Abstract
13-cis-retinoic acid (RA) is typically used in post-remission maintenance therapy in patients with neuroblastoma. However, side effects and recurrence are often observed. We investigated the use of miRNAs as a strategy to replace RA as promoters of differentiation. miR-124 was identified as the top candidate in a functional screen. Genomic target analysis indicated that repression of a network of transcription factors could be mediating most of miR-124’s effect in driving differentiation. To advance miR-124 mimic use in therapy and better define its mechanism of action, a high-throughput siRNA morphological screen focusing on its transcription factor targets was conducted and ELF4 was identified as a leading candidate for miR-124 repression. By altering its expression levels, we showed that ELF4 maintains neuroblastoma in an undifferentiated state and promotes proliferation. Moreover, ELF4 transgenic expression was able to counteract the neurogenic effect of miR-124 in neuroblastoma cells. With RNA-seq, we established the main role of ELF4 to be regulation of cell cycle progression, specifically through the DREAM complex. Interestingly, several cell cycle genes activated by ELF4 are repressed by miR-124, suggesting that they might form a TF-miRNA regulatory loop. Finally, we showed that high ELF4 expression is often observed in neuroblastomas and is associated with poor survival.
Introduction
Neuroblastoma is the most common extracranial solid tumor among infants younger than 12 months, and is responsible for 7% of childhood cancers and 15% of cancer-related childhood deaths (1). These tumors arise from neural crest cell precursors of the sympathetic nervous system that fail to differentiate into neurons (1,2). Induction of malignant cells to differentiate into mature cells through the use of 13-cis-retinoic acid (RA) has been a mainstay treatment for post-remission maintenance therapy in patients with neuroblastoma (2). Although this therapy has drastically improved patient survival, it is often accompanied by side effects and high rates of recurrence (3). We have previously evaluated miRNA mimics as potential alternatives to RA treatment in a high-throughput screen, and identified miR-124 as one of the strongest inducers of differentiation (4).
miR-124 is a neuron-enriched, highly conserved miRNA which ranks as the most highly expressed miRNA in the human brain (5). miR-124 dysregulation has been implicated in a variety of neurological disorders and cancers with neuronal origin (5). miR-124 is defined as a tumor suppressor miRNA and is typically absent or down-regulated in tumors, very likely due to promoter hyper-methylation (6). miR-124 tumor suppressive functions include inhibition of proliferation, regulation of cell cycle genes such as CDK4 (7), and inhibition of self-renewal, migration and invasion through regulation of SCP1, PTPN12, ROCK1, Twist, and SNAI2 (5). In neuroblastoma, low miR-124 expression is associated with an undifferentiated state (8).
miR-124 expression levels increase during neural stem cell (NSC) differentiation (9) and ectopic expression enhances neuronal differentiation of mouse neural stem cells and decreases proliferation, expression of stem cell markers and growth and self-renewal of neurospheres (9). In a prior study to understand how miR-124 induces differentiation, we blocked its function with antagomiRs to determine how this treatment suppressed critical changes in gene expression during neurogenesis. Gene ontology analysis of 910 miR-124 targets identified in this study indicated transcription factors as one of the most highly enriched terms (9). This finding supports the concept of miRNA-transcription factor (TF) networks as critical players in cell fate determination (9,10). miRNA-TF networks are essential for a wide range of processes, such as embryogenesis, hematopoiesis, myogenesis, and macrophage differentiation (11). Furthermore dysregulation of miRNA-TF networks has been observed in a variety of cancers (12).
We hypothesized that the transcription factors targeted by miR-124 are critical to maintaining the undifferentiated state of neuroblastoma cells. Ectopic expression of miR-124 in neuroblastoma cells would decrease their expression levels, allowing cells to turn on a differentiation program. To advance the use of miR-124 mimics in neuroblastoma therapy and establish its mechanism of action, we evaluated miR-124 targeted transcription factors to identify the ones contributing the most to the undifferentiated state and proliferation. ELF4 was the top hit in our functional screen and was selected for further analysis. Characterization of ELF4 impact on gene expression by RNA-seq identified cell cycle regulation as a main route of action. ELF4 knockdown affected CDK2/4/5/6 expression along with various cyclins. Interestingly, a comparative analysis suggested that miR-124 and ELF4 form a feed-forward loop, where they antagonize one another via their opposing regulatory roles in the expression of shared target genes. Finally, we determined that ELF4 expression could potentially serve as a prognostic marker as its levels in neuroblastoma tumors are predictive of patient survival. Taken together, these findings support a role for miR-124 mimics in neuroblastoma differentiation therapy.
Materials and Methods
Cell Lines and Transfection
Neuroblastoma cell lines BE(2)-C, CHP-212, SH-SY5Y, SK-N-AS and SK-N-DZ were obtained from the American Type Culture Collection. KELLY and NGP cells were were a gift from Dr. Raymond Stallings and were obtained originally from the European Collection of Animal Cell Cultures (Porton Down, UK) and Cancer Therapy and Research Centre, San Antonio, TX respectively. Cells were grown in DMEM/F12 (GIBCO) supplemented with 10% fetal bovine serum (GIBCO) and 1% pen/strep (GIBCO). HeLa cells were obtained from the American Type Culture Collection and were cultured in RPMI-1640 (GIBCO) supplemented with 10% FBS and 1% pen/strep. 293T cells were obtained from the American Type Culture Collection and were cultured in DMEM-High Glucose (HyClone) supplemented with 10% FBS and 1% pen/strep. Cells were passaged no more than fifteen times and were tested for mycoplasma contamination using DAPI staining (2 μg/mL) (Thermo Fisher). All cells were authenticated using short tandem repeat profiling.
SMARTpool siRNAs against the transcription factors were obtained from Dharmacon (Supplementary Table 1). Neuroblastoma cells (BE(2)-C, CHP-212, KELLY, NGP, SH-SY5Y, SK-N-AS, and SK-N-DZ) were reverse transfected into 96-wells with siRNAs using RNAiMAX (Thermo Fisher).
Quantification of Neurite Outgrowth
96 hours after transfection, cells were imaged on an IncuCyte ZOOM Imaging System (Essen Bioscience) at 10X or 20X magnification. Neurite outgrowth was quantified and a neurite definition for each cell line was created using the NeuroTrack software module (Essen BioScience). Knockdown efficiency for each siRNA was validated at 48 hours by qRT-PCR and decreases in mRNA expression of 85% or more were observed (qPCR method described below).
MTS Assay
Neuroblastoma cells were plated into 96-well plates and transfected as described above. After culturing for 120 hours, quantification of viable cells was assessed using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega).
Cell Proliferation Assay
Neuroblastoma cells were plated into 96-well plates, transfected as described above, and placed into the IncuCyte ZOOM Imaging System (Essen BioScience). Cell confluence was monitored periodically using the Confluence Processing analysis tool (Essen BioScience). Cell growth curves were generated by plotting cell confluence as a function of time.
Ki67 Quantification Assay
Neuroblastoma cells were plated into 96-well plates and transfected as described above. After 120 hours, cells were fixed in 4% paraformaldehyde (Sigma-Aldrich) for 15 minutes at room temperature. Cells were permeabilized and blocked with blocking buffer (1X PBS/5% BSA/0.3% Triton™ X-100 (Sigma-Aldrich)) for 1 hour at room temperature. Cells were then incubated with Ki67 antibody (Abcam: ab15580) overnight at a dilution of 1:150. Cells were then washed three times with PBS and incubated with a Cy™−3 anti-rabbit secondary antibody (Jackson ImmunoResearch: 711-165-152) at a concentration of 1:500 for 1 hour at room temperature. Following three washes with PBS, cells were incubated in 2 μg/mL DAPI (Thermo Fisher) for 5 mins at room temperature. Cells were then manually counted utilizing an epifluorescence microscope. For each replicate, 500 cells were counted, and Ki67 positive cells were determined as the number of positive cells divided by the total number of cells (DAPI positive).
Caspase-3/−7 Assay
Neuroblastoma cells were plated into 96-well opaque plates and transfected as described above. After 48 hours, cells were assessed for Caspase-3/−7 activity utilizing the Caspase-Glo® 3/7 Assay (Promega).
mRNA Expression
Total RNA was isolated from neuroblastoma cells treated with specific conditions (e.g. siRNA against TFs, siControl/siELF4, miRNA Control/miR-124) with TRIzol (Thermo Fisher). cDNA was synthesized using a High-Capacity cDNA Synthesis Kit (Thermo Fisher). Primer pairs and TaqMan probes used for knockdown validation and gene expression quantification are listed in Supplementary Table 1. PowerUp SYBR Green Master Mix and TaqMan Master Mix were used for qRT-PCR (Thermo Fisher) GAPDH was utilized as a reference gene. The delta-delta Ct method was used to compare mRNA levels between different conditions (13).
Plasmid and Lentiviral Expression Vectors
A clone containing the ELF4 3’UTR was obtained from the DNASU Plasmid Depository (HsUT00698332). The 3’UTR sequence was PCR amplified and ligated into the pmirGLO vector (Promega). Another clone was prepared by deleting the predicted miR-124 binding site. Deletion of the binding site was performed using the QuikChange II Site-Directed Mutagenesis Kit (Agilent) with the following primer pair: CTA TGC GTG TTT CCA GCA GTT TTT CTA ATA AAA TCA GTT TAT and ATA AAC TGA TTT TAT TAG AAA AAC TGC TGG AAA CAC GCA TAG,
A clone containing the ELF4 open reading frame was obtained from Vigene (Catalog # CH815249), PCR amplified, and cloned into pUltra (a gift from Malcolm Moore: Addgene plasmid #24129) using the EcoRI and BamHI restriction sites. The ligated product was then transformed into Stbl3 cells (ThermoFisher). Lentivirus was prepared and titered as described previously (14).
Luciferase Assay
HeLa and 293T cells were plated into 96-well plates, grown for 16 hours, and transfected using Lipofectamine 3000 (Thermo Fisher) with the ELF4 3’UTR construct along with miR-124 mimic or control oligos. Firefly luciferase luminescence was measured 48 hours later using the Dual-Glo® Luciferase Assay System (Promega). Sea pansy luminescence was measured as a transfection control.
RNA-seq Analysis
To identify the transcriptional impact of ELF4, BE(2)-C cells were treated in triplicate with either siELF4 or siControl. 48 hours later, RNA was isolated with TRIzol (ThermoFisher). Samples were sequenced using poly-A selected mRNA at the GCCRI Genome Sequencing Facility (UTHSCSA). Reads were aligned to the human genome (UCSC version hg19) using TopHat. Feature Counts was used to count reads mapping to genes and differential gene expression analysis was performed using DESeq2 with default parameters (15). Differentially expressed genes can be found in Supplementary Table 2. Complete sequencing data can be found at (GEO: GSE125772).
Gene Set Enrichment Analysis and Network Interaction Assessment
Significant differentially expressed genes were identified at an FDR-corrected threshold of p < 0.0001 and log2 fold-change ≥ 0.5 or ≤ −0.5. Enriched gene ontologies and gene sets were identified through Protein Analysis Through Evolutionary Relationships (PANTHER; http://pantherdb.org) and the Molecular Signatures Database v6.2 (MSigDB; http://software.broadinstitute.org/gsea/msigdb/index.jsp). Enriched gene ontologies or gene sets of interest were further investigated by examining their interactions based on experimental data, co-occurrence, literature and database information, using the STRING database (https://string-db.org). Only interactions with medium (0.4) scores and above were used. Interactions were clustered by MCL clustering with an inflation parameter of 3. Graphics depicting the interactions between the genes identified were generated with Cytoscape 3.70 (https://cytoscape.org). To identify druggable targets, the Drug Gene Interaction Database (DGIdb) was used (http://www.dgidb.org). In addition to MSigDB, Enrichr was used to identify gene-sets from various databases (http://amp.pharm.mssm.edu/Enrichr).
Analysis of ELF4 and miR-124 Antagonism
Neuroblastoma cells [BE(2)-C, KELLY and SK-N-DZ] were infected with a MOI of 10 of ELF4 or control lentiviral expression vectors. 72 hours after transduction, cells were reverse transfected with either miR-124 mimic or control, and neurite outgrowth was assessed as described above. RNA was collected 48 hours after miRNA mimic transfection and qRT-PCR was performed as described in the mRNA Expression section to assess impact on co-targeted genes.
Analysis of Patient Survival
Patient survival data was obtained from the R2 genomics analysis and visualization platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Neuroblastoma patients in the Versteeg and Seeger cohorts were divided into high and low groups based on median untransformed ELF4 expression (probe: 31845_at) (16,17). Survival analysis was performed using the log-rank (Mantel-Cox) test as implemented in GraphPad Prism. Univariate and multivariate analyses were performed utilizing a Cox proportional hazards regression model as implemented in the R package ‘survival’ (https://CRAN.R-project.org/package=survival). For the Versteeg cohort the following variables were utilized: ELF4 expression, stage, age, MYCN status, and gender.
Statistical Analysis
The siRNA screen was performed utilizing technical triplicates for each of the assays (MTS, neurite outgrowth, Ki67 staining, and Caspase-3/−7 activity). Differences in proliferation were identified utilizing multiple t-testing adjusted with a Bonferroni correction. Significant differences were identified using a Student’s t-test with a Holm-Sidak correction for multiple testing, with a corrected p value of 0.05 considered statistically significant. qRT-PCR measurements were performed with biological and technical triplicates, with differences in expression assessed by Student’s t-test. Luciferase assays were performed with biological and technical triplicates, with differences in activity assessed by Student’s t-test. Antagonism and synergy were assessed using the Bliss independence model (18) using two-way ANOVA with a Tukey’s range test for multiple comparisons. Overlap between sets of miRNA targets was assessed by hypergeometric distribution test. RNA-seq analysis also employed biological triplicates.
Results
miR-124 regulated transcription factors contribute to neuroblastoma proliferation and undifferentiated state
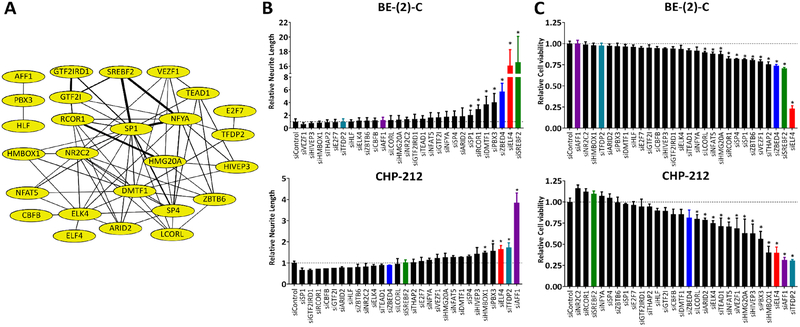
Neuroblastoma cells can be differentiated into post-mitotic neurons through the use of retinoic acid. We and others have shown that similar changes can be obtained through transfection of mimics of miR-124 (4,8). Our previous analyses suggest that miR-124 promotes differentiation at least in part by repressing transcription factors implicated in the maintenance of stem cell phenotypes (9). To identify the transcription factors responsible for maintaining the undifferentiated state and functioning as mediators of the observed effects of miR-124 in the process of neuronal differentiation, we conducted a functional screen with a selected group of 28 highly interconnected transcription factors (Fig. 1A).
Figure 1. miR-124 regulated transcription factors contribute to neuroblastoma proliferation and maintenance of the undifferentiated state.
(A) Transcription factor network regulated by miR-124 (built with STRING). (B-C) BE(2)-C and CHP-212 cells were reverse transfected with siRNAs against each transcription factor. 120 h later: (B) Neurite length was measured utilizing the IncuCyte Neurotrack software module; (C) Quantification of viable cells was assessed by MTS assay. Statistical significance of observed differences was determined by Student’s t-test with a Holm-Šídák correction for multiple comparisons.
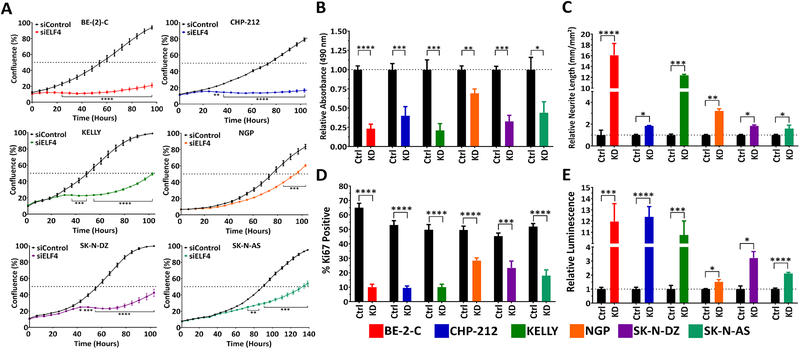
A well-recognized marker of neuroblastoma cell differentiation in vitro is neurite outgrowth. We have previously conducted high-content screens based on this morphological indicator to identify inducers of neuroblastoma differentiation (4). To analyze the network of selected transcription factors, we knocked down each one individually in BE(2)-C and CHP-212 cells via siRNA transfection. Knockdown efficiency was measured by qRT-PCR (Supplementary Fig. S1). Quantification of neurite outgrowth was done with the InCucyte Zoom Neurotrack module (Fig. 1B). In addition to assessing morphological changes, we used an MTS assay to examine if loss of any transcription factor had an impact on the number of viable cells (Fig. 1C). Of the 28 transcription factors screened, knock-down of ELF4 resulted in a consistent effect in all analyses performed, and was selected for further analysis. ELF4 knock-down in six different neuroblastoma cell lines [BE(2)-C, CHP-212, KELLY, SK-N-DZ, NGP, and SK-N-AS] had a significant effect on proliferation, Ki67 positivity, Caspase-3/−7 activity, viability, morphology, and differentiation (Fig. 2, 3).
Figure 2. ELF4 is critical for neuroblastoma cell proliferation and reduction of intracellular levels of ELF4 induces differentiation.
(A) Neuroblastoma cell lines were reverse transfected with siRNAs (siELF4 and control) and their proliferation was monitored by live-cell imaging (IncuCyte). (B) Knockdown of ELF4 with siRNA results in a significant decrease in number of viable cells (MTS assay, 120 h post-transfection). (C) Silencing of ELF4 results in differentiation (neurite outgrowth, 120 h post-transfection). (D) ELF4 knockdown significantly diminishes the percentage of Ki67 positive cells (Ki67 quantification, 120 h post-transfection). (E) Loss of ELF4 induces apoptosis (Caspase-3/−7 assay, 48 h post-transfection). Statistical significance of observed differences was determined by Student’s t-test. Proliferation data was adjusted for multiple testing using a Bonferroni correction. * = p<0.05, ** = p<0.01, *** = p<0.001, **** = p<0.0001.
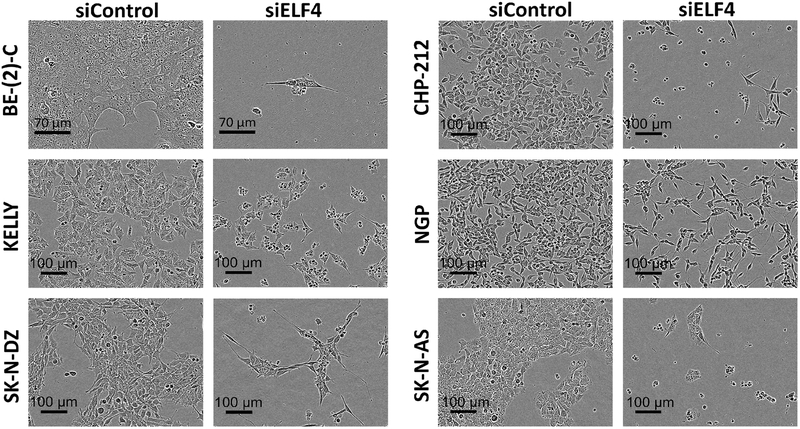
Figure 3. Morphological changes of neuroblastoma cells following ELF4 knockdown.
Images of neuroblastoma cells 120 h after reverse transfection with siRNAs.
Since we originally identified ELF4 as a target of miR-124 in mouse NSCs (9), we sought to validate this regulation in human cells. miR-124 mimic transfection in BE(2)-C, CHP-212, KELLY, and SK-N-DZ cells resulted in a significant downregulation of ELF4 mRNA (Fig. S2A). Next, we conducted a luciferase assay in two different cell lines to demonstrate a direct and specific interaction of miR-124 with ELF4. The 3’UTR of ELF4 was cloned into a dual luciferase vector. Another construct was prepared with a deletion of the miR-124 binding site (Fig. S2B, C). When the ELF4 3’ UTR reporter vector was co-transfected with miR-124 mimics, a decrease in luciferase activity was observed in comparison to control. miR-124 mimic co-transfection with the ELF4 3’ UTR reporter from which the miR-124 target site had been deleted showed no decrease in luciferase activity (Fig. S2B).
ELF4 ectopic expression antagonizes miR-124 effect on differentiation of neuroblastoma cells
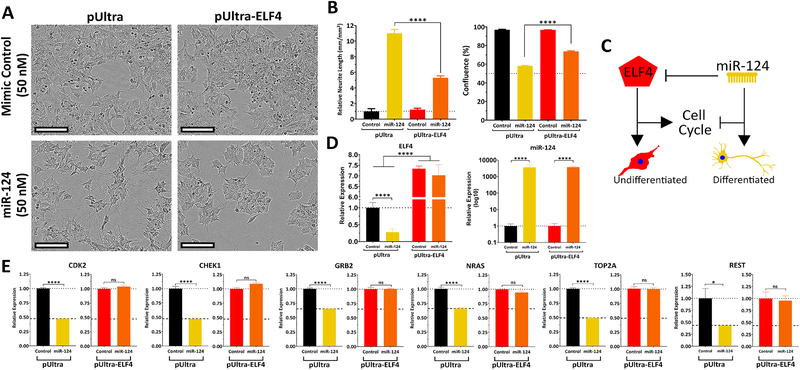
To corroborate ELF4 as a critical target of miR-124, we tested if transgenic expression of ELF4 could counteract the effect of miR-124 on differentiation. We transduced neuroblastoma cells (BE(2)-C, SK-N-DZ, and KELLY) with either control lentivirus or one expressing the open reading frame of ELF4. Infected cells were later transfected with miR-124 or control mimics. miR-124 transfection induced strong differentiation in cells infected with pUltra (control). The effect of miR-124 on neurite outgrowth and proliferation was partially neutralized by the ectopic expression of ELF4 (Fig. 4A–C, S3 and S4). Based on these results, we propose a model for the interaction between ELF4 and miR-124 in the regulation of neuroblastoma differentiation (Fig. 4C). To assess this antagonism model further, we examined mRNA levels of several genes predicted to be regulated by miR-124 and ELF4 in opposing directions. We observed that when miR-124 mimics and ELF4 expression vector were co-transfected, ELF4 counteracted miR-124 repressive effect on several of its target genes – CDK2, CHEK1, GRB2, NRAS, TOP2A, and REST (Fig. 4D, E, S3, S4) – supporting the phenotypic observations.
Figure 4. ELF4 ectopic expression antagonizes the effect of miR-124 on differentiation of neuroblastoma cells.
(A-B) BE(2)-C cells were first infected with ELF4-expressing lentivirus or control then reverse transfected with miRNA mimics (miR-124 or control). 120 hours later, the impact on differentiation and confluence was measured. (A) Morphology of transfected cells (scale bar = 100 μm). (B) Quantification of neurite outgrowth of treated cells. (C) Confluence of treated cells. (D) Proposed model of ELF4 and miR-124 antagonism. (E) ELF4 and miR-124 expression levels in SK-N-BE(2)-C transfected cells. (F) Expression analysis by qRT-PCR of miR-124 and ELF4-overexpressing shared targets in co-transfected cells were transfected with miRNA mimics; 48 hours later RNA was isolated and qRT-PCR was used to measure expression of a select group of co-targeted genes. Statistical significance of observed changes in neurite outgrowth was determined by a two-way ANOVA with Tukey’s range test for multiple comparisons. A Student’s t-test was used to assess differences in expression. * = p<0.05, **** = p<0.0001.
High ELF4 expression correlates with poor prognosis in neuroblastoma patients
The impact of ELF4 on neuroblastoma cell proliferation and maintenance of an undifferentiated phenotype suggests that high ELF4 expression could contribute to neuroblastoma development and response to therapy. We stratified the neuroblastoma patient cohort described by Molenaar, et al. (16) based on median ELF4 expression and found that patients with high tumor ELF4 expression show poor survival rates relative to those with low ELF4 levels, (Fig. S5A and B). In a multivariate analysis featuring additional variables (stage, age, gender and MYCN status), ELF4 was found to be an independent risk factor (p=0.00601) (Supplementary Table 3). In another cohort featuring metastatic non-MYCN amplified tumors (17), high ELF4 expression correlated with poor relapse-free survival (Fig. S5C,D and Supplementary Table 3), indicating that ELF4 expression can potentially predict poor outcome.
Synergistic effect of ALK and ELF4 inhibition on cell proliferation
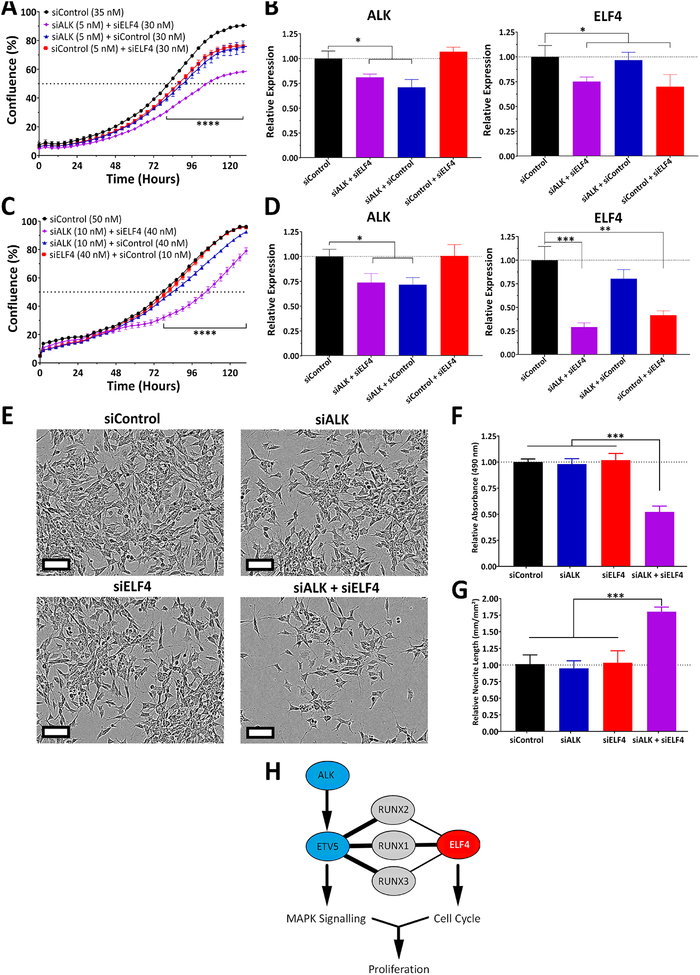
We assessed the importance of ELF4 in multiple neuroblastoma cell lines. In SH-SY5Y cells, ELF4 knockdown had little impact on proliferation or differentiation. Initially we predicted a MYCN dependency, but SK-N-AS cells, which are not MYCN amplified, responded well to ELF4 knockdown. In addition, ELF4 expression strongly correlates with poor relapse-free survival in a MYCN-non-amplified cohort (Fig. S5C and D). Therefore, we hypothesized that a different mechanism was driving resistance. ALK mutations are a frequent cause of germline neuroblastoma, and SH-SY5Y cells carry the F1174L mutation (19). Several groups have demonstrated efficacy of ALK inhibition in inhibiting neuroblastoma cell proliferation. More importantly, inhibitors of ALK and CDK4/6 have been found to work synergistically (19). Taking into account the RNA-seq results that suggest that ELF4 directly or indirectly regulates cell cycle progression genes, we decided to perform dual knockdowns of ALK and ELF4. Low levels of ALK or ELF4 knockdown (29% and 30%, respectively) had minor effects on proliferation of SH-SY5Y cells, but when both ALK and ELF4 were partially knocked down, the effect on proliferation and differentiation was greater (Fig. 5A–B, E–G). To further confirm this synergistic effect, we performed the dual knockdown (29% decrease in ALK and 60% decrease in ELF4) in KELLY cells, which carry the same ALK mutation (F1174L). The results were similar to what we observed in SH-SY5Y cells (Fig. 5C–D, and S6). Inhibition of both ALK and ELF4 resulted in a synergistic effect, with a combination index (CI) of 0.82 for SH-SY5Y and 0.28 for KELLY at 120 hours, based on the Bliss independence model (18). ALK, a receptor tyrosine kinase, acts upstream of the MAPK signaling cascade, partially functioning through the transcription factor ETV5 (20). ETV5, like ELF4, is an ETS-family member, and both have been shown to interact with members of the Runt-related transcription factor family (RUNX) as identified by STRING. We propose that this interaction explains how the combined reduction in expression of ALK and ELF4 leads to a strong reduction in cell proliferation (Fig. 5H).
Figure 5. Synergy between ALK and ELF4 inhibition on NB cell proliferation.
(A) SH-SY5Y cells were reverse transfected with siRNAs (siALK, siELF4 or control) and their proliferation was monitored with live-cell imaging (IncuCyte). (B) mRNA levels of ALK and ELF4 48 h after transfection. (C) KELLY cells were reverse transfected with siRNAs (siALK, siELF4 or control) and their proliferation was monitored with live-cell imaging. (D) mRNA levels of ALK and ELF4 48 h after transfection. (E) Morphological changes observed. (F) Quantification of the number of viable cells (MTS assay, 120 hours after transfection). (G) Quantification of differentiation (neurite outgrowth, 120 hours after transfection). (H) Proposed model of ALK and ELF4 synergy, based on STRING protein-protein interactions. Statistical significance of observed changes was determined by Student’s t-test with a nominal significance threshold of p = 0.05. * = p<0.05, ** = p<0.01, *** = p<0.001.
ELF4 regulates a network of cell cycle genes
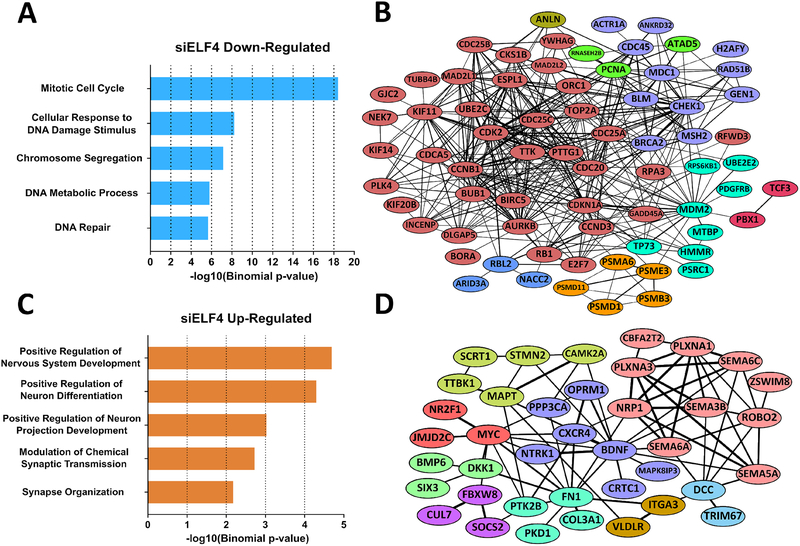
To determine the genes and pathways regulated by ELF4, we performed an RNA-seq analysis on BE(2)-C cells following ELF4 knockdown. 940 genes showed a decrease in expression, while 722 genes were up-regulated (Supplementary Table 4). A subset of differentially expressed genes were validated with qRT-PCR in three cell lines (BE(2)-C, SK-N-DZ, and KELLY) (Fig. S7). To determine the extent to which the differentially regulated genes are preferentially associated with specific pathways or biological functions, we performed a gene-set enrichment analysis using PANTHER and the molecular signatures database (MSigDB). Down-regulated genes are highly enriched in cell cycle pathways and complexes involved in cell cycle progression. These down-regulated genes possess many protein-protein interactions between them and include well-recognized cell cycle genes such as CDK2/5/16, cyclins A2/B1/D3/F/T1, CDC20, PCNA, and MCM2. On the other hand, the set of up-regulated genes shows enrichment of differentiation and neurogenesis ontology terms, supporting the observation that loss of ELF4 induces differentiation (Fig. 6 C and D, Supplementary Table 4). The upregulated genes include doublecortin, NTRK1, NTNG1/−2, NRP1, and ROBO2, all of which are involved in neuronal differentiation as well as peripheral nervous system maturation (21–24). In fact, high NTRK1 expression is a positive predictor of neuroblastoma patient survival as well as differentiation ability (25).
Figure 6. ELF4 regulates a network of cell cycle genes.
(A) Gene ontology analysis of siELF4-downregulated genes with PANTHER reveals that ELF4 targets are preferentially associated with cell cycle pathways. (B) Cell cycle-related genes (GO: Regulation of Mitotic Cell Cycle) regulated by ELF4 form a highly connected network according to STRING. Link thickness is based on the confidence of the interaction. Nodes color-coded based on MCL clustering. (C) siELF4-upregulated genes are highly associated with neuronal processes (PANTHER), supporting the observation that ELF4 loss induces differentiation and that differentiation requires cell cycle exit. (D) Upregulated genes (GO: Neurogenesis) linked to neuronal function form a highly connected network according to STRING. Link thickness is based on the confidence of the interaction. Nodes color-coded based on MCL clustering.
ELF4 and miR-124 have opposite regulatory impact on cell cycle related genes
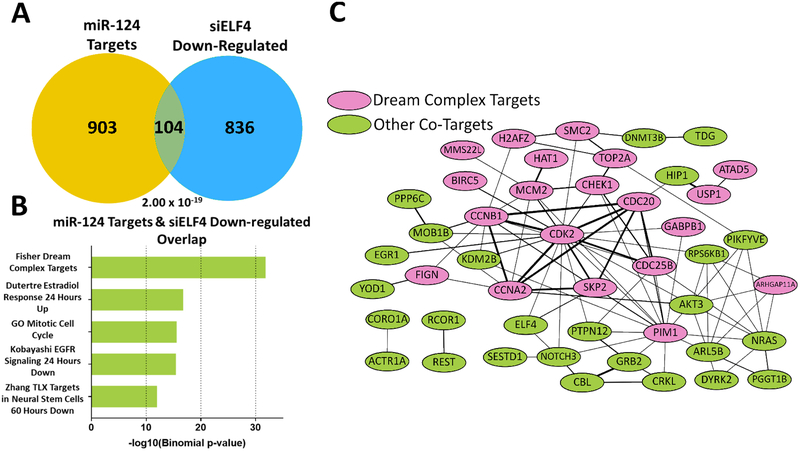
miRNAs and transcription factors interact in the context of complex networks that modulate a variety of biological processes, including development and tumorigenesis (10,26). We were curious as to whether the miR-124-ELF4 interaction goes beyond the impact of miR-124 impact on ELF4 expression. More specifically, we wanted to know if their antagonistic effect on neuronal differentiation could block ELF4 activity in two ways: 1) by direct repression, and 2) by decreasing the expression of ELF4 target genes. To evaluate this possible feed-forward loop, we first generated a list of miR-124 targets by combining miR-124 targets identified by our group and high confidence validated targets from miRTarBase (9,27) (Supplementary Table 5). We then compared this list (1,007 genes) to the list of genes displaying a decrease in expression upon ELF4 knockdown (940 genes). We found a statistically significant overlap of 104 genes based on a hypergeometric distribution test (p = 2.00 × 10−19) (Fig. 7A, Supplementary Table 6). Gene ontology analysis with MSigDb revealed an enrichment in cell cycle regulation (Fig. 7B, Supplementary Table 7). One of the resulting gene sets featured targets of the DREAM complex. This complex is essential for maintenance of quiescence by regulating the transition from both G1/S and G2/M phases. DREAM complex dysregulation has been implicated in a variety of cancers, including neuroblastoma, where the complex is reactivated by MYCN and FOXM1 to promote cell cycle progression (28). Protein interaction analysis showed that genes affected by both miR-124 and ELF4 are highly connected, forming a dense network (Fig. 7C). Finally, in our antagonism experiments, we found that ELF4 overexpression prevents miR-124 from downregulating several co-target genes, including CDK2, CHEK1, GRB2, NRAS, TOP2A and REST (Fig. 4E, S4, S5). These results indicate that failure to downregulate ELF4 diminishes the ability of miR-124 to downregulate target genes found in the overlap. Taken together, these results suggest that miR-124 and ELF4 form a miRNA-transcription factor regulatory loop in cell fate decisions; however further analysis is required to characterize this interaction completely.
Figure 7. miR-124 and ELF4 regulate a common set of targets in opposing directions.
(A) miR-124 targets and siELF4-downregulated genes overlap significantly. Statistical significance was determined by hypergeometric distribution test. (B) MSigDb analysis of the 104 overlapping genes reveals highly enriched gene sets centered on cell cycle regulation (e.g. Fisher Dream Complex, GO Cell Cycle). (C) Protein-protein interactions between the co-targeted genes reveals a highly interconnected network. DREAM Complex targets (pink) account for a large majority of the overlapped genes (34/104). Link thickness is based on the confidence of the interaction (STRING).
Discussion
ELF4 transcriptional targets and pathways
Differentiation is an essential therapeutic approach for treating neuroblastoma, but not all children respond to the agents currently in use. Regulators of the undifferentiated phenotype can offer insight on differentiation therapy resistance. We identified ELF4 as a critical target of the pro-neurogenic miR-124 and as a novel driver of neuroblastoma’s undifferentiated state and proliferation. RNA-seq data implicated ELF4 in cell cycle progression, as it potentially regulates, either directly or indirectly, CDK2, CDK4, CDK5, and CDK6, in addition to other cell cycle genes. In support of this observation, Miyazaki, et al. (29) showed that ELF4 activity is highest in the G1 phase and that ELF4 is directly regulated through phosphorylation by the cyclin A2-CDK2 complex, both of which are found to be down-regulated when ELF4 is knocked down. These findings also suggest the existence of a positive regulatory loop between ELF4 and the cyclin A2-CDK2 complex.
Increased expression of ELF4 has been proposed to contribute to tumorigenesis via its critical role in cell cycle progression. Genetic ablation of ELF4 resulted in decreased proliferation of glioma-initiating cells (30). Several reports describe a close relationship between ELF4 and the tumor suppressor p53. ELF4 promotes the expression of MDM2, a repressor of p53, enabling transformation (31). MDM2 also increases MYCN mRNA stability and translation (32). In addition, ELF4 expression is tightly controlled by the p53-MDM2-E2F1 signaling axis (33). In relapsed neuroblastoma, a high frequency of p53/MDM2/p14ARF pathway alterations is observed (34). Due to its relationship to this pathway, ELF4 may be implicated in relapses as well.
ELF4 is critical to stem cell proliferation. In ELF4−/− mice, hematopoietic stem cells have reduced proliferation and become stuck in a quiescent state (35). Interestingly, our gene-set enrichment analysis (GSEA) of siELF4-downregulated genes identified an enrichment of DREAM complex targets. As this complex regulates quiescence transitions, our data supports the phenotype observed in ELF4−/− mice (36).
ALK and ELF4 relationship
We found an interesting mechanism by which the receptor tyrosine kinase ALK may compensate for ELF4 loss. Not only are ALK mutations a common driver of familial neuroblastoma, but ALK is often dysregulated in non-mutated neuroblastomas (37). In six of the seven cell lines we assessed, ELF4 knockdown affected proliferation, independent of MYCN amplification status. However, knockdown of ELF4 in SH-SY5Y cells produced a partial inhibition of cell proliferation. We hypothesized that ALK mutation in these cells could compensate for the negative impact of ELF4 knockdown on CDKs and cyclins, which was confirmed by dual knock-down experiments. This finding is supported by a study showing the synergistic effect of dual ALK and CDK4/6 inhibition on neuroblastoma cell proliferation (19). ALK-ELF4 compensation was verified in a MYCN-amplified cell line, suggesting this relationship is MYCN independent.
miR-124 and ELF4 transcription factor network
Our work highlights the importance of miRNA-transcription factor crosstalk in tumorigenesis. While miR-124 targets are distributed across several different biological functions, it appears that targeting transcription factors, as seen here with ELF4, is critical for driving major phenotype changes. miR-124 and ELF4 potentially share a significant number of target genes. Analysis of these targets using gene-set enrichment (GSEA) revealed the DREAM complex and cell cycle as important pathways. The DREAM complex tightly regulates the transition between G0 and G1 phases of the cell cycle, in addition to regulating gene expression through G1/S and G2/M phases (38). This complex is responsible for maintaining quiescence, and following ELF4 knockdown or miR-124 overexpression, the downregulation of DREAM complex target genes appears to be relevant for terminal differentiation of neuroblastoma cells.
Another enriched target gene set is regulated by estrogen (Dutertre Estradiol Response, ID: M2156). Estrogen signaling has been implicated in neuroblastoma proliferation and phenotype (39). Moreover, estrogen receptor α was found to enhance the transcriptional activity of ETS-1, an ELF4 family member, in neuroblastoma cells (40). Analysis of siELF4-downregulated genes with the gene-set enrichment program Enrichr revealed that 219 out of the 940 genes are also responsive to the phytoestrogen coumestrol, based on the Drug Signatures Database (DSigDB). Although the relationship between estrogen signaling and ELF4 is not well defined, it appears that both miR-124 and ELF4 target several components of estrogen signaling.
Genes regulated by miR-124 and ELF4 have druggable potential
Analysis of the 104 antagonistically regulated genes via the Drug Gene Interaction Database identified numerous genes with druggable domains. In fact, five genes (NRAS, PDE10A, SLC12A2, RPS6KB1, and TOP2A) have FDA-approved drugs that either directly or indirectly target them. More importantly, expression of several of these genes strongly correlates with patient survival. NRAS, for example, is upstream of the MEK signaling cascade, which is dysregulated in neuroblastoma (41). MEK inhibitors, such as cobimetinib, have demonstrated efficacy against neuroblastomas (42). TOP2A, a topoisomerase, has been shown to be important for response to topoisomerase inhibitor doxorubicin (43), a front-line agent against high-risk neuroblastoma (44). SLC12A2, also known as NKCC1, is a Na+, K+, and Cl− co-transporter that can be inhibited with furosemide or bumetanide. This transporter was shown to be critical in EGF and FGF signaling (45). Furthermore, in normal neural progenitor cells, NKCC1 knockdown results in decreased proliferation, suggesting that this co-transporter is important in cell cycle progression in neural stem cell populations (46). More importantly, it was found that overexpression of NKCC1 could induce transformation in mouse fibroblasts (47). RPS6KB1, also known as p70-S6 kinase, is an important downstream target of mTOR. Inhibition of mTOR in neuroblastoma has demonstrated anti-proliferative effects (48). PDE10A, a phosphodiesterase, can be inhibited by dipyridamole, which prevents neuroblastoma cell growth (49). Finally, preclinical assessment of other co-targeted genes, such as CDK2, also has also revealed promising leads for treating neuroblastoma (50).
In conclusion, our work establishes the transcription factor ELF4 as a target of neurogenic miRNA miR-124 and as a critical regulator of neuroblastoma cell identity, cell cycle and proliferation. We also showed the potential of ELF4 as a predictor of patient survival. Finally, we demonstrated that miR-124 can induce differentiation through regulation of transcription factors and corroborated the importance of miRNA-transcription factor crosstalk in cancer-relevant phenotypes.
Supplementary Material
Implications:
miR-124 induces neuroblastoma differentiation partially through the downregulation of transcription factor ELF4, which drives neuroblastoma proliferation and its undifferentiated phenotype.
Acknowledgments
This work was supported by two grants from the Owens Foundation to LOFP and EK, by NIH grant CA198648 (VRI), and by the IIMS-CTSA (UTHSCSA). AK was supported by NIH Supplement 2R01 HG006015 and the Greehey Foundation. We also thank the Biomedical Research Computing Facility and the Texas Advanced Computing Center (TACC) at UT Austin for the use of computational facilities.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Park JR, Eggert A, Caron H. Neuroblastoma: biology, prognosis, and treatment. Elsevier. 2008; [DOI] [PubMed] [Google Scholar]
- 2.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. [DOI] [PubMed] [Google Scholar]
- 3.Matthay KK, Villablanca JG, Seeger RC. Treatment of High-Risk Neuroblastoma with Intensive Chemotherapy, Radiotherapy, Autologous Bone Marrow Transplantation, and 13-cis-Retinoic Acid. Mass Medical Soc. 1999; [DOI] [PubMed] [Google Scholar]
- 4.Zhao Z, Ma X, Hsiao T-H, Lin G, Kosti A, Yu X, et al. A high-content morphological screen identifies novel microRNAs that regulate neuroblastoma cell differentiation. Oncotarget. 2014;5:2499–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Luo Z-M, Guo X-M, Su D-F, Liu X. An updated role of microRNA-124 in central nervous system disorders: a review. Front Cell Neurosci. 2015;9:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong KY, So CC, Loong F, Chung LP, Lam WWL, Liang R, et al. Epigenetic inactivation of the miR-124–1 in haematological malignancies. PLoS One. 2011;6:e19027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng X, Ma L, Wu M, Zhang G, Jin C, Guo Y, et al. miR-124 radiosensitizes human glioma cells by targeting CDK4. J Neurooncol. 2013;114:263–274. [DOI] [PubMed] [Google Scholar]
- 8.Samaraweera L, Grandinetti KB, Huang R, Spengler BA, Ross RA. MicroRNAs define distinct human neuroblastoma cell phenotypes and regulate their differentiation and tumorigenicity. BMC Cancer. 2014;14:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos MCT, Tegge AN, Correa BR, Mahesula S, Kohnke LQ, Qiao M, et al. miR-124, −128, and −137 Orchestrate Neural Differentiation by Acting on Overlapping Gene Sets Containing a Highly Connected Transcription Factor Network. Stem Cells. 2016;34:220–232. [DOI] [PubMed] [Google Scholar]
- 10.Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, et al. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288:77–87. [DOI] [PubMed] [Google Scholar]
- 12.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719–732. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 14.Kutner RH, Zhang X-Y, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4:495–505. [DOI] [PubMed] [Google Scholar]
- 15.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- 16.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. [DOI] [PubMed] [Google Scholar]
- 17.Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98:1193–1203. [DOI] [PubMed] [Google Scholar]
- 18.Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3:e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood AC, Krytska K, Ryles HT, Infarinato NR, Sano R, Hansel TD, et al. Dual ALK and CDK4/6 Inhibition Demonstrates Synergy against Neuroblastoma. Clin Cancer Res. 2017;23:2856–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Delisle L, Pierre-Eugène C, Louis-Brennetot C, Surdez D, Raynal V, Baulande S, et al. Activated ALK signals through the ERK-ETV5-RET pathway to drive neuroblastoma oncogenesis. Oncogene. 2018;37:1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown JP, Couillard-Després S, Cooper-Kuhn CM, Winkler J, Aigner L, Kuhn HG. Transient expression of doublecortin during adult neurogenesis. J Comp Neurol. 2003;467:1–10. [DOI] [PubMed] [Google Scholar]
- 22.Shiau CE, Bronner-Fraser M. N-cadherin acts in concert with Slit1-Robo2 signaling in regulating aggregation of placode-derived cranial sensory neurons. Development. 2009;136:4155–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maden CH, Gomes J, Schwarz Q, Davidson K, Tinker A, Ruhrberg C. NRP1 and NRP2 cooperate to regulate gangliogenesis, axon guidance and target innervation in the sympathetic nervous system. Dev Biol. 2012;369:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz Q, Maden CH, Vieira JM, Ruhrberg C. Neuropilin 1 signaling guides neural crest cells to coordinate pathway choice with cell specification. Proc Natl Acad Sci USA. 2009;106:6164–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimada H, Nakagawa A, Peters J, Wang H, Wakamatsu PK, Lukens JN, et al. TrkA expression in peripheral neuroblastic tumors: prognostic significance and biological relevance. Cancer. 2004;101:1873–1881. [DOI] [PubMed] [Google Scholar]
- 26.Martinez NJ, Walhout AJM. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31:435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou C-H, Shrestha S, Yang C-D, Chang N-W, Lin Y-L, Liao K-W, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Decaesteker B, Denecker G, Van Neste C, Dolman EM, Van Loocke W, Gartlgruber M, et al. TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat Commun. 2018;9:4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazaki Y, Boccuni P, Mao S, Zhang J, Erdjument-Bromage H, Tempst P, et al. Cyclin A-dependent phosphorylation of the ETS-related protein, MEF, restricts its activity to the G1 phase of the cell cycle. J Biol Chem. 2001;276:40528–40536. [DOI] [PubMed] [Google Scholar]
- 30.Bazzoli E, Pulvirenti T, Oberstadt MC, Perna F, Wee B, Schultz N, et al. MEF promotes stemness in the pathogenesis of gliomas. Cell Stem Cell. 2012;11:836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sashida G, Liu Y, Elf S, Miyata Y, Ohyashiki K, Izumi M, et al. ELF4/MEF activates MDM2 expression and blocks oncogene-induced p16 activation to promote transformation. Mol Cell Biol. 2009;29:3687–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu L, Zhang H, He J, Li J, Huang M, Zhou M. MDM2 regulates MYCN mRNA stabilization and translation in human neuroblastoma cells. Oncogene. 2012;31:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suico MA, Fukuda R, Miyakita R, Koyama K, Taura M, Shuto T, et al. The transcription factor MEF/Elf4 is dually modulated by p53-MDM2 axis and MEF-MDM2 autoregulatory mechanism. J Biol Chem. 2014;289:26143–26154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carr-Wilkinson J, O’Toole K, Wood KM, Challen CC, Baker AG, Board JR, et al. High Frequency of p53/MDM2/p14ARF Pathway Abnormalities in Relapsed Neuroblastoma. Clin Cancer Res. 2010;16:1108–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacorazza HD, Yamada T, Liu Y, Miyata Y, Sivina M, Nunes J, et al. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell. 2006;9:175–187. [DOI] [PubMed] [Google Scholar]
- 36.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. [DOI] [PubMed] [Google Scholar]
- 37.Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciana P, Ghisletti S, Mussi P, Eberini I, Vegeto E, Maggi A. Estrogen receptor alpha, a molecular switch converting transforming growth factor-alpha-mediated proliferation into differentiation in neuroblastoma cells. J Biol Chem. 2003;278:31737–31744. [DOI] [PubMed] [Google Scholar]
- 40.Cao P, Feng F, Dong G, Yu C, Feng S, Song E, et al. Estrogen receptor α enhances the transcriptional activity of ETS-1 and promotes the proliferation, migration and invasion of neuroblastoma cell in a ligand dependent manner. BMC Cancer. 2015;15:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka T, Higashi M, Kimura K, Wakao J, Fumino S, Iehara T, et al. MEK inhibitors as a novel therapy for neuroblastoma: Their in vitro effects and predicting their efficacy. J Pediatr Surg. 2016;51:2074–2079. [DOI] [PubMed] [Google Scholar]
- 42.Singh A, Ruan Y, Tippett T, Narendran A. Targeted inhibition of MEK1 by cobimetinib leads to differentiation and apoptosis in neuroblastoma cells. J Exp Clin Cancer Res. 2015;34:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, et al. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci USA. 2008;105:9053–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kushner BH, LaQuaglia MP, Bonilla MA, Lindsley K, Rosenfield N, Yeh S, et al. Highly effective induction therapy for stage 4 neuroblastoma in children over 1 year of age. J Clin Oncol. 1994;12:2607–2613. [DOI] [PubMed] [Google Scholar]
- 45.Panet R, Eliash M, Atlan H. Na+/K+/Cl− cotransporter activates MAP-kinase cascade downstream to protein kinase C, and upstream to MEK. J Cell Physiol. 2006;206:578–585. [DOI] [PubMed] [Google Scholar]
- 46.Young SZ, Taylor MM, Wu S, Ikeda-Matsuo Y, Kubera C, Bordey A. NKCC1 knockdown decreases neuron production through GABA(A)-regulated neural progenitor proliferation and delays dendrite development. J Neurosci. 2012;32:13630–13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panet R, Marcus M, Atlan H. Overexpression of the Na(+)/K(+)/Cl(−) cotransporter gene induces cell proliferation and phenotypic transformation in mouse fibroblasts. J Cell Physiol. 2000;182:109–118. [DOI] [PubMed] [Google Scholar]
- 48.Johnsen JI, Segerström L, Orrego A, Elfman L, Henriksson M, Kågedal B, et al. Inhibitors of mammalian target of rapamycin downregulate MYCN protein expression and inhibit neuroblastoma growth in vitro and in vivo. Oncogene. 2008;27:2910–2922. [DOI] [PubMed] [Google Scholar]
- 49.Tebbi CK, Chervinsky D, Baker RM. Modulation of drug resistance in homoharringtonine-resistant C-1300 neuroblastoma cells with cyclosporine A and dipyridamole. J Cell Physiol. 1991;148:464–471. [DOI] [PubMed] [Google Scholar]
- 50.Dolman MEM, Poon E, Ebus ME, den Hartog IJM, van Noesel CJM, Jamin Y, et al. Cyclin-Dependent Kinase Inhibitor AT7519 as a Potential Drug for MYCN-Dependent Neuroblastoma. Clin Cancer Res. 2015;21:5100–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.