Abstract
Inhibition of ribonucleotide reductase (RNR), the rate-limiting enzyme in the synthesis of deoxyribonucleotides, causes DNA replication stress and activates the ATR-CHK1 pathway. Notably, a number of different cancers, including Ewing sarcoma tumors, are sensitive to the combination of RNR and ATR-CHK1 inhibitors. However, multiple, overlapping mechanisms are reported to underlie the toxicity of ATR-CHK1 inhibitors, both as single-agents and in combination with RNR inhibitors, toward cancer cells. Here, we identified a feedback loop in Ewing sarcoma cells in which inhibition of the ATR-CHK1 pathway depletes RRM2, the small subunit of RNR, and exacerbates the DNA replication stress and DNA damage caused by RNR inhibitors. Mechanistically, we identified that the inhibition of ATR-CHK1 activates CDK2, which targets RRM2 for degradation via the proteasome. Similarly, activation of CDK2 by inhibition or knockdown of the WEE1 kinase also depletes RRM2 and causes DNA damage and apoptosis. Moreover, we show that the concurrent inhibition of ATR and WEE1 has a synergistic effect in Ewing sarcoma cells. Overall, our results provide novel insight into the response to DNA replication stress, as well as a rationale for targeting the ATR, CHK1, and WEE1 pathways, in Ewing sarcoma tumors.
INTRODUCTION
Ewing sarcoma is a bone and soft tissue sarcoma that is caused by a chromosomal translocation that fuses the EWSR1 gene to members of the ETS family of transcription factors, most frequently FLI1 (1). The EWS-FLI1 oncogene is an attractive therapeutic target in Ewing sarcoma tumors because it is required for tumorigenesis and specific for tumor cells (1). Directly targeting EWS-FLI1, though, has proven to be challenging and the standard treatment for Ewing sarcoma, which has changed very little in the past two decades, consists of dose-intensified, cytotoxic chemotherapy in combination with surgery and radiation (2). However, an alternative approach to directly inhibiting EWS-FLI1 function is to target unique vulnerabilities incurred by the oncogene. For example, Ewing sarcoma cells exhibit elevated levels of endogenous DNA replication stress and are sensitive to inhibitors of ribonucleotide reductase (RNR), the rate limiting enzyme in the synthesis of deoxyribonucleotides (3–5). Ewing sarcoma cells are also dependent on the ataxia telangiectasia and rad3-related protein (ATR) and checkpoint kinase 1 (CHK1) pathway, which plays a key role in orchestrating the cellular response to DNA replication stress, for survival (3,4,6).
Ewing sarcoma tumors are sensitive in vitro and in vivo to ATR and CHK1 inhibitors, both as single agents and in combination with other drugs (3,4,6–10). Notably, ATR-CHK1 inhibitors are also reported to sensitize a range of other tumor types to DNA-damaging agents and, in some cases, elicit single agent cytotoxicity (11). For example, Lowery et al. recently showed that the CHK1 inhibitor prexasertib has antitumor effects as both a monotherapy and in combination with chemotherapy in multiple preclinical models of pediatric cancers, including malignant rhabdoid tumors, rhabdomyosarcoma, neuroblastoma, and osteosarcoma (8). The ATR-CHK1 pathway, when activated by DNA replication stress, orchestrates a multifaceted response that arrests cell cycle progression, suppresses origin firing, stabilizes replication forks, and promotes fork repair and restart (12). However, ATR and CHK1 also have critical and unique functions outside of S phase and the response to DNA replication stress. For example, ATR and/or CHK1 regulate chromosome segregation, the S/G2 checkpoint, the G2/M transition, double-strand DNA break repair, and the response to osmotic and mechanical stress (13–17). Consequently, the effects of inhibiting ATR or CHK1 are variable and multiple mechanisms are reported to underlie the selective toxicity of ATR-CHK1 inhibitors toward cancer cells (18).
In the current study, we identified that the inhibition of the ATR-CHK1 pathway in Ewing sarcoma cells experiencing DNA replication stress leads to the aberrant activation of CDK2 and cell death. Similarly, activation of CDK2 by inhibiting the WEE1 kinase with AZD1775, or knockdown of WEE1 with siRNA, also causes DNA damage and apoptosis. Moreover, from a mechanistic standpoint, we show that active CDK2 targets ribonucleotide reductase M2 (RRM2), the small subunit of ribonucleotide reductase (RNR), for degradation. Notably, RRM2 is required for DNA replication and DNA damage repair. Thus, we describe a novel feedback loop in Ewing sarcoma cells in which the inhibition of the ATR-CHK1 or WEE1 pathways during DNA replication stress, due to inhibition of RRM2 or other causes, leads to the aberrant activation of CDK2, degradation of RRM2, enhanced DNA replication stress, increased DNA damage, and apoptosis.
MATERIALS AND METHODS
Cell lines and culture
Cell lines were maintained at 37° C in a 5% CO2 atmosphere. The A673, TC32, TC71, and EW8 cell lines were kindly provided by Dr. Kimberly Stegmaier (Dana-Farber Cancer Institute, Boston, MA). The BJ-tert, HEK-293T, RPE-tert, and U2OS cell lines were obtained from ATCC. Cells were cultured as previously described(6,10). Cell lines were authenticated by DNA fingerprinting using the short tandem repeat (STR) method and used within 5–10 passages of thawing.
Chemical compounds
Chemical compounds were purchased from Selleckchem (LY2603618), ThermoFisher Scientific (puromycin, doxycycline, and geneticin), and MedChemExpress (AZD1775, AZD6738, RO-3306, roscovitine, VX-970, NSC663284, and GDC-0575).
Thymidine double block
Cells were treated for 18 h overnight with thymidine (2 mM). The thymidine was then removed by washing the cells with pre-warmed 1x PBS. Fresh medium was then added and the cells were incubated for 9 h in a tissue culture incubator at 37 °C. The cells were then treated with a second round of thymidine (2 mM) for another 18 h at 37 °C.
Cell cycle analysis
Cell cycle analysis was performed in duplicate using the Click-iT EdU-488 kit for flow cytometry (ThermoFisher Scientific). Cells were labeled with EdU for two hours and analysis was performed according to the manufacturer’s instructions. Flow cytometry was performed on a Becton Dickinson LSR II instrument.
Cell cycle analysis with EdU-647 labeling
The doxycycline-inducible shRRM1 and shRRM2 cells, which express green fluorescent protein, were labeled with EdU-647 using the Click-iT EdU-647 kit for flow cytometry (ThermoFisher Scientific). Cells were labeled with EdU-647 for two hours and analysis was performed on a Becton Dickinson FACS Aria instrument. DNA content was measured using the Hoescht stain.
Cell viability
Cell proliferation was measured using the resazurin (AlamarBlue) fluorescence and Cell-Titer-Glo luminescence assays as previously described (6,10). Drug synergy testing was performed by treating cells for 72 h with drug combinations of up to 1000 nM AZD6738 and 500 nM AZD1775. Survival was assayed by Cell-Titer-Glo and each experiment was repeated at least two times. The data were analyzed using SynergyFinder (https://synergyfinder.fimm.fi) (19). For the dose response experiment, approximately 5 × 104 cells were plated per well of a 96-well plate in the presence or absence of doxycycline. Cells were treated with a range of AZD1775 concentrations for 72 hours. Fluorescence readings were obtained after adding the AlamarBlue reagent (Sigma) using a FLUOstar Omega microplate reader (BMG Labtech). IC50 values were then calculated using log-transformed and normalized data (GraphPad Prism 8.01).
siRNA transfection
Cells (1.5–3 × 105) were plated one day prior to transfection in six-well plates. Cells were transfected with siRNA using Lipofectamine RNAiMax (Thermo Fisher Scientific) as previously described (5,6,10). siCHK1, siCDC25A, siCDK1, siCDK2, and siWEE1 were SMARTpool ON-TARGETplus reagents (GE Dharmacon). siRRM2 (siRRM2_R2B) was described previously (5). siControl was purchased from Cell Signaling Technology (#6568).
Doxycycline-inducible shRRM1 and shRRM2
shERWOOD UltramiR Lentiviral Inducible shRNA plasmids targeting RRM1 (TLHSU2300–6240) and RRM2 (TLHSU2300–6241) were obtained from Transomic Technologies (Huntsville, AL). Lentivirus was produced by transfecting HEK-293T cells with the shRNA plasmid and packaging plasmids (psPAX2 and pMD2.G) according to the FuGENE 6 (Roche) protocol. For the lentiviral transduction, Ewing sarcoma cells were incubated with 2 mL of virus and 6 mg/mL of polybrene (Sigma-Aldrich) for 12–16 hours. Cells were selected in 1 μg/mL puromycin 48 hours after transduction.
Doxycycline-inducible CDK1-AF and CDK2-AF
Plasmids were synthesized by VectorBuilder (Shenandoah, TX). The full-length CDK1 and CDK2 cDNAs, modified to convert Thr13 and Tyr14 to Ala13 and Phe14, were synthesized and inserted into lentiviral vectors downstream of the TRE3G doxycycline-inducible promoter (20). N-terminal FLAG and V5 tags were added to CDK1 and CDK2, respectively. The pLVX-EF1a-Tet3G vector (Clontech) was used to expresses the Tet-On 3G transactivator protein from the human EF1 alpha promoter. Lentivirus was prepared as described above. EW8 cells were sequentially infected and selected with geneticin 500 μg/mL (pLVX-EF1a-Tet3G) and puromycin 1 μg/mL (CDK1-AF and CDK2-AF).
Doxycycline-inducible TO-FLAG-RRM2-T33A
The full length RRM2 cDNA, with a Threonine-33 to Alanine-33 mutation and a FLAG-tag, was obtained as a gene block (IDT; Coralville, IA) and inserted into the Lenti-X-Tet-One vector (Clontech). After verification by sequencing, the plasmid was used to make lentivirus, as described above.
Immunoblotting
Immunoblots were performed as previously described (10). Protein loading for the immunoblots was normalized using cell number. Antibodies to the following proteins were used in the immunoblots: phospho-Histone-139 H2A.X (Cell Signaling, #9718, 1:1000), phospho-Chk1–345 ( Cell Signaling, #2348, 1:1000), Chk1 (Cell Signaling, #2360, 1:1000), PARP (Cell Signaling, #9532, 1:1000), cleaved caspase-3 (Cell Signaling, #9664, 1:1000), RRM1 (Cell Signaling, #8637, 1:1000), RRM2 (Santa Cruz, #398294, 1:500), Actin (Cell Signaling, #4970, 1:1000), CDK1 (Cell Signaling, #9116, 1:1000), CDK2 (Millipore, #05–596, 1:1000), P-CDK1/2 (Cell Signaling, #4539, 1:1000), Lamin A/C (Cell Signaling, #2032, 1:1000), P-Lamin A/C (Ser22) (Cell Signaling, #13448, 1:1000), WEE1 (Cell Signaling, #13084, 1:1000), V5 (Abcam, ab9116, 1:2000), FLAG (Sigma, F1804, 1:1000), and tubulin (Proteintech, 66031–1, 1:2000).
Phos-tag Gel Electrophoresis
Protein lysates were collected using RIPA buffer (Sigma, #R0276) containing cOmplete EDTA-free protease inhibitor cocktail (Sigma, #11873580001). An aliquot of lysate was removed from the sample and treated, according to the manufacturer’s instructions, with lambda phosphatase (Lambda PP; New England BioLabs, #P0753S) to serve as a positive control. Electrophoresis was then performed using precast Supersep Phos-tag 12.5% gels (Fisher Scientific) at 90V for 1.5 h. Gels were then washed three times in transfer buffer containing EDTA followed by one wash in transfer buffer without EDTA. Proteins were then transferred to polyvinylidene difluoride membranes at 100V for 1 h. Antibodies are described above. A sample of the protein lysate was also evaluated using standard polyacrylamide gel electrophoresis, which does not separate the unphosphorylated and phosphorylated CDK1 and CDK2 proteins, to ensure that the levels of total CDK1 and CDK2 were unchanged by the drug treatment.
γH2AX flow cytometry
Cells (3 × 105 cells/well) were plated in a 6-well plate and allowed to adhere overnight. The cells were then treated with drugs, or vehicle, as described. Cells were labeled with EdU-488 for 2 h using the Click-iT EdU-488 kit for flow cytometry (ThermoFisher Scientific). Flow cytometry for γH2AX and EdU-488 were then performed as previously described (10).
Caspase-3/7 Activation
Caspase-3/7 activation was measured using the Casp-Glo 3/7 Luminescence assay (Promega), according to the manufacturer’s instructions. The fold increase in caspase-3/7 activity was calculated by comparing drug to vehicle (DMSO) treated cells.
RT-qPCR
Total RNA was extracted using RNAeaasy kit following the manufacturer’s instructions. 1 μg of total RNA was reverse-transcribed into first-strand cDNA using random hexamer primers and the SuperScript III Reverse Transcriptase (Invitrogen). RT-qPCR was performed on the ViiA 7 Real-Time PCR System (Life Technologies) using SYBR Select Master Mix (Life Technologies). RT-qPCR was performed for RRM1 (5’-GCCAGGATCGCTGTCTCTAAC-3’), RRM2 (5’-CACGGAGCCGAAAACTAAAGC-3’), and WEE1 (5’-AACAAGGATCTCCAGTCCACA-3’). Reactions were performed in triplicate and gene expression was normalized to actin.
Statistical Analysis
Student’s t-test was used to calculate P-values for the comparison of two groups and analyses for more than two groups were conducted with a one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparisons test. Statistical analyses were conducted using GraphPad Prism 8.01.
RESULTS
Inhibition of the ATR-CHK1 pathway in Ewing sarcoma cells in S phase causes DNA damage and apoptosis
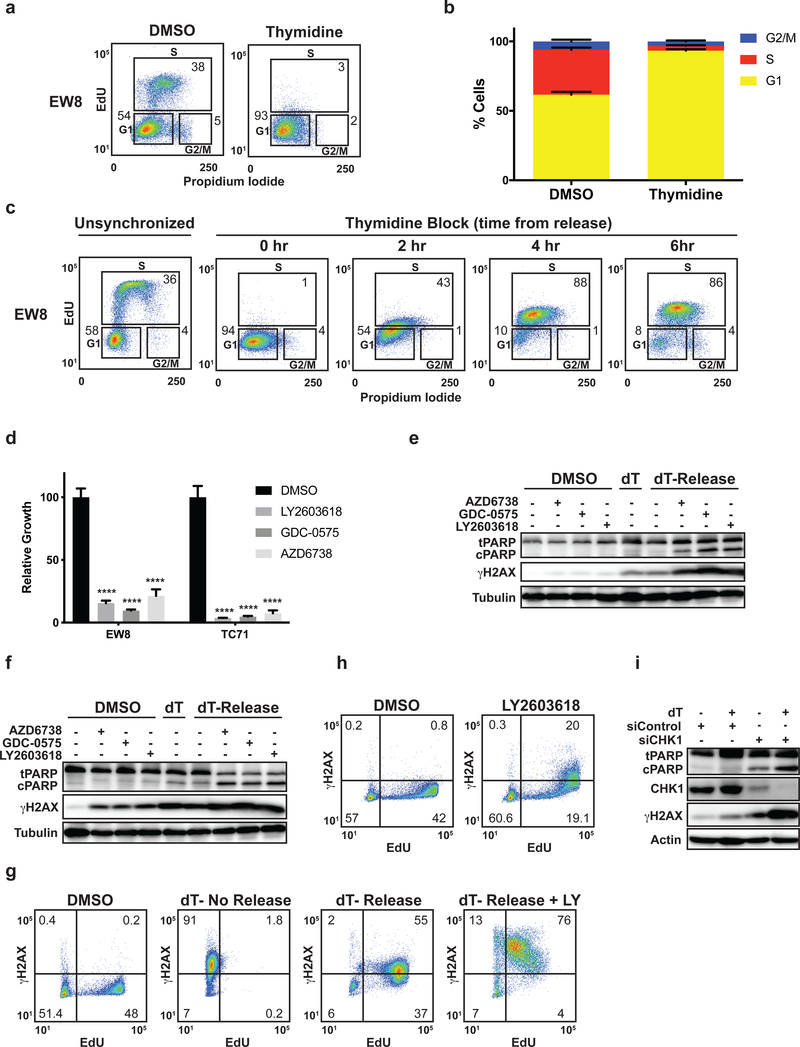
In order to delineate the cell-cycle dependent effects of inhibition of the ATR-CHK1 pathway in Ewing sarcoma, we synchronized cells at the G1/S border using a double thymidine block. We chose to synchronize the cells using thymidine, as opposed to other methods, because a thymidine double block 1) arrests cells at the G1/S border and 2) causes DNA replication stress and DNA damage by inhibiting ribonucleotide reductase (RNR) (21–23). Based on our previous work demonstrating synergy between ATR-CHK1 inhibitors and drugs that cause DNA replication stress, we hypothesized that the induction of DNA damage and/or replication stress by thymidine would recapitulate, in part, the effects of gemcitabine and other inhibitors of RNR (6). Figures 1A and 1B show that the treatment of multiple Ewing sarcoma cells lines with a double thymidine block causes accumulation of cells at the G1/S border. Removal of the thymidine after the double block then results in the orderly progression of cells through S phase (Figure 1C and Supplementary Figure 1A). Next, to determine whether ATR-CHK1 signaling is required for cell survival in early S phase, we released Ewing sarcoma cells from a double thymidine double block in the presence of two different CHK1 inhibitors (LY2603618 and GDC-0575), an ATR inhibitor (AZD6738), or vehicle (24–26). The ATR and CHK1 inhibitors were removed after a 4 h incubation, while the cells were still in S phase (Figure 1C). Cell growth was then quantified 24 h after drug removal using Cell-Titer-Glo. Figure 1D shows that treatment with the ATR and CHK1 inhibitors significantly reduced cell growth after release from the thymidine block. However, in contrast to these thymidine-treated cells, the treatment of unsynchronized cells with CHK1 or ATR inhibitors for 4 h had a minimal impact on cell growth (Supplementary Figure 1B).
Figure 1.
Inhibition of the ATR-CHK1 pathway in Ewing sarcoma cells in S phase causes DNA damage and apoptosis. (A) Representative cell cycle analysis (EdU and propidium iodide) plot for Ewing sarcoma cells (EW8) synchronized at the G1/S border using a thymidine double block. (B) Summary of cell cycle analysis for four additional Ewing sarcoma cell lines that were subjected to a thymidine double block. (C) EW8 cells were arrested at the G1/S border using a thymidine double block and then released into S phase. Cell cycle analysis with EdU and propidium iodide was performed at the indicated time points after release from the block. Results are representative of two independent experiments. (D) EW8 and TC71 cells were released from a thymidine double block in the presence of LY2603618 (1 μM), GDC-0575 (1 μM), AZD6738 (1 μM), or DMSO for 4 h. Drugs were then removed and cell viability was quantified 24 h later using Cell-Titer-Glo. (E, F) EW8 (E) and TC71 (F) cells were released from a thymidine double block in the presence of drugs, as described in (D), for 4 h. Cell lysates were then collected for immunoblotting. (G) EW8 cells were released from a thymidine double block in the presence of LY2603618 or DMSO for 4 h. Cells were then labeled with EdU and fixed for flow cytometry for γH2AX. Results are representative of two independent experiments. (H) Unsynchronized EW8 cells were treated with LY2603618 or DMSO for 4 h, pulse-labeled with EdU, and fixed for flow cytometry for γH2AX. Results are representative of three independent experiments. (I) EW8 cells were released from a thymidine double block 24 h after transfection with siRNA targeting CHK1. Cellular lysates were collected 4 h after the cells were released from the block. **** indicates P < 0.0001.
Figures 1E and 1F show that inhibition of ATR or CHK1 in cells released from a thymidine double block results in phosphorylation of H2AX (γH2AX), a marker of DNA damage, and cleavage of PARP, a marker of apoptosis. In addition, we also observed that thymidine, even in the absence of an ATR or CHK1 inhibitor, induced phosphorylation of H2AX in Ewing sarcoma cells, consistent with reports of thymidine causing replication stress and DNA damage in other cell types (21–23). In particular, the TC71 cells were more sensitive than the EW8 cells to thymidine as a single agent. However, the phosphorylation of H2AX was enhanced when the thymidine double block was followed by treatment with an ATR or CHK1 inhibitor. Similar results were obtained with the TC32 cell line (Supplementary Figure 1C). Flow cytometry was also used to quantify the phosphorylation of H2AX in EW8 cells (Figure 1G), as well as the additional Ewing sarcoma cell lines (TC71 and TC32) (Supplementary Figure 1D). We also treated unsynchronized cells with a CHK1 inhibitor for 4 h and pulse-labeled the cells with EdU. Figure 1H shows that a CHK1 inhibitor, even in the absence of thymidine treatment or a DNA replication stress inducer, caused DNA damage (γH2AX) in cells with active DNA replication. The effect of the CHK1 inhibitor as a single agent, however, was diminished relative to the combination of thymidine, which causes replication stress, and the CHK1 inhibitor. Finally, to ensure that the effects of the ATR and CHK1 inhibitors were on-target we used a siRNA, validated in our previous work, to knockdown CHK1 in cells treated with a double thymidine block (5). Similar to the results using the small-molecule inhibitors, Figure 1I shows that siCHK1, but not siControl, caused phosphorylation of H2AX and cleavage of PARP.
Inhibition of the ATR-CHK1 pathway in Ewing sarcoma cells in S phase activates CDK1/2
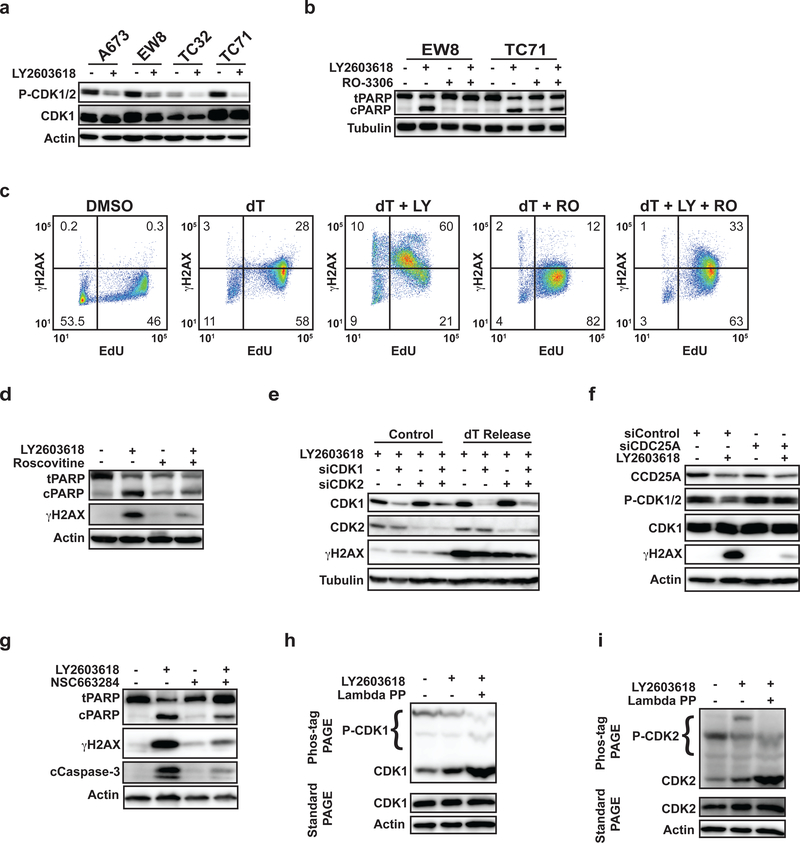
CDK1 and CDK2 are critical mediators of cell cycle progression that are regulated by the ATR-CHK1-CDC25A pathway (27,28). In the setting of DNA replication stress, ATR-CHK1 negatively regulates CDC25A, which de-phosphorylates and activates CDK1/2, to restrain cell cycle progression and promote DNA damage repair. Figure 2A shows that the release of Ewing sarcoma cells from a thymidine double block in the presence of a CHK1 inhibitor results in an increase in the unphosphorylated, or activated, forms of CDK1 and/or CDK2. Notably, the phosphorylation-specific antibody for CDK1 is not specific for CDK1 and also detects the phosphorylated form of CDK2 (29). In contrast, the treatment of several non-Ewing sarcoma cell lines, including BJ-tert, RPE-tert, and U2OS, with a thymidine block followed by a CHK1 inhibitor did not alter the phosphorylation of CDK1/2 (Supplementary Figure 2A). Next, EW8 and TC71 cells were released from a thymidine double block in the presence of a CHK1 inhibitor (LY2603618), the CDK1/2 inhibitor RO-3306, or the combination of LY2603618 and RO-3306 (30). In a similar experiment, EW8 cells were also pulse-labeled with EdU and fixed for flow cytometry for γH2AX. Figures 2B and 2C show that inhibition of CDK1/2 with RO-3306 reduced both the cleavage of PARP and the phosphorylation of H2AX, respectively. Treatment with RO-3306 also blocked morphologic changes suggestive of apoptosis and cell death (Supplementary Figure 2B). Similar results were obtained with the TC32 cell line (Supplementary Figure 2C). Roscovitine, an additional inhibitor of CDK1/2, also reduced the cleavage of PARP and phosphorylation of H2AX caused by the CHK1 inhibitor (Figure 2D) (31). Next, we used siRNA to knockdown CDK1, CDK2, and the combination of CDK1 and CDK2. Supplementary Figure 2D shows that the knockdown of CDK1 and CDK2 did not impair the entry of cells into S phase at 24 h after siRNA transfection (29). However, Figure 2E shows that the knockdown of CDK1 and CDK2 decreased the phosphorylation of H2AX in cells released from a thymidine double block in the presence of a CHK1 inhibitor. Figure 2F shows that the siRNA-mediated knockdown of CDC25A, which is regulated by ATR-CHK1 and de-phosphorylates CDK1/2, blocked the activation of CDK1/2 and significantly reduced the phosphorylation of H2AX caused by treatment of the cells with a CHK1 inhibitor. Similarly, treatment of EW8 cells with NSC663284, an inhibitor of CDC25A, also reduced phosphorylation of H2AX and cleavage of PARP (Figure 2G). Finally, as phospho-CDK1 and phospho-CDK2 antibodies are cross-reactive and not specific to phospho-CDK1 or phospho-CDK2, we used phos-tag polyacrylamide electrophoresis to discern activation of CDK1 versus CDK2 (29). Figures 2H and 2I show that treatment of EW8 cells with LY2603618 after release from a thymidine block increased the unphosphorylated, or activated, forms of both CDK1 and CDK2. Of note, a sample of the protein lysate was also evaluated using standard polyacrylamide gel electrophoresis to ensure that the levels of total CDK1 and CDK2 were unaltered by the drug treatment.
Figure 2.
Inhibition of the ATR-CHK1 pathway in Ewing sarcoma cells in S phase activates CDK1/2. (A) Multiple Ewing sarcoma cell lines were released from a thymidine double block in the presence of LY2603618 (1 μM) or DMSO. Cellular lysates were harvested 4 h after release from the block and probed for CDK1/2 phosphorylation by immunoblotting. (B) EW8 and TC71 cells were released from a thymidine double block in the presence of LY2603618 (1 μM), RO-3306 (10 μM), or the combination of LY2603618 and RO-3306. Cellular lysates were collected for immunoblotting 4 h after release from the block. (C) EW8 cells were released from a thymidine double block in the presence of LY2603618 (1 μM), RO-3306 (10 μM), or the combination of LY2603618 and RO-3306 for 4 h. Cells were then labeled with EdU and fixed for flow cytometry for γH2AX. Results are representative of two independent experiments. (D) EW8 cells were released from a thymidine double block in the presence of LY2603618 (1 μM), Roscovitine (10 μM), or the combination of LY2603618 and Roscovitine. Cellular lysates were collected for immunoblotting 4 h after release from the block. (E) EW8 cells were released from a thymidine double block 24 h after transfection with siRNA targeting CDK1, CDK2, or CDK1 and CDK2. Cellular lysates were collected 4 h after the cells were released from the block in the presence of LY2603618 and probed for CDK1, CDK2, and γH2AX. (F) EW8 cells were released from a thymidine double block 24 h after transfection with siRNA targeting CDC25A. Cellular lysates were collected 4 h after the cells were released from the block in the presence of LY2603618 and probed for CDC25A, CDK1, P-CDK1/2, and gH2AX. (G) EW8 cells were released from a thymidine double block in the presence of LY2603618 (1 μM), NSC663284 (10 μM), or the combination of LY2603618 and NSC663284. Cellular lysates were collected for immunoblotting 4 h after release from the block. (H, I) EW8 cells were released from a thymidine double block in the presence of LY2603618 and then cellular lysates were collected 4 h after release. Electrophoresis was performed using phos-tag gels and the gels were probed with CDK1 (H) and CDK2 (I) antibodies. As a control, an aliquot of protein lysate was treated with lambda phosphatase. The same lysates were also evaluated using standard polyacrylamide gel electrophoresis, which does not separate the unphosphorylated and phosphorylated CDK1 and CDK2 proteins, to ensure that the levels of total CDK1 and CDK2 are not altered by drug treatment.
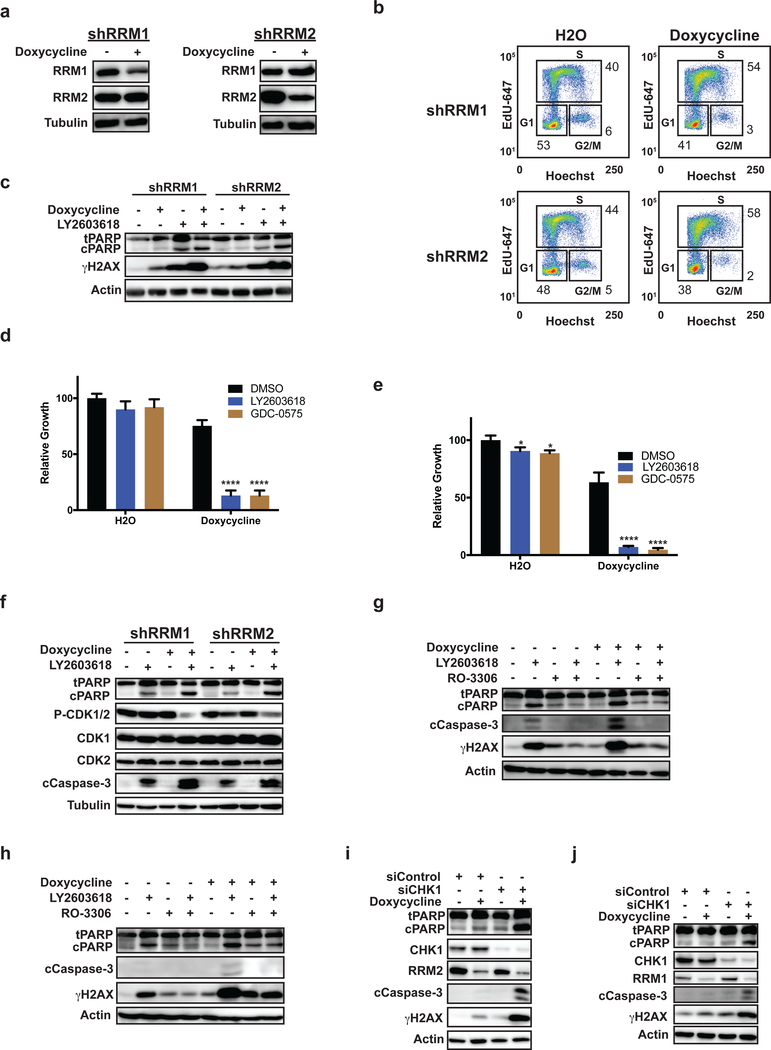
Inhibition of CHK1 in Ewing sarcoma cells with shRNA-mediated knockdown of RRM1 or RRM2 activates CDK1/2 and causes apoptosis
Thymidine causes DNA replication stress and, as shown above, increased the DNA damage and apoptosis caused by ATR and CHK1 inhibitors (21–23). To complement the thymidine studies, we also used a genetic approach to partially reduce the levels of the RRM1 and RRM2 subunits of ribonucleotide reductase (RNR), the rate-limiting enzyme in the synthesis of deoxyribonucleotides (dNTPs), and impair DNA replication. In a previous publication, we showed that the knockdown of RRM1 or RRM2 using potent siRNAs caused replication stress, activation of ATR-CHK1 signaling, apoptosis, and cell death in Ewing sarcoma cells (5). However, in the current work, we used doxycycline-inducible shRNA to more modestly reduce the levels of RRM1 or RRM2 (Figure 3A). Figure 3B shows that this partial knockdown of RRM1 or RRM2 increased the percentage of cells in S phase, but did not arrest the cells. Reducing RRM1 and RRM2 levels also caused a mild increase in the phosphorylation of H2AX, but no significant cleavage of PARP (Figure 3C). However, treatment of these knockdown cells with CHK1 inhibitors for 4 h caused an increase in phosphorylation of H2AX, cleavage of PARP, and a significant reduction in cell viability compared to the parental cells (Figure 3C–E and Supplementary Figure 3A–B). Next, we treated the shRRM1 and shRRM2 knockdown cells with a CHK1 inhibitor and assessed, by immunoblotting, for activation of CDK1/2. Figure 3F shows that treatment of the cells with a CHK1 inhibitor, in the presence of doxycycline and knockdown of RRM1 or RRM2, caused activation of CDK1/2, as well as cleavage of PARP and caspase-3. However, treatment of these cells lines with a CDK1/2 inhibitor, RO-3306, in combination with the CHK1 inhibitor reduced the phosphorylation of H2AX and decreased cleavage of PARP and caspase-3, similar to the results obtained using a thymidine block (Figure 3G–H). Figures 3I and 3J show that knockdown of CHK1 in the EW8-shRRM2 and EW8-shRRM1 cells using siRNA caused phosphorylation of H2AX, cleavage of PARP, and cleavage of caspase-3. We also generated an additional cell line with doxycycline-inducible shRNA knockdown of RRM1. Supplementary Figures 3C–E show that the treatment of TC71-shRRM1 cells with a CHK1 inhibitor resulted in cleavage of PARP, phosphorylation of H2AX, and activation of CDK1/2, similar to the results obtained with the EW8 cells.
Figure 3.
Inhibition of CHK1 in Ewing sarcoma cells with shRNA-mediated knockdown of RRM1 or RRM2 activates CDK1/2 and causes apoptosis. (A) EW8 cell lines with doxycycline-inducible, shRNA-mediated knockdown of RRM1 and RRM2 were treated with doxycycline or vehicle for 48 h. Cellular lysates were then collected for immunoblotting for RRM1 and RRM2. (B) The EW8-shRRM1 and EW8-shRRM2 cell lines were treated with doxycycline or vehicle for 48 h, fixed, and subjected to cell cycle analysis using EdU and Hoechst. Results are representative of two independent experiments. (C) EW8-shRRM1 and EW8-shRRM2 cells were treated with doxycycline or vehicle for 24 h and then treated with LY2603618 (1 μm) for 4 h. Cellular lysates were then collected for immunoblotting for cleaved and total PARP and γH2AX. (D, E) The EW8-shRRM1 (D) and EW8-shRRM2 (E) cell lines were treated with doxycycline, LY2603618, or GDC-0575 as described in (C). LY2603618 and GDC-0575 were removed after 4 h and cell growth was quantified 48 h later using Cell-Titer-Glo. (F) The EW8-shRRM1 and EW8-shRRM2 cell lines were treated with doxycycline and LY2603618 as described in (C). LY2603618 was removed after 4 h and cellular lysates were then collected for immunoblotting. (G, H) EW8-shRRM1 (G) and EW8-shRRM2 (H) cells, grown in the presence of doxycycline or vehicle, were treated with LY2603618, RO-3306, or the combination of LY2603618 and RO-3306 for 4 h. Cellular lysates were then collected for immunoblotting for cleaved and total PARP, γH2AX, and cleaved caspase-3. (I) EW8-shRRM2 cells, grown in the presence of doxycycline or vehicle, were transfected with siRNA targeting CHK1. Cellular lysates were collected for immunoblotting 24 h after transfection. (J) EW8-shRRM1 cells, grown in the presence of doxycycline or vehicle, were treated with LY2603618 for 4 h. Cellular lysates were then harvested for immunoblotting 24 h after transfection. **** indicates P < 0.0001.
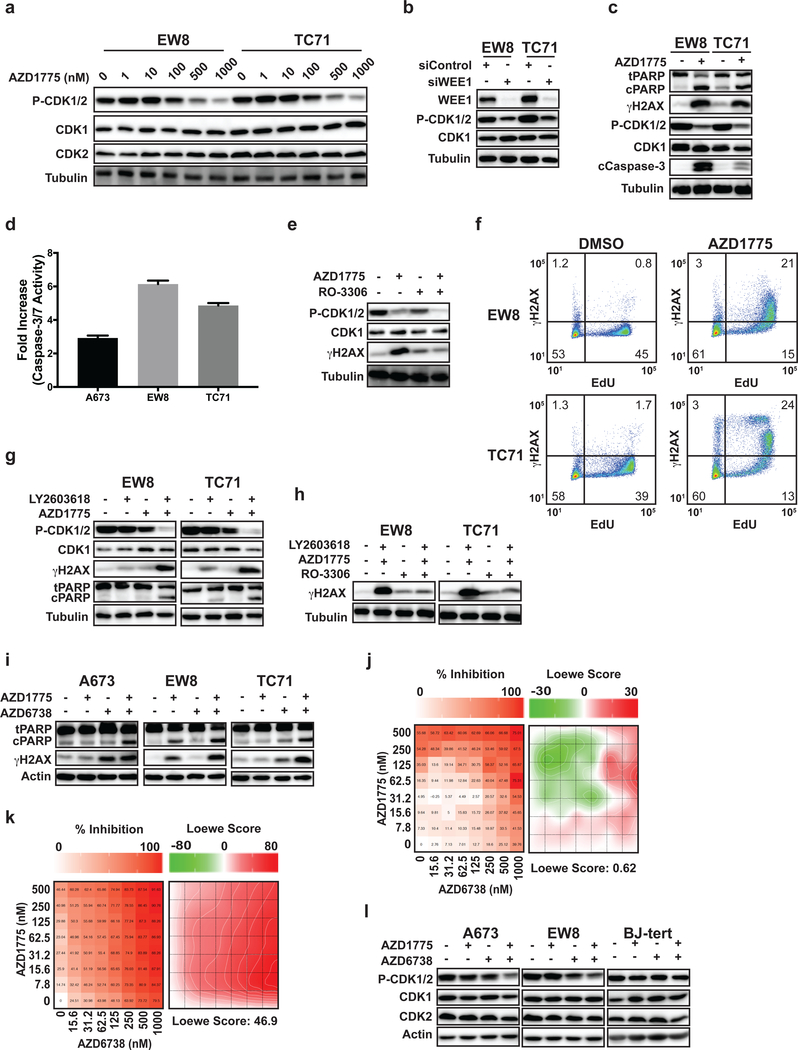
Inhibition of WEE1 kinase with AZD1775 activates CDK1/2 and causes apoptosis in Ewing sarcoma cells
The WEE1 kinase phosphorylates and inactivates CDK1/2 (32). Figure 4A shows that treatment of EW8 and TC71 cells with the WEE1 kinase inhibitor AZD1775 caused a dose-dependent activation, or decreased phosphorylation, of CDK1/2. Similarly, knockdown of WEE1 using siRNA caused activation of CDK1/2 (Figure 4B). In addition, treatment with AZD1775 also caused cleavage of PARP, cleavage of caspase-3, activation of caspase 3/7 as assessed using a luminescence assay, and phosphorylation of H2AX (Figure 4C–D). However, the phosphorylation of H2AX induced by AZD1775 was blocked by concurrent treatment of the cells with the CDK1/2 inhibitor RO-3306 (Figure 4E). Next, we treated Ewing sarcoma cells with AZD1775 and then pulse-labeled the cells with EdU to identify cells with active DNA replication. Figure 4F shows that the DNA damage induced by AZD1775, as assessed by staining for γH2AX, occurred in cells with active DNA replication.
Figure 4.
Inhibition of WEE1 kinase with AZD1775 activates CDK1/2 and causes apoptosis in Ewing sarcoma cells. (A) EW8 and TC71 cells were treated with different doses of AZD1775 (0–1000 nM) for 6 h. Cellular lysates were then harvested for immunoblotting for CDK1 and P-CDK1/2. (B) EW8 and TC71 were transfected with siRNA targeting WEE1. Cellular lysates were harvested 24 h after transfection and immunoblotting was performed for CDK1 and P-CDK1/2. (C) EW8 and TC71 were treated with AZD1775 (500 nM) for 18 h. Cellular lysates were then collected for immunoblotting for cleaved and total PARP, γH2AX, CDK1, P-CDK1/2, and cleaved caspase-3. (D) EW8, TC71, and A673 cells were treated with AZD1775 (500 nM) for 18 h and then caspase-3/7 activation was quantified using Casp-Glo 3/7 Luminescence assay. (E) EW8 cells were treated with AZD1775 (500 nM), RO-3306 (10 μM), or the combination of AZD1775 and RO-3306 for 6 h. Cellular lysates were then collected for immunoblotting for γH2AX, CDK1, and P-CDK1/2. (F) EW8 and TC71 cells were treated with AZD1775 (500 nM) for 6 h. Cells were then labeled with EdU and fixed for flow cytometry for γH2AX. Results are representative of two independent experiments. (G) EW8 and TC71 cells were treated with AZD1775 (100 nM), LY2603618 (100 nM), or the combination of AZD1775 and LY2603618 for 18 h. Cellular lysates were then collected for immunoblotting for gH2AX, CDK1, P-CDK1/2, and cleaved and total PARP. (H) EW8 and TC71 cells were treated for 6 h with AZD1775 (100 nM), LY2603618 (100 nM), RO-3306 (10 μM) or the combination of AZD1775 and LY2603618 with RO-3306. Cellular lysates were then collected for immunoblotting for γH2AX. (I) A673, EW8, and TC71 cells were treated with AZD1775 (100 nM), the ATR inhibitor AZD6738 (100 nM), or the combination of AZD1775 and AZD6738 for 18 h. Cellular lysates were then collected for immunoblotting for cleaved and total PARP and γH2AX. (J, K) BJ-tert (J) and A673 (K) cells were treated for 72 h with a combination of up to 1000 nM AZD6738 and 500 nM AZD1775. Survival was assayed by Cell-Titer-Glo and each experiment was repeated 2 times. Color bars indicate % inhibition normalized to untreated cells. Loewe matrix plots for drug cooperativity are also shown. (L) A673, EW8, and BJ-tert cells were treated with AZD1775 (100 nM), AZD6738 (100 nM), and the combination of AZD1775 (100 nM) and AZD6738 (100 nM). Cellular lysates were collected for immunoblotting 24 h after the drugs were added.
We then treated Ewing sarcoma cells with low doses of AZD1775 and a CHK1 inhibitor, LY2603618. Figure 4G demonstrates that the combination of the drugs, but neither drug as a single agent (at low concentrations), caused activation of CDK1/2, cleavage of PARP, and phosphorylation of H2AX. The combination of the drugs did not alter the cell cycle, though (Supplementary Figure 4A–B). Furthermore, as shown in Figure 4H, the addition of the CDK1/2 inhibitor RO-3306 to the combination of the WEE1 and CHK1 inhibitors significantly reduced phosphorylation of H2AX. Inhibition of ATR in combination with inhibition of WEE1 also caused cleavage of PARP and phosphorylation of H2AX (Figure 4I). Next, BJ-tert cells and multiple Ewing sarcoma cell lines were incubated with different concentrations of inhibitors of WEE1 (AZD1775) and ATR (AZD6738) for 72 h before quantifying the surviving cells with Cell-Titer-Glo. Figure 4J shows that the drug combination did not cause synergistic toxicity (Loewe Score 0.62) with the BJ-tert cells, consistent with reports that this drug combination is selective for cancer cells and causes minimal systemic toxicity in vivo (33). In contrast, the combination of AZD1775 and AZD6738 was highly synergistic with multiple Ewing sarcoma cell lines, as shown in a representative Loewe synergy matrix plot (Loewe Score 46.9) (Figure 4K) and summarized for multiple cell lines (Table 1) (Supplementary Figure 4C–E) (19). AZD1775 also showed synergy (Loewe Score 44.9) with an additional ATR inhibitor, VX-970, in TC71 cells (Supplementary Figure 5) (34). Finally, the combination of AZD1775 and AZD6738 activated CDK1/2 in the A673 and EW8, but not BJ-tert, cells (Figure 4L).
Table 1.
Average synergy scores for drug combinations calculated using the Loewe Additivity and Bliss Independence methods.
| Cell Line | Drug 1 | Drug 2 | Loewe Synergy Score | Bliss Synergy Score |
|---|---|---|---|---|
| BJ-tert | AZD1775 | AZD6738 | 0.6 | 0.4 |
| EW8 | AZD1775 | AZD6738 | 34.8 | 10.8 |
| TC71 | AZD1775 | AZD6738 | 21.3 | 6.8 |
| TC32 | AZD1775 | AZD6738 | 27.7 | 9.1 |
| A673 | AZD1775 | AZD6738 | 46.9 | 11.7 |
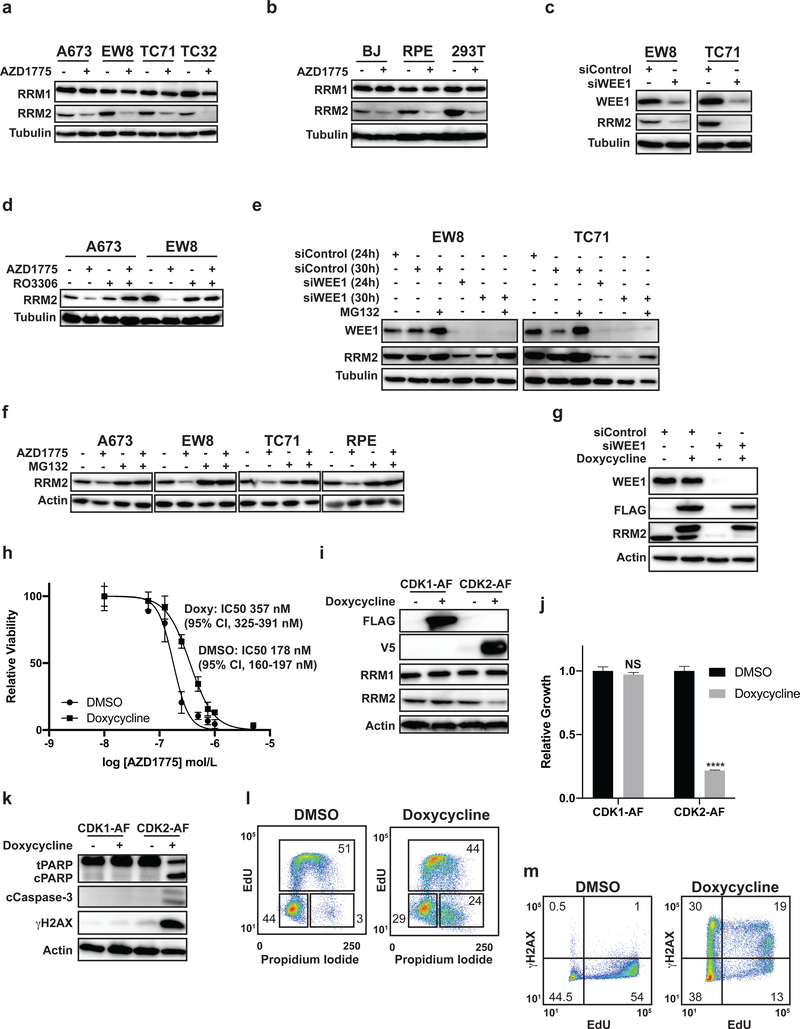
CDK2 activation in Ewing sarcoma cells causes RRM2 degradation, DNA damage, and apoptosis
In a previous publication, and Supplementary Figure 6A, we showed that knockdown or inhibition of RRM2 caused apoptosis in Ewing sarcoma cells (5). RRM2, which is required for DNA replication and the repair of DNA damage, is phosphorylated on Threonine-33 by CDK1/2 in G2 phase after completion of DNA synthesis and, thereby, targeted for ubiquitin-mediated proteolysis (35,36). Figure 5A shows that treatment of four Ewing sarcoma cell lines with AZD1775 caused a reduction in RRM2 protein levels. Similar results were obtained with three non-Ewing sarcoma cell lines, suggesting the effect of AZD1775 on RRM2 is not specific to Ewing sarcoma cells (Figure 5B) (36). Using a genetic approach, we also reduced WEE1 levels with siRNA, which activates CDK1/2 (Figure 4B), and identified a comparable reduction in RRM2 levels (Figure 5C). Next, we treated Ewing sarcoma cells with AZD1775 in combination with the CDK1/2 inhibitor RO-3306. As shown in Figure 5D, inhibition of CDK1/2 with RO-3306 blocked the reduction in RRM2 levels. We then used siRNA to reduce the levels of WEE1 and assessed whether inhibition of the proteasome with MG132 would rescue RRM2 levels. Figure 5E shows that MG132 increased the levels of RRM2 in EW8 and TC71 cells after knockdown of WEE1. Similar results were obtained when AZD1775 was combined with MG132 in A673, EW8, TC71, and RPE cells (Figure 5F). In addition, we did not observe a significant effect of siWEE1 or AZD1775 on RRM2 mRNA levels, as assessed using qPCR (Supplementary Figure 6B–E). Next, we expressed a doxycycline-inducible FLAG-RRM2 transgene (TO-FLAG-RRM2-T33A) with a T33A mutation, which was previously shown to be resistant to the ubiquitin-mediated-proteolysis caused by inhibition of WEE1, in a Ewing sarcoma cell line (36). Figure 5G shows that FLAG-RRM2-T33A, compared to the endogenous RRM2 protein, is resistant to the degradation caused by knockdown of WEE1. Furthermore, the doxycycline-inducible expression of FLAG-RRM2-T33A also significantly reduced the sensitivity of cells to the WEE1 inhibitor AZD1775 (IC50 178 nM versus 357 nM) (Figure 5H).
Figure 5.
CDK2 activation in Ewing sarcoma cells promotes RRM2 degradation. (A) Four Ewing sarcoma cell lines were treated with AZD1775 (500 nM) for 8 h. Cellular lysates were then harvested for immunoblotting for RRM1 and RRM2. (B) Three non-Ewing sarcoma cell lines were treated with AZD1775 as described in (A). (C) EW8 and TC71 cells were transfected with siRNA targeting WEE1. Cellular lysates were then collected 24 h after transfection for immunoblotting for WEE1 and RRM2. (D) EW8 and TC71 cells were treated with AZD1775 (500 nM), RO-3306 (10 μM), or the combination of AZD1775 and RO-3306 for 6 h. Cellular lysates were then harvested for immunoblotting for RRM2. (E) EW8 and TC71 cells were transfected with siWEE1 or siControl. MG132 (1 μM) was added 24 h after transfection. Cellular lysates were collected at 24 and 30 h after transfection for immunoblotting for RRM2 and WEE1. (F) A673, EW8, TC71, and RPE-tert cells were treated with AZD1775 (500 nM), MG132 (1 μM), or the combination of AZD1775 (500 nM) and MG132 (1 μM) for 6 h. Lysates were then collected for immunoblotting. (G) TO-FLAG-RRM2-T33A cells were treated with doxycycline in combination with siControl or siWEE1 for 24 h. Cellular lysates were then collected for immunoblotting. (H) Dose response curves for TO-FLAG-RRM2-T33A cells treated with different concentrations of AZD1775 in the presence or absence of doxycycline. Cell viability was assessed 72 h after drug addition using the AlamarBlue assay. Error bars represent the mean ± SD of three technical replicates. The results are representative of two independent experiments. (I) FLAG-CDK1-AF and V5-CDK2-AF cell lines were treated with doxycycline for 24 h and then cellular lysates were harvested for immunoblotting for FLAG, V5, RRM1, and RRM2. (J) FLAG-CDK1-AF and V5-CDK2-AF cell lines were treated with doxycycline for 72 h and then cell growth was quantified by Cell-Titer-Glo. (K) FLAG-CDK1-AF and V5-CDK2-AF cell lines were treated with doxycycline for 24 h and then cellular lysates were harvested for immunoblotting for cleaved PARP, cleaved caspase-3, and γH2AX. (L) Cell cycle analysis (EdU and propidium iodide) plot for V5-CDK2-AF cells treated with doxycycline for 24 h. Results are representative of two independent experiments. (M) V5-CDK2-AF cells were treated with doxycycline for 24 h and then pulse-labeled with EdU and fixed for flow cytometry for γH2AX. Results are representative of two independent experiments.**** indicates P < 0.0001.
Earlier studies did not distinguish whether CDK1 or CDK2 regulates RRM2 degradation (36). In addition, phos-tag gel electrophoresis (Figures 2H and 2I) showed activation of both CDK1 and CDK2 in Ewing sarcoma cells. Consequently, in order to identify the functional consequences of activation of CDK1 or CDK2, we generated constitutively active mutants of CDK1 and CDK2 which substitute alanine for threonine at position 14 and phenylalanine for tyrosine at position 15 (20,37–39). The FLAG-CDK1-AF and V5-CDK2-AF constructs were cloned into doxycycline-inducible vectors and expression of the tagged proteins in EW8 cells were observed when cells were treated with doxycycline (Figure 5I). Notably, expression of constitutively active CDK2-AF, but not CDK1-AF, caused a reduction in the level of the RRM2 protein (Figure 5I). Figures 5J and 5K show that the inducible expression of CDK2-AF also significantly reduced cell viability and increased markers of apoptosis. In addition, expression of CDK2-AF, but not CDK1-AF (Supplementary Figure 7A–B), reduced DNA replication, as expected based on the reduction of levels of RRM2, and increased phosphorylation of H2AX (Figure 5L–M). Despite the lack of effect of CDK1-AF on RRM2 levels or cell viability, we did observe increased phosphorylation of the CDK1 target lamin A/C, demonstrating that CDK1-AF is functional (Supplementary Figure 7C).
DISCUSSION
Ewing sarcoma cells exhibit elevated levels of endogenous DNA replication stress, which may, in part, explain the sensitivity of this cancer to RNR, ATR, and CHK1 inhibitors (3,4). Notably, both haploinsufficiency of EWSR1 and the EWS-FLI1 oncogene are reported to contribute to DNA replication stress and DNA damage in Ewing sarcoma tumors. For example, Gorthi et al. recently showed that depletion of EWSR1 increased the formation of R-loops, which are DNA-RNA hybrids that impair the progression of the DNA replication machinery, and activated ATR in Ewing sarcoma tumors (3). In other work, depletion of EWSR1 was also reported to impact the splicing of genes involved in DNA repair and genotoxic stress signaling, which suggests the role of EWSR1 in replication stress may be multifactorial (40). Similarly, the EWS-FLI oncogene is reported to regulate multiple aspects of the cellular response to genotoxic stress, as reviewed by Ghosal et al. (41). For example, SLFN11 is a direct transcriptional target of EWS-FLI1 and overexpression of SLFN11 sensitizes cancer cells to a wide range of DNA-targeted therapies by blocking replication fork progression (42,43). In addition, Stewart et al. showed that Ewing sarcoma cells are defective in DNA damage repair and downregulate multiple repair genes, including BRCA1, GEN1, and ATM (44). Furthermore, germline sequencing of patients with Ewing sarcoma tumors has identified a significant enrichment for mutations in genes involved in DNA damage repair (45). Overall, these studies suggest that the etiology of the elevated levels of endogenous DNA replication stress in Ewing sarcoma cells may involve multiple mechanisms.
Similarly, a number of different mechanisms have been reported to underlie the toxicity of ATR-CHK1 inhibitors, both as single agents and in combination with other drugs, toward cancer cells (11–13,18). These different mechanisms are likely related, in part, to the diverse functions and the multiple downstream targets of the ATR and CHK1 kinases. In this work, we identified that inhibition of the ATR-CHK1 and WEE1 pathways in Ewing sarcoma cells causes activation of CDK1 and CDK2. In addition, inhibition or siRNA-mediated knockdown of CDK1/2 reduced DNA damage and apoptosis. Furthermore, using expression of constitutively active CDK1 and CDK2 mutants, we also identified that the aberrant activation of CDK2 leads to the degradation of RRM2 and, thereby, generates a feedback loop which exacerbates DNA replication stress, increases DNA damage, and induces apoptosis (Figure 6). However, although overexpression of constitutively active CDK1 did not deplete RRM2 or induce DNA damage in Ewing sarcoma cells, we did note that knockdown of CDK1 with siRNA modestly reduced DNA damage in the setting of replication stress (Figure 2G) and, therefore, we cannot fully rule out a contribution from CDK1 in mediating the effects of inhibition of the ATR-CHK1 pathway.
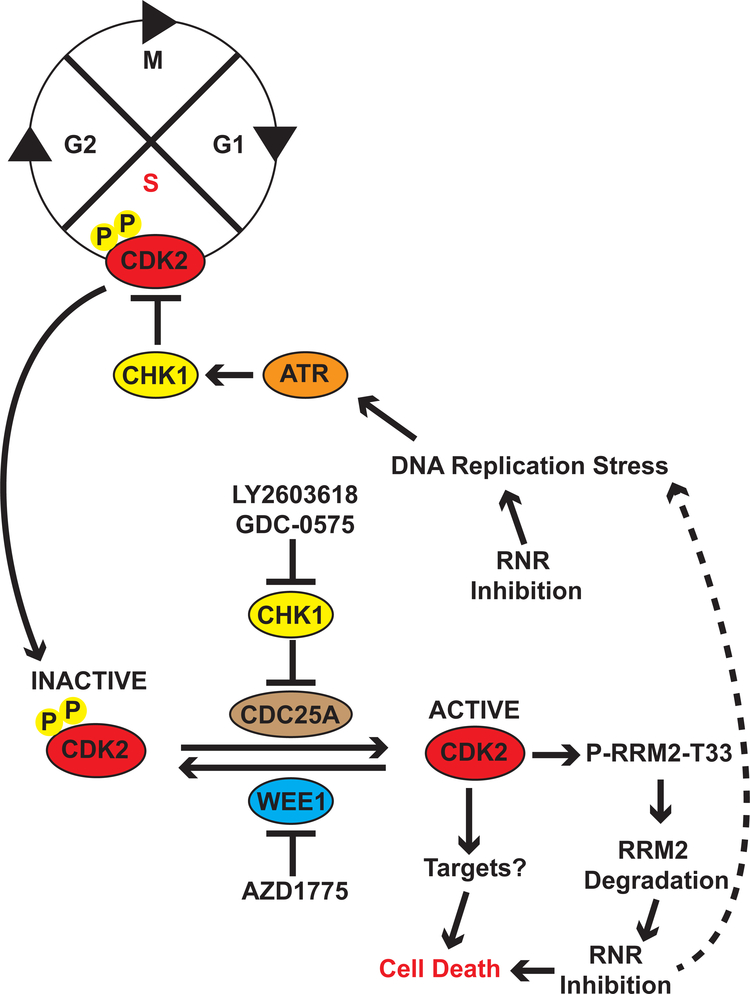
Figure 6.
Proposed mechanism for the regulation of RRM2 levels by the ATR-CHK1 and WEE1 pathways in Ewing sarcoma cells. Inhibition of the ATR-CHK1 and WEE1 pathways in Ewing sarcoma cells experiencing DNA replication stress, caused by inhibition of RRM1, RRM2, or other mechanisms, leads to the aberrant activation of CDK2. Active CDK2, in turn, phosphorylates and targets RRM2 for degradation via the proteasome. This reduction in the level of the RRM2 protein caused by activation of CDK2 generates a feedback loop which exacerbates DNA replication stress, increases DNA damage, and induces apoptosis.
The classical view is that CDK2 regulates entry into S phase and the initiation of DNA replication, while entry into M phase requires the activity of CDK1 (27,46). However, there is significant functional overlap between these two kinases and the aberrant activation of both CDK1 and CDK2 have been reported to cause DNA damage and toxicity. For example, Szmyd et al. showed that the monoallelic expression of CDK1-AF is embryonic lethal in mice and causes an S phase arrest with associated phosphorylation of H2AX and activation of the DNA damage checkpoint (47). Similarly, Hughes et al. showed that expression of CDK2-AF in HCT116 cells caused premature entry into S phase, abnormal DNA replication, and sensitivity to replication stress (48). Additional reports have also shown that CHK1 inhibitors, as single agents or in combination with gemcitabine, cause CDK2-dependent toxicity (49,50). On the other hand, Garcia et al. showed that the expression of the combination of CDK1-AF and CDK2-AF, but neither construct alone, enhanced the DNA damage and apoptosis caused by cytarabine in leukemia cells (51). Overall, these studies suggest that activation of CDK1 and CDK2, as well as the downstream impact of active CDK1 and CDK2, is likely to depend on multiple factors, including tumor type, the phase of the cell cycle, and the presence of replication stress.
Notably, from a mechanistic standpoint, we identified that the activation of CDK2 promotes the ubiquitin-mediated proteolysis of RRM2, which is the small subunit of ribonucleotide reductase (RNR). RNR catalyzes the rate-limiting step in the synthesis of deoxyribonucleotides from ribonucleotides and inhibition of RRM2, or the RRM1 subunit of RNR, depletes nucleotides and causes DNA replication stress and DNA damage (52). The levels of RRM2 peak during S phase and RRM2 is normally degraded during G2 phase (35). However, in Ewing sarcoma cells, we identified that the aberrant activation of CDK2, caused by inhibiting ATR, CHK1, or WEE1, leads to the proteolysis of RRM2. Notably, inhibition or knockdown of RRM2 causes DNA replication stress, DNA damage, and apoptosis in Ewing sarcoma tumors (5,6). Consequently, inhibition of the ATR-CHK1 pathway, or the WEE1 kinase, in Ewing sarcoma cells experiencing replication stress generates a feedback loop that depletes RRM2, exacerbates replication stress, and increases DNA damage. Finally, although we identified RRM2 as one target of active CDK2, we also note that CDK2 phosphorylates numerous substrates and we expect that the toxicity caused by the aberrant activation of CDK2 in Ewing sarcoma cells is dependent on multiple downstream targets and not solely RRM2 (53,54).
We also found that the inhibition of CHK1 and WEE1 are not equivalent, which is consistent with reports in the literature (33,55–61). For example, the WEE1 inhibitor AZD1775 was able to activate CDK1/2 in both Ewing and non-Ewing sarcoma lines (Figure 5A–B). In contrast, inhibition of CHK1 activated CDK1/2 in the Ewing sarcoma cell lines, but not the control cell lines that we tested (Figure 2A and Supplementary Figure 2A). We hypothesize that this reflects the distinct substrates and functions of the ATR, CHK1, and WEE1 kinases. Notably, although ATR, CHK1, and WEE1 all function in the DNA replication stress pathway, the functions of these kinases are non-redundant and, in some cases, dependent on cell type (33,55–61). For example, WEE1 directly phosphorylates CDK1/2 while CHK1 indirectly regulates phosphorylation of CDK1/2 via the phosphatase CDC25A (Figure 2F and 2G). Consequently, we hypothesize that this direct versus indirect regulation may contribute to the difference between the inhibitors. In addition, WEE1 inhibitors block the phosphorylation of both cyclin-bound and cyclin-unbound CDK2 (62–64). In contrast, CDC25A can dephosphorylate CDK2 only when CDK2 is bound to a cyclin (62–64). Furthermore, both WEE1 and CHK1 inhibitors cause DNA replication stress via regulation of origin licensing and differences in the degree of replication stress caused by these drugs could impact the activation of CDK2 (18,65). Consequently, defining the distinct roles and functions of ATR, CHK1, and WEE1 inhibitors in Ewing sarcoma tumors will be a focus of future investigation.
In summary, we identified a novel feedback loop in Ewing sarcoma cells in which the inhibition of the ATR-CHK1 pathway, or the WEE1 kinase, during DNA replication stress leads to the aberrant activation of CDK2, degradation of RRM2, enhanced DNA replication stress, increased DNA damage, and apoptosis. In other tumor types, inhibition of CHK1 has also been reported to activate CDK2, but it is unclear whether this also results in depletion of RRM2 and exacerbation of DNA replication stress as the downstream impact of active CDK2 appears variable between tumor types (49,63). However, in future work, we plan to investigate this feedback loop in other cell types, as well as identify additional downstream targets of active CDK2. Overall, our work provides novel mechanistic insight into the causes and consequences of DNA replication stress and generates further support for targeting the RNR, ATR-CHK1, and WEE1 pathways in Ewing sarcoma tumors.
Supplementary Material
Implication.
Targeting the ATR, CHK1, and WEE1 kinases in Ewing sarcoma cells activates CDK2 and increases DNA replication stress by promoting the proteasome-mediated degradation of RRM2.
ACKNOWLEDGMENTS
DJG is supported by a University of Iowa Dance Marathon Award, a Holden Comprehensive Cancer Center Sarcoma Multidisciplinary Oncology Group Seed Grant, a University of Iowa Oberley Seed Grant, St. Baldrick’s Research Foundation, Aiming for a Cure Foundation, The Matt Morrell and Natalie Sanchez Pediatric Cancer Research Foundation, and NIH Grant R37-CA217910. The authors would also like to acknowledge use of the University of Iowa Flow Cytometry Core Facility (NIH/NCI P30CA086862)
Footnotes
The authors declare no potential conflicts of interest
REFERENCES
- 1.Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5. [DOI] [PubMed] [Google Scholar]
- 2.Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley M-C, et al. Ewing sarcoma: current management and future approaches through collaboration. J Clin Oncol. 2015;33:3036–46. [DOI] [PubMed] [Google Scholar]
- 3.Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature. 2018;555:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nieto-Soler M, Morgado-Palacin I, Lafarga V, Lecona E, Murga M, Callen E, et al. Efficacy of ATR inhibitors as single agents in Ewing sarcoma. Oncotarget. 2016;7:58759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goss KL, Gordon DJ. Gene expression signature based screening identifies ribonucleotide reductase as a candidate therapeutic target in Ewing sarcoma. Oncotarget. 2016;7:63003–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goss KL, Koppenhafer SL, Harmoney KM, Terry WW, Gordon DJ. Inhibition of CHK1 sensitizes Ewing sarcoma cells to the ribonucleotide reductase inhibitor gemcitabine. Oncotarget. 2017;8:87016–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowery CD, VanWye AB, Dowless M, Blosser W, Falcon BL, Stewart J, et al. The checkpoint kinase 1 inhibitor prexasertib induces regression of preclinical models of human neuroblastoma. Clin Cancer Res. 2017;23:4354–63. [DOI] [PubMed] [Google Scholar]
- 8.Lowery CD, Dowless M, Renschler M, Blosser W, VanWye AB, Stephens JR, et al. Broad spectrum activity of the checkpoint kinase 1 inhibitor prexasertib as a single agent or chemopotentiator across a range of preclinical pediatric tumor models. Clin Cancer Res. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henssen AG, Reed C, Jiang E, Garcia HD, von Stebut J, MacArthur IC, et al. Therapeutic targeting of PGBD5-induced DNA repair dependency in pediatric solid tumors. Sci Transl Med. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koppenhafer SL, Goss KL, Terry WW, Gordon DJ. mTORC1/2 and Protein Translation Regulate Levels of CHK1 and the Sensitivity to CHK1 Inhibitors in Ewing Sarcoma Cells. Mol Cancer Ther. 2018;17:2676–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rundle S, Bradbury A, Drew Y, Curtin NJ. Targeting the ATR-CHK1 Axis in Cancer Therapy. Cancers (Basel). 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol. 2017;18:622–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krämer A, Mailand N, Lukas C, Syljuåsen RG, Wilkinson CJ, Nigg EA, et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–91. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Mazzanti M, Mistrik M, Kosar M, Beznoussenko GV, Mironov AA, et al. ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell. 2014;158:633–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeche L, Nguyen HD, Buisson R, Zou L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science. 2018;359:108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, et al. An intrinsic S/G2 checkpoint enforced by ATR. Science. 2018;361:806–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobbelstein M, Sørensen CS. Exploiting replicative stress to treat cancer. Nat Rev Drug Discov. 2015;14:405–23. [DOI] [PubMed] [Google Scholar]
- 19.Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics. 2017;33:2413–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai D, Gu Y, Morgan DO. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurose A, Tanaka T, Huang X, Traganos F, Darzynkiewicz Z. Synchronization in the cell cycle by inhibitors of DNA replication induces histone H2AX phosphorylation: an indication of DNA damage. Cell Prolif. 2006;39:231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halicka D, Zhao H, Li J, Garcia J, Podhorecka M, Darzynkiewicz Z. DNA Damage Response Resulting from Replication Stress Induced by Synchronization of Cells by Inhibitors of DNA Replication: Analysis by Flow Cytometry. Methods Mol Biol. 2017;1524:107–19. [DOI] [PubMed] [Google Scholar]
- 23.Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M. ATM is required for the cellular response to thymidine induced replication fork stress. Hum Mol Genet. 2004;13:2937–45. [DOI] [PubMed] [Google Scholar]
- 24.King C, Diaz H, Barnard D, Barda D, Clawson D, Blosser W, et al. Characterization and preclinical development of LY2603618: a selective and potent Chk1 inhibitor. Invest New Drugs. 2014;32:213–26. [DOI] [PubMed] [Google Scholar]
- 25.Di Tullio A, Rouault-Pierre K, Abarrategi A, Mian S, Grey W, Gribben J, et al. The combination of CHK1 inhibitor with G-CSF overrides cytarabine resistance in human acute myeloid leukemia. Nat Commun. 2017;8:1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Checkley S, MacCallum L, Yates J, Jasper P, Luo H, Tolsma J, et al. Bridging the gap between in vitro and in vivo: Dose and schedule predictions for the ATR inhibitor AZD6738. Sci Rep. 2015;5:13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–6. [DOI] [PubMed] [Google Scholar]
- 28.Sørensen CS, Syljuåsen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 2012;40:477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakurikar N, Eastman A. Critical reanalysis of the methods that discriminate the activity of CDK2 from CDK1. Cell Cycle. 2016;15:1184–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103:10660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N, et al. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem. 1997;243:527–36. [DOI] [PubMed] [Google Scholar]
- 32.Matheson CJ, Backos DS, Reigan P. Targeting WEE1 kinase in cancer. Trends Pharmacol Sci. 2016;37:872–81. [DOI] [PubMed] [Google Scholar]
- 33.Bukhari AB, Lewis CW, Pearce JJ, Luong D, Chan GK, Gamper AM. Inhibiting Wee1 and ATR kinases produces tumor-selective synthetic lethality and suppresses metastasis. J Clin Invest. 2019;129:1329–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall AB, Newsome D, Wang Y, Boucher DM, Eustace B, Gu Y, et al. Potentiation of tumor responses to DNA damaging therapy by the selective ATR inhibitor VX-970. Oncotarget. 2014;5:5674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Angiolella V, Donato V, Forrester FM, Jeong Y-T, Pellacani C, Kudo Y, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfister SX, Markkanen E, Jiang Y, Sarkar S, Woodcock M, Orlando G, et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell. 2015;28:557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu B, Mitra J, van den Heuvel S, Enders GH. S and G2 phase roles for Cdk2 revealed by inducible expression of a dominant-negative mutant in human cells. Mol Cell Biol. 2001;21:2755–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin P, Gu Y, Morgan DO. Role of inhibitory CDC2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol. 1996;134:963–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ford JB, Baturin D, Burleson TM, Van Linden AA, Kim Y-M, Porter CC. AZD1775 sensitizes T cell acute lymphoblastic leukemia cells to cytarabine by promoting apoptosis over DNA repair. Oncotarget. 2015;6:28001–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paronetto MP, Miñana B, Valcárcel J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol Cell. 2011;43:353–68. [DOI] [PubMed] [Google Scholar]
- 41.Ghosal G, Yustein JT. EWS-FLI1 Regulates Genotoxic Stress Response in Ewing Sarcoma. J Cancer Biol Res. 2015;3:1063–70. [Google Scholar]
- 42.Tang S-W, Bilke S, Cao L, Murai J, Sousa FG, Yamade M, et al. SLFN11 Is a Transcriptional Target of EWS-FLI1 and a Determinant of Drug Response in Ewing Sarcoma. Clin Cancer Res. 2015;21:4184–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci USA. 2012;109:15030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart E, Goshorn R, Bradley C, Griffiths LM, Benavente C, Twarog NR, et al. Targeting the DNA repair pathway in Ewing sarcoma. Cell Rep. 2014;9:829–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brohl AS, Patidar R, Turner CE, Wen X, Song YK, Wei JS, et al. Frequent inactivating germline mutations in DNA repair genes in patients with Ewing sarcoma. Genet Med. 2017;19:955–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szmyd R, Niska-Blakie J, Diril MK, Renck Nunes P, Tzelepis K, Lacroix A, et al. Premature activation of Cdk1 leads to mitotic events in S phase and embryonic lethality. Oncogene. 2019;38:998–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes BT, Sidorova J, Swanger J, Monnat RJ, Clurman BE. Essential role for Cdk2 inhibitory phosphorylation during replication stress revealed by a human Cdk2 knockin mutation. Proc Natl Acad Sci USA. 2013;110:8954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren NJH, Eastman A. Inhibition of checkpoint kinase 1 following gemcitabine-mediated S phase arrest results in CDC7- and CDK2-dependent replication catastrophe. J Biol Chem. 2019;294:1763–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gadhikar MA, Zhang J, Shen L, Rao X, Wang J, Zhao M, et al. CDKN2A/p16 Deletion in Head and Neck Cancer Cells Is Associated with CDK2 Activation, Replication Stress, and Vulnerability to CHK1 Inhibition. Cancer Res. 2018;78:781–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia TB, Fosmire SP, Porter CC. Increased activity of both CDK1 and CDK2 is necessary for the combinatorial activity of WEE1 inhibition and cytarabine. Leuk Res. 2018;64:30–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aye Y, Li M, Long MJC, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34:2011–21. [DOI] [PubMed] [Google Scholar]
- 53.Chi Y, Welcker M, Hizli AA, Posakony JJ, Aebersold R, Clurman BE. Identification of CDK2 substrates in human cell lysates. Genome Biol. 2008;9:R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P. CDK substrate phosphorylation and ordering the cell cycle. Cell. 2016;167:1750–1761.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buisson R, Boisvert JL, Benes CH, Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol Cell. 2015;59:1011–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanjiv K, Hagenkort A, Calderón-Montaño JM, Koolmeister T, Reaper PM, Mortusewicz O, et al. Cancer-Specific Synthetic Lethality between ATR and CHK1 Kinase Activities. Cell Rep. 2016;14:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koh S-B, Wallez Y, Dunlop CR, Bernaldo de Quirós Fernández S, Bapiro TE, Richards FM, et al. Mechanistic Distinctions between CHK1 and WEE1 Inhibition Guide the Scheduling of Triple Therapy with Gemcitabine. Cancer Res. 2018;78:3054–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wayne J, Brooks T, Massey AJ. Inhibition of Chk1 with the small molecule inhibitor V158411 induces DNA damage and cell death in an unperturbed S-phase. Oncotarget. 2016;7:85033–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung S, Vail PJ, Witkiewicz AK, Knudsen ES. Coordinately targeting cell cycle checkpoint functions in integrated models of pancreatic cancer. Clin Cancer Res. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilsker D, Chung JH, Pradilla I, Petermann E, Helleday T, Bunz F. Targeted mutations in the ATR pathway define agent-specific requirements for cancer cell growth and survival. Mol Cancer Ther. 2012;11:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilsker D, Bunz F. Loss of ataxia telangiectasia mutated- and Rad3-related function potentiates the effects of chemotherapeutic drugs on cancer cell survival. Mol Cancer Ther. 2007;6:1406–13. [DOI] [PubMed] [Google Scholar]
- 62.Coulonval K, Bockstaele L, Paternot S, Roger PP. Phosphorylations of cyclin-dependent kinase 2 revisited using two-dimensional gel electrophoresis. J Biol Chem. 2003;278:52052–60. [DOI] [PubMed] [Google Scholar]
- 63.Sakurikar N, Thompson R, Montano R, Eastman A. A subset of cancer cell lines is acutely sensitive to the Chk1 inhibitor MK-8776 as monotherapy due to CDK2 activation in S phase. Oncotarget. 2016;7:1380–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warren NJH, Donahue KL, Eastman A. Differential Sensitivity to CDK2 Inhibition Discriminates the Molecular Mechanisms of CHK1 Inhibitors as Monotherapy or in Combination with the Topoisomerase I Inhibitor SN38. ACS Pharmacol Transl Sci. 2019;2:168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ubhi T, Brown GW. Exploiting DNA replication stress for cancer treatment. Cancer Res. 2019;79:1730–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.