Abstract
To cure a patient’s cancer is to eradicate invasive cells from the ecosystem of the body. However, the ecological complexity of this challenge is not well understood. Here we show how results from eradications of invasive mammalian species from islands—one of the few contexts in which invasive species have been regularly cleared—inform new research directions for treating cancer. We first summarize the epidemiological characteristics of island invader eradications and cancer treatments by analyzing recent datasets from the Database of Invasive Island Species Eradications and The Cancer Genome Atlas, detailing the superior successes of island eradication projects. Next, we compare how genetic and environmental factors impact success in each system. These comparisons illuminate a number of promising cancer research and treatment directions, such as heterogeneity engineering as motivated by gene drives and adaptive therapy; multi-scale analyses of how population heterogeneity potentiates treatment resistance; and application of ecological data mining techniques to high-throughput cancer data. We anticipate that interdisciplinary comparisons between tumor progression and invasive species would inspire development of novel paradigms to cure cancer.
Introduction
Improvements in DNA and RNA sequencing technology have transformed our knowledge of cancer [1,2], yet they have also revealed a daunting ecological complexity. The cancer ecosystem includes genetically heterogeneous cancer cells and localized microenvironments in a web of interactions among tumor, immune, and stromal cells [3,4], and this ecosystem interfaces with the greater environment of the patient’s body. The complexity of the cancer ecosystem has made it difficult to prioritize cancer cell features for targeted treatment. To solve this challenge, studies of pest animal ecosystems can provide valuable insights. Invasive mammalian species, particularly rats, have caused serious ecological damage and extinguished native species on islands around the world, and hundreds of projects to eradicate these invaders have been attempted globally [5]. Notably, the Predator-Free New Zealand movement has set a goal of eliminating rats from the country’s archipelago by 2050 in order to save native fauna from this efficient predator and allow ecosystem recovery [6]. Despite the enormous differences in scale, we argue that a systems approach comparing island restoration to cancer therapy will elucidate factors that influence the success of different treatments.
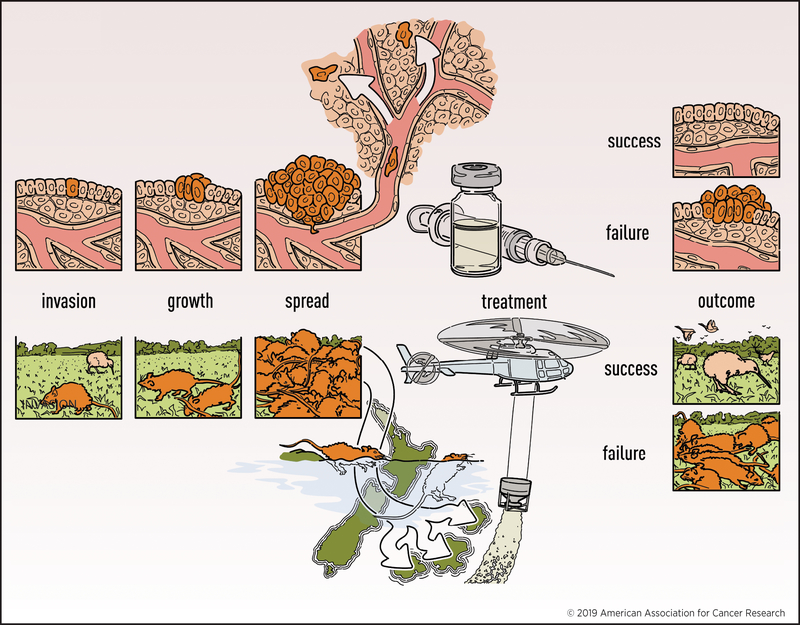
Epidemiologically, invasive species and cancers share similar properties [7,8] in terms of invasion, growth, spread, treatment, and outcome, as shown in Fig. 1. The isolated naive environments of islands have made them key systems for studying and repairing ecologies disrupted by invasive species. Invading rats typically arrive on islands as small populations or even single pregnant individuals via boats or by swimming, analogous to cancers developing within the body from a single tumor cell. For both invasive species and cancers, these initial populations then grow until they are detected, at which point action must be taken as soon as possible to halt their expansion.
Fig 1. Comparison of invasive species and cancer.
Cancers and invasive species are systems with analogous processes for invasion, growth, spread, treatment, and outcome.
Epidemiological comparison of invasive species and cancer
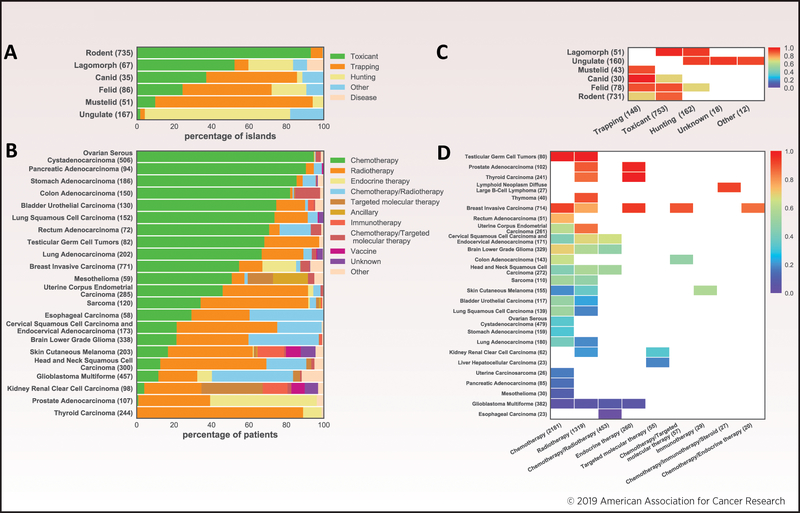
Multiple methods have been deployed to eradicate invasive species and to treat cancer. We have assessed the prevalence of common strategies (Fig. 2A, 2B, and Supplementary Table S1) by analyzing the Database of Island Invasive Species Eradications (DIISE) [9] and The Cancer Genome Atlas (TCGA) [1]. Statistics for the six most frequently targeted mammalian clades in the DIISE, i.e. rodent, lagomorph (rabbit and hare), canid (dog and fox), felid (cat), mustelid (mongoose and weasel), and ungulate (hoofed mammals), are shown in Fig. 2A. These eradication projects are undertaken based on available funding [10] and socio-legal capacity, making uninhabited islands the preferred target [11]. The most prevalent eradication strategies are trapping, hunting, and toxicant, with toxicant being the most used method (67% of all attempts). Rodents are the most frequently targeted group (735 eradications, 64% of all attempts), with most cases treated by toxicant (94% of attempts). Similarly, we analyzed 22 tumor types from TCGA (minimum cohort size of 50 patients), and stratified them by treatment strategy. Common non-surgical cancer treatments include chemotherapy, radiotherapy, immunotherapy, and targeted molecular therapy (Fig. 2B), with chemotherapy and radiotherapy being the most prevalent cancer treatments.
Fig 2. Prevalence of strategies for invasive species eradication and cancer treatment and their success rates.
(A) Percentage of islands that received specific primary eradication strategies, stratified by invasive mammalian species. (B) Percentage of patients that received specific initial treatments, stratified by cancer type. (C) Success rates for different primary treatments. Species are ordered by the mean of their success rates across all treatments. Only eradication cases with at least 10 islands were considered. (D) Five-year survival rate (Kaplan-Meier estimation) for cancer types as a function of initial treatment. Cancer types are ordered by the mean of their survival rates across all treatments. Data in panels (A) and (C) are from the Database of Island Invasive Species Eradications, with the number of islands in each dataset shown in parentheses. Data in panels (B) and (D) are from The Cancer Genome Atlas (TCGA) through TumorPortal [1], with the number of patients in each dataset shown in parentheses.
There are parallels between the strategies for eradicating invasive species from islands [5] and treating cancer in the human body [4]. For example, rodent toxicants are broadly administered either aerially or via ground-based devices, analogous to the treatment of cancer by chemotherapy through the bloodstream. Hunting is similar to cancer immunotherapy, as each method targets elusive quarry in a highly specific fashion. Animal trapping and cancer targeted molecular therapy are each moderately specific and complement broad-spectrum methods. Moreover, in each system it is vital that eradication methods avoid side effects that would be detrimental to native fauna [12] or patient health.
The success rates of common strategies for invasive species eradication and cancer treatment are summarized in Fig. 2C and 2D. Success rates have been high for all invasive species–treatment combinations (Fig. 2C), with a median success rate of 91% (confidence interval 80–97%, Jeffreys binomial proportion test at 95% confidence). Some species–treatment combinations have been particularly successful, e.g. canid–trapping has had a 100% success rate (CI 86–100%) and even the least successful have worked well, i.e. felid–hunting has had a 69% success rate (CI 44–87%). These eradications can be confidently ascertained by surveying islands several generations after treatment, e.g. two years for rats, providing ample time for populations to rebound from small numbers [13]. For comparison, cancer treatment success rates are shown in Fig. 2D. The median of the five-year survival rate for combinations of cancer type–primary nonsurgical treatment is 52% (CI 35–65%; beta product procedure at 95% confidence [14]). Testicular germ cell tumors treated with chemotherapy have the best outcome, as 100% of patients survive (CI 88–100%), while esophageal carcinoma treated with chemotherapy/radiotherapy has the worst outcome, as 0% of patients survive. Some cancer types have high survival rates across multiple different treatments, notably testicular germ cell tumors, prostate adenocarcinoma, thyroid carcinoma, and breast invasive carcinoma. Conversely, others have low survival regardless of the treatment used, e.g. glioblastoma multiforme has a survival rate <8% for every primary therapy used. Overall, eradication success rates are higher and more stable across different island invasive species, whereas success rates are lower and vary more across cancers.
Factors impacting invasive species and cancer eradication
It is informative to compare and contrast the factors that affect the success of eradication and treatment in these systems. Here we highlight several aspects, namely: distinctiveness between the invader and native species; effectiveness of common treatments across diverse invaders; intrapopulation heterogeneity of the invader; and environmental interactions (Table 1). We have chosen these factors because current measurements allow them to be quantitatively compared, though more comprehensive descriptions of each system can be found elsewhere [4,15].
Table 1:
Comparison of key parallel factors for invasive mammalian species eradication and cancer treatment, and the relative impact of each factor on treatment success.
| Invasive mammals | Cancer | Relative impact on treatment success | |
|---|---|---|---|
| Distinctiveness between host/invader | High: Millions of coding point mutations between invader and native species | Low: <1000 coding point mutations between tumor and normal cells | Lower treatment specificity and success against cancer than treatments against mammalian island invaders. |
| Genetic diversity among similarly treated species | Genetic differences are high: millions of coding mutations between different species. | Genetic differences are low: <100,000 coding germline differences between patients and <1000 coding mutations between patient tumor / germline. | Treatment response varies little between mammalian species but varies much between different cancer types. |
| Invader intrapopulation heterogeneity | Low: small populations (~103 for rats on most islands) and low mutation rates | High: large number of cells (~109) and high mutation rates | Cancers have more opportunities for evolution of resistance. |
| Invader environment interactions | Decoupling of invader genetics and ecological environment | Primary tumor genetics evolve in a fixed microenvironment, followed by decoupling at metastasis. | Microenvironment may be more targetable in cancer, especially for primary tumors. Rats are robust across island environments. |
Distinctiveness between the invader and native species is critical to enable specific eradication of the invader with minimal impact to the host ecosystem. At the genetic level, there is greater divergence between invasive mammals and the native species they threaten, which are typically birds, than there is between a patient’s cancer cells and normal cells. Rats and birds have millions of coding sequence differences [16] while cancer and normal cells differ by only 10–1000 somatic coding point mutations [17]. This makes treatment specificity easier to achieve for rat invasions, and indeed anticoagulant/citric acid cycle toxicant strategies have often succeeded in controlling or eradicating rats while permitting native bird species to re-expand [18]. Optimization for feeding preferences, which may derive from genetic differences, has been important to this specificity, e.g. use of compressed cereal bait matrices with green coloring reduces toxicant appeal to most birds. Meanwhile, non-specific side effects limit most cancer treatments.
The effectiveness of common treatments across diverse invaders differs substantially between invasive mammals and cancers. A wide range of invasive mammalian species have similar responses to the most commonly applied toxicants despite tens of millions of years of evolutionary divergence, as exemplified by the species listed in Fig. 2. On the other hand, cancers have more variable responses to chemotherapy and radiotherapy, which are the most common treatments, even though any two tumors have comparatively low evolutionary divergence. Cancers from different patients are distinguished by the germline differences between the patients (~105 coding mutations) and the cancer somatic mutations (~103 coding mutations), which sum to fewer mutations than distinguish mammals and birds. Thus, interpatient tumor diversity does not preclude successful pan-cancer treatments. The greater constraint is that such pan-cancer effectiveness must be achieved with minimal impact on normal cells.
Intrapopulation genetic heterogeneity is fundamental to survival for both island invaders and treated cancer cells. Cancers are made up of multiple subclones with distinct mutations that may confer resistance to treatment [19], and high intratumoral heterogeneity provides more opportunities for resistance. Invasive rodents are subject to the same dynamics but evolve resistance less frequently than cancers do [20]. This may be due to the low population sizes of rodents, as there are on average 103 rats on islands for which exterminations have been attempted [21] compared to the 109 cells in a 1 cm3 neoplasm [22]. Consistent with this trend, eradication attempts on larger islands have had lower success rates (82%) than on smaller islands (91%) (Fisher’s exact test P = 4.7⨉10−4, DIISE database), but both small and large islands have better outcome rates than median cancer survival. Rapid mutation rates also potentiate resistance in cancer, as the cancer mutation rate (10−7/bp/generation) is higher than that for rats (10−8/bp/generation) [16], in addition to cancer cells having much shorter generation times.
Environmental interactions also distinguish invading species and cancers. An underlying reason is that cancers usually originate in the tissues in which they develop, whereas invasive species immigrate to naive islands. As a result, cancer cells–like native cells–have strong interactions with the local microenvironment. For example, fibroblasts influence chemoresistance of cancer cells, and the immune system has major impacts on patient survival [23]. Environmental factors play less of a role for invasive rats than in cancer, as rats are known for their ability to live in diverse biomes. This is also true in the treatment context, as both tropical and temperate islands have rat eradication success rates >80% [15]. Nevertheless, some environmental factors have been found that reduce the success of rat eradication efforts, notably higher temperatures, presence of local agriculture, and presence of species that eat bait (land crabs) or provide alternative food sources (coconut palms). These environmental considerations are also related to cancer metastasis, in which cancer cells grow in the new microenvironment of a different organ. Likewise, rats can swim to other islands more than 1km away. However, nearby islands are not inherently different in environment than the original island, unlike metastasis to different organs. As a result, spatial dissemination of rats is less associated with differential treatment resistance than cancer metastasis.
Opportunities for future research
Here we discuss potential research directions motivated by these interdisciplinary comparisons.
Heterogeneity engineering
A promising new cancer approach that combines aspects of invasive species studies and cancer genetics is “heterogeneity engineering,” which we define to describe engineering of population heterogeneity to induce desired growth behaviors. In the invasive species field this concept has been commonly considered for gene drives, which are CRISPR/cas9 constructs engineered into an organism’s DNA in order to propagate the construct through the population [24–26]. Gene drives are being proposed to introduce maladaptive genetic traits with the goal of population eradication. The CRISPR/cas9 elements enable the construct to duplicate itself into the homologous chromosome of its genomic locus, allowing it to rapidly spread in a sexually reproducing population. An early gene drive concept has been to include a sex-linked infertility gene within the construct, which has the effect of limiting population reproduction while still allowing the construct to propagate. Recently, safety concerns about single locus drives have led to consideration of time-dependent multi-component genotype engineering via gene drive “daisy chains” [27]. Analogous ideas have been less considered in the cancer field, but could have similar value, e.g. to create tumor populations that grow rapidly but have specified genetic weaknesses that can later be targeted; or to synchronize multiclonal and microenvironmental dynamics to enhance immunotherapy.
Although gene drives cannot be applied to cancer cells because of their lack of sexual reproduction, the broader concept of heterogeneity engineering may still be useful. For example, cancer adaptive therapy is a version of heterogeneity engineering without genome editing that treats a tumor as a mixture of two subpopulations. In cancer adaptive therapy, treatment is timed to target a fast-growing subpopulation and then re-applied when the fast-growing subpopulation re-expands. This is analogous to adaptive ecosystem management, which denotes regular monitoring and episodic ‘knockdown’ of the invasive population, used to control invasive mammalian pests [28]. The success of cancer adaptive therapy in recent clinical trials [29,30], as well as analogous subclonal dynamics in xenograft studies [31], shows how time-dependent control of intratumoral heterogeneity can be useful for treatment outcomes. This utility arises because the competitive behaviors among subclones enable manipulation of tumor growth. We hypothesize that it would be possible to combine genetic manipulation and adaptive therapy concepts by removing some tumor cells from a patient and then genetically engineering them to outcompete other cancer cells while minimizing their ability to evolve resistance. Such cells could then be re-inserted into the tumor to replace the extant tumor cells, followed by a hyper-effective drug treatment to kill the engineered cells. Although this scenario may seem complex, competitive population replacement has been successful in fecal transplant treatment of gut bacterial dysbiosis [32], and the replacement populations can still be susceptible to antibiotic treatment.
Adaptive therapy is just one realization of heterogeneity engineering, and there are likely to be more powerful systems of multiple interacting subpopulations whose dynamics could each be triggered by external agents. Sophisticated phenotypes for cancer suppression are potentially possible, e.g. in which tradeoffs between immune evasion and chemotherapy resistance are sharpened, or particular cancer subpopulations are favored in the presence of specific immune cell types. Heterogeneity engineering of cancer by CRISPR/cas9 may also eventually be achievable directly in vivo for patients [33]. Cancer cell specificity will be a key issue, and this topic can benefit from recent discussions of how to achieve cas9 sequence specificity for invader-specific targeting in the invasive species literature [27]. Comprehensive targeting of every cancer cell will also be challenging, and therefore new strategies that take into account subclonal competition will likely need to be integrated.
Population scale issues in evolution of resistance
The effects of intratumoral population dynamics on resistance remain poorly understood, aside from early concepts such as larger populations providing more opportunities for resistance [34,35] and some tumors being more treatable after a bottleneck [36]. Because resistance is common in cancer but rare for rat eradications, comparing their population dynamics can provide insights. Typical tumor and rat populations differ in scale, but the Predator-Free New Zealand project [37] aims to remove all 108–109 rats from the country’s 26 million hectares of land [21], a population size similar to the cells in an initially detectable neoplasm. Other recently developed cancer models cover sizes from a few cells (microfluidics [38]) to thousands of cells (organoids [39]) to billions of cells (xenografts [40]). These models enable comparisons to a range of rat population sizes, which could illuminate fundamental questions in cancer biology such as how the propensity for resistance is related to the size of the tumor. This relationship affects strategies for early detection and treatment. While this relationship can be studied empirically in cancer experimental systems, basic understanding may require mathematical modeling. Aspects related to cancer spatial dynamics can be difficult to investigate because of the challenge of tracking individual cells in vivo for long periods of time. Rats, on the other hand, can be individually tagged and monitored, and therefore mathematical models of rat spatial dynamics may benefit analogous models for cancer. Population scale comparisons to other systems may also be illuminating. For example, immunoediting is a resistance mechanism of cancer to T-cells [41], but it imposes a fitness cost that is not well quantified. Characterizations of immunoediting as a function of population size in microbial or viral infections [42] may help quantify the likelihood of immuno-resistance during cancer growth or under immunotherapy. Such quantifications of fitness cost could be used to design adaptive therapy strategies [43] for cancers containing immunotherapy-resistant subclones.
Big data approaches to the tumor microenvironment
Methods to quantify the environment are rapidly developing in both cancer and macroscopic ecological systems, enabling potent data mining approaches. In the Predator-Free New Zealand project, invasive rats are being studied across islands with diverse latitudinal, rural, and urban biomes through quantifications such as temperature, rainfall, species tracking, remote sensing [44], and metagenomic DNA sampling [45]. Analogously, cancers are now studied across diverse tissue types, and the local microenvironment is quantified through measures such as MRI, immunostaining, cell sorting, and expression profiling. In each system, these rich datasets can be mined to reveal relationships among environment variables and invader growth. For example, random forests have been used to predict rat population growth [15] and ecological databases [46] provide powerful resources for study by algorithmic approaches such as ensemble regression, classification trees, and association rule mining [47]. Development of big data approaches for cancer histopathology, immunohistochemistry, and expression data is a burgeoning field [48,49]. These approaches provide a new way to answer the pressing question of what tumor microenvironmental features are important to response [50].
Recent efforts in both fields have focused on advances in data collection. In cancer, image collections from TCGA have recently become available and these have been mined to build classifiers for outputs such as mutation status [51] or cancer subtype [48]. While many hospitals have additional slide collections, these are not openly available -- greater sharing could have a profound effect on the field. In ecological systems, new eco-sensing data collection efforts are actively being developed to forecast biodiversity [52]. This is applicable but not limited to invasive species ecology. For example, neural networks have been applied to high resolution camera images to assist in the identification of biological features such as coral reefs [53], a problem analogous to the identification of tumor regions with specific phenotypes. Approaches to determine features predictive of outcome are actively being developed in both fields. Natural geographic variations (e.g. low altitude vs. high altitude) assist in this analysis for ecological studies, just as stromal and vascular features may be helpful in cancer studies. Such approaches may clarify the spatial scales at which treatment resistance occurs in tumors, and they will be increasingly important as new technologies such as spatial transcriptomics [54] and imaging mass cytometry [55] become prevalent.
Conclusion
Cancer and invasive mammalian species are, despite the differences in scale, essentially similar in their processes of invasion, growth, and treatment. Key distinctions include the higher success of invasive mammal eradication projects, the low genetic divergence between cancer and normal cells, the high genetic divergence among clades of similarly treatable mammals, the greater intrapopulation heterogeneity of cancers, and the closer link between genetics and environment in cancers.
Promising future directions suggested by these comparisons include: heterogeneity engineering, multi-scale systems comparisons of population dynamics, and adaptation of image data mining approaches from ecological studies to cancer. Inter-system comparisons may help clarify parameter ranges for which heterogeneity engineering approaches will be effective [56]. It is promising that there have been recent calls to expand clinical trial testing of cancer adaptive therapy and treatment-switching [57]. The results of such studies would be highly informative for development of two-population heterogeneity engineering and shed light on when higher order population engineering might be most necessary. In organismal ecology studies, population engineering is just a part of the broader topic of eco-engineering, which may include environmental control as well. Large scale imaging approaches at a variety of scales provide the raw data for selecting what environmental perturbations may be useful in concert with heterogeneity engineering, and the feature analysis techniques being developed for eco-sensing projects should be applicable to the cancer problem.
Our discussions here could be developed in many ways. Firstly, we have focused mainly on the analogy between mammalian island invaders and cancer. Some strengths of this analogy are the genomic similarities between invasive mammals and human cancer cells, as well as the fact that both systems are invasions into closed systems. Many other invasive species systems can provide further insights, though they lack that particular combination of strengths. Examples include island invasions by insects, invasions of pests into agricultural fields, and microbial invasions into the human body [58]. Secondly, these discussions have emphasized genetic measures, but epigenetic, tissue-level, or organismal-level phenotypes will all be important. Thirdly, the survival data we have presented do not account for cancer survival / eradication success without treatment (e.g. spontaneous remission or animals naturally dying out). Further data collection will be necessary to take this into account, as they are not available in DIISE or TCGA.
Despite these caveats and the seeming differences across fields, invasive mammal eradication in New Zealand and cancer cure have many congruences in genetic and ecological complexity, positioning them as valuable parallel systems for developing improved eradication approaches. We believe that comparative studies of these and other invasive systems have great potential for developing new cancer treatment paradigms, and we hope that these perspectives will encourage others to develop insights across interdisciplinary divides.
Supplementary Material
Acknowledgements
We thank medical illustrator Matt Wimsatt for assistance with Figure 1. We also thank Razelle Kurzrock and Sheng Li for helpful discussions. JHC acknowledges support from the National Cancer Institute under award numbers R01CA230031, R21CA191848, U24 CA224067, and P30CA034196. JCR acknowledges support from the New Zealand Ministry of Business Innovation and Employment (MBIE) under award numbers RDF-UOA1404 (Rutherford Discovery Fellowship from the Royal Society of New Zealand) and 1617–44-003 (BioHeritage National Science Challenge from Manaaki Whenua-Landcare Research).
Footnotes
Conflict of Interest
The authors declare no potential conflicts of interest.
Reference
- 1.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357. doi: 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- 3.Maley CC, Aktipis A, Graham TA, Sottoriva A, Boddy AM, Janiszewska M, et al. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer. 2017;17: 605–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144: 646–674. [DOI] [PubMed] [Google Scholar]
- 5.Russell JC, Meyer J-Y, Holmes ND, Pagad S. Invasive alien species on islands: impacts, distribution, interactions and management. Environ Conserv. 2017;44: 359–370. [Google Scholar]
- 6.Russell JC, Innes JG, Brown PH, Byrom AE. Predator-Free New Zealand: Conservation Country. Bioscience. 2015;65: 520–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pienta KJ, McGregor N, Axelrod R, Axelrod DE. Ecological therapy for cancer: defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Transl Oncol. 2008;1: 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amend SR, Pienta KJ. Ecology meets cancer biology: the cancer swamp promotes the lethal cancer phenotype. Oncotarget. 2015;6: 9669–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DIISE [Internet]. The Database of Island Invasive Species Eradications, developed by Island Conservation, Coastal Conservation Action Laboratory UCSC, IUCN SSC Invasive Species Specialist Group, University of Auckland and Landcare Research New Zealand. 2015. Available: http://diise.islandconservation.org/
- 10.Russell JC, Meyer J-Y, Holmes ND, Pagad S. Invasive alien species on islands: impacts, distribution, interactions and management. Environ Conserv. Cambridge University Press; 2017;44: 359–370. [Google Scholar]
- 11.Glen AS, Atkinson R, Campbell KJ, Hagen E, Holmes ND, Keitt BS, et al. Eradicating multiple invasive species on inhabited islands: the next big step in island restoration? Biol Invasions. Springer Netherlands; 2013;15: 2589–2603. [Google Scholar]
- 12.Courchamp F, Chapuis J-L, Pascal M. Mammal invaders on islands: impact, control and control impact. Biol Rev Camb Philos Soc. 2003;78: 347–383. [DOI] [PubMed] [Google Scholar]
- 13.Samaniego-Herrera A, Anderson DP, Parkes JP, Aguirre-Muñoz A. Rapid assessment of rat eradication after aerial baiting. J Appl Ecol. 2013;50: 1415–1421. [Google Scholar]
- 14.Fay MP, Brittain EH, Proschan MA. Pointwise confidence intervals for a survival distribution with small samples or heavy censoring. Biostatistics. 2013;14: 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes ND, Griffiths R, Pott M, Alifano A, Will D, Wegmann AS, et al. Factors associated with rodent eradication failure. Biol Conserv. 2015;185: 8–16. [Google Scholar]
- 16.Lynch M Evolution of the mutation rate. Trends Genet. 2010;26: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349: 1483–1489. [DOI] [PubMed] [Google Scholar]
- 18.Croll DA, Newton KM, McKown M, Holmes N, Williams JC, Young HS, et al. Passive recovery of an island bird community after rodent eradication. Biol Invasions. 2015;18: 703–715. [Google Scholar]
- 19.Morgillo F, Della Corte CM, Fasano M, Ciardiello F. Mechanisms of resistance to EGFR-targeted drugs: lung cancer. ESMO Open. 2016;1: e000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prescott CV, Buckle AP, George Gibbings J, Allan ENW, Stuart AM. Anticoagulant resistance in Norway rats (Rattus norvegicusBerk.) in Kent – a VKORC1 single nucleotide polymorphism, tyrosine139phenylalanine, new to the UK. Int J Pest Manage. 2010;57: 61–65. [Google Scholar]
- 21.Wilson DJ, Efford MG, Brown SJ, Williamson JF, McElrea GJ. Estimating density of ship rats in New Zealand forests by capture- mark-recapture trapping. N Z J Ecol. 2007;31: 47–59. [Google Scholar]
- 22.Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW. Cell Growth and Size Homeostasis in Proliferating Animal Cells. Science. American Association for the Advancement of Science; 2009;325: 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10 GPR77 Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell. 2018;172: 841–856.e16. [DOI] [PubMed] [Google Scholar]
- 24.Serr ME. Towards a Genetic Approach to Invasive Rodent Eradications: Assessing Reproductive Competitiveness Between Wild-derived and Laboratory Mice. North Carolina State University; 2019. [Google Scholar]
- 25.Harvey-Samuel TO, Campbell KJ, Edgington M, Alphey L. Trialling gene drives to control invasive species: what, where and how? Island invasives: scaling up to meet the challenge. by: IUCN, Gland, Switzerland; 2019; 618. [Google Scholar]
- 26.Campbell KJ, Saah JR, Brown PR, Godwin J, Gould F, Howald GR, et al. A potential new tool for the toolbox: assessing gene drives for eradicating invasive rodent populations. Island invasives: scaling up to meet the challenge. by: IUCN, Gland, Switzerland; 2019; 6. [Google Scholar]
- 27.Esvelt KM, Gemmell NJ. Conservation demands safe gene drive. PLoS Biol. 2017;15: e2003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans JM, Wilkie AC, Burkhardt J. Adaptive Management of Nonnative Species: Moving Beyond the “Either-Or” Through Experimental Pluralism. J Agric Environ Ethics. 2008;21: 521–539. [Google Scholar]
- 29.Zhang J, Cunningham JJ, Brown JS, Gatenby RA. Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat Commun. 2017;8: 1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West JB, Dinh MN, Brown JS, Zhang J, Anderson AR, Gatenby RA. Multidrug Cancer Therapy in Metastatic Castrate-Resistant Prostate Cancer: An Evolution-Based Strategy. Clin Cancer Res. 2019; doi: 10.1158/1078-0432.CCR-19-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H, Kumar P, Menghi F, Noorbakhsh J, Cerveira E, Ryan M, et al. High-resolution deconstruction of evolution induced by chemotherapy treatments in breast cancer xenografts. Sci Rep. 2018;8: 17937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits LP, Bouter KEC, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic Potential of Fecal Microbiota Transplantation [Internet]. Gastroenterology. 2013. pp. 946–953. doi: 10.1053/j.gastro.2013.08.058 [DOI] [PubMed] [Google Scholar]
- 33.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noorbakhsh J, Kim H, Namburi S, Chuang J. Distribution-based measures of tumor heterogeneity are sensitive to mutation calling and lack strong clinical predictive power. Scientific Reports. 2018;In press. doi: 10.1101/248435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor Evolution as a Therapeutic Target. Cancer Discov. 2017; doi: 10.1158/2159-8290.CD-17-0343 [DOI] [PubMed] [Google Scholar]
- 36.Kim T, Tyndel MS, Kim HJ, Ahn J-S, Choi SH, Park HJ, et al. Spectrum of somatic mutation dynamics in chronic myeloid leukemia following tyrosine kinase inhibitor therapy. Blood. 2017;129: 38–47. [DOI] [PubMed] [Google Scholar]
- 37.Pennisi E New Zealand’s “mind-blowing” goal: Rat-free by 2050. Science. 2016; doi: 10.1126/science.aag0692 [DOI] [Google Scholar]
- 38.Boussommier-Calleja A, Li R, Chen MB, Wong SC, Kamm RD. Microfluidics: A new tool for modeling cancer-immune interactions. Trends Cancer Res. 2016;2: 6–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clevers H Modeling Development and Disease with Organoids. Cell. 2016;165: 1586–1597. [DOI] [PubMed] [Google Scholar]
- 40.Dobrolecki LE, Airhart SD, Alferez DG, Aparicio S, Behbod F, Bentires-Alj M, et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016;35: 547–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology. Wiley-Blackwell; 2007;121: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philip JR Goulder BDW. HIV and HLA Class I: an evolving relationship. Immunity. NIH Public Access; 2012;37: 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ibrahim-Hashim A, Robertson-Tessi M, Enriquez-Navas PM, Damaghi M, Balagurunathan Y, Wojtkowiak JW, et al. Defining Cancer Subpopulations by Adaptive Strategies Rather Than Molecular Properties Provides Novel Insights into Intratumoral Evolution. Cancer Res. 2017;77: 2242–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell JC, Stanley MC. An overview of introduced predator management in inhabited landscapes. Pac Conserv Biol. CSIRO PUBLISHING; 2018;24: 371–378. [Google Scholar]
- 45.Creer S, Deiner K, Frey S, Porazinska D, Taberlet P, Kelley Thomas W, et al. The ecologist’s field guide to sequence-based identification of biodiversity. Methods Ecol Evol. 2016;7: 1008–1018. [Google Scholar]
- 46.Ecological Data Wiki [Internet]. [cited 8 Feb 2019]. Available: https://ecologicaldata.org/
- 47.LaDeau SL, Han BA, Rosi-Marshall EJ, Weathers KC. The Next Decade of Big Data in Ecosystem Science. Ecosystems. 2016;20: 274–283. [Google Scholar]
- 48.Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I. Deep Convolutional Neural Networks Enable Discrimination of Heterogeneous Digital Pathology Images. EBioMedicine. 2018;27: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ash J, Darnell G, Munro D, Engelhardt B. Joint analysis of gene expression levels and histological images identifies genes associated with tissue morphology [Internet]. 2018. doi: 10.1101/458711 [DOI] [PMC free article] [PubMed]
- 50.Pitt JM, Marabelle A, Eggermont A, Soria J-C, Kroemer G, Zitvogel L. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27: 1482–1492. [DOI] [PubMed] [Google Scholar]
- 51.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning [Internet]. Nature Medicine. 2018. pp. 1559–1567. doi: 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson CB. Biodiversity monitoring, earth observations and the ecology of scale. Ecol Lett. 2018;21: 1572–1585. [DOI] [PubMed] [Google Scholar]
- 53.Brodrick PG, Davies AB, Asner GP. Uncovering Ecological Patterns with Convolutional Neural Networks. Trends Ecol Evol. 2019; doi: 10.1016/j.tree.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 54.Eng C-HL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature. Nature Publishing Group; 2019;568: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11: 417–422. [DOI] [PubMed] [Google Scholar]
- 56.West J, You L, Brown J, Newton PK, Anderson ARA. Towards multi-drug adaptive therapy [Internet]. bioRxiv. 2018. p. 476507. doi: 10.1101/476507 [DOI] [Google Scholar]
- 57.McCoach CE, Bivona TG. Engineering Multidimensional Evolutionary Forces to Combat Cancer. Cancer Discov. AACR; 2019; Available: http://cancerdiscovery.aacrjournals.org/content/9/5/587.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glickman MS, Sawyers CL. Converting cancer therapies into cures: lessons from infectious diseases. Cell. 2012;148: 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.