that drive cancer growth and invasiveness. The AP-2γ gene signature predicts outcome of patients with HER2+ breast cancer and pathway analysis predicts that subsets of patients will respond to drugs that target the MAPK or retinoic acid pathways.
Keywords: Breast Cancer, HER2-positive, TFAP2C, AP-2γ, CDH5, CDKN1A, MAPK13, Retinoic Acid, Transcription
Introduction
The AP-2 transcription factor family plays a critical role in establishing clinically relevant patterns of gene expression in breast cancer. Expression of ERα (encoded by the ESR1 gene) is transcriptionally regulated in breast cancer (1), and AP-2γ (encoded by the TFAP2C gene) was identified as one of the key factors controlling ESR1/ERα gene transcription (2–6). In addition, AP-2γ regulates the expression of many other genes associated with the luminal breast cancer phenotype (4,6,7) and collaborates with FOXA1 and ERα in the transcriptional regulation of genes in hormone-responsive breast cancer (8). Within ERα-positive breast cancers, high levels of AP-2γ expression have been associated with poor patient survival and resistance to hormonal therapy (9,10). Knockdown of TFAP2C in ERα-positive breast cancer cell lines repressed expression of ERα and other markers of luminal breast cancer and induced markers associated with epithelial-mesenchymal transition (EMT) and the cancer stem cell phenotype (11). The highly homologous AP-2 family member, AP-2α (encoded by the TFAP2A gene), was shown to regulate the expression of HER2 (12), which may function through several regulatory regions (13). Subsequent work suggested that AP-2γ contributes to HER2 gene regulation (14,15), and an enhancer element has been described in the HER2 gene that is activated by AP-2γ (16).
The HER2+ breast cancer subtype lacks ERα and progesterone receptor (PgR) expression with amplified HER2 expression and has a worse clinical course compared to the luminal breast cancer subtypes. However, even within the HER2+ subtype of breast tumors, there is a high degree of heterogeneity (17). Detailed genetic analysis of HER2+ breast cancers identified a subset displaying an addiction to TFAP2C gene amplification (18). For example, knockdown of TFAP2C in SKBR3 HER2+ breast cancer cells reduced cell growth, and this dependency was associated with amplification of the TFAP2C gene locus. In contrast, other HER2-amplified cell lines failed to demonstrate reduced growth with knockdown of TFAP2C, suggesting a significant diversity in the pattern of genes regulated by AP-2γ in HER2+ breast cancer. We sought to examine the regulation of genes by AP-2γ in HER2+ breast cancers with the goals of characterizing differential AP-2γ target genes and defining pathways of gene regulation by AP-2γ that modulate cancer growth and progression.
MATERIALS AND METHODS
Cell Culture
Cell lines HCC1954, SKBR3, MCF-7, HCC202, HCC1569, and MDA-MB-453 were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA), used at low (<10) passage number without further authentication or mycoplasma testing, and propagated in the appropriate medium as recommended by the manufacturer. For experiments with all-trans-retinoic acid (ATRA) (Sigma, USA, Cat#302–79-4), cells were grown in red-free media containing 10% charcoal stripped fetal bovine serum (Gibco, Cat#F6765) to remove endogenous steroids. Cells were subsequently treated for 48 hours with concentrations 0.1 and 0.5 μM of ATRA or DMSO as a negative control.
Gene Knockdown
Cells were transfected using small interfering RNA (siRNA) directed towards non-targeting #1 (NT Thermo Fisher Scientific, ID:4390843), TFAP2C (TFS, ID:107041), CDH5 (TFS, ID:10509), CDKN1A/p21 (TFS, ID: HSS173521), and MAPK13 (TFS, ID: VHS40525) with lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, USA, Cat# 13778150), accordingly a manufacturer’s instruction. After 72 to 96 hours of incubation, cells were immediately analyzed or used in subsequent experiments. Cell clones of HCC1954 lines with stable knockdown of TFAP2C (Sigma, USA, Cat#TRCN0000019745) and negative control (Sigma, Cat#SHC002) were generated using lentivirus-mediated shRNA cassette as described previously (11).
Expression Analysis
RNA: mRNA from cell lysates were obtained from cell lines using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA, Cat#74104) and converted to cDNA by polymerase chain reaction (qPCR) using random hexamers (Thermo Fisher Scientific) method. Using the delta-delta CT method of quantitative PCR (qPCR), relative gene expression was calculated using TaqMan primers to TFAP2C (TFS, Cat#4331182, ID: Hs00231476_m1), CDH5 (TFS, Cat#4331182, ID: Hs00901465_m1), CDKN1A (TFS, Cat#4331182, ID: Hs00355782_m1), MAPK11 (TFS, Cat#4331182, ID: Hs00177101_m1), MAPK12 (TFS, Cat#4331182, ID: Hs00268060_m1), MAPK13 (TFS, Cat#4331182, ID: Hs00234085_m1), and MAPK14 (TFS, Cat#4331182, ID: Hs01051152_m1) with GAPDH (TFS, Cat#4331182, ID: Hs02758991_g1) and 18s rRNA subunit (TFS, Cat#4331182, ID: Hs03003631_g1) used as an endogenous control. Western blots: protein was isolated in RIPA lysis buffer (Millipore, Cat#20–188), supplemented with protease inhibitor (Roche, Cat#11836170001) and PhosSTOP (Roche, Cat#4906845001). Primary antibodies were used according to the manufacturer’s recommendations: AP-2γ 1:700 (Abcam, Cat#ab76007), VE-cadherin 1:1000 (Cell Signaling Technology, Cat#2500), p21 1:700 (CST, Cat#2946), p38 MAPK 1:850 (CST, Cat#8690), p38δ/MAPK13 1:1000 (CST, Cat#2308), HER2/ErbB2 1:700 (CST, Cat#2165), and ERα 1:700 (Millipore, Cat#04–820). GAPDH 1:2000 (Santa Cruz, Cat#sc-32233) was used as a loading control. Secondary antibodies were used according to manufacturer specification: anti-rabbit HRP 1:5000 (CST, Cat#7074), anti-mouse HRP 1:2000 (CST, Cat#7076), and anti-goat 1:2000 (TFS, Cat#31402). Protein was visualized with SuperSignal West Dura extended duration substrate (TFS, Cat#34075) and SuperSignal West Femto maximum sensitivity substrate (TSF, Cat#34095).
RNA-seq
Experimental setup and analyses were performed in accordance to ENCODE Guidelines and Best Practices for RNA-seq. TFAP2C knockdown was completed on HCC1954, SKBR3 and HCC1569 cell lines (biologic triplicates) using 96-hour siRNA transfection. RNA (100–200 ng/μL) was harvested, knockdown confirmed with RT-PCR and protein western blot, frozen, and sent to the University of Nebraska for further processing. The RNA quality was confirmed by the receiving facility and was subsequently sequenced (technical replicates specified by facility) with specifications for gene differentiation (50 base pair, single-end reads). Sequencing depth was adequate (~50 million reads). Lentiviral shRNA knockdown of TFAP2C (Sigma, Cat# TRCN0000019745) in HCC1954 was completed with RNA being sent to the same facility for sequencing (50 base pair, paired-end reads; ~75 million reads). The RNA-seq analysis of the raw data was performed utilizing the Galaxy web platform at usegalaxy.org with the built-in tools: Bowtie2 for mapping and Cuffdiff for differential gene expression analysis, as recommended. For gene expression comparisons, genes with significant expression changes as determined by Cuffdiff data analysis were included. RNA-seq data is available at the GEO database (National Center for Biotechnology Information, Bethesda, MD, USA) under accession number GSE126898.
ChIP-seq
ChIP-seq was accomplished with AP-2γ antibody (Santa Cruz, Cat#sc-12762) as previously described (7). ChIP-seq data is available in the GEO database (National Center for Biotechnology Information, Bethesda, MD, USA) under accession number GSE126898.
Cell Viability Assay
Cells were plated on 96 well plates in technical quadruplicates. Ninety-six hours after siRNA transfection, cells were incubated with MTT (Thermo Fisher Scientific, Cat#M6494)) according to manufacturer’s recommended. Measurements were obtained on an Infinity 200 Pro (Tecan: Switzerland) plate reader at an absorbance of 570 nm. For cell counting, cells were plated on 96 well plates in technical quintuplicate. Ninety-six hours after siRNA transfection, cells were dyed with trypan blue and counted using a hemacytometer.
Invasion Assay
Invasion assay was performed using 24-well Transwell polycarbonate membranes (8.0-μm pores; Corning Inc., Corning, NY, USA, Cat#353097), coated with 100μl Matrigel for 30 min. at 37° C, washed with 500 μl PBS, and 1 × 106 cells plated in serum-free media 96 hours after siRNA transfection. Cells were loaded into an invasion chamber with Growth Factor Reduced Matrigel (Corning Cat#354230) diluted 1:10 in appropriate medium. After the 24-hour incubation, migrated cells were fixed and stained with 0.5% crystal violet for 20 minutes. The cells that had penetrated through the membrane were quantified under a microscope at ×40 magnification.
Regression Analysis and Model
Cox-regression analysis was performed on expression values for 41-gene, 16-gene and 3-gene panels from the Yau (19) dataset. Expression values were obtained from the Xena cancer browser (20). Cox-regression was performed using the “survival” package (version 2.38) in R (21). The Cox coefficients were used to determine “recurrence scores” (RS) as described by Paik (22). Using the mean RS to separate patients into “low RS” and “high RS”, we generated Kaplan-Meier (KM) survival curves to evaluate if a combination of 41-gene, 16-gene, 3-gene panels or “significant” (p-value ≤ 0.05) genes from the Cox-regression conferred similar hazard or protection. The “survminer” package (version 0.4.3) in R was used for KM plots (23).
Statistical Analysis
Parametric data analysis and graphs were completed using Student’s t-test with GraphPad Prism 8.
RESULTS
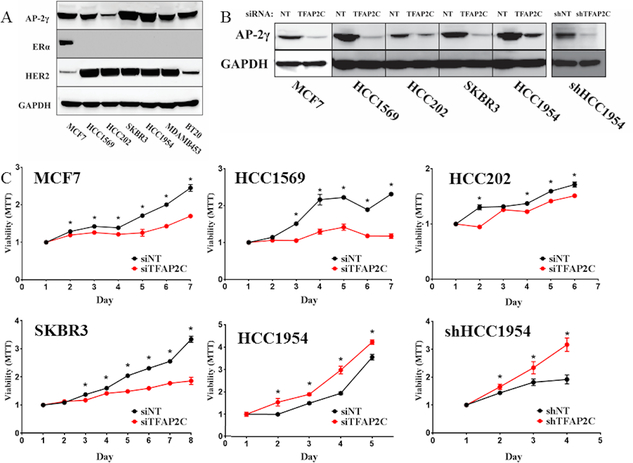
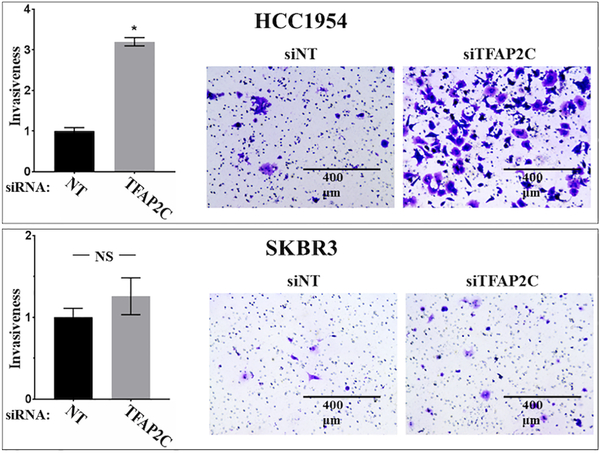
As a first step to examining the role of AP-2γ in HER2+ breast cancer, a panel of breast cancer cell lines was screened for the expression of AP-2γ with focus on HER2+ breast cell lines (Figure 1A). HCC1569, HCC202, SKBR3, HCC1954, and MDA-MB-453 were all confirmed to be ERα-negative with amplified expression of HER2. All of these cell lines expressed AP-2γ protein, though HCC202 had a significantly reduced level of expression. TFAP2C was knocked down in a panel of breast cancer cell lines using siRNA compared to a non-targeting (NT) siRNA (Figure 1B) and was found to repress cell growth in MCF-7, HCC202, HCC1569, and SKBR3 (Figure 1C). Although the effect on proliferation was significant in HCC202, the decrease in proliferation was modest, possibly due to the relatively low expression of AP-2γ. The findings in SKBR3 were consistent with previously published data (18); however, knockdown of TFAP2C increased proliferation in HCC1954, consistent with previously published data that knockdown of TFAP2C in HCC1954 failed to repress proliferation (18). To confirm this finding, stable knockdown of TFAP2C was performed in HCC1954 using a lentiviral delivered shRNA, which significantly reduced expression of AP-2γ (Figure 1B). HCC1954 cells with stable knockdown of TFAP2C (shHCC1954) were confirmed to have a significantly increased proliferative rate compared to cells with a non-targeting shRNA (Figure 1C). Previous studies in lung cancer reported that knockdown of TFAP2C enhanced cancer cell migration and invasion (24). Whereas knockdown of TFAP2C increased invasiveness in HCC1954, invasiveness was not significantly altered in SKBR3 (Figure 2). These findings demonstrate that AP-2γ has differential effects on cell growth and invasiveness in HCC1954 and SKBR3 cell lines.
Figure 1. Proliferative Response with Knockdown of TFAP2C.
A. A panel of breast cancer cell lines was screened for expression of AP-2γ, ERα and HER2. B. Knockdown of TFAP2C with siRNA and stable knockdown of TFAP2C with shRNA in HCC1954 cells. C. Proliferative response to knockdown of TFAP2C using MTT assay in cell lines indicated.
Figure 2. Invasion Assay in HCC1954 and SKBR3 with TFAP2C Knockdown.
Graph showing relative cell invasiveness after knockdown of TFAP2C compared to non-targeting (NT) siRNA in HCC1954 (top) and SKBR3 (bottom). Panels to right show examples of cell invasion assay with knockdown of NT and TFAP2C.
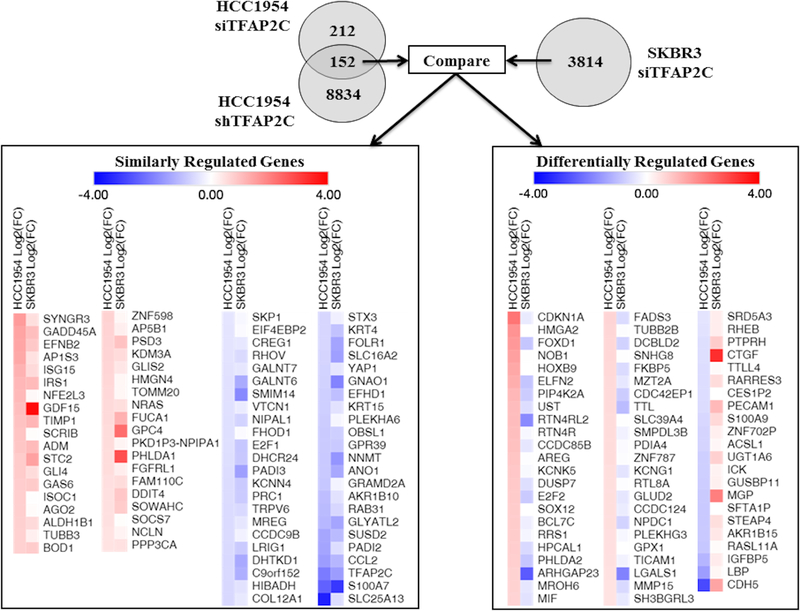
To identify the key AP-2γ target genes mediating cell growth and invasiveness, RNA-seq was used to characterize significant changes in gene expression with knockdown of TFAP2C. By RNA-seq analysis, knockdown of TFAP2C in HCC1954 with siRNA (compared to NT siRNA) identified 364 genes with significantly altered expression (Figure 3). To further create specificity for AP-2γ-regulated genes and reduce possible “off-target” effects, RNA-seq analysis was performed comparing expression in shHCC1954 with shTFAP2C vs. shNT; this analysis identified 8,986 genes with significantly altered expression. A comparison of the two data sets yielded 152 AP-2γ target genes that were consistently altered with knockdown of TFAP2C by siRNA and shRNA in HCC1954 cells. Because HCC1954 demonstrated opposite growth regulation and invasiveness with knockdown of TFAP2C compared to SKBR3, we hypothesized that the AP-2γ target genes responsible for growth and invasion would be differentially regulated with knockdown of TFAP2C in HCC1954 versus SKBR3. Hence, RNA-seq analysis was performed in SKBR3 after knockdown of TFAP2C with siRNA; in this analysis, a total of 3814 genes were significantly altered. The pattern of expression for the 152 AP-2γ target genes identified in HCC1954 was subsequently compared to expression changes in SKBR3. Of note, only 79 of the 152 TFAP2C target genes in HCC1954 were found to change expression significantly in the RNA-seq data set from SKBR-3. However, important physiologic differences could be due to genes that display significant changes in HCC1954 only. When examining the pattern of expression for all 152 AP-2γ target genes in HCC1954 and SKBR3, 84 genes were noted to change in the same direction (Similarly Regulated Genes, left panel of Figure 3). Within the set of similarly regulated genes, 38 genes increased and 46 genes decreased with knockdown of TFAP2C. Particularly reassuring was the finding that expression of the TFAP2C gene demonstrated the third most significant decrease in expression in both cell lines. Differential regulation was found for 68 genes; 46 genes had increased expression in HCC1954 and decreased in SKBR3, whereas, 22 genes had decreased expression in HCC1954 and increased in SKBR3.
Figure 3. Summary of RNA-seq Data with Knockdown of TFAP2C.
Significant changes in gene expression were first compared in HCC1954 with siRNA or shRNA stable knockdown. The 152 genes with significant and consistent changes were compared to knockdown of TFAP2C in SKBR3 cells with siRNA. Similarly regulated genes (on left) and differentially regulated genes (on right) are summarized with intensity of color indicating direction and magnitude of gene expression changes.
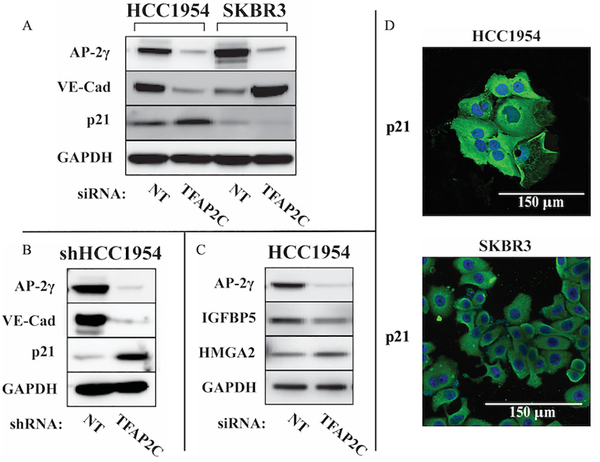
More detailed analysis was focused on the 68 AP-2γ target genes that changed in different directions (Differentially Regulated Genes, right panel, Figure 3) in HCC1954 compared to SKBR3 with knockdown of TFAP2C. Selected Western blots were performed to demonstrate changes in protein levels that confirmed the RNA-seq data (Figure 4 A–C). Within the data set for HCC1954, CDKN1A had the greatest increase in expression and CDH5 had the greatest decrease. Western blots confirmed that vascular endothelial cadherin (VE-cadherin) (encoded by the CDH5 gene) decreased in HCC1954 and increased in SKBR3, whereas, p21 (encoded by the CDKN1A gene) increased in HCC1954 and decreased in SKBR3 with knockdown of TFAP2C. Identical changes in VE-cadherin and p21 were found in shHCC1954 cells with TFAP2C knockdown compared to shNT (Figure 4B). Additional Western blots for IGFBP5 and HMGA2 with knockdown of TFAP2C in HCC1954 cells further confirmed the validity of the RNA-seq results (Figure 4C); these proteins were not consistently identified in protein extracts from SKBR3 (data not shown). Immunofluorescence staining with anti-p21 confirmed cytoplasmic localization (Figure 4D), which is consistent with previous findings in HER2-positive cell lines (25).
Figure 4. Expression of Proteins of Differentially Regulated AP-2γ-Responsive Genes.
A. Western blots for AP-2γ, VE-cadherin and p21 in HCC1954 and SKBR3 with knockdown of TFAP2C compared to NT. B. Western blots for protein expression in HCC1954 with stable knockdown comparing shRNA for NT vs. TFAP2C. C. Select Western blots of additional AP-2γ-responsive genes in HCC1954. D. Immunofluorescence staining for p21 in HCC1954 and SKBR3 demonstrates cytoplasmic localization.
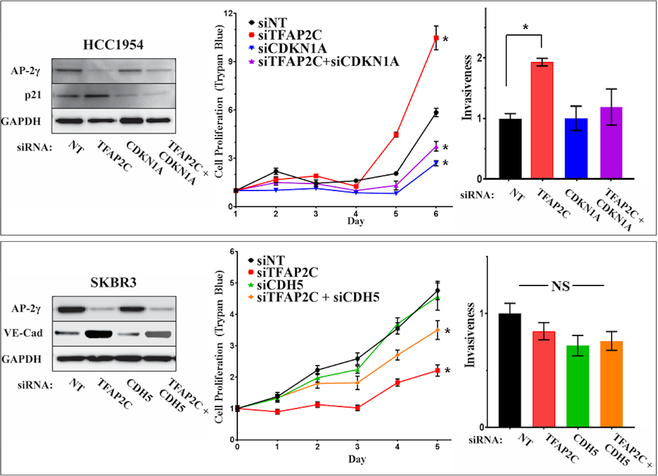
Further attempts were made to establish the role of specific AP-2γ target genes in regulating cell growth and invasion. Since CDKN1A was induced by knockdown of TFAP2C in HCC1954, TFAP2C was knocked down with or without co-knockdown of CDKN1A and proliferation was assessed by counting viable cells (Figure 5). Knockdown of CDKN1A reduced cell growth and partially reversed the growth stimulatory effects of TFAP2C knockdown. Assessment of proliferation in parallel experiments using MTT assay gave identical results (Figure S1A). Since knockdown of TFAP2C in HCC1954 was associated with increased cell invasiveness, the role of p21 in altering invasiveness was also assessed. Knockdown of CDKN1A alone did not alter invasiveness; however, knockdown of CDKN1A reversed the increase in invasiveness noted with knockdown of TFAP2C (Figure 5). In parallel experiments in SKBR3 cells, CDH5 was knocked down with and without co-knockdown of TFAP2C. Interestingly, knockdown of CDH5 alone had no significant effect on cell proliferation (Figure 5) and similar findings were demonstrated when proliferation was assessed by MTT assay (Figure S1B). However, knockdown of CDH5 partially rescued the reduction in cell proliferation induced by knockdown of TFAP2C. This finding suggests that part of the growth suppressive effects induced in SKBR3 through loss of AP-2γ is mediated by CDH5 up-regulation. Knockdown of TFAP2C with co-knockdown of CDH5 in SKBR-3 confirmed no significant effect on invasion, though there was a slight reduction in invasiveness with knockdown of CDH5 that failed to reach statistical significance (p=0.12) (Figure 5). These findings support the conclusion that regulation of CDH5 and CDKN1A contribute to alterations of proliferation and invasiveness induced by knockdown of TFAP2C.
Figure 5. Response of Proliferation and Invasiveness to CDKN1A and CDH5.
HCC1954 cells (top panels) with knockdown of TFAP2C, CDKN1A or both; western blots confirm knockdown of proteins (left panel); proliferation determined by counting viable cells (trypan blue) (middle panel); bar graph showing relative invasiveness in HCC1954 with knockdown of TFAP2C with or without knockdown of CDKN1A (right panel). Parallel experiments in SKBR3 cells (bottom panels) with knockdown of TFAP2C, CDH5 or both; western blots confirm knockdown (left panel); proliferation measured by viable cell counts (trypan blue) (middle panel); bar graph shows relative cell invasiveness (right panel).
Mechanisms of Gene Regulation by TFAP2C
Since AP-2γ is a transcription factor, many of the changes in gene expression identified are thought to occur through altered transcription of AP-2γ target genes. The findings, however, may be due to genes that are both primary gene targets and secondary gene targets regulated through intermediary factor(s). ChIP-seq was performed in SKBR3 and HCC1954 to define potential differences in AP-2γ genomic occupancy. Most of the similarly regulated genes have patterns of occupancy that indicate primary targeting by AP-2γ in both SKBR3 and HCC1954. For example, the regulatory regions for S100A7, CCL2, KRT15, and GDF15 demonstrate comparable patterns of AP-2γ occupancy (Figure S2). In evaluating the 68 differentially expressed genes, 31 (46%) had differential patterns of occupancy that could explain differences in regulation by TFAP2C knockdown. Examples of genes with differential occupancy of AP-2γ include E2F2, RTN4R, RASL11A and UST (Figure S3). These differences in patterns of occupancy could be due to variations in epigenetic chromatin structure at regulatory regions that block AP-2γ occupancy, as previously demonstrated for the GPX1 gene (26). On the other hand, 37 (54%) of the differentially regulated genes had patterns of AP-2γ occupancy that were comparable. For example, the regulatory regions of CDKN1A, CDH5, IGFBP5, and S100A9 all have similar patterns of AP-2γ occupancy in HCC1954 and SKBR3 (Figure S4). Although there were examples of slight differences in the comparative peak height in certain cases, the differences in patterns of occupancy were unlikely to account for differences in expression. Hence, differences in occupancy alone would not explain differential response to TFAP2C knockdown. To determine additional regulatory nodes that may be involved in the transcriptional pathways differentially regulated by AP-2γ in HER2+ cell lines, the differentially regulated AP-2γ target genes were analyzed using the Ingenuity Pathway Analysis software (Figures S5 & S6). Two regulatory nodes were identified as potentially contributing to differential expression of the AP-2γ target genes: p38/MAPK and retinoic acid (Figure S5A & S6).
Regulation by p38δ/MAPK13
p38/MAPKs are a group of mitogen-activated protein kinases that regulate responses to inflammation, oxidative stress and several oncogenic properties involving tumor progression including angiogenesis, invasion and metastasis. The p38/MAPK family is composed of four isoforms encoded by separate genes: p38α/MAPK14, p38β/MAPK11, p38γ/MAPK12, and p38δ/MAPK13 (27). To determine which isoform may be involved in differential response to AP-2γ, the expression of all four isoforms were examined in HCC1954 and SKBR3 with knockdown of TFAP2C (Figure S5B). Interestingly, the expression of MAPK13 increased in HCC1954 and decreased in SKBR3 with TFAP2C knockdown, whereas, the other p38/MAPKs demonstrated no significant change (Figure S5B).
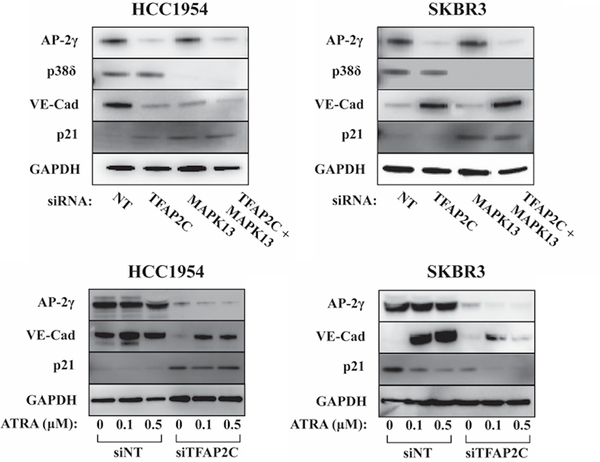
In HCC1954 knockdown of MAPK13 repressed expression of VE-cadherin and increased that of p21, and these effects were comparable to the effects of TFAP2C knockdown (Figure 6). These data suggest that effects on CDH5 and CDKN1A expression with TFAP2C knockdown may be mediated through repression of MAPK13 in HCC1954 cells. Conversely, knockdown of MAPK13 in SKBR3 cells failed to demonstrate any effect on VE-cadherin expression; similar to effects in HCC1954, knockdown of MAPK13 induced p21 expression in SKBR3, but this effect was opposite to knockdown of TFAP2C (Figure 6). Overall, the data support the conclusion that in HCC1954, p38δ represents a regulatory node that contributes to the regulation of CDH5 and CDKN1A with knockdown of TFAP2C. However, in SKBR3, knockdown of MAPK13 does not reproduce the effect of TFAP2C knockdown indicating that either p38δ/MAPK13 is not involved or p38δ activity is up-regulated by knockdown of TFAP2C. In either case, activity of p38δ appears to be a candidate regulatory node contributing to the differential expression of CDH5 and CDKN1A in HER2+ cell lines.
Figure 6. Expression of VE-cadherin and p21 in Response to MAPK13 and ATRA.
Western blots for AP-2γ, p38δ, VE-cadherin, p21 and GAPDH showing response in HCC1954 and SKBR3 cells with knockdown of TFAP2C with or without co-knockdown of MAPK13 (top panels). Western blots showing changes of expression of AP-2γ, VE-cadherin, p21 and GAPDH in HCC1954 and SKBR3 with knockdown using NT vs. TFAP2C siRNA with increasing concentration of ATRA at 0, 0.1 and 0.5 μM (bottom panels).
Regulation by Retinoic Acid/ATRA
To examine the potential role of retinoic acid signaling, the effect of all-trans retinoic acid (ATRA) was examined in the HER2+ cell lines with and without knockdown of TFAP2C (Figure 6). In SKBR3, ATRA induced VE-cadherin levels by 35-fold and repressed p21 expression, responses that were similar to the effects of TFAP2C knockdown, which significantly blunted the response to ATRA treatment. In HCC1954 ATRA had insignificant effects on VE-cadherin and p21 expression, though with TFAP2C knockdown, there was a modest induction of VE-cadherin from the lower baseline expression. Therefore, SKBR3 cells were more responsive to ATRA with significant changes in the expression of both VE-cadherin and p21 that mirrored the effects of TFAP2C knockdown. Retinoic acid response in HCC1954 was negligible in the presence of TFAP2C expression. The findings suggest that p38δ is an important regulatory node for the AP-2γ pathway in HCC1954 cells and that retinoic acid is a more important regulatory node in SKBR3 cells. Differences in the involvement of these two regulatory nodes may account for differential effects of TFAP2C knockdown.
Use of the Gene Signature to Predict Outcome in HER2 Breast Cancer
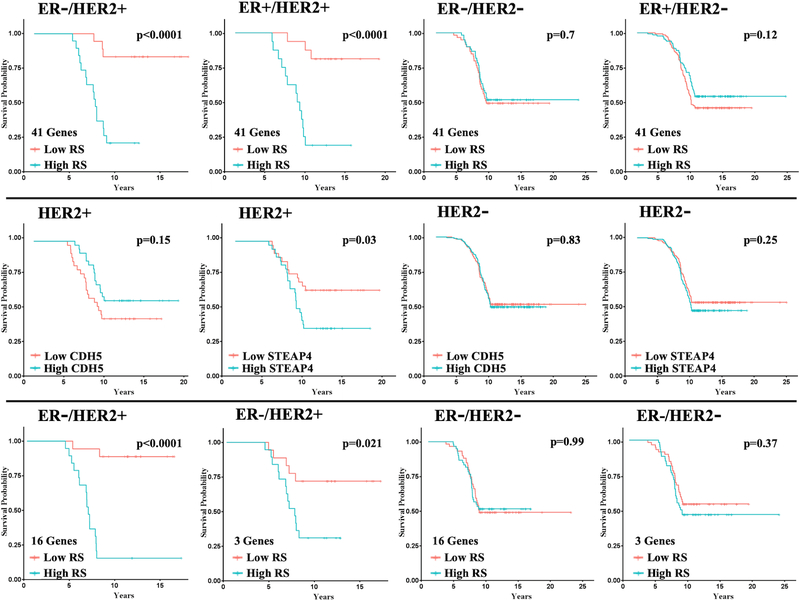
The set of genes differentially expressed with knockdown of TFAP2C comparing HCC1954 and SKBR3 are implicated in the growth and invasiveness of HER2+ breast cancer. The clinical importance of the gene signature based on the differentially expressed genes was analyzed by examining an association between the expression of the gene set and outcome in patients with breast cancer. Regression analysis was used to establish a correlation between expression of the differentially regulated genes and outcome using datasets with gene expression data linked to clinical outcome. The Yau et al. (19) dataset had data for 69 HER2+ breast cancer patients with distant metastasis-free survival (DMFS) that included expression data for 41 of the 68 genes in our gene signature. Using regression analysis, there were 10 genes that demonstrated hazard ratios (HRs) with statistical significance (Figure S7), with 7 genes associated with a low HR (ACSL1, CDH5, DCBLD2, FOXD1, KCNK5, MGP, RHEB) and three genes associated with a high HR (CDC42EP1, HOXB9, STEAP4). A model was developed to create a recurrence score (RS) using the expression of all 41 genes with high or low RS based on values above or below the median (Supplemental Data, Table 1). There was a striking difference in DMFS comparing high vs. low RS in ER-/HER2+ tumors (Figure 7). When the model was applied to other tumor subtypes, patients with ER+/HER2+ tumors also demonstrated a significant predictive capacity for high vs. low RS. However, when applied to HER2- tumor subtypes, the model failed to demonstrate significant differences for high vs. low RS (Figure 7). Although the regression model was developed based on the expression of the gene set, the expression of individual genes (e.g., CDH5 and STEAP4) was examined based on relative gene expression level above (high) or below (low) the median. As predicted by the model, high CDH5 expression was associated with better outcome, whereas, high STEAP4 expression was associated with worse DMFS in patients with HER2+ tumors (Figure 7). On the other hand, no differences in DMFS were found in patients with HER2- tumors (Figure 7).
Figure 7. Kaplan-Meier Plots for DMFS Based on Recurrence Score Model.
Top Row: Using the model with the 41-gene panel, Kaplan-Meier plots for distant metastasis-free survival (DMFS) for patients with each tumor subtypes as shown. For ER-/HER2+ n=37, ER+/HER2+ n=32, ER-/HER2- n=119, ER+/HER2- n=266.
Middle Row: Examples of Kaplan-Meier plots for DMFS for individual genes, CDH5 and STEAP4, for HER2+ and HER2- tumors (with any ER status).
Bottom Row: Kaplan-Meier plots for DMFS using the model with 16-gene and 3-gene panels for ER-/HER2+ and ER-/HER2- patients, as indicated.
In an attempt to further reduce the number of genes in the regression model, AP-2γ target genes were identified in a third HER2+ breast cancer line, HCC1569. As described above, siRNA was used to knockdown TFAP2C expression compared to NT siRNA and RNA-seq analysis was used to identify significant changes in gene expression. Genes with significant changes in expression with TFAP2C knockdown in HCC1569 were compared to the 68 differentially expressed TFAP2C target genes; 32 of the 68 AP-2γ target genes were found to be significantly altered in HCC1569. Of these 32 genes, 16 genes were contained within the 41 genes available in the Yau et al. dataset. A regression model was built based on these 16 AP-2γ target genes (Supplemental Data, Table S2). The RS based on the 16-gene model was highly predictive of DMFS in ER-/HER2+ patients (Figure 7, bottom row); however, there was no difference in DMFS in ER-/HER2- patients. Regression analysis indicated that three genes (ICK, KCNG1 and STEAP4) in the 16-gene panel were independently significant (Table S2). A model built using these three genes was also highly predictive of DMFS in ER-/HER2+ patients but not in ER-/HER2- patients. These findings provide additional evidence that the AP-2γ target genes identified in the HER2+ breast cancer model as likely involved in growth and invasion are predictive of outcome in patients with HER2+ breast cancer.
DISCUSSION
Several previous studies of breast cancer have developed gene signatures that are predictive of outcome and in some cases can predict response to chemotherapy (22,28). In addition, molecular profiling has been able to select patients that are less likely to benefit from adjuvant chemotherapy, thus avoiding unnecessary treatment (28,29). Many of the gene signature models developed have been focused on ERα-positive breast cancer. In some cases, gene signature models that perform well in ERα-positive/HER2-positive patients do not have predictive capacity in the ERα-negative/HER2-positive breast cancer subtype (30). The HER2+ breast cancer subtype has a poor prognosis relative to other intrinsic breast cancer subtypes (31). Hence, development of molecular profiles for the HER2+ breast cancer subtype will help to stratify patients more likely to benefit from additional therapy and will further define pathways that can be targeted by directed therapy. The current study is unique from several perspectives. Previous gene expression signatures were developed by using unsupervised hierarchical clustering algorithms to define similarities in patterns of expression to create associations with clinical phenotypes. The current model was developed by defining a gene signature based on a set of genes likely to be associated with growth and invasion in HER2+ breast cancer cell lines. Furthermore, other models in common use are more appropriate for ERα-positive tumors, whereas, the current model was developed specifically for application to the HER2+ breast cancer subtype. The associations between outcome in patients with the HER2+ breast cancer subtype and the gene expression signature described herein support the clinical relevance of the gene signature and the likelihood that the identified genes drive cancer cell growth and invasiveness.
Studies examining cell growth and invasiveness indicate that VE-cadherin/CDH5 and p21/CDKN1A influence the growth and progression of HER2+ breast cancer. Within the normal mammary gland, CDH5 is expressed in the basal cell lineage within the quiescent mammary stem cell population that demonstrate multipotency and the capacity to develop mammary ducts (32). Studies of the role of VE-cadherin in cancer have resulted in conflicting findings suggesting that VE-cadherin may have opposing effects depending upon the cancer type. In melanoma VE-cadherin is overexpressed in aggressive subtypes and is involved in angiogenesis and formation of vascular-like networks mimicking embryonic vascular networks (33,34). Epithelial-mesenchymal transition (EMT) is associated with “cadherin switching” characterized by down-regulation of E-cadherin/CDH1 and up-regulation of both N-cadherin/CDH2 and VE-cadherin/CDH5 (33,35). High expression of VE-cadherin/CDH5 was reported to be associated with recurrence and the development of metastatic disease in breast cancer (36) and gastric cancer (37). However, knockdown of CDH5 in MCF-7 luminal breast cancer cells increased cell invasiveness suggesting that VE-cadherin suppresses metastatic potential (38). Our data supports the finding that VE-cadherin/CDH5 reduces cell growth and invasiveness in the HER2+ breast cancer subtype.
P21 is a cyclin-dependent kinase inhibitor and induction of p21 normally leads to cell cycle arrest and inhibition of proliferation (39). However, p21 can also inhibit apoptosis and, within certain contexts, p21 can paradoxically promote cell proliferation and tumor growth (40,41). The paradoxical effects of p21 promoting cell proliferation, tumorigenicity and invasiveness has been attributed to either its subcellular localization or deficiency of p53 (42,43). Our findings that knockdown of CDKN1A repressed cell growth in certain HER2+ breast cancer cells were corroborated by published experiments in AU565 cells (44). The potential role of p21 inducing cell proliferation and invasiveness in breast cancer is further supported by clinical studies showing that high p21 expression is associated with high tumor grade and advanced stage in breast cancer patients (45). Also consistent with our findings, the worst 5-year survival rate was associated with high HER2 expression and cytoplasmic p21 localization (46).
The differential effects of AP-2γ in HCC1954 and SKBR3 were mirrored by a divergence in response to key regulatory pathways. In HCC1954 cells, knockdown of TFAP2C and MAPK13 induced identical changes in the expression of CDH5 and CDKN1A. Emerging data indicates an important role for MAPK13/ p38δ in several physiologic processes and oncogenesis. Studies in different cancer models have uncovered both oncogenic and tumor suppressor functions of p38δ dependent on the cell type. In breast cancer, overexpression of p38δ is associated with a worse prognosis, and knockdown of MAPK13 in MCF-7 and MDA-MB-231 breast cancer cells reduced proliferation and decreased cell detachment (47). High expression of MAPK13/p38δ was identified in a variety of cancer cell types, including gynecological cancers where expression of p38δ was required to maintain the cancer stem cell (CSC)/tumor initiating cell (TIC) population (48). In contrast, the opposite findings were described in non-small cell lung cancer where p38δ repressed cancer stem cell markers and inhibited the tumor cell initiating ability (49). In the current study, knockdown of MAPK13 in HCC1954 cells induced alterations in CDH5 and CDKN1A expression that mirrored changes with knockdown of TFAP2C, supporting a tumor inhibitory role in this cell line.
The retinoic acid pathway was identified as an important regulatory node that demonstrated differential activation in HER2+ breast cancer cell lines. In SKBR-3 cells, ATRA induced CDH5 and repressed CDKN1A, which mirrored effects of TFAP2C knockdown. Furthermore, the effects of ATRA were significantly dependent upon AP-2γ, since knockdown of TFAP2C blunted the effects of ATRA. The current findings are in agreement with previous studies reporting that ATRA reduced proliferation and invasiveness of SKBR3 and MCF-7 cells (50,51). In SKBR3 cells, ATRA activates a mammary epithelial differentiation program that decreased migration of cells with increased formation of adherens junctions involving up-regulation of CDH5; whereas, HCC1954 cells are relatively resistant to the action of ATRA (52). The current findings demonstrate that response to ATRA in SKBR3 cells is dependent upon AP-2γ, consistent with earlier findings that AP-2γ regulates genes in the retinoic acid response pathway in MCF-7 cells (7).
In conclusion, TFAP2C knockdown in the HER2+ breast cancer subtype demonstrated cell-specific differences that were characterized by a set of differentially regulated AP-2γ target genes, which are involved in cell proliferation and invasion. Retinoic acid and MAPK13/ p38δ represent two regulatory nodes that demonstrated differential responses that mirror knockdown of TFAP2C. These findings suggest that HER2+ breast cancers may be characterized by their sensitivity to retinoic acid and MAPK inhibitors, which may offer new therapeutic options for different subsets of patients with HER2+ breast cancer. Further work will be needed to define patterns of expression that might predict response to either retinoic acid or MAPK inhibitor.
Supplementary Material
Implications.
A set of genes regulated by AP-2γ in HER2+ breast cancer that drive proliferation and invasion were identified and provided a gene signature that is predictive of outcome in HER2+ breast cancer.
ACKNOWLEDGEMENTS
This work was supported by the NIH grants R01CA183702 (PI: R.J.W.), T32CA148062 (P.I.: R.J.W.), P30CA08686218S6, (PI: G.J.W.) and by a generous gift from the Kristen Olewine Milke Breast Cancer Research Fund. V.T.W., B.K., K.K. and A.C.B. were supported by the NIH grant T32CA148062.
Footnotes
Conflict of Interest: The authors state no conflicts of interest.
REFERENCES
- 1.Weigel RJ, deConinck EC. Transcriptional control of estrogen receptor in estrogen receptor-negative breast carcinoma. Cancer research 1993;53(15):3472–4. [PubMed] [Google Scholar]
- 2.deConinck EC, McPherson LA, Weigel RJ. Transcriptional regulation of estrogen receptor in breast carcinomas. Mol Cell Biol 1995;15(4):2191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McPherson LA, Baichwal VR, Weigel RJ. Identification of ERF-1 as a member of the AP2 transcription factor family. Proceedings of the National Academy of Sciences of the United States of America 1997;94(9):4342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodfield GW, Horan AD, Chen Y, Weigel RJ. TFAP2C controls hormone response in breast cancer cells through multiple pathways of estrogen signaling. Cancer research 2007;67(18):8439–43. [DOI] [PubMed] [Google Scholar]
- 5.Kang HJ, Lee MH, Kang HL, Kim SH, Ahn JR, Na H, et al. Differential Regulation of Estrogen Receptor alpha Expression in Breast Cancer Cells by Metastasis-Associated Protein 1. Cancer research 2014. doi 10.1158/0008-5472.CAN-13-2020. [DOI] [PubMed] [Google Scholar]
- 6.Bogachek MV, Chen Y, Kulak MV, Woodfield GW, Cyr AR, Park JM, et al. Sumoylation pathway is required to maintain the Basal breast cancer subtype. Cancer cell 2014;25(6):748–61 doi 10.1016/j.ccr.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodfield GW, Chen Y, Bair TB, Domann FE, Weigel RJ. Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes, chromosomes & cancer 2010;49(10):948–62 doi 10.1002/gcc.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, et al. AP-2gamma regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. The EMBO journal 2011;30(13):2569–81 doi 10.1038/emboj.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gee JM, Eloranta JJ, Ibbitt JC, Robertson JF, Ellis IO, Williams T, et al. Overexpression of TFAP2C in invasive breast cancer correlates with a poorer response to anti-hormone therapy and reduced patient survival. J Pathol 2009;217(1):32–41. [DOI] [PubMed] [Google Scholar]
- 10.Perkins SM, Bales C, Vladislav T, Althouse S, Miller KD, Sandusky G, et al. TFAP2C expression in breast cancer: correlation with overall survival beyond 10 years of initial diagnosis. Breast Cancer Res Treat 2015;152(3):519–31 doi 10.1007/s10549-015-3492-2. [DOI] [PubMed] [Google Scholar]
- 11.Cyr AR, Kulak MV, Park JM, Bogachek MV, Spanheimer PM, Woodfield GW, et al. TFAP2C governs the luminal epithelial phenotype in mammary development and carcinogenesis. Oncogene 2015;34(4):436–44 doi 10.1038/onc.2013.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosher JM, Williams T, Hurst HC. The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proceedings of the National Academy of Sciences of the United States of America 1995;92(3):744–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delacroix L, Begon D, Chatel G, Jackers P, Winkler R. Distal ERBB2 promoter fragment displays specific transcriptional and nuclear binding activities in ERBB2 overexpressing breast cancer cells. DNA Cell Biol 2005;24(9):582–94 doi 10.1089/dna.2005.24.582. [DOI] [PubMed] [Google Scholar]
- 14.Ailan H, Xiangwen X, Daolong R, Lu G, Xiaofeng D, Xi Q, et al. Identification of target genes of transcription factor activator protein 2 gamma in breast cancer cells. BMC cancer 2009;9:279 doi 1471-2407-9-279 [pii] 10.1186/1471-2407-9-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nolens G, Pignon JC, Koopmansch B, Elmoualij B, Zorzi W, De Pauw E, et al. Ku proteins interact with activator protein-2 transcription factors and contribute to ERBB2 overexpression in breast cancer cell lines. Breast Cancer Res 2009;11(6):R83 doi 10.1186/bcr2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Kulak MV, Borcherding N, Maina PK, Zhang W, Weigel RJ, et al. A novel HER2 gene body enhancer contributes to HER2 expression. Oncogene 2018;37(5):687–94 doi 10.1038/onc.2017.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol 2011;5(1):5–23 doi 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiu KK, Wetterskog D, Mackay A, Natrajan R, Lambros M, Sims D, et al. Integrative molecular and functional profiling of ERBB2-amplified breast cancers identifies new genetic dependencies. Oncogene 2014;33(5):619–31 doi 10.1038/onc.2012.625. [DOI] [PubMed] [Google Scholar]
- 19.Yau C, Esserman L, Moore DH, Waldman F, Sninsky J, Benz CC. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res 2010;12(5):R85 doi 10.1186/bcr2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klonowska K, Czubak K, Wojciechowska M, Handschuh L, Zmienko A, Figlerowicz M, et al. Oncogenomic portals for the visualization and analysis of genome-wide cancer data. Oncotarget 2016;7(1):176–92 doi 10.18632/oncotarget.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Therneau TM. A Package for Survival Analysis in S. version 2.38. 2015. [Google Scholar]
- 22.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351(27):2817–26. Epub 004 Dec 10. [DOI] [PubMed] [Google Scholar]
- 23.Kassambara A, Kosinski M. Survminer: Drawing Survival Curves using ggplot2. 2018. [Google Scholar]
- 24.Chang TH, Tsai MF, Gow CH, Wu SG, Liu YN, Chang YL, et al. Upregulation of microRNA-137 expression by Slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer letters 2017;402:190–202 doi 10.1016/j.canlet.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nature cell biology 2001;3(3):245–52 doi 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 26.Kulak MV, Cyr AR, Woodfield GW, Bogachek M, Spanheimer PM, Li T, et al. Transcriptional regulation of the GPX1 gene by TFAP2C and aberrant CpG methylation in human breast cancer. Oncogene 2013;32(34):4043–51 doi 10.1038/onc.2012.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuenda A, Sanz-Ezquerro JJ. p38gamma and p38delta: From Spectators to Key Physiological Players. Trends in biochemical sciences 2017;42(6):431–42 doi 10.1016/j.tibs.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med 2016;375(8):717–29 doi 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 29.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N Engl J Med 2015;373(21):2005–14 doi 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Risi E, Grilli A, Migliaccio I, Biagioni C, McCartney A, Guarducci C, et al. A gene expression signature of Retinoblastoma loss-of-function predicts resistance to neoadjuvant chemotherapy in ER-positive/HER2-positive breast cancer patients. Breast Cancer Res Treat 2018;170(2):329–41 doi 10.1007/s10549-018-4766-2. [DOI] [PubMed] [Google Scholar]
- 31.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America 2001;98(19):10869–74 doi 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H, Miao Z, Zhang X, Chan UI, Su SM, Guo S, et al. Single-cell RNA-Seq reveals cell heterogeneity and hierarchy within mouse mammary epithelia. J Biol Chem 2018;293(22):8315–29 doi 10.1074/jbc.RA118.002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol 2009;1(6):a003129 doi 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendrix MJ, Seftor EA, Meltzer PS, Gardner LM, Hess AR, Kirschmann DA, et al. Expression and functional significance of VE-cadherin in aggressive human melanoma cells: role in vasculogenic mimicry. Proceedings of the National Academy of Sciences of the United States of America 2001;98(14):8018–23 doi 10.1073/pnas.131209798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labelle M, Schnittler HJ, Aust DE, Friedrich K, Baretton G, Vestweber D, et al. Vascular endothelial cadherin promotes breast cancer progression via transforming growth factor beta signaling. Cancer research 2008;68(5):1388–97 doi 10.1158/0008-5472.CAN-07-2706. [DOI] [PubMed] [Google Scholar]
- 36.Fry SA, Robertson CE, Swann R, Dwek MV. Cadherin-5: a biomarker for metastatic breast cancer with optimum efficacy in oestrogen receptor-positive breast cancers with vascular invasion. British journal of cancer 2016;114(9):1019–26 doi 10.1038/bjc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inokuchi M, Higuchi K, Takagi Y, Tanioka T, Nakagawa M, Gokita K, et al. Cadherin 5 Is a Significant Risk Factor for Hematogenous Recurrence and a Prognostic Factor in Locally Advanced Gastric Cancer. Anticancer research 2017;37(12):6807–13 doi 10.21873/anticanres.12141. [DOI] [PubMed] [Google Scholar]
- 38.Zheng L, Zhang Z, Zhang S, Guo Q, Zhang F, Gao L, et al. RNA Binding Protein RNPC1 Inhibits Breast Cancer Cell Metastasis via Activating STARD13-Correlated ceRNA Network. Mol Pharm 2018;15(6):2123–32 doi 10.1021/acs.molpharmaceut.7b01123. [DOI] [PubMed] [Google Scholar]
- 39.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature 1993;366(6456):701–4 doi 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 40.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nature reviews Cancer 2009;9(6):400–14 doi 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer research 2005;65(10):3980–5 doi 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 42.Zohny SF, Al-Malki AL, Zamzami MA, Choudhry H. p21(Waf1/Cip1): its paradoxical effect in the regulation of breast cancer. Breast cancer 2018. doi 10.1007/s12282-018-0913-1. [DOI] [PubMed] [Google Scholar]
- 43.Georgakilas AG, Martin OA, Bonner WM. p21: A Two-Faced Genome Guardian. Trends Mol Med 2017;23(4):310–9 doi 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Chiang CT, Way TD, Lin JK. Sensitizing HER2-overexpressing cancer cells to luteolin-induced apoptosis through suppressing p21(WAF1/CIP1) expression with rapamycin. Mol Cancer Ther 2007;6(7):2127–38 doi 10.1158/1535-7163.MCT-07-0107. [DOI] [PubMed] [Google Scholar]
- 45.Zohny SF, Baothman OA, El-Shinawi M, Al-Malki AL, Zamzami MA, Choudhry H. The KIP/CIP family members p21^{Waf1/Cip1} and p57^{Kip2} as diagnostic markers for breast cancer. Cancer Biomark 2017;18(4):413–23 doi 10.3233/CBM-160308. [DOI] [PubMed] [Google Scholar]
- 46.Xia W, Chen JS, Zhou X, Sun PR, Lee DF, Liao Y, et al. Phosphorylation/cytoplasmic localization of p21Cip1/WAF1 is associated with HER2/neu overexpression and provides a novel combination predictor for poor prognosis in breast cancer patients. Clin Cancer Res 2004;10(11):3815–24 doi 10.1158/1078-0432.CCR-03-0527. [DOI] [PubMed] [Google Scholar]
- 47.Wada M, Canals D, Adada M, Coant N, Salama MF, Helke KL, et al. P38 delta MAPK promotes breast cancer progression and lung metastasis by enhancing cell proliferation and cell detachment. Oncogene 2017;36(47):6649–57 doi 10.1038/onc.2017.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasuda K, Hirohashi Y, Kuroda T, Takaya A, Kubo T, Kanaseki T, et al. MAPK13 is preferentially expressed in gynecological cancer stem cells and has a role in the tumor-initiation. Biochem Biophys Res Commun 2016;472(4):643–7 doi 10.1016/j.bbrc.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 49.Fang Y, Wang J, Wang G, Zhou C, Wang P, Zhao S, et al. Inactivation of p38 MAPK contributes to stem cell-like properties of non-small cell lung cancer. Oncotarget 2017;8(16):26702–17 doi 10.18632/oncotarget.15804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanderhoeven F, Redondo AL, Martinez AL, Vargas-Roig LM, Sanchez AM, Flamini MI. Synergistic antitumor activity by combining trastuzumab with retinoic acid in HER2 positive human breast cancer cells. Oncotarget 2018;9(41):26527–42 doi 10.18632/oncotarget.25480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin G, Zhu S, Wu Y, Song C, Wang W, Zhang Y, et al. omega-3 free fatty acids and all-trans retinoic acid synergistically induce growth inhibition of three subtypes of breast cancer cell lines. Sci Rep 2017;7(1):2929 doi 10.1038/s41598-017-03231-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Zanetti A, Affatato R, Centritto F, Fratelli M, Kurosaki M, Barzago MM, et al. All-trans-retinoic Acid Modulates the Plasticity and Inhibits the Motility of Breast Cancer Cells: ROLE OF NOTCH1 AND TRANSFORMING GROWTH FACTOR (TGFbeta). J Biol Chem 2015;290(29):17690–709 doi 10.1074/jbc.M115.638510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.