Abstract
Publication bias, more generally termed as small-study effect, is a major threat to the validity of meta-analyses. Most meta-analysts rely on the p-values from statistical tests to make a binary decision about the presence or absence of small-study effects. Measures are available to quantify small-study effects’ magnitude, but the current literature lacks clear rules to help evidence users in judging whether such effects are minimal or substantial. This article aims to provide rules of thumb for interpreting the measures. We use six measures to evaluate small-study effects in 29,932 meta-analyses from the Cochrane Database of Systematic Reviews. They include Egger’s regression intercept and the skewness under both the fixed-effect and random-effects settings, the proportion of suppressed studies, and the relative change of the estimated overall result due to small-study effects. The cutoffs for different extents of small-study effects are determined based on the quantiles in these distributions. We present the empirical distributions of the six measures and propose a rough guide to interpret the measures’ magnitude. The proposed rules of thumb may help evidence users grade the certainty in evidence as impacted by small-study effects.
Keywords: Cochrane Database of Systematic Reviews, meta-analysis, publication bias, small-study effect, systematic review
Introduction
Meta-analyses are powerful tools to combine and compare information from multiple sources and provide the most comprehensive evidence for decision-making. They have been frequently applied to facilitate evidence-based medicine, and innovative approaches have been increasingly developed to meet contemporary needs of decision makers and overcome various challenges.1,2 An essential difficulty in meta-analyses is to reduce potential bias from individual studies, as well as from the process of combining them. The PRISMA statement guides the conduct and reporting of the systematic review process3,4, the Cochrane Collaboration provides instructions on assessing the risk of bias of individual studies5,6, and the GRADE Working Group has established approaches to rating the certainty of evidence.7,8 Most efforts aim to avoid unwarranted strong inference based on misleading or low-quality evidence.9 Nevertheless, compared with handling bias within studies, it is more challenging to effectively detect or even correct publication bias. Studies with statistically significant results or results in certain directions may be more likely published10–13, which can seriously bias meta-analytic conclusions.
An ideal method to remedy publication bias is to retrieve unpublished studies from various sources besides published journal articles. These sources include clinical trial registries, drug or device approving agencies, scientific conference abstracts and proceedings.14 However, this method is not always feasible. For example, the unpublished sources may not be available, and the retrieval process may be cumbersome and time-consuming. Also, unpublished databases have not been peer-reviewed, and their quality is not guaranteed. Even if some unpublished data were incorporated into a meta-analysis, the likelihood of publication bias remains. Therefore, when using such non-statistical methods to deal with publication bias, meta-analysts need to pursue a tradeoff between time, effort, costs, and the importance of unpublished data.15
Consequently, in addition to searching for unpublished databases, statistical methods have been popular to assess publication bias.16–18 These include Egger’s regression19, the trim-and-fill method20, and a recently proposed skewness of the collected studies.21 Meta-analysts usually determine the presence or absence of publication bias based on these tests’ significance, while the bias direction and magnitude can also deliver critical information.22 Although Egger et al.19 suggested to measure the bias direction and magnitude using their proposed regression’s intercept, most meta-analysts reported only this regression test’s p-value, not the intercept itself. Such a measure has been insufficiently reported primarily because researchers lack an intuitive guide to interpret the measure’s magnitude.
Of note, most statistical methods, including Egger’s regression, are designed based on examining the funnel plot’s asymmetry. Such asymmetry is sometimes termed as small-study effect, rather than publication bias, to remind meta-analysts that publication bias may not be the only cause of the asymmetry.18 Many other factors may also lead to an asymmetrical funnel plot (e.g., due to heterogeneity or simply by chance). Consequently, we use the term small-study effects throughout this article to more accurately describe the statistical methods’ results. We cautiously note that the statistical methods may only assist the assessment of small-study effects, rather than ascertaining publication bias. The funnel plot’s asymmetry should be carefully interpreted on a case-by-case basis from both statistical and clinical perspective, e.g., using the comprehensive guidelines provided by Sterne et al.18
This article summarizes six measures for small-study effects and presents their empirical distributions based on many meta-analyses in the Cochrane Database of Systematic Reviews (CDSR). We provide rules of thumb to quantitatively help decision makers evaluate the magnitude of small-study effects.
Methods
Data Collection
The Cochrane Collaboration provides a large collection of systematic reviews in healthcare. We searched for all reviews in the CDSR from 2003 Issue 1 to 2018 Issue 5 and downloaded their data on 27 May 2018 via the R package “RCurl”. Each Cochrane review corresponded to a distinct healthcare-related topic and usually contained multiple meta-analyses on different outcomes and/or treatment comparisons. We pooled all meta-analyses from all reviews and classified them into two groups based on their outcomes (binary and non-binary). For meta-analyses with binary outcomes, regardless the effect sizes used in the original Cochrane reviews, we assessed their small-study effects based on (log) odds ratios so that the associated measures could be more consistent across meta-analyses. For meta-analyses with non-binary outcomes, we used the originally reported effect sizes (e.g., mean differences, standardized mean differences, rate ratios), because it was impossible to transform them into a common type of effect size using the available data from the CDSR. Finally, it was difficult to justify small-study effects in meta-analyses with few studies16,23, so we only considered meta-analyses with at least five studies.
Meta-analyses in the same Cochrane review often shared some common studies, so their results were possibly correlated. In addition to the full database as collected above, we considered a reduced database by selecting the largest meta-analysis (with respect to the number of studies) from each Cochrane review. The meta-analyses in the reduced database could be considered independent, and they were used as a sensitivity analysis.
Measures for Small-Study Effects
We measured small-study effects using six methods briefly described in Table 1 for each Cochrane meta-analysis. The first four measures were based on Egger’s regression. We considered the regression intercept (originally introduced by Egger et al.19) and the recently proposed skewness of the regression errors.21 Egger’s regression was originally described under the fixed-effect (FE) setting.24 However, heterogeneity often existed between studies, and it was commonly modeled using random effects (RE) with additive between-study variances.25,26 We considered the regression intercept and the skewness under both the FE and RE settings, denoted as and , and and , respectively. Under the RE setting, we first used the test to examine the significance of heterogeneity. If its p-value <0.05, the between-study variance was estimated using the DerSimonian–Laird method27; otherwise, it was set to zero.
Table 1.
Statistical methods for assessing small-study effects.
| Method | Description |
|---|---|
| Funnel plot | Presenting the study-specific effect size against its standard error (or the inverse of standard error). It is roughly symmetrical around the overall effect size if no small-study effects appear. |
| Regression test | SND = + precision + error. Under the fixed-effect setting, SND (standard normal deviate) = and precision =; under the random-effects setting, SND = and precision =. Here, y and s are the study-specific effect size and its standard error within studies, respectively, and is between-study variance due to heterogeneity. It tests for whether = 0. |
| Regression intercept ( or ) | An estimate of the intercept of the regression test under the fixed-effect () or random-effects () setting. |
| Skewness ( or ) | An estimate of the skewness of the study-specific errors of the regression test under the fixed-effect () or random-effects () setting. |
| Trim-and-fill method | Estimating the suppressed studies and thus correcting small-study effects based on funnel plot’s asymmetry. |
| Proportion of suppressed studies () | , where n is the number of studies in the original meta-analysis, and is the estimated number of suppressed studies using the trim-and-fill method. |
| Relative change of overall result by incorporating imputed suppressed studies () | , where is the estimated overall result in the original meta-analysis of published studies, and is that after incorporating imputed suppressed studies using the trim-and-fill method. |
The other two measures were based on the trim-and-fill method20, which could use the observed studies in a meta-analysis to impute the suppressed studies and thus adjust for small-study effects. We estimated the number of suppressed studies , and the overall effect sizes before and after correcting small-study effects, denoted as and , respectively. Consequently, small-study effects were measured using the proportion of suppressed studies, , and the relative change of the estimated overall effect size, .14
All six measures were theoretically zero when no small-study effects presented. The five measures except can be positive or negative; their signs indicated small-study effects’ direction, and their absolute values implied the effects’ magnitude. Positive regression intercepts or skewness indicated that studies with more negative results (i.e., on the funnel plot’s left side) tended to be suppressed; negative ones indicated that suppressed studies tended to be in the positive direction. When using , determining the small-study effects’ direction depended on the overall effect size’s direction. Moreover, unlike the above five measures, lied within 0–100%; it informed only small-study effects’ magnitude, not their direction.
Deriving Rules of Thumb for Small-Study Effects’ Magnitudes
The six measures’ empirical distributions were obtained using the Cochrane meta-analyses. We determined the cutoffs for different magnitudes of small-study effects based primarily on the quantiles in these distributions. We roughly classified the magnitudes into four levels, i.e., unimportant, moderate, substantial, and considerable; each level contained roughly 30% of the distribution. Also, we permitted the levels’ ranges to overlap, because the importance of small-study effects depended on many factors (e.g., disease outcome) and strict cutoffs without overlapped ranges might be misleading. Also, we rounded the selected quantiles to be in simple forms (with few digits after the decimal point) so that they could be easily summarized and applied. These labels of magnitudes and the approach of overlapping categories have been similarly used with the statistic for heterogeneity.6,28
Results
We obtained 18,562 eligible meta-analyses with binary outcomes and 11,370 ones with non-binary outcomes. The reduced database contained 1960 and 1342 meta-analyses with binary and non-binary outcomes, respectively. Figure 1 presents the flow chart of the selection process.
Figure 1.
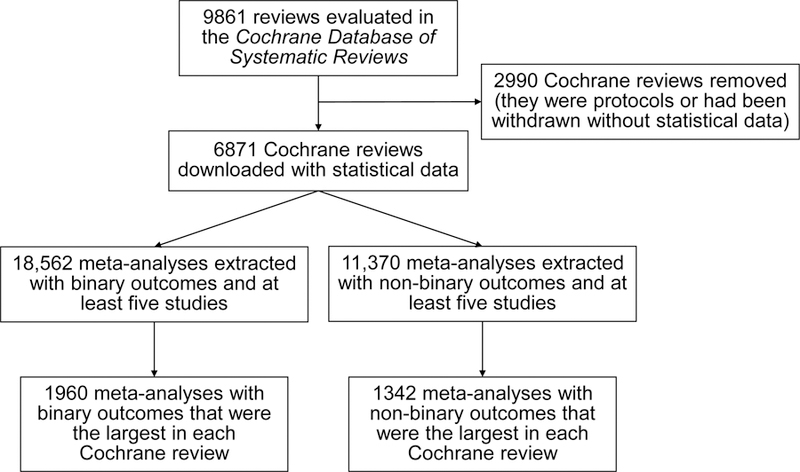
Flow chart of selecting the Cochrane meta-analyses.
Meta-Analyses with Binary Outcomes
Figures S1 and S2 in the Supplementary Material present the empirical distributions of measures for small-study effects and their absolute values (except ) among meta-analyses with binary outcomes. The vertical dashed lines depict the null value (zero) of no small-study effects. The distributions of Egger’s regression intercept and the skewness were approximately symmetrical around zero. The averages of the regression intercepts and were −0.14 and −0.16 with standard deviations (SDs) 2.04 and 3.30, respectively; both had a median around −0.14. Also, 55.2% and 55.0% meta-analyses had negative and , respectively, and 44.8% and 45% meta-analyses had positive ones. The regression intercept was extreme in a few meta-analyses. Specifically, 279 (1.5%) and 254 (1.4%) meta-analyses had less than −4 and greater than 4, respectively. Using the regression intercepts under the RE setting, more meta-analyses had extreme measures: was less than −4 in 560 (3.0%) meta-analyses and was greater than 4 in 495 (2.7%) meta-analyses.
Compared with the regression intercept, the skewness was more concentrated and symmetrical around zero. The averages of both and were close to zero with SDs around 0.50; their medians were also near zero. Moreover, 49.9% and 49.8% of the meta-analyses had negative and , respectively, and 50.1% and 50.2% had positive ones. Only 14 and 11 (< 0.1%) meta-analyses had and greater than 2 in absolute magnitude.
When calculating and , the trim-and-fill algorithm did not converge in one meta-analysis with binary outcomes. Also, it did not identify any suppressed studies in 6099 (32.9%) meta-analyses; thus, their and were exactly zero and their empirical distributions had high frequencies massed at zero. The average of was 13.6% with SD 11.6%; its median was 14.3%. The was less than 40% in all meta-analyses. The average of was 57.0% with a huge SD 8087.9%, and its median was zero. The was negative in 28.9% meta-analyses and was positive in 38.2% meta-analyses. The empirical distribution of the absolute had a decreasing trend, which was similar to those of the absolute regression intercept and skewness, while its frequency at zero was much higher. The empirical distribution of did not have an obvious decreasing trend as the measure increased.
Meta-Analyses with Non-Binary Outcomes
Figures S3 and S4 in the Supplementary Material present the measures’ empirical distributions among meta-analyses with non-binary outcomes. Compared with those with binary outcomes, Figure S3 indicates that more meta-analyses had extreme measures for small-study effects, especially when using the regression intercept. The averages of and were −0.26 and −0.40 with SDs 2.74 and 17.89, respectively; their medians were around −0.24. The large SD of was due to its distribution’s long tails. Many meta-analyses had extreme regression intercepts. Moreover, 56.2% and 55.8% meta-analyses had negative and , respectively, and 43.8% and 44.1% had positive ones. In one meta-analysis, all studies had equal effect sizes, so all measures for small-study effects were exactly zero.
The skewness was mostly lied between −2 and 2. The and had averages around −0.04 with SDs 0.64 and 0.61, respectively, and their medians were around −0.03. The and were negative in 52.4% and 52.5% meta-analyses and were positive in 47.6% and 47.4% ones, respectively. Only 1.1% and 0.8% meta-analyses had and greater than 2 in absolute magnitude. These proportions were slightly greater than those with binary outcomes.
The trim-and-fill method did not identify any suppressed studies in 5168 (45.5%) meta-analyses, whose and were zero. The had an average 11.0% with SD 11.6%, and its median was 10.0%. It was also less than 40% in all meta-analyses as in those with binary outcomes. The average of was 4.4% with SD 838.6%, and its median was 0%. The was negative in 26.1% and positive in 28.4% meta-analyses, respectively.
Rules of Thumb for Magnitudes of Small-Study Effects
Table 2 provides rules of thumb for interpreting small-study effects’ magnitudes based on the measures’ empirical distributions. In absolute magnitude, small-study effects might be unimportant if the regression intercepts and , the skewness and , the proportion of suppressed studies , and the relative change were less than 0.6, 0.2, 4%, and 2%, respectively. Around 25% to 45% meta-analyses with either binary or non-binary outcomes had measures within these ranges. The and generally indicated unimportant small-study effects in more meta-analyses than the other measures. If the six measures’ absolute values were within 0.4–1, 0.15–0.4, 2%–18%, and 1%–25% accordingly, small-study effects might be moderate; around 20% to 35% Cochrane meta-analyses had measures within the ranges. Small-study effects might be substantial if the absolute measures were within 0.8–2, 0.35–0.7, 16%–26%, and 20%–80% accordingly, and might be considerable if they were at least 1.8, 0.65, 24%, and 75% accordingly. The regression intercepts and skewness indicated substantial small-study effects in around 30% to 35% meta-analyses, and considerable small-study effects in around 20% to 40% ones. The and implied substantial and considerable small-study effects in less meta-analyses than the other measures.
Table 2.
Proposed rules of thumb for assessing small-study effects’ magnitudes.
| Measure range and proportion of Cochrane meta-analyses contained in the range among the full/reduced database with binary/non-binary outcomes |
||||||
|---|---|---|---|---|---|---|
| Measure | Database | Outcome | Unimportant small-study effects | Moderate small-study effects | Substantial small-study effects | Considerable small-study effects |
| Full | Binary | 39.6% | 31.2% | 34.3% | 18.7% | |
| Reduced | Binary | 38.5% | 31.1% | 35.4% | 18.7% | |
| Full | Non-binary | 28.5% | 24.7% | 34.0% | 32.8% | |
| Reduced | Non-binary | 26.7% | 24.1% | 35.5% | 33.9% | |
| Full | Binary | 32.0% | 35.3% | 30.7% | 18.8% | |
| Reduced | Binary | 32.2% | 32.9% | 29.9% | 21.0% | |
| Full | Non-binary | 28.8% | 31.1% | 30.2% | 25.7% | |
| Reduced | Non-binary | 26.8% | 29.8% | 30.8% | 27.9% | |
| Full | Binary | 38.8% | 30.2% | 32.2% | 21.8% | |
| Reduced | Binary | 37.0% | 29.9% | 33.4% | 22.8% | |
| Full | Non-binary | 27.8% | 23.3% | 29.9% | 38.2% | |
| Reduced | Non-binary | 25.4% | 21.9% | 31.0% | 40.5% | |
| Full | Binary | 32.3% | 35.2% | 30.7% | 18.7% | |
| Reduced | Binary | 33.0% | 32.8% | 30.1% | 19.9% | |
| Full | Non-binary | 29.9% | 31.7% | 30.5% | 24.2% | |
| Reduced | Non-binary | 27.8% | 31.5% | 31.4% | 26.2% | |
| Full | Binary | 33.2% | 31.1% | 26.0% | 24.6% | |
| Reduced | Binary | 31.8% | 31.9% | 25.8% | 23.2% | |
| Full | Non-binary | 45.7% | 25.3% | 22.3% | 19.6% | |
| Reduced | Non-binary | 42.9% | 28.0% | 23.9% | 17.7% | |
| Full | Binary | 34.0% | 28.8% | 26.3% | 17.3% | |
| Reduced | Binary | 32.0% | 30.4% | 28.5% | 16.1% | |
| Full | Non-binary | 47.2% | 26.2% | 21.7% | 10.5% | |
| Reduced | Non-binary | 44.2% | 29.1% | 21.9% | 10.6% | |
Sensitivity Analysis
Figures S5–S8 in the Supplementary Material show the measures’ histograms among the reduced database. The shapes of the histograms were similar to their counterparts in Figures S1–S4 using the full database.
Discussion
Main Findings
We have presented rules of thumb for interpreting measures for small-study effects based on their empirical distributions among many Cochrane meta-analyses. The results can aid decision makers who are tasked with appraising evidence and determining the certainty warranted by the evidence. Using the GRADE approach, small-study effects lead to rating down certainty. Using the proposed rules of thumb, the certainty in evidence to support a decision may not need to be rated down if small-study effects’ magnitude was small or if the effects’ direction was inconsistent with exaggerated study results.22
As Egger’s regression test is often considered more statistically powerful than the trim-and-fill method in many situations,16 the regression intercept and skewness may be preferred measures than the trim-and-fill-based and , although the latter two may be more straightforward than the former ones. Also, despites its intuitive interpretation as the proportion of suppressed studies, has several drawbacks. First, it accounts only for the number of suppressed studies, not their weights in the estimated overall result. If the suppressed studies are small, small-study effects may have little influence on the meta-analysis, and might exaggerate small-study effects. Second, does not take continuous values like other measures; instead, it must be fractions. If a meta-analysis has five studies, is at least 16.7% (1/6 × 100%) even when only one suppressed study is identified.
Moreover, the regression intercept might be extreme, possibly because of outliers, while the skewness was mostly within a reasonable range. Under the RE setting, the regression intercept was more extreme in some meta-analyses. This may be due to poor between-study variance estimates affected by small-study effects.29
Limitations
This article has several limitations. First, many limitations are inherent to the available statistical methods for small-study effects. These methods are often underpowered and require strong assumptions. For example, the trim-and-fill method may perform poorly in the presence of high heterogeneity or when its assumption about suppressed studies is seriously violated.30,31 In such cases, meta-analysts may avoid using and to quantify small-study effects. Also, Egger’s regression may have inflated false positive rates in certain cases.17 The inflation is due to the intrinsic association between effect sizes and their within-study variances; such effect sizes include odds ratios, risk ratios, risk differences, as well as standardized mean differences.17,32–37 These issues need to be carefully taken into account when exploring publication bias. Although alternative methods may control false positive rates better than Egger’s regression in some cases17, they may be also seriously underpowered and not applicable to generic effect sizes. As an illustrative article for interpreting small-study effects’ magnitude based on empirical evidence, we did not comprehensively consider all available alternative methods; assessing the measures for small-study effects based on the alternatives will be one of our future studies.
Second, our analysis was restricted to the Cochrane meta-analyses, which focused on healthcare-related topics. However, many meta-analyses were performed to investigate diverse (e.g., ecological and educational) topics.2 Therefore, our findings may not be directly generalized to those meta-analyses.
Third, we considered Cochrane meta-analyses containing at least five studies, because it was difficult to justify small-study effects in very small meta-analyses. However, it is infeasible to establish a widely-accepted eligibility criterion on the number of studies for appropriately assessing small-study effects. The cutoff may be some other values, say ten.18
Finally, although we classified small-study effects’ magnitudes into four levels, their true importance relates to the disease outcome’s type and its context. For example, unimportant increase in mortality may be important in certain meta-analyses. Furthermore, our classifications were based roughly on the quantiles of the measures’ empirical distributions; however, no gold standard of small-study effects can be feasibly applied to examine their true magnitudes in all meta-analyses in the large database. Therefore, the proposed rules of thumb may serve as auxiliary assessment of small-study effects; to ascertain such effects or publication bias, meta-analysts should carefully investigate individual studies on a case-by-case basis.18
Supplementary Material
References
- 1.Sutton AJ, Higgins JPT. Recent developments in meta-analysis. Statistics in Medicine 2008;27(5):625–50. [DOI] [PubMed] [Google Scholar]
- 2.Gurevitch J, Koricheva J, Nakagawa S, Stewart G. Meta-analysis and the science of research synthesis. Nature 2018;555:175–82. [DOI] [PubMed] [Google Scholar]
- 3.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLOS Medicine 2009;6(7):e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stewart LA, Clarke M, Rovers M, et al. Preferred reporting items for a systematic review and meta-analysis of individual participant data: the PRISMA-IPD statement. JAMA 2015;313(16):1657–65. [DOI] [PubMed] [Google Scholar]
- 5.Higgins JPT, Altman DG, Gøtzsche PC, et al. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011;343:d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions Chichester, UK: John Wiley & Sons; 2008. [Google Scholar]
- 7.Guyatt GH, Oxman AD, Vist G, et al. GRADE guidelines: 4. Rating the quality of evidence—study limitations (risk of bias). Journal of Clinical Epidemiology 2011;64(4):407–15. [DOI] [PubMed] [Google Scholar]
- 8.Hultcrantz M, Rind D, Akl EA, et al. The GRADE Working Group clarifies the construct of certainty of evidence. Journal of Clinical Epidemiology 2017;87:4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonner A, Alexander PE, Brignardello-Petersen R, et al. Applying GRADE to a network meta-analysis of antidepressants led to more conservative conclusions. Journal of Clinical Epidemiology 2018;102:87–98. [DOI] [PubMed] [Google Scholar]
- 10.Chavalarias D, Wallach JD, Li AHT, Ioannidis JPA. Evolution of reporting p values in the biomedical literature, 1990–2015. JAMA 2016;315(11):1141–48. [DOI] [PubMed] [Google Scholar]
- 11.Cristea IA, Ioannidis JPA. P values in display items are ubiquitous and almost invariably significant: A survey of top science journals. PLOS ONE 2018;13(5):e0197440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dwan K, Altman DG, Arnaiz JA, et al. Systematic review of the empirical evidence of study publication bias and outcome reporting bias. PLOS ONE 2008;3(8):e3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickersin K The existence of publication bias and risk factors for its occurrence. JAMA 1990;263(10):1385–89. [PubMed] [Google Scholar]
- 14.Eyding D, Lelgemann M, Grouven U, et al. Reboxetine for acute treatment of major depression: systematic review and meta-analysis of published and unpublished placebo and selective serotonin reuptake inhibitor controlled trials. BMJ 2010;341:c4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmucker CM, Blümle A, Schell LK, et al. Systematic review finds that study data not published in full text articles have unclear impact on meta-analyses results in medical research. PLOS ONE 2017;12(4):e0176210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Z-C, Zhou X-H, He J. Statistical methods for dealing with publication bias in meta-analysis. Statistics in Medicine 2015;34(2):343–60. [DOI] [PubMed] [Google Scholar]
- 17.Peters JL, Sutton AJ, Jones DR, et al. Comparison of two methods to detect publication bias in meta-analysis. JAMA 2006;295(6):676–80. [DOI] [PubMed] [Google Scholar]
- 18.Sterne JAC, Sutton AJ, Ioannidis JPA, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011;343:d4002. [DOI] [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315(7109):629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duval S, Tweedie R. A nonparametric “trim and fill” method of accounting for publication bias in meta-analysis. Journal of the American Statistical Association 2000;95(449):89–98. [Google Scholar]
- 21.Lin L, Chu H. Quantifying publication bias in meta-analysis. Biometrics 2018;74(3):785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murad MH, Chu H, Lin L, Wang Z. The effect of publication bias magnitude and direction on the certainty in evidence. BMJ Evidence-Based Medicine 2018;23(3):84–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins JPT, Thompson SG, Spiegelhalter DJ. A re-evaluation of random-effects meta-analysis. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2009;172(1):137–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno SG, Sutton AJ, Ades AE, et al. Assessment of regression-based methods to adjust for publication bias through a comprehensive simulation study. BMC Medical Research Methodology 2009;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins JPT. Commentary: heterogeneity in meta-analysis should be expected and appropriately quantified. International Journal of Epidemiology 2008;37(5):1158–60. [DOI] [PubMed] [Google Scholar]
- 26.Riley RD, Higgins JPT, Deeks JJ. Interpretation of random effects meta-analyses. BMJ 2011;342:d549. [DOI] [PubMed] [Google Scholar]
- 27.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials 1986;7(3):177–88. [DOI] [PubMed] [Google Scholar]
- 28.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327(7414):557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson D The implications of publication bias for meta-analysis’ other parameter. Statistics in Medicine 2006;25(17):2911–21. [DOI] [PubMed] [Google Scholar]
- 30.Terrin N, Schmid CH, Lau J, Olkin I. Adjusting for publication bias in the presence of heterogeneity. Statistics in Medicine 2003;22(13):2113–26. [DOI] [PubMed] [Google Scholar]
- 31.Peters JL, Sutton AJ, Jones DR, et al. Performance of the trim and fill method in the presence of publication bias and between-study heterogeneity. Statistics in Medicine 2007;26(25):4544–62. [DOI] [PubMed] [Google Scholar]
- 32.Macaskill P, Walter SD, Irwig L. A comparison of methods to detect publication bias in meta-analysis. Statistics in Medicine 2001;20(4):641–54. [DOI] [PubMed] [Google Scholar]
- 33.Lin L Bias caused by sampling error in meta-analysis with small sample sizes. PLOS ONE 2018;13(9):e0204056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwetsloot P-P, Van Der Naald M, Sena ES, et al. Standardized mean differences cause funnel plot distortion in publication bias assessments. eLife 2017;6:e24260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamman EA, Pappalardo P, Bence JR, et al. Bias in meta-analyses using Hedges’ d. Ecosphere 2018;9(9):e02419. [Google Scholar]
- 36.Doncaster CP, Spake R. Correction for bias in meta-analysis of little-replicated studies. Methods in Ecology and Evolution 2018;9(3):634–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pustejovsky JE, Rodgers MA. Testing for funnel plot asymmetry of standardized mean differences. Research Synthesis Methods 2019;10(1):57–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


