Abstract
Centrosome amplification (CA), or a numerical increase in centrosomes, is common in human cancers, particularly those with high-risk features. We have discovered that cells with CA have an increased burden of autophagy, a catabolic process whereby autophagosomes engulf damaged organelles and proteins and deliver these contents to the lysosome for degradation and subsequent recycling. Cells with CA demonstrate an accumulation of autophagosomes. We evaluated the alternative hypotheses that CA alters autophagy by modulating microtubule networks and impairing trafficking versus altering lysosome clustering and organization versus chromosome missegregation-induced proteotoxic stress. Using LC3 reporter assays and autophagosome tracking experiments, we demonstrate that CA causes an accumulation of autophagosomes by interfering with autophagosome trafficking. To establish if this was a druggable weakness, we tested autophagy inhibitors in our cell models of CA. Cells with CA are sensitized to chemical and genetic autophagy inhibition. Taken together, our results suggest that autophagy is disrupted by CA and sensitizes cells to inhibition of autophagy. These findings suggest a novel precision medicine strategy, whereby CA increases reliance on autophagy and serves as a biomarker for autophagy inhibitors in high-risk cancers.
Keywords: centrosome, centrosome amplification, PLK4, autophagy, chloroquine
INTRODUCTION
Autophagy, or macroautophagy, is a catabolic process whereby autophagosomes engulf damaged organelles and protein aggregates and deliver these contents to the lysosome for degradation (1,2). Autophagy is activated under various cellular stress conditions such as starvation and oxidative stress. In cancer, autophagy has been reported to be both a tumor suppressor and promoter; the prevailing view is that autophagy is a tumor suppressive pathway and inhibits tumor initiation, but once the tumor has developed, autophagy provides nutrients for the growing tumor and is pro-tumorigenic (3). As such, there are a number of ongoing clinical trials evaluating autophagy inhibitors, such as hydroxychloroquine, in advanced and drug-resistant cancers.
Autophagy is a complex, multi-step process during which the following major events happen in sequence: (1) sequestering of cytoplasmic contents by an isolation membrane or phagophore; (2) closure of the isolation membrane to form a double-membrane autophagosome; (3) fusion of the autophagosome with the lysosome to create the autolysosome; and (4) degradation of autophagic cargo by lysosomal enzymes (4–6). The cytoskeleton, particularly the microtubule cytoskeleton, is crucial for autophagy (7). Movement of endolysosomal organelles requires coupling to microtubule motors and actin, with long-range transport mostly dependent on microtubule motors (e.g. dynein, kinesins) and short-range transport mostly dependent on actin and myosin (8). Further, autophagosome movements depend on intact microtubules to bring them into the proximity of lysosomes, which cluster near the centrosome, the major microtubule organizing center in animal cells (6). When microtubules are disrupted with depolymerizing drugs such as nocodazole and vinblastine, autophagosomes accumulate, suggesting impaired trafficking (9–13). These observations suggest that centrosomes may play a key role in regulating autophagy.
Centrosome amplification (CA), or a numerical increase in centrosomes, is reported in virtually all human cancer types (14–16). CA promotes error-prone mitoses with multipolar spindles and merotelic microtubule-kinetochore attachments (17), high-grade phenotypes (15), invasiveness (18,19), and ultimately worse clinical outcomes (15). Given that CA is very specific to cancer cells, it represents a potential therapeutic target for cancer therapy. Here, we report that CA disrupts autophagy by interfering with autophagosome trafficking. Critically, cells with CA have enhanced sensitivity to inhibitors of autophagy. This suggests that CA may be a potential biomarker for clinical development of autophagy inhibitors in cancer.
MATERIALS AND METHODS
Cell culture
All cell lines were propagated at 37 °C in 5% CO2. RPE1 cells were grown in a 1:1 mixture of DMEM and Ham’s F-12 medium supplemented with 2.5 mm l-glutamine, with 10% fetal bovine serum and 100 units/ml penicillin-streptomycin. MCF10A (non-transformed breast line) and two MCF10A-derivative cell lines, DCIS and Ca1d (20), were grown in 1:1 DMEM and Ham’s F-12 supplemented with 2.5 mm l-glutamine, horse serum, EGF, hydrocortisone, cholera toxin, and 100 units/mL penicillin-streptomycin. Doxycycline-inducible PLK4 cell lines (RPE1 and MCF10A) (18) were used to model centrosome amplification after doxycycline at 2 μg/mL. HeLa and Phoenix cells were cultured in DMEM high glucose medium with 10% fetal bovine serum and 100 units/ml penicillin-streptomycin. DLD1 cells were cultured in RPMI 1640 medium with 10% fetal bovine serum and 100 units/ml penicillin-streptomycin. Tetraploid DLD1 cells were generated as previously described (21). Briefly DLD1 cells were treated with blebbistatin to induce cytokinesis failure, subcloning by limiting dilution, and screening and validating clones using propidium iodide staining and flow cytometric detection.
RPE1 and MCF10A cell lines were obtained from ATCC in 2005 and validated by karyotype in 2012. DCIS and Cald cells were provided by Dr. Patricia Keely (University of Wisconsin). Doxycycline-inducible RPE1 and MCF10A cells were obtained from Dr. David Pellman (Harvard) and validated by assessing centrioles before/after addition of doxycycline. HeLa and Phoenix packaging cells were obtained from ATCC in 2005. DLD1 cells were obtained from ATCC in 2016. Mycoplasma testing was performed on all cell lines using R&D Systems Mycoprobe Mycoplasma Detection Kit with help from the UW Small Molecule Screening and Synthesis Facility.
The LC3B autophagic flux reporter assay used pBABE-puro-mCherry-EGFP-LC3B, a gift from Jay Debnath (Addgene plasmid # 22418)(22). Retrovirus was generated by transfecting a T25 of Phoenix cells with 2 μg of the retroviral vector and 1 μg pVSV-G pantropic envelope vector using Fugene transfection reagent (Promega). Phoenix cells were given fresh media 24 hours after transfection, and this supernatant was collected 48 hours after transfection and used to transduce RPE and MCF10A cells. Polybrene was added to a concentration of 10 μg/mL immediately prior to infecting target cells. Cells were selected with puromycin for 5 days, and monoclonal cell lines were isolated and propagated.
ATG5 depletion was performed using shRNAs generated from pLKO.1-TRC cloning vector, a gift from David Root (Addgene plasmid #10878). The following sequences were used: #1 (5’-CCAGATATTCTGGAATGGAAA-3’), #2 (5’-CCTTTCATTCAGAAGCTGTTT-3’). Non-hairpin insert pLKO.1-TRC control vector, a gift from David Root (Addgene plasmid #10879), was used as a control. Lentivirus was generated by transfecting a T25 of 293T cells with 2 μg pLKO.1 lentiviral vector, 1 μg pVSV-G pantropic envelope vector, and 1 μg psPAX2 packaging vector using Fugene (Promega). Cells were given fresh media 24 hours after transfection, and supernatant containing lentivirus was collected 48 hours after transfection and added to RPE target cells with 10 μg/mL polybrene. Cells were given fresh media the next day, selected with puromycin for 5 days, then allowed to recover in fresh media before using in experiments. ATG5 gene expression was assessed by qRT-PCR.
Cellular senescence was assayed using a pH-dependent β-galactosidase staining kit (Cell Signaling Technology) according to the manufacturer’s instructions.
Chemicals used in this study include chloroquine (Fisher, ICN19391910, 100 μm), aphidicolin (Fisher, BP615, 5 μM), doxycycline (Fisher, BP26531, 2 μg/mL), nocodazole (Sigma, M1404, 0.2 μg/ml), 3-MB-PP1 (Toronto Research Chemicals, 10 μM), bafilomycin A1 (Fisher, NC9686929, 100nM), puromycin (Invivogen, 10 μg/mL), polybrene (Millipore, TR-1003-G, 10 μg/mL), blebbistatin (Tocris, 176010R), doxorubicin (Fisher, 15910101), paclitaxel (Fisher, P3456), vincristine (Acros, 203440050), thapsigargin (Fisher, AC328570010), tunicamycin (MP Biomedicals, ICN15002801), BI-2536 (Selleck Chemicals, S1109), BI-6727 (Selleck Chemicals, S2235).
Immunofluorescence
Cells were seeded on sterilized glass coverslips in 24-well plates and fixed with 4% paraformaldehyde or 100% ice-cold methanol for 15 minutes. Methanol was used when analyzing centrioles. Fixed cells were then blocked for 30 minutes in 3% bovine serum albumin (BSA) and 0.1% Triton X-100 in PBS (PBSTx + BSA). Primary antibodies were incubated in PBSTx + BSA for at least 1 hour at room temperature in a humidified chamber and washed three times in PBSTx, followed by secondary antibody (Alexa fluor-conjugated, Invitrogen, 1:350) incubation in PBSTx + BSA for 30 minutes at room temperature and one wash with PBSTx. Cells were counterstained with DAPI and mounted on glass slides with Prolong Gold antifade medium (Invitrogen). Image acquisition was performed on a Nikon Eclipse Ti inverted microscope equipped with: 10x, 20x, 40x, 60x, and 100x objectives; a temperature-controlled motorized stage with 5% CO2 support (In Vivo Scientific); and Hamamatsu ORCA Flash 4.0 camera. For images displayed in the figures, optical sections were taken at 0.2-μm intervals and deconvolved using Nikon Elements. Where appropriate, the observer was blinded to treatment condition during image acquisition and analysis. Images were processed and analyzed using Nikon Elements.
Antibodies utilized for immunofluorescence include: p62/SQSTM1 (Santa Cruz, SC-28359, 1:1000), alpha tubulin (Abcam ab4074, 1:5000), lysosomal-associated membrane protein-1 (LAMP-1; H4A3, deposited to the DSHB by August, J.T. / Hildreth, J.E.K.; 1:500), LAMP-2 (H4B4, deposited to the DSHB by August, J.T. / Hildreth, J.E.K.; 1:500), LC3B (Cell Signaling Technology, 2775, 1:500), pericentrin (Abcam, ab4448, 1:1000), centrin (Millipore, 04–1624, 1:500), and gamma tubulin (Abcam, ab27074, 1:1000). Alexa fluor-conjugated secondary antibodies were used (Invitrogen, 1:350).
Breast cancer tissue microarray analysis
The breast cancer tissue microarray (TMA) used in this analysis has been described previously (15). Briefly, samples were obtained from primary breast tumor blocks obtained at time of surgery for stage I-III breast cancer patients seen at the University of Wisconsin Carbone Cancer Center under an IRB-approved protocol. The TMA contains three 0.6mm punch biopsies of 5 μm thickness from each patient’s tumor (n = 379 breast cancers) and 15 normal breast controls from mammoplasty specimens.
Breast cancer TMA slides were deparaffinized by heating at 60° C for 25 minutes and incubation in xylene (2 × 10 minutes), and rehydrated in serial ethanol dilutions. Antigen retrieval was performed using a microwave for 7 minutes with citrate buffer (Citra Plus, Biogenex, HK080–5K). Blocking was done for 30 minutes in 10% bovine serum albumin (BSA) in TBS. Tissues were probed with anti-pan cytokeratin (Abcam, ab7753, 1:500), anti-pericentrin (Abcam, ab4448, 1:500) and anti-p62 (Santa Cruz, SC-28359, 1:500) antibodies diluted in 5% BSA and TBS for one hour at room temperature. Tyramide signal amplification (Perkin Elmer) was used for signal amplification; this method employs HRP-conjugated secondary antibodies, tyramide-conjugated biotin, and streptavidin-conjugated fluorophores. Slides were then incubated with anti-mouse (Abcam, ab7068) and anti-rabbit secondary antibodies (Abcam, ab) for 1 hour at room temperature. Slides were washed 3 times with TBST after primary and secondary antibody incubations. Slides were counterstained for DNA with 4’,6-diamidino-2-phenylindole (DAPI) and mounted with ProLong Diamond antifade reagent (Life Technologies).
To quantify protein expression, the TMA slides were imaged using a Vectra automated quantitative pathology imaging system (PerkinElmer Life Sciences) with a 40x objective. Tissue images were segmented and scored using inForm (version 1.4.0). We quantified cytoplasmic pericentrin and p62 in cytokeratin-positive cells.
Flow cytometry
For cell cycle and DNA content analyses, cells were harvested, washed with PBS, fixed in cold 70% EtOH for at least 24 hours, washed once in PBS, and resuspended in PBS with 0.5 mg/mL RNase A and 50 mg/mL propidium iodide. Samples were incubated at 4° C overnight and analyzed on a flow cytometer (FACSCalibur, BD Biosciences).
For apoptosis staining, cells were collected, centrifuged, resuspended in Annexin V binding buffer, and stained with FITC Annexin V and propidium iodide, per manufacturer’s protocol (BD Biosciences). Cells positive for both Annexin V and propidium iodide were considered apoptotic.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
RNA was isolated from cells using the RNeasy Micro Kit (Qiagen, Valencia, CA), and converted to cDNA using the Quantitect Reverse Transcription Kit (Qiagen). Primer sequences are shown in Supplemental Table 1 (23–25). Primers were used at a final concentration of 0.67 μM each, and 100 ng cDNA was used per reaction. A StepOne Plus (Applied Biosystems) real-time PCR thermal cycler was used for amplification with iQ SYBR Green Supermix (Bioard) per manufacturer’s protocol. Quantification of autophagy genes was normalized to three housekeeping genes (RRN18S, GAPDH and ACTB). The ΔΔCT method was employed to calculate the fold change in expression (26).
Immunoblotting
Cells were lysed in buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 0.5% NP-40, 10% glycerol) containing phosphatase inhibitors (10 mm sodium pyrophosphate, 5 mM β-glycerol phosphate, 50 mm NaF, 0.3 mm Na3VO4), 1 mM PMSF, protease inhibitor cocktail (Thermo Scientific), and 1 mm dithiothreitol. Samples heated in SDS buffer. Proteins were separated by SDS-PAGE (15% bis-acrylamide gels, 37.5:1 acrylamide:bis-acrylamide), transferred to PVDF membrane (Millipore), and blocked for at least 30 minutes in 5% milk and 0.1% Tween 20 in Tris-buffered saline, pH 7.4 (TBST + milk). Membranes were incubated at 4° C overnight with primary antibodies diluted in TBST + 5% milk, washed three times with TBST, and incubated for 30 minutes at room temperature in secondary antibodies conjugated to horseradish peroxidase in TBST + 5% milk. Membranes were washed and developed with luminol/peroxide (Millipore) and visualized with a BioRad ChemiDoc. LC3-I is more labile and more sensitive to freezing-thawing and to degradation in SDS sample buffer than LC3-II (1), so fresh samples were used and not subjected to repeated freeze-thaw cycles.
Antibodies utilized for immunoblotting include: LC3B (CST, 4108, 1:1000), p62/SQSTM1 (Santa Cruz, SC-28359, 1:1000), actin (DSHB, JLA20, 1:1000), and tubulin (Proteintech, 11224–1-AP, 1:5000). HRP-conjugated secondary antibodies were used (Jackson, 1:7500). Relative intensities of bands were calculated using ImageJ from scanned images and normalized to their respective β-actin intensity.
Cell Proliferation Assays
Doxycycline-inducible cell lines were pre-treated with doxycycline for 24 hours to induce CA, then harvested for drug screening. 1000 cells in 150 μL media were plated per well in 96 well plates. Cells were allowed to attach overnight, then chloroquine was added the next morning in 50 μL. After 5 days of incubation in the presence of chloroquine and doxycycline, 50μL of Cell Titer Glo (Promega) was added to each well, incubated for 15 minutes at room temperature, and luminescence was analyzed using an Enspire Plate Reader (Perkin Elmer) at the UW Small Molecule Screening and Synthesis Facility. From each experimental luminescence value, we subtracted the luminescence value of wells with media plus Cell Titer Glo (indicating background luminescence without cells) and normalized to the luminescence value of untreated cells within each condition. This screen was replicated using Vita-Orange Cell Viability Reagent (Biotool) in 96-well plates (same protocol as above), as well as crystal violet staining in 24-well plates. For crystal violet staining, cells were pre-treated with doxycycline for 24 hours, then harvested and plated in 24-well plates at a density of 5000 cells/well. Chloroquine was added the next day, and cells were incubated for 5 days before staining with crystal violet.
Autophagosome Trafficking
RPE PLK4 WT and ΔC cells were transfected with pGFP-LC3 (Addgene; plasmid #21073) using Mirius LT1 transfection reagent (Mirius Bio), plated on the cover slip of 35mm dishes (MatTek), and treated with 2 μg/mL doxycycline for 24 hours (if indicated). Live imaging of autophagosome traffic and autophagic flux was performed as previously described (27). Live, transfected cells were rapidly imaged (2000ms per Z-stack, 50 times) using: the Revolution XD spinning-disk microscopy system equipped with Yokogawa CSU-X1 confocal spinning disk head; Nikon Eclipse Ti inverted microscope surrounded by an Okolab cage incubator; iXon x3 897 EM-CCD camera; Andor laser combiner with four solid-state lasers at 405, 488, 561 and 640 nm and corresponding band-pass filter sets (Sutter); and ASI motorized stage with piezo-Z for rapid Z-stack acquisition as previously described (27,28). Andor IQ2 software was used for image acquisition and Imaris X64 (Bitplane) for image analysis. Spots module was used to obtain speed and track displacement length of autophagosomes.
Statistics
Statistical evaluations were performed using Prism software (GraphPad). Two-tailed t-tests were used for comparing two groups, and one-way ANOVA and Tukey multiple comparison tests were used for comparing more than two groups. P-values <0.05 were considered significant for all tests, and designations are made in the figures for statistical significance. Unless otherwise stated, data are presented as means ± SEM of three or more independent experiments, with at least three replicates per condition per experiment.
RESULTS
Centrosome amplification disrupts autophagy
CA is specific to cancer cells and therefore represents an attractive therapeutic target (17). Based on findings that microtubule disruption can impair autophagy, we hypothesized that CA would cause aberrations in autophagocytic load and trafficking and thereby alter autophagy. To test this, we utilized two cell line models of CA, which mimic the biology of cancer cells. These RPE1 and MCF10A cell lines utilize a doxycycline-inducible PLK4 transgene (labeled PLK4 WT) (18,29). As a control for doxycycline treatment and for increased kinase activity in the cell, we utilized a doxycycline-inducible truncated construct (amino acids 1–608 of PLK4), which lacks a crucial C-terminal localization domain (labeled ΔC) and does not result in CA when overexpressed (Supplemental Figure 1A–B) (18,29).
To assess autophagy in these models of CA, we first measured the autophagy markers p62 and LC3B by immunofluorescence. CA increased p62 foci and the expression of both p62 and LC3B (Figure 1A–D), suggesting that there is a block in the autophagy pathway resulting in buildup of autophagosomes and autophagic cargo. Chloroquine, which is known to inhibit lysosome acidification and results in autophagosome accumulation, was used as a positive control.
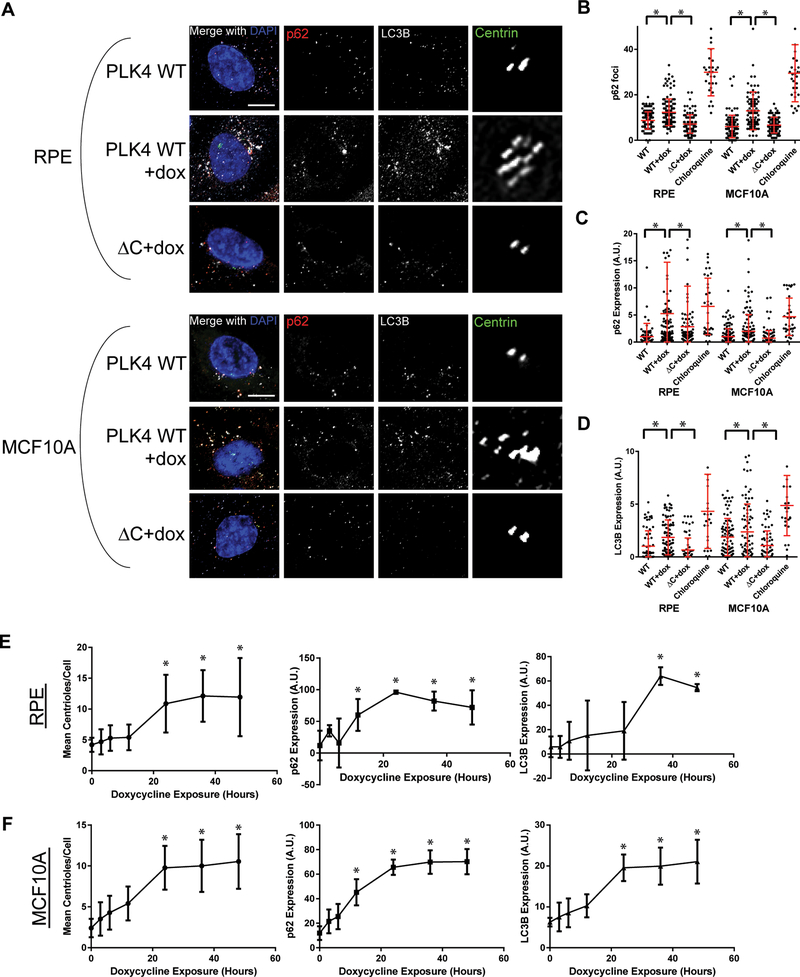
Figure 1: Centrosome amplification disrupts autophagy.
(A) Micrographs of fixed cells probed for autophagososomes (p62/SQSTM1, red; LC3B, white) and centrioles (centrin, green) in a doxycycline-inducible PLK4 overexpression system. The PLK4 WT cell line overexpresses full length PLK4 when treated with doxycycline, while the ΔC cell line overexpresses a truncated form of PLK4 (the first 608 amino acids, lacking the C-terminal localization domain) and does not result in centrosome amplification. The centrin panel shows enlargement of the centrosome. Scale bar = 5 μm. (B) Quantification of p62 immunofluorescence. Chloroquine, an autophagy inhibitor, was used as a positive control. (C) Quantification of p62 foci exceeding a predefined threshold in the indicated cell lines pictured in panel A. In panels B, C, and D, dots represent individual cells. To delineate individual cells and to ensure we assessed only intracellular puncta, we co-stained for α-tubulin. (D) Quantification of LC3B immunofluorescence. (E-F) RPE PLK4 WT (E) and MCF10A PLK4 WT (F) cells were treated with doxycycline and fixed after 3, 6, 12, 24, 36, and 48 hours. Centrioles (centrin foci), p62 expression, and LC3B expression were quantified. Dots represent at least 30 cells with bars indicating SD. Throughout the figure, bars represent means ± SEM from at least 3 independent experiments. *P value < 0.05 with correction for multiple comparisons.
To demonstrate that these observations are not simply due to increasing PLK4 expression or activity in the cell, we evaluated the timing of CA versus the development of p62 and LC3B accumulation in cells. Although doxycycline is expected to induce PLK4 expression in a matter of hours (Supplemental Figure 1C), increases in p62 and LC3B expression required at least 24 hours and occur in concert with the timing of CA (Figure 1E–F). To further confirm that these observations were not specific to PLK4, we also induced CA by overexpressing STIL. As expected, STIL overexpression-induced CA disrupts autophagy, as assessed by p62 and LC3B quantitative immunofluorescence (Supplemental Figure 2A–D). As a third method of inducing CA, we generated tetraploid cells by cytokinesis failure (21). Concordantly, tetraploid cells with CA demonstrated an increase in p62 and LC3B expression (Supplemental Figure 2E–K). There is heterogeneity in centriole number in our tetraploid cells; when comparing tetraploid cells with CA to tetraploid cells without CA, we observe significantly greater p62 and LC3B expression in tetraploid cells with CA (Supplemental Figure 2H–I). We conclude that CA, regardless of how it is generated, alters the autophagy pathway in human cells, as detected by multiple distinct measures.
To assess the potential reversibility of this phenotype, we treated multiple centrosome-amplified cancer cell lines with the PLK4 inhibitor centrinone B to reduce centrinone number (Supplemental Figure 3).(30) Some of these cell lines demonstrate a significant reduction in p62 and LC3B expression after centrinone B treatment (e.g. Cal51, HeLa), while others are not affected (e.g. MDA-MB-231, MDA-MB-468), suggesting there may be other contributors to autophagy defects in these cancer cell lines.
Centrosome amplification causes autophagosome accumulation
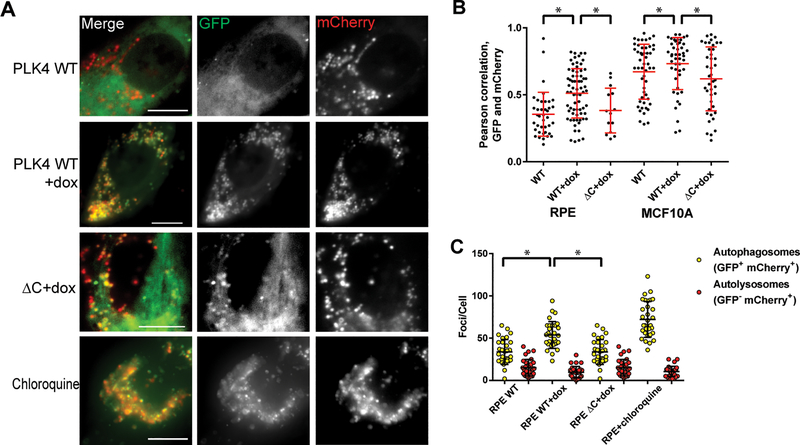
To determine which part of the autophagy pathway is disrupted by CA, we utilized an LC3B reporter construct, in which LC3B is fused with both mCherry and GFP, whose fluorescent properties change as a result of changes in pH (Figure 2A). This reporter enables simultaneous estimation of both the induction of autophagy and flux through autophagic compartments. The GFP signal is quenched by the acidic and/or proteolytic conditions of the lysosome lumen, whereas mCherry is more stable with changes in pH (31,32). Therefore, colocalization of both GFP and mCherry fluorescence (yellow foci) indicates a compartment that has not fused with a lysosome, such as the phagophore or an autophagosome. In contrast, mCherry signal without GFP (red foci) corresponds to an autolysosome. Therefore ratios of GFP to mCherry are a measure of autophagic flux. Firstly, we find that CA (PLK4 WT+dox conditions) increases the Pearson correlation of GFP and mCherry channels (Figure 2B), consistent with an increase in non-acidified autophagosomes. Secondly, we quantified the number of GFP+ mCherry+ foci and GFP- mCherry+ foci. Autophagic flux is increased when both yellow and red puncta are increased, while autophagic flux is blocked when only yellow puncta are increased without an accompanying increase of red puncta in cells (33). As expected, chloroquine increased the number of yellow puncta without a significant increase in red puncta (Figure 2C). CA resulted in a similar increase in the number of yellow puncta without a significant increase in red puncta compared to controls (Figure 2C). We conclude that CA results in an accumulation of autophagosomes.
Figure 2: Centrosome amplification causes an accumulation of autophagosomes.
(A) Representative immunofluorescent micrographs of cells expressing an LC3B reporter, in which LC3B is fused to both GFP and mCherry. In acidic environments (i.e. the lysosome), the GFP signal is quenched. Scale bar = 5 μm. (B) Quantification of Pearson correlations between GFP and mCherry expression. Higher values indicate more GFP-mCherry overlap, indicating more autophagosomes. (C) Quantification of yellow and red foci in RPE cells. Yellow foci represent autophagosomes, while red foci represent autolysosomes. Dots represent individual cells. Bars represent means ± SEM. *P value < 0.05.
In addition, we assessed the expression of autophagy-related genes by qRT-PCR. Cells with CA exhibited greater expression of the autophagy related genes ATG5, BECN1/Beclin-1, and p62/SQSTM1 (Supplemental Figure 4A). These data suggest a compensatory increase in autophagy to counteract an autophagy defect in cells with CA, as has been seen in other models of autophagy inhibition (34). We also assessed LC3B-II/I expression by western blotting but did not observe a significant change with CA (Supplemental Figure 4B–D); this assay may not be as sensitive as the other assays we have performed.
Centrosome amplification correlates with p62 expression in human breast cancer cell lines and primary breast cancer tissues
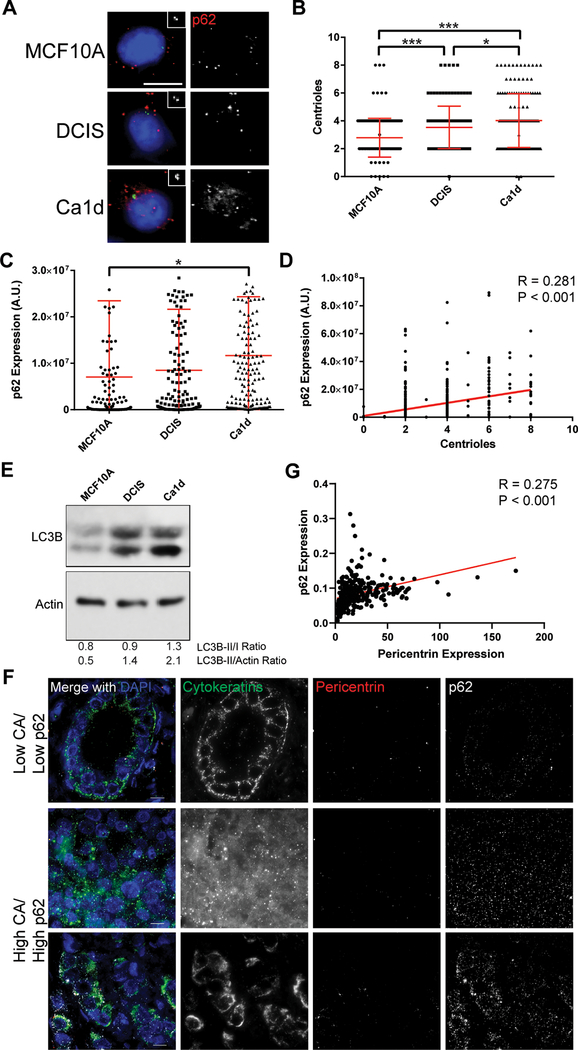
To assess whether our findings were consistent in different models and in a more cancer-relevant system, we utilized a syngeneic cell line panel, consisting of MCF10A (non-transformed, immortalized human epithelial cell line) and two derivative cell lines, DCIS (mimics the biology of breast ductal carcinoma in situ) and Ca1d (mimics the biology of invasive breast carcinoma). We assessed centrioles and autophagy by immunofluorescent staining of centrin and p62, respectively (Figure 3A). We find an increase in centrioles in DCIS compared to MCF10A and an even greater increase in centrioles in Ca1d compared to the two other cell lines (Figure 3B). A similar trend was observed in p62 expression (Figure 3C). Further, centrioles and p62 expression were significantly correlated after combining the data from all 3 cell lines (Figure 3D). Consistently, we find an increase in LC3-II/I ratio from MCF10A to DCIS to Ca1d Figure 3E). We conclude that CA correlates with autophagy in an independent breast cancer cell line panel.
Figure 3: Centrosome amplification correlates with p62 in breast cancer models.
(A) Representative images of MCF10A, DCIS, and Ca1d cell lines, representing benign breast epithelium, ductal carcinoma in situ, and invasive ductal carcinoma, respectively. Insets show enlargements of the centrioles (centrin staining). Blue = DNA/DAPI, green = centrin, red = p62. Scale bar = 10 μm. (B) Correlation of centrioles with p62 expression, quantified from immunofluorescence images. Pearson r = 0.281, P value < 0.001. Data are combined from all 3 cell lines shown in panel A. (C) Quantification of centrioles in each of the 3 cell lines. (D) Quantification of p62 expression in each of the 3 cell lines. ANOVA P value = 0.013, Tukey’s multiple comparisons test demonstrated significant difference between MCF10A and Ca1d (P < 0.015). Dots represent individual cells. Bars represent means ± SD. *P value < 0.05, ***P< 0.001. (E) Western blotting for LC3B and actin loading control. (F) Representative images of primary breast tumors from the breast tissue microarray (TMA) analysis. Tissues were stained for pericentrin (to mark centrosomes and assess centrosome amplification), p62 (to mark autophagosomes and assess autophagy), pan-cytokeratins (to identify epithelial cancer cells), and DAPI (to label nuclei). (G) Correlation of p62 expression with pericentrin expression in the breast TMA. Each dot represents the average of over 300 cells for one tumor.
To further assess the relationship between CA and autophagy in human cancer, we utilized a cohort of nearly 400 primary human breast cancers. We probed for centrosomes (pericentrin) and p62 to assess the correlation of CA with autophagy in human cancer (Figure 3F). We find a significant positive correlation between pericentrin and p62 in this cohort (Figure 3G). We conclude that CA correlates with autophagy dysregulation in primary human breast cancer.
Centrosome amplification-induced autophagy defect is due to disrupted autophagosome trafficking
We hypothesized three non-mutually exclusive mechanisms for the autophagy defect (Figure 4A). We first hypothesized that supernumerary centrosomes could generate increased lysosomal hubs, impairing autophagosome processing. However, there was no evident increase in lysosomal hubs as measure by total or peri-centrosomal LAMP1 expression (Supplemental Figure 5); LAMP1 is a major lysosomal membrane glycoprotein that is responsible for maintain lysosome structure and function. We next considered the role of aberrant mitosis on autophagosomes in post-mitotic progeny. Previously, aneuploidy and chromosome missegregation induced by inhibition of the mitotic checkpoint kinase MPS1 have been shown to disrupt autophagy (34). This is thought to be due to an aneuploidy-induced gene imbalance, causing altered protein complex stoichiometry, increased proteotoxic stress, and accumulation of autophagic products in lysosomes (35,36). CA is known to cause chromosome missegregation via multipolar spindles, merotelic kinetochore attachments, and lagging chromosomes (37). To determine if our observed effect of CA on autophagy is dependent on mitosis and chromosome missegregation events, we arrested cells at the G1/S transition with aphidicolin (Supplemental Figure 4), then treated with doxycycline for 24 hours to induce CA; cells arrested with aphidicolin (Supplemental Figure 6A) are still able to duplicate and amplify centrosomes (Supplemental Figure 6B–C and ref (38)). We compared the change in p62 or LC3B expression between aphidicolin-arrested and non-arrested conditions. We observe a milder increase in p62 and LC3B in cells with CA and treated with aphidicolin compared to untreated cells with CA. (Supplemental Figure 6D–E); however, the statistically significant difference in p62 and LC3B seen between CA (WT+dox) and non-CA controls (WT or ΔC+dox conditions) is maintained in the aphidicolin-treated conditions (Supplemental Figure 6D–E). We conclude mitosis and/or chromosome missegregation may be minor contributors to the observed effects of CA on autophagy.
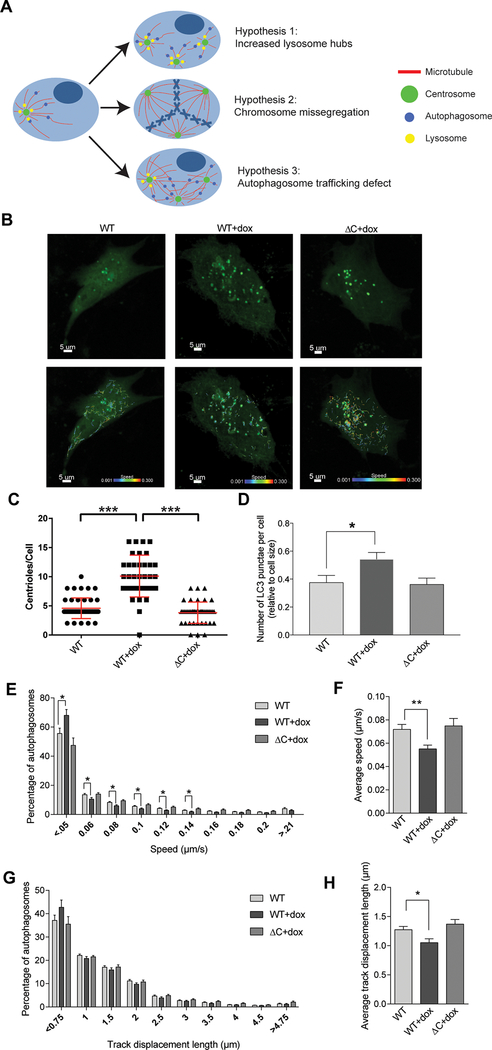
Figure 4: Centrosome amplification impairs autophagosome trafficking.
(A) Diagrammatic representation of three potential hypotheses for disrupted autophagy in cells with centrosome amplification. In the first scenario, centrosome amplification results to a disordered array of microtubules, thereby decreasing the efficiency of transportation of autophagic vesicles in the cell. In the second scenario, an increase in centrosomes generates an increase in lysosomes hubs, as lysosomes tend to cluster around centrosomes. In the third scenario, centrosome amplification causes chromosome missegregation, resulting in aneuploidy and therefore altered stoichiometry of protein complexes, increasing the cell’s dependence on autophagy. (B) Cells were plated in glass-bottom chamber slides, treated with doxycycline (if indicated), and transduced with baculovirus encoding LC3-GFP to label autophagosomes. Timelapse imaging was used to measure autophagosome trafficking. Representative images of RPE cells are shown both with and without the autophagosome traces. Scale bar = 5 μm. Colored lines on the bottom images correlate with autophagosome speed (color of line) and distance (length of line). (C) Following imaging, cells were fixed and stained to assess CA, which is quantified in this panel by counting centrioles (centrin foci). Dots represent individual cells. (D) Quantification of the number of autophagosomes (LC3B punctae) per cell. (E) Histogram demonstrating the spread of autophagosome trafficking speed by condition. (F) Average autophagosome speed for each condition. (G) Histogram demonstrating the spread of autophagosome track displacement lengths by condition. (H) Average autophagosome track displacement length for each condition. Bars represent means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. NS = not significant. ANOVA and t test with Tukey’s correction for multiple comparisons were utilized. RPE cells were utilized for all experiments in this figure.
Lastly, we tested the hypothesis that CA disrupts trafficking of autophagosomes, likely from the known impact on microtubule networks (18,39). Movement of autophagosomes to lysosomes is dependent on microtubules (9–13), and microtubule disruption with nocodazole and paclitaxel inhibit autophagy, as evaluated by increased p62 and LC3B expression, increased LC3B-II/I ratios, and autophagosome accumulation (Supplemental Figure 7). Additionally, CA can disrupt microtubules and cause increased microtubule nucleation (40). To test our hypothesis, we measured autophagosome trafficking by live-cell imaging. Cells were plated in glass-bottom chambers, treated with doxycycline, and transduced with baculovirus expressing LC3B-GFP (Figure 4B). Following each experiment, we fixed the cells and stained in situ to confirm CA in the PLK4 WT+doxycycline condition (Figure 4C). Consistent with our previous findings, CA caused an increase in the number of autophagosomes (Figure 4D). Furthermore, CA reduced autophagosome trafficking speed (Figure 4E–F) and track displacement (Figure 4G–H).
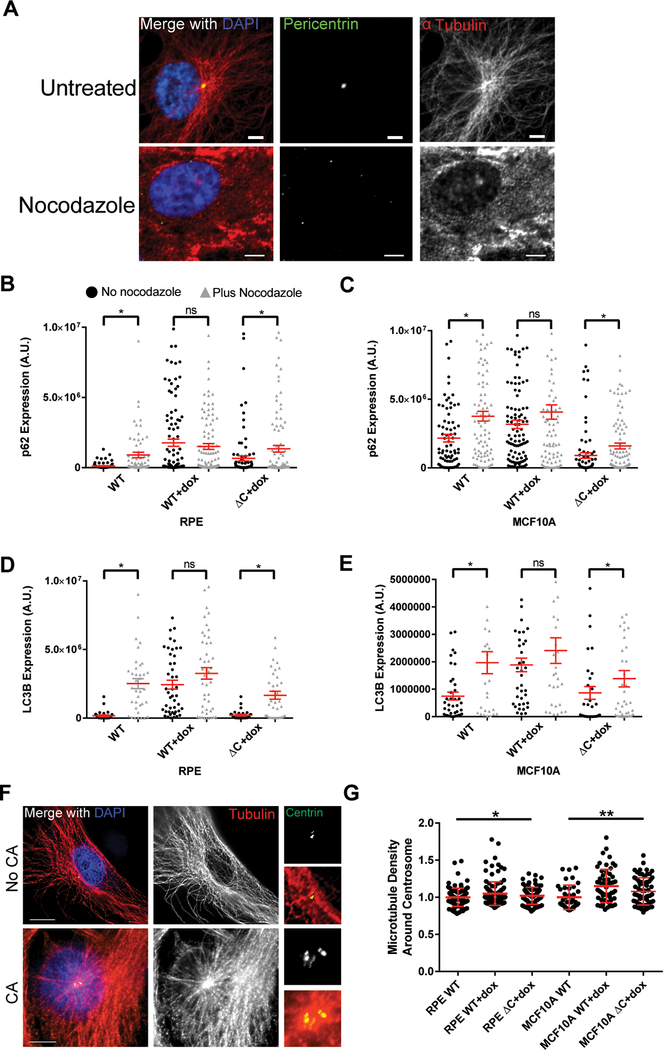
To assess whether altered autophagosome trafficking in cells with CA is due to disrupted microtubule networks, we compared p62 and LC3B expression before and after acute microtubule disruption with nocodazole (Figure 5). If the mechanism by which CA inhibits autophagy is due to microtubule disruption, then we would expect no significant increase in autophagosomes (assessed by p62 and LC3B immunofluorescence) after cells with CA are treated with nocodazole. Conversely, if the mechanism is not due to microtubule disruption, then nocodazole treatment should further disrupt autophagy and increase autophagosomes. We find that nocodazole significantly increases p62 and LC3B expression in controls, as expected, but does not significantly increase p62 and LC3B in cells with CA (Figure 5). Based on these data, we conclude that the CA-induced autophagy defect depends on disruption of microtubules.
Figure 5: Centrosome amplification disrupts microtubule nucleation.
(A) Immunofluorescent images demonstrating nocodazole-induced microtubule depolymerization. Cells were treated with 0.2 μg/mL nocodazole for 2 hours before fixation. (B-E) Quantification of p62 (B-C) and LC3B (D-E) in RPE (B,D) and MCF10A (C,E) cell lines. Cells were first pre-treated with doxycycline to induce CA for 24 hours, then treated with 0.2 μg/mL nocodazole for 2 hours. Black circles indicate cells not treated nocodazole, while gray triangles indicate cells treated with nocodazole. (F) Representative images of microtubule networks emanating from the centrosome(s) in cells with normal or extra centrosomes. Smaller images on the right are enlargements of the centrosome. Scale bars = 10 μm. (G) Quantitative immunofluorescence was used to quantify microtubule density around the centrosome. Bars represent means ± SEM. *P value < 0.05. NS = not significant. T tests were used for comparisons in B-E and ANOVA was used for G.
Centrosome amplification sensitizes to inhibition of autophagy
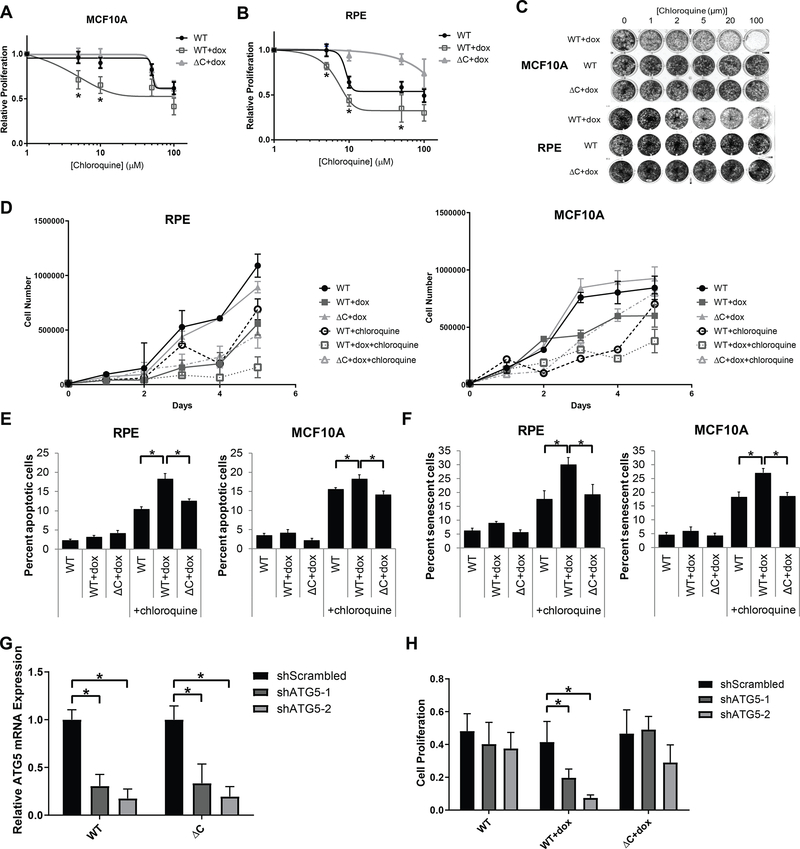
The role of autophagy in cancer has been somewhat unclear and controversial. Most data support the conclusion that autophagy is a tumor-suppressive pathway, but that after a tumor has initiated, autophagy helps the tumor progress. As such, chloroquine and its derivative hydroxychloroquine, FDA-approved drugs for non-oncologic indications, are currently being investigated for cancer treatment. Therefore, the effect of CA on autophagy could have clinical implications. Because cells with CA display an accumulation of autophagosomes, we hypothesized that they are more dependent on autophagy for survival and are more sensitive to inhibition of autophagy. We assessed cell viability in the RPE-1 and MCF10A models of CA treated with chloroquine. We also screened a panel of other drugs in these cell lines, finding that cells with CA appear more resistant to anti-mitotic drugs, such as PLK1 inhibitors and vinca alkaloids (Supplemental Figure 8); this finding is likely due to the slower proliferative rate of cells with CA (41) and is consistent with previous reports (42). In both cell lines, cells with CA were more sensitive to chloroquine, as assessed by Cell Titer Glo viability assays (Figure 6A–B), crystal violet staining (Figure 6C), and cell counts (Figure 6D). We then assessed the mechanism of reduced viability by testing the hypotheses that chloroquine increases either apoptosis or senescence to a greater extent in cells with CA versus controls. Our data demonstrate a significantly greater rate of both apoptosis (Figure 6E) and senescence (Figure 6F) in cells with CA (PLK4 WT+dox conditions) versus controls (PLK4 WT and ΔC+dox).
Figure 6: Centrosome amplification sensitizes cells to chloroquine.
(A-B) Cells were pre-treated with doxycycline for 24 hours to induce centrosome amplification, then 1000 cells per well were plated in 96 well plates. Chloroquine was added the next day at the indicated concentrations, then proliferation was assessed 3 days later. Curves were compared by logistic regression and extra sum-of-squares F test. For MCF10A (A), P = 0.03; for RPE (B), P = 0.04. In addition, asterisks display significant differences (P value < 0.05) from two-way ANOVA tests. (C) 5000 cells per well were plated in 24 well plates. Doxycycline was added the next day, followed by chloroquine at the in dicated concentration the following day. The plates were stained with crystal violet 5 days after the addition of chloroquine. (D) Cell counts were performed using cells treated with 10 μM chloroquine for the indicated time points. (E) Apoptosis assays were performed by staining cells with propidium iodide and anti-Annexin V-FITC after 5 days of treatment with 10 μM chloroquine. Cells staining positive for both propidium iodide and Annexin V were considered to be apoptotic. (F) Senescence assays were performed by assessing β-galactosidase activity after 5 days of treatment with 10 μM chloroquine. (G) RPE cells were transduced with retrovirus expressing shRNA targeting two different sequences of ATG5 or a scrambled shRNA control. ATG5 knockdown efficiency was assessed by qRT-PCR. (H) Proliferation was assessed 3 days after addition of doxycycline. Bars represent means ± SEM. *P value < 0.05.
Lastly, we assessed sensitivity to genetic autophagy inhibition by depleting ATG5, a crucial autophagy gene (Figure 6G). We find that cells with CA (PLK4 WT+dox) are more sensitive to ATG5 depletion compared to controls (PLK4 WT and ΔC+dox; Figure 6H).We conclude that cells with CA are more sensitive to autophagy inhibition, and that the mechanism of reduced viability with chloroquine treatment involves both apoptosis and senescence.
DISCUSSION
CA is common in cancer, particularly in higher risk cancers, and has a number of effects on cell physiology, such as chromosome missegregation, increased microtubule nucleation, and cellular invasiveness (15,17–19). As CA is very specific to cancer cells, it represents a potentially attractive therapeutic target. Therefore, it is important to continue to determine the consequences of CA that render cancer exquisitely sensitive to particular interventions. Herein we have reported a novel precision medicine strategy, whereby cells with CA demonstrate disrupted autophagy and are more susceptible to inhibition of autophagy. This may indicate that CA is a potential biomarker for treatment with autophagy inhibitors. We demonstrate that the mechanism of CA-induced autophagy disruption is predominantly dependent on interference of autophagosome trafficking.
Autophagy occurs in the following stages: formation of phagophores around damaged proteins and organelles, maturation into autophagosomes, targeting and trafficking of autophagosomes to lysosomes, formation of autolysosomes by fusion of autophagosomes and lysosomes, and degradation of the autophagic cargo within the lysosomes for recycling and future use. Past evidence has pointed to the necessity of cytoskeletal elements, particularly microtubules, in autophagy. However, the exact role of microtubules remains unclear due to contradictory studies. Stable microtubules appear to be important for early stages of autophagy, as vinblastine and nocodazole inhibit fusion of autophagosomes with endosomes (9). Further, autophagosome trafficking to lysosomes for autolysosome formation depends on stable microtubules, as taxol-mediated microtubule stabilization does not affect autophagosome and lysosome fusion (9), while microtubule depolymerization with nocodazole or vinca alkaloids causes an increase in autophagosomes (9–13) and LC3-II levels (9,11,43). Our data presented herein also follow this paradigm, as microtubule disruption with either nocodazole or paclitaxel inhibits autophagy, as assessed by p62 and LC3B immunofluorescence, LC3B tandem tag reporter assay, and LC3B western blotting. Further, our data demonstrate that gain of centrosomes increases microtubule density and alters trafficking of autophagosomes. Taking all these data into account, we conclude that trafficking of autophagosomes along centrosome-organized microtubule networks towards lysosomes is necessary to allow for efficient autophagosome-lysosome fusion and clearance of autophagic cargo.
In addition to disruption of microtubules, a minor mechanism of CA-induced autophagy alterations may be aberrant cell divisions. Cells with CA are more likely to undergo chromosome missegregation (37), often resulting in aneuploid progeny. In eukaryotic cells, changes in gene copy number largely lead to a proportional change in the amount of protein produced. As a result, aneuploid cells have more imbalances in protein complex stoichiometry and generate more misfolded and aggregated proteins that accumulate in lysosomes and are difficult to degrade (34,44,45). Previous work demonstrates that the accumulated autophagosomal proteins are not efficiently cleared within lysosomes, but that lysosomal functions in aneuploidy cells remain intact (34). It remains unclear why these proteins are not efficiently cleared. Our data also confirm that lysosome function is unaltered by CA and that the proteasome pathway does not compensate for this increased need for protein degradation (data not shown). Additionally, our study suggests that the increased autophagic cargo created by chromosome missegragation due to CA is not efficiently cleared because CA disrupts autophagosome trafficking. Further work is necessary to fully assess the reason for inefficient clearance of autophagic cargo in aneuploid cells without CA.
Recent work has suggested the importance of the centrosome for autophagy. Specifically, the centrosome regulates autophagosome formation by facilitating transport of GABARAP, another ATG8 family member similar to LC3, to the forming autophagosome membrane (46). Additional work has suggested that autophagy plays a role in maintaining the proper number of centrosomes via degradation of CEP63 (47). Our work adds to this growing body of knowledge suggesting that the centrosome contributes to autophagy homeostasis, and that aberrancy of centrosome number disrupts autophagy.
The role of autophagy in cancer has been controversial, but the prevailing view had been that autophagy acts as a tumor suppressor initially but helps the tumor progress once it has already developed (48). Evidence for autophagy being a tumor suppressor comes from the observation of increased tumorigenesis in mice with allelic loss of Becn1, a crucial autophagy gene (49,50); however, it was later determined that BECN1 is codependent on deletion of the adjacent BRCA1 gene, suggesting that loss of BRCA1 was driving tumorigenesis. As the tumor progresses, tumor cells become dependent on autophagy for survival, so autophagy is pro-tumorigenic in later stages of cancer (48). This is substantiated by observations that loss of function of core autophagy genes is uncommon in cancer, whereas mutations that activate autophagy have been identified (51). Further, immunohistochemical analyses demonstrate that more aggressive tumors express lower levels of p62 and LC3B, consistent with increased autophagy (52). There are several ongoing clinical trials of chloroquine in advanced cancer, and second-generation agents specific to autophagic machinery are in development (e.g. ULK1 and ATG4 inhibitors). Heretofore, clinical trials involving autophagy inhibitors have been largely unsuccessful, but do show activity in a subset of patients (53). One reason for this is that the most commonly used agents, chloroquine and hydroxychloroquine, are pleiotropic compounds and affect processes other than autophagy, such as the cellular redox state and mitochondrial function. Another potential reason for the failure of autophagy inhibitors is that we have not been able to adequately identify the subset of patients that will benefit. Our work suggests that the use of CA as a biomarker for autophagy inhibitors could increase the success of autophagy inhibition as a therapeutic strategy for cancer.
There are some limitations to this study. First, most experiments were performed with one model of CA (i.e. PLK4 overexpression). However, key portions of our findings were validated in additional models including STIL overexpression, cytokinesis failure, and models of breast cancer progression which harbor CA, lending credence to our findings. Next, we have not demonstrated the correlation between CA and autophagy in human cancer samples. A potential problem with using CA as a biomarker is that analysis of centrosomes in human cancer patients may not realistically translate well to clinical practice, as this method is time- and labor-intensive. Another potential challenge relates to the use of CA as a therapeutic target. While CA is specific to cancer cells, it is usually only present in a fraction of the cells in a tumor and may be a transient phenotype; for example, we previously reported that an average of 23% of breast cancer cells have CA (15). However, these and other data are based on immunohistochemical staining of tissue sections, which may underestimate the number of centrosomes via sectioning artifact. Lastly, our experiments using centrinone B to inhibit PLK4 in cancer cell lines resulted in a significant reduction in p62 and LC3B in some cell lines (e.g. Cal51, HeLa) but not others (e.g. MDA-MB-231, MDA-MB-468). This suggests a broader theme and limitation of this work: there are likely many contributors to autophagy defects in human cancer other than CA.
In conclusion, we found that CA disrupts autophagosome trafficking, resulting in autophagosome accumulation. Further, CA confers a vulnerability and renders cells more susceptible to autophagy inhibition. These results suggest a rationale for precisely targeting autophagy in high-risk cancers with CA.
Supplementary Material
IMPLICATIONS.
Our study suggests that centrosome amplification could be used as a predictive biomarker for treatment with autophagy inhibitors.
Acknowledgements
The authors thank: David Pellman, Jun Wan, Beth Weaver, Patricia Keely, and Peiman Hematti for reagents and use of equipment; Alka Choudhary for exceptional technical assistance; the University of Wisconsin Translational Initiatives in Pathology (TRIP) Laboratory; the University of Wisconsin Small Molecule Screening and Synthesis Facility; the University of Wisconsin Carbone Cancer Center Flow Cytometry Laboratory. Research reported in this publication was supported by the following NIH awards: F30CA203271 to RAD, R01 GM097245 to MEB, R01 EY023299 to AL, University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520. RAD has also been supported by the University of Wisconsin Medical Scientist Training Program (T32GM008692) and University of Wisconsin ICTR TL1 training program (supported under NIH awards UL1TR000427 and TL1TR000429). The content is the responsibility of the authors and does not necessarily represent the views of the NIH.
Financial Support: This work is supported by the following NIH grants: F30CA203271, R01 GM097245, P30 CA014520, T32GM008692, UL1TR000427, TL1TR000429, and R01 EY023299. The content is the responsibility of the authors and does not necessarily represent the views of the NIH.
Abbreviations
- CA
centrosome amplification
- DCIS
ductal carcinoma in situ
- LC3
microtubule associated protein 1 light chain 3
- PLK4
Polo-like kinase 4
- WT
wild type
Footnotes
Conflicts of Interest: The authors declare no conflicts of interests.
REFERENCES
- 1.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007;8:931–7 [DOI] [PubMed] [Google Scholar]
- 3.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol 2011;8:528–39 [DOI] [PubMed] [Google Scholar]
- 4.Seglen PO, Bohley P. Autophagy and other vacuolar protein degradation mechanisms. Experientia 1992;48:158–72 [DOI] [PubMed] [Google Scholar]
- 5.Amaya C, Fader CM, Colombo MI. Autophagy and proteins involved in vesicular trafficking. FEBS Lett 2015;589:3343–53 [DOI] [PubMed] [Google Scholar]
- 6.Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 2006;281:36303–16 [DOI] [PubMed] [Google Scholar]
- 7.Mackeh R, Perdiz D, Lorin S, Codogno P, Poüs C. Autophagy and microtubules - new story, old players. J Cell Sci 2013;126:1071–80 [DOI] [PubMed] [Google Scholar]
- 8.Bonifacino JS, Neefjes J. Moving and positioning the endolysosomal system. Curr Opin Cell Biol 2017;47:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köchl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006;7:129–45 [DOI] [PubMed] [Google Scholar]
- 10.Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA. Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol 1992;152:458–66 [DOI] [PubMed] [Google Scholar]
- 11.Fengsrud M, Roos N, Berg T, Liou W, Slot JW, Seglen PO. Ultrastructural and immunocytochemical characterization of autophagic vacuoles in isolated hepatocytes: effects of vinblastine and asparagine on vacuole distributions. Exp Cell Res 1995;221:504–19 [DOI] [PubMed] [Google Scholar]
- 12.Punnonen EL, Reunanen H. Effects of vinblastine, leucine, and histidine, and 3-methyladenine on autophagy in Ehrlich ascites cells. Exp Mol Pathol 1990;52:87–97 [DOI] [PubMed] [Google Scholar]
- 13.Seglen PO, Berg TO, Blankson H, Fengsrud M, Holen I, Strømhaug PE. Structural aspects of autophagy. Adv Exp Med Biol 1996;389:103–11 [DOI] [PubMed] [Google Scholar]
- 14.Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci 2011;7:1122–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denu RA, Zasadil LM, Kanugh C, Laffin J, Weaver BA, Burkard ME. Centrosome amplification induces high grade features and is prognostic of worse outcomes in breast cancer. BMC Cancer 2016;16:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denu RA, Shabbir M, Nihal M, Singh CK, Longley BJ, Burkard ME, et al. Centriole Overduplication is the Predominant Mechanism Leading to Centrosome Amplification in Melanoma. Mol Cancer Res 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc Lond B Biol Sci 2014;369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, et al. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 2014;510:167–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pannu V, Mittal K, Cantuaria G, Reid MD, Li X, Donthamsetty S, et al. Rampant centrosome amplification underlies more aggressive disease course of triple negative breast cancers. Oncotarget 2015;6:10487–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat 2001;65:101–10 [DOI] [PubMed] [Google Scholar]
- 21.Choudhary A, Zachek B, Lera RF, Zasadil LM, Lasek A, Denu RA, et al. Identification of Selective Lead Compounds for Treatment of High-Ploidy Breast Cancer. Mol Cancer Ther 2016;15:48–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 2009;10:173–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chinnadurai R, Copland IB, Ng S, Garcia M, Prasad M, Arafat D, et al. Mesenchymal Stromal Cells Derived From Crohn’s Patients Deploy Indoleamine 2,3-dioxygenase-mediated Immune Suppression, Independent of Autophagy. Mol Ther 2015;23:1248–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toepfer N, Childress C, Parikh A, Rukstalis D, Yang W. Atorvastatin induces autophagy in prostate cancer PC3 cells through activation of LC3 transcription. Cancer Biol Ther 2011;12:691–9 [DOI] [PubMed] [Google Scholar]
- 25.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 2010;38:D792–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
- 27.Toops KA, Tan LX, Jiang Z, Radu RA, Lakkaraju A. Cholesterol-mediated activation of acid sphingomyelinase disrupts autophagy in the retinal pigment epithelium. Mol Biol Cell 2015;26:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toops KA, Tan LX, Lakkaraju A. A detailed three-step protocol for live imaging of intracellular traffic in polarized primary porcine RPE monolayers. Exp Eye Res 2014;124:74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci 2010;123:2163–9 [DOI] [PubMed] [Google Scholar]
- 30.Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, et al. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007;3:452–60 [DOI] [PubMed] [Google Scholar]
- 32.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131–45 [DOI] [PubMed] [Google Scholar]
- 33.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010;140:313–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santaguida S, Vasile E, White E, Amon A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev 2015;29:2010–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santaguida S, Amon A. Aneuploidy triggers a TFEB-mediated lysosomal stress response. Autophagy 2015;11:2383–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 2015;16:473–85 [DOI] [PubMed] [Google Scholar]
- 37.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009;460:278–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol 1995;130:105–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci U S A 1998;95:2950–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A 2002;99:1978–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, et al. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev 2012;26:2684–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konotop G, Bausch E, Nagai T, Turchinovich A, Becker N, Benner A, et al. Pharmacological Inhibition of Centrosome Clustering by Slingshot-Mediated Cofilin Activation and Actin Cortex Destabilization. Cancer Res 2016 [DOI] [PubMed] [Google Scholar]
- 43.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev 2012;26:2696–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stingele S, Stoehr G, Storchova Z. Activation of autophagy in cells with abnormal karyotype. Autophagy 2013;9:246–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joachim J, Razi M, Judith D, Wirth M, Calamita E, Encheva V, et al. Centriolar Satellites Control GABARAP Ubiquitination and GABARAP-Mediated Autophagy. Curr Biol 2017;27:2123–36.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe Y, Honda S, Konishi A, Arakawa S, Murohashi M, Yamaguchi H, et al. Autophagy controls centrosome number by degrading Cep63. Nat Commun 2016;7:13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White E The role for autophagy in cancer. J Clin Invest 2015;125:42–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003;112:1809–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003;100:15077–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev 2016;30:1913–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams O, Dislich B, Berezowska S, Schläfli AM, Seiler CA, Kroell D, et al. Prognostic relevance of autophagy markers LC3B and p62 in esophageal adenocarcinomas. Oncotarget 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chude CI, Amaravadi RK. Targeting Autophagy in Cancer: Update on Clinical Trials and Novel Inhibitors. Int J Mol Sci 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.