Abstract
Integrins are transmembrane proteins that mediate cell adhesion to the extracellular matrix. Integrin α11 (ITGA11) is not expressed in normal alveolar epithelial cells and is a known receptor for collagen. While integrin α11β1 overexpression in the tumor stroma has been associated with tumor growth and metastatic potential of non–small cell lung cancer (NSCLC), little is known about the role of ITGA11 in tumor cells. Thus, we examined the RNA expression of ITGA11 by quantitative RT‐PCR in 80 samples collected from NSCLC patients who had undergone surgical resection and analyzed the clinical outcomes. We found that high expression of ITGA11 was associated with lower recurrence‐free survival in all NSCLC patients (P = 0.043) and in stage I NSCLC patients (P = 0.049). These results were consistent with in silico analyses of the Cancer Genome Atlas database. We also analyzed cell proliferation, migration and invasion capacity in lung cancer cell lines after overexpression of ITGA11. Overexpression of ITGA11 in lung cancer cell lines had little effect on cell proliferation but resulted in increased migration and invasion capacity. Our findings suggest that ITGA11 plays a significant role in cancer migration and invasion, leading to higher recurrence. ITGA11 expression may be a predictor of poor prognosis in patients with surgically resected NSCLC.
Keywords: integrin α11, invasion, migration, non–small cell lung cancer, postoperative recurrence
High expression of integrin α11 (ITGA11) in non–small cell lung cancer was associated with higher cancer stage and postoperative recurrence. Our findings in human cell lines suggest that ITGA11 plays a significant role in cancer migration and invasion, leading to higher recurrence. ITGA11 expression may be a predictor of poor prognosis in patients with surgically resected non–small cell lung cancer.

1. INTRODUCTION
Lung cancer is the most common cause of cancer‐related death worldwide,1, 2 and non–small cell lung cancer (NSCLC) is the major histological type.3 While patients with early stage NSCLC undergo surgery with curative intent, prognosis is poor for patients with recurrent or advanced lung cancer.4 Although treatments such as molecular targeted drugs and immune checkpoint inhibitors improve the outcome of unresectable or recurrent NSCLC, they cannot cure lung cancer.5, 6
Integrins are transmembrane proteins that mediate cell adhesion to the extracellular matrix (ECM). Integrins are heterodimeric glycoproteins composed of α and β subunits.7 Integrin α chains are classified into four main categories according to the different ligands they recognize, including laminin and collagen.8 The interaction of integrins with their ligands trigger cellular responses such as cell adhesion, cell survival, differentiation and migration.9 The expression and activity of integrins vary among different organs and between normal and tumor tissue. While normal alveolar epithelial cells express laminin‐binding integrins such as integrin α3β1, α6β1 and α6β4,10, 11 the expression of integrin receptors change after malignant transformation. Integrin α5, a receptor for fibronectin, is generally not found in normal lung tissue, but it is expressed in a considerable fraction of lung cancers.12, 13 In addition, aberrant expression of integrins in cancer is reported to promote tumor progression and metastasis.12, 14 Overexpression of integrin α5 in node negative NSCLC is associated with decreased survival.12 Similarly, expression of αvβ6 in NSCLC is associated with poor outcome.14
Integrin α11 (ITGA11) dimerize with β1‐subunit and function as collagen receptors. Integrin α11β1 is shown to be expressed on embryonic fibroblasts or myofibroblasts.15, 16, 17 In addition, several studies demonstrated that the expression of integrin α11β1 was controlled by transforming growth factor‐beta (TGF‐β), and ITGA11 regulated embryonic mesenchymal cell differentiation on the collagen matrix.18, 19 Integrins enable cells to interact with the ECM and function as an important mediator of epithelial‐mesenchymal transition (EMT).20 The expression of ITGA11 is reported to be upregulated by EMT in several tumor models,21, 22 and it may affect tumor progression. Whereas integrin α11β1 overexpression in the tumor stroma is associated with tumor growth and metastatic potential of NSCLC,23 little is known about the role of ITGA11 in tumor cells in cell growth and metastatic capacity. The aim of the present study was to determine whether and how aberrant ITGA11 expression in lung cancer leads to worse outcomes.
2. MATERIAL AND METHODS
2.1. Patients and samples
We collected 80 resected non–small cell lung carcinoma (NSCLC) samples between January 2007 and December 2010 at The University of Tokyo Hospital (Tokyo, Japan), and reviewed medical records of all patients. All diagnoses were pathologically confirmed, and staging was determined according to the 7th edition of the Union for International Cancer Control (UICC) tumor‐node‐metastasis (TNM) staging classification. The following parameters were recorded and analyzed: age, sex, disease stage, smoking status, lymphatic invasion (ly), vascular invasion (v) and EGFR status. Recurrence‐free survival was measured from the time of surgery to disease recurrence or death. This project was approved by the Institutional Review Board of The University of Tokyo Hospital (Tokyo, Japan), and all patients enrolled in the research gave written informed consent.
2.2. Integrin expression of lung cancer samples
Total RNA was isolated using RNAiso Plus (Takara Bio, Shiga, Japan). Using SuperScript III (Thermo Fisher Scientific), 1 μg of total RNA was reverse‐transcribed into cDNA. Optimized primers of targeting genes including integrin α11 (ITGA11) and RNA polymerase II subunit A (POLR2A) were designed using the Primer Analysis Software (OLIGO; Molecular Biology Insights). cDNA was amplified using Thunderbird SYBR qPCR Mix (Toyobo) and the following primer sets: ITGA11 (F, CCTGTGGCCAGGGTTCAC; R, CCCACGACCAGCCACTTATT) and POLR2A (F, GAAGGCCAAGCAGGACGTAA; R, GCAGAGGAGCCAGTCTTGTC). The primer sets (final concentration for each primer, 400 nmol/L) were used in a final volume of 16 μL per well. The thermal profile used for quantitative RT‐PCR was 95°C for 1 minute, 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. Dissociation curves were obtained after the last PCR cycle. Background‐corrected fluorescence data were analyzed using the 7500 Fast Real‐Time PCR System and 7500 software v2.3 (Thermo Fisher Scientific). The relative expression level of each sample was determined after normalization to POLR2A using the ΔΔCt method. The cycle number difference (ΔCt) between ITGA11 and POLR2A was calculated for each replicate. Relative target gene expression values were calculated using the mean ΔCt of three replicates.
2.3. The cancer genome atlas database
RNA‐sequencing (RNA‐seq) data of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) from the Cancer Genome Atlas (TCGA) database were obtained (https://cancergenome.nih.gov/). The expression level of ITGA11 was determined by fragments per kilobase of exon per million mapped fragments (FPKM) from HTSeq pipeline (GDC.h38 GENCODE v22, data status as of June 1, 2016). We excluded cases lacking clinical information. A total of 994 cases with LUAD or LUSC were analyzed. The datasets used in this study are listed in Table S1.
2.4. Lung cancer cell lines
Lung cancer cell lines (Calu‐1, H23, H441) were obtained from the American Type Culture Collection and the Health Science Research Resources Bank. These cell lines were cultured according to each manufacturer’s protocol. Total RNA was isolated using RNAiso. ITGA11 expression of each cell line was measured using quantitative RT‐PCR, revealing that the expression level of ITGA11 was high in Calu‐1 and low in H23 and H441.
2.5. Lentiviral production and infection
Full‐length cDNA of ITGA11 was amplified from reverse‐transcribed cDNA of Calu‐1 cells (F, ATGGACCTGCCCAGGG; R, agccTCACTCCAGCACTTTG) and inserted into pGEM‐T Easy vector according to the manufacturer’s instructions (Promega). cDNA of ITGA11 was subcloned into the CSII‐CMV‐MCS‐IRES2‐Bsd vector (RIKEN BioResource Research Center) between the NhelI and EcoRI restriction enzyme sites with FLAG tag. The blank vector or CSII‐CMV‐GFP‐IRES2‐Bsd vector was used as control for lentiviral infection. These plasmids were transfected into H23 and H441 cell lines using HilyMax (Dojindo). Transfected cells were selected in RPMI 1640 medium with 10 µg/mL of blasticidin for 5 days. Overexpression of ITGA11 in transfected cells was checked by quantitative RT‐PCR.
2.6. 2.6 Evaluation of the expression of epithelial‐mesenchymal transition‐related molecules
Cells were plated at 1 × 105 cells/well in 6‐well plates with or without collagen coating for 48 hours in RPMI 1640 medium containing 10% FBS. Total RNA was isolated using RNAiso and cDNA was synthesized. The following primer sets were used to detect the gene expression of TGF‐β1, SNAI1, ZEB1, E‐cadherin and vimentin: TGF‐β1 (F, TTTTGATGTCACCGGAGTTG; R, GGCGCTAAGGCGAAAG), SNAI1 (F, ACTATGCCGCGCTCTTTCC; R, TGGGGTTGAGGATCTCCG), ZEB1 (F, TACAAACATCACCTAAAAGAGCAC; R, GTCGCCCATTCACAGGTATC), E‐cadherin (F, CACAGCAGAACTAACACACGG; R, CACACACGCTGACCTCTAAG) and vimentin (F, AGTACCGGAGACAGGTGCAG; R, GCTTCAACGGCAAAGTTCTC). Gene expression of each sample was measured using quantitative RT‐PCR with normalization to POLR2A.
2.7. Evaluation of cell size using ImageJ
Cell size was calculated with ImageJ software (National Institutes of Health). We plated 1 × 105 cells in 6‐well plates with or without collagen coating for 24 hours in RPMI 1640 medium containing 0.5% FBS. The cell size of each condition was evaluated from the average area of 50 cells.
2.8. Cell proliferation assay
Cell proliferation assay was performed with a Cell Counting Kit 8 (CCK‐8) assay (Promega). Cells were plated in 96‐well plates with or without collagen coating at 5000 cells/well and cultured at 37°C and 5% CO2. To detect the cell growth, 10 μL of CCK‐8 solution was added to each well every 24 hours according to the instructions of the manufacturer. We also performed cell proliferation assays with 6‐well plates with or without collagen coating. Cells were plated at 1 × 105 cells/well and counted every 24 hours with a cell counter. Each sample was analyzed in triplicate.
2.9. Cell migration assay
Cell migration capacity was measured by migration assay with 2‐well culture inserts (ibidi, Martinsried, Germany). The culture inserts were placed into 24‐well plates with or without collagen coating, and H23 cells (35 000 cells/well) and H441 cells (50 000 cells/well) were incubated in the culture insert wells for 24 hours. Then we removed the culture inserts and added RPMI 1640 medium containing 10% FBS to the plates. Changes in gap area were evaluated with ImageJ software (National Institutes of Health) at different time points, and the migration rate was calculated according to the ratio of the gap area.
2.10. Cell invasion assay
Cell invasion capacity was measured by Cultrex Cell Migration Assay Kit (Trevigen). H23 cells (50 000 cells/well) suspended in RPMI 1640 containing 0.5% FBS were seeded onto the top chamber of a 96‐well plate with 8‐μm pores, with or without collagen coating, and RPMI 1640 containing 10% FBS was added into the bottom chamber. After incubation for 48 hours, cells migrating to the bottom chamber were quantified according to the manufacturer’s instructions. Invasion capacity was calculated as the ratio of cells invading through collagen‐coated pores to cells migrating through normal pores.
2.11. Statistical analysis
Statistical analysis was carried out using SPSS software (SPSS). χ 2‐precision tests were used to analyze pathological data. One‐way ANOVA and Tukey post‐hoc analysis was used for multiple comparisons of mRNA expression levels. The Kaplan‐Meier method was used to analyze recurrence‐free survival, and the log‐rank test was used to examine any differences. Differences were considered significant when the P‐value was <0.05. All experiments were carried out at least three times.
3. RESULTS
3.1. Clinical characteristics of non–small cell lung carcinoma patients
The clinical characteristics of 80 patients with NSCLC are summarized in Table 1. Among 80 patients, 36 were female (45%), 37 were never smokers (46%) and the median age was 68 (range, 36‐83). A total of 73 were adenocarcinomas (91%) and 7 were squamous cell carcinomas (9%). Because we analyzed patients with resectable lung cancer, the frequency of advanced stage was low (8/80, 10%). Vascular invasion was positive in 25 patients (31%) and lymphatic invasion was positive in 15 patients (19%). A total of 28 patients (35%) had EGFR gene mutations, 16 patients (20%) had wild‐type EGFR and 36 patients (45%) had unknown EGFR mutation status.
Table 1.
Clinical characteristics of 80 non–small cell lung cancer (NSCLC) patients who had undergone surgical resection
| N = 80 | % | |
|---|---|---|
| Age (median, y) | 68 (36‐83) | |
| Female | 36 | 45 |
| Never smoker | 37 | 46 |
| Histologic type | ||
| Adenocarcinoma | 73 | 91 |
| Squamous carcinoma | 7 | 9 |
| Stage | ||
| I | 57 | 71 |
| II | 15 | 19 |
| III/IV | 8 | 10 |
| Vascular invasion | ||
| Negative | 56 | 70 |
| Positive | 24 | 30 |
| Lymphatic invasion | ||
| Negative | 65 | 81 |
| Positive | 15 | 19 |
| EGFR mutation | ||
| Wild type | 16 | 20 |
| Mutated | 29 | 36 |
| Unknown | 35 | 44 |
EGFR, epidermal growth factor receptor.
3.2. High expression of integrin α11 was associated with higher stage and lower recurrence‐free survival
We investigated mRNA expression of ITGA11 in 80 NSCLC patients and found that high expression of ITGA11 in lung cancer was associated with higher stage (Figure 1A). In contrast, expression of ITGA11 was not related to tumor size, vascular invasion or lymphatic invasion (Figure 1B‐D). In addition, expression of ITGA11 was not related to EGFR mutation status (P = 0.563).
Figure 1.
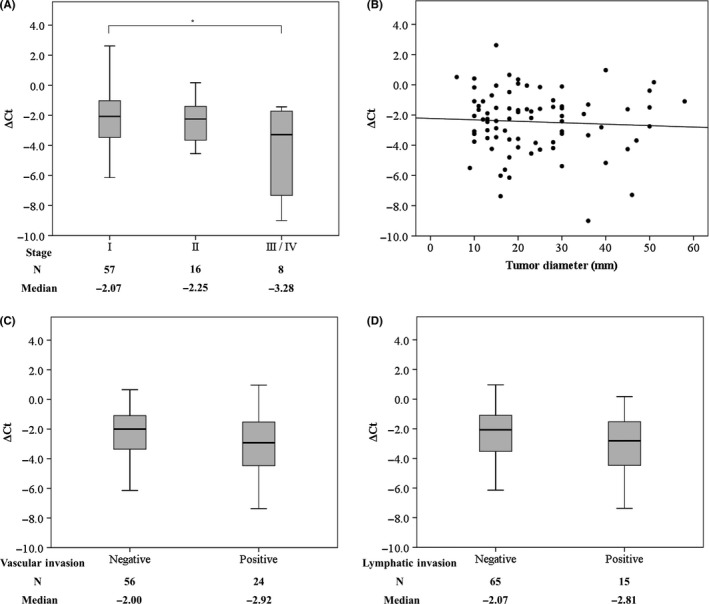
High expression of integrin α11 (ITGA11) was associated with higher stages. Expression of ITGA11 in 80 non–small cell lung cancer (NSCLC) patients was analyzed by quantitative RT‐PCR. RNA polymerase II subunit A (POLR2A) was used as internal control. A, Expression of ITGA11 and clinical stage. B, Relationship between the expression of ITGA11 and tumor diameter. Correlation coefficient Rs = 0.009. C, D, Expression of ITGA11 and vascular or lymphatic invasion. Experiments were carried out three times. *P < 0.05
Next, using the Kaplan‐Meier method, we analyzed the relationship between the expression level of ITGA11 and recurrence‐free survival. The optimal cutoff for ΔCt was calculated as −1.6 by receiver operating characteristic curve analysis (area under the curve 0.67; specificity 0.895, sensitivity 0.293). According to the cutoff value, we classified 54 patients as having high expression and 26 patients as having low expression; no significant characteristic differences were seen between the two groups. Kaplan‐Meier analysis showed that high expression of ITGA11 was associated with lower recurrence‐free survival (P = 0.043, Figure 2A). In addition, we investigated recurrence‐free survival of 57 patients with stage I NSCLC using the same method and found 37 patients with high expression and 20 patients with low expression. In stage I patients, high expression of ITGA11 was also associated with lower recurrence‐free survival (P = 0.049, Figure 2B).
Figure 2.
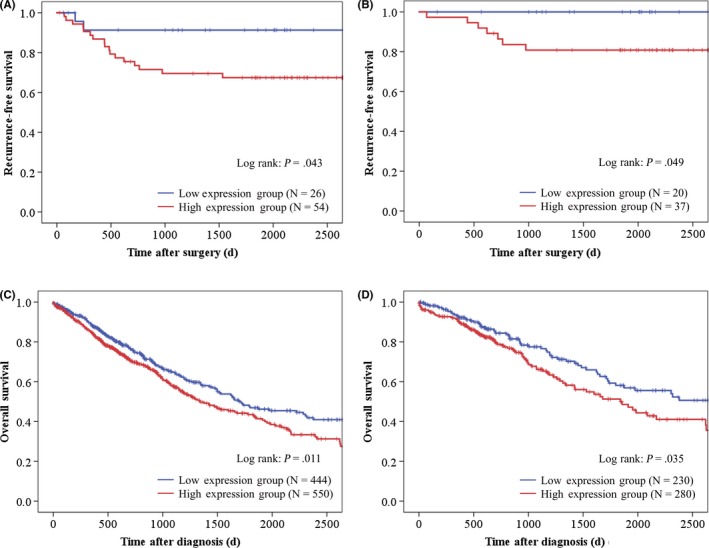
Kaplan‐Meier analysis of NSCLC patients according to expression levels of integrin α11 (ITGA11). A, B, Recurrence‐free survival of all patients with non–small cell lung cancer (NSCLC) (A) or patients with stage I NSCLC (B) in the present study. C, D, Overall survival of all patients with NCSLC (C) or patients with stage I NSCLC (D) in the Cancer Genome Atlas database
3.3. High expression of integrin α11 was associated with lower overall survival in the Cancer Genome Atlas database
We analyzed the expression level of ITGA11 in the lung adenocarcinoma and lung squamous cell carcinoma samples from the TCGA database. We assigned patients to high and low expression groups according to the expression level of ITGA11 as described above. The optimal cutoff value of FPKM was calculated as −1.895, and we classified 550 patients as having high expression and 444 patients as having low expression. Kaplan‐Meier analysis showed that high expression of ITGA11 was associated with lower overall survival (P = 0.011, Figure 2C). We also analyzed the overall survival of 510 patients with stage I NSCLC using the same method (280 patients with high expression and 230 patients with low expression). In stage I patients from the TCGA database, high expression of ITGA11 was also associated with lower overall survival (P = 0.035, Figure 2D).
3.4. Overexpression of integrin α11 leads to bigger cell size but has little effect on cell proliferation
To analyze the function of ITGA11 in lung cancer cells, we overexpressed ITGA11 in H23 cells or H441 cells with lentiviral vectors. ITGA11 overexpression was confirmed by RT‐PCR. Next, we analyzed the expression of EMT‐related molecules and found that there was no significant relationship between expression of ITGA11 and TGF‐β1, SNAI1, ZEB1, E‐cadherin or vimentin when ITGA11 was overexpressed in H23 cells on collagen‐coated plates (Figure 3). Expression levels of these molecules were also not significantly different in H23 cells when plated without collagen coating or in H441 cells with or without collagen‐coated plates (data not shown). While no morphological changes were seen in ITGA11 overexpressed cells, ITGA11 overexpression led to larger cell size in H23 cells on plates with or without collagen coating (Figure 4A). We investigated cell proliferation using Cell Counting Kit‐8 and by simple cell counting (Figure 4B,C). In H23 and H441 cells that overexpressed ITGA11, the cell number was similar to controls regardless of collagen coating. Simple cell counting of either cell in 6‐well plates with or without collagen coating resulted in similar counts.
Figure 3.
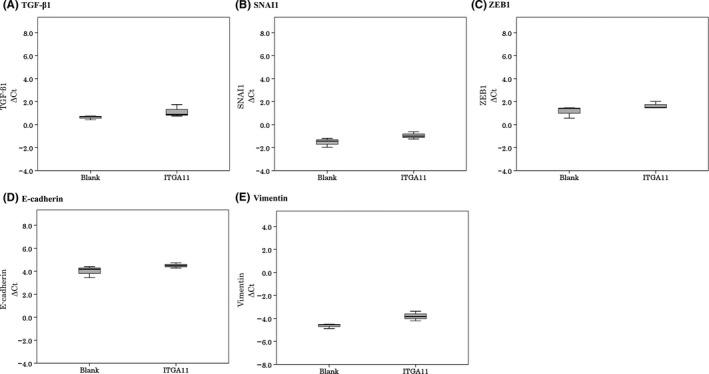
Epithelial‐mesenchymal transition‐related molecules in lung cancer cell lines. The expression levels of TGF‐β1 (A), SNAI1 (B), ZEB1 (C), E‐cadherin (D) and vimentin (E) in H23 cells are shown after overexpression of integrin α11 (ITGA11) and plated on collagen coating plates. There was no significant relationship between expression of ITGA11 and EMT‐related molecules. Experiments were carried out three times
Figure 4.
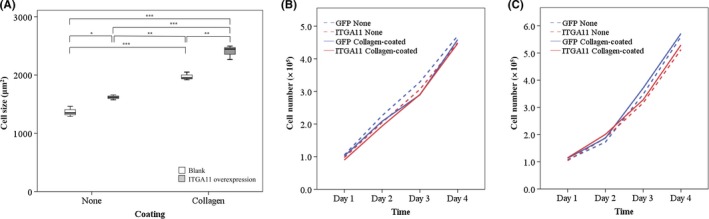
Overexpression of integrin α11 (ITGA11) in lung cancer cell lines. A, Overexpression of ITGA11 leads to larger size in H23 cells with or without collagen coating. The average cell area of 50 cells was measured. Experiments were carried out three times. *P < 0.05; **P < 0.01; ***P < 0.001. B, C, Simple cell counting of H23 (B) and H441 (C) cells that overexpressed ITGA11. Experiments were carried out three times in 6‐well plates with or without collagen coating. The lines are shown as mean experimental data, and no significant difference was found
3.5. Overexpression of integrin α11 leads to higher migration and invasion capacity
We evaluated the migration capacity in H23 cells or H441 cells after overexpression of ITGA11. While the migration rate of both cell lines was similar on plastic plates, the migration rate of both cell lines overexpressing ITGA11 was significantly higher on collagen‐coated plates compared with the control cells (Figure 5A,B). Subsequently, we performed an invasion assay with H23 cells. We analyzed the ratio of H23 cells invading through collagen‐coated filters and cells migrating through normal filters. H23 cells overexpressing ITGA11 had significantly higher invasion capacity compared with control cells (Figure 5C).
Figure 5.
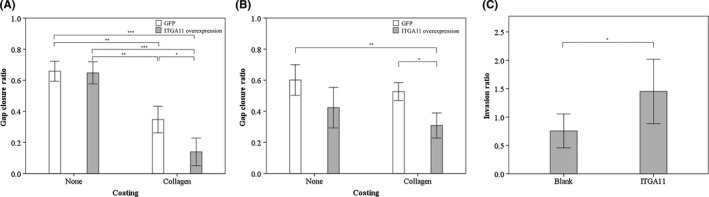
Overexpression of integrin α11 (ITGA11) leads to higher migration and invasion capacity in lung cancer cell lines. A, B, Migration assay of H23 (A, n = 4) and H441 (B, n = 7) cells overexpressing ITGA11. C, Invasion assay of H23 cells overexpressing ITGA11 (n = 10). The vertical axis indicates the ratio of cells invading through collagen‐coated filter and cells migrating through normal filter. Data represent the mean ± SE. *P < 0.05; **P < 0.01; ***P < 0.001
4. DISCUSSION
Our study demonstrated that high expression of ITGA11 was associated with advanced stage and lower recurrence‐free survival in patients with NSCLC. This was consistent with in silico analyses of the TCGA dataset. Furthermore, functional analyses showed that lung cancer cells overexpressing ITGA11 resulted in increased migration and invasion capacity. Thus, we conclude that higher expression of ITGA11 is associated with higher postoperative recurrence in NSCLC through increased migration and invasion. Our results indicate that ITGA11 expression may be a predictor of poor prognosis in patients with surgically resected NSCLC and ITGA11 may be a therapeutic target for NSCLC.
Integrins play important roles in tumor progression and metastatic processes in various types of human cancer. Aberrant integrin expression in cancer cells changes adhesion capacity to their surrounding microenvironment and promotes tumor progression.24 Previous studies revealed that the expression of integrin α2β1 enhances cell migration and proliferation on the collagen matrix in gastric cancer and pancreatic cancer cells.25, 26 ITGA11 was expressed in metastatic malignant melanomas and high mRNA levels of the collagen receptor integrins including α1, α2 and α11 were correlated with shorter survival.27 In this study, we showed high ITGA11 expression in primary NSCLC correlated with poor prognosis using our clinical samples and in silico analysis. Poor prognosis was also shown in patients with stage I lung cancer with high expression of ITGA11.
Integrin α11 affects cell spreading, motility and collagen remodeling in mesenchymal cells.15, 28 The lack of ITGA11 chain in fibroblasts was reported to reduce cell attachment and cell spread on collagen I, which was consistent with our results of larger size in ITGA11‐overexpressed cells. Moreover, knockdown of ITGA11 in myofibroblasts has previously been shown to strongly inhibit their migration.17 Integrin α11 in cancer‐associated stromal fibroblasts (CAF) has been reported to promote tumor progression in NSCLC by affecting the collagen stiffness of the tumor stroma.23 In pancreatic ductal adenocarcinoma, ITGA11 in pancreatic stellate cells (PSC) regulates differentiation of PSC into CAF and induces migration of tumor cells.29 In addition, ITGA11 in CAF interacts with PDGFR‐β to promote CAF‐induced tumor cell invasion in breast cancer.30 However, little is known about the role of ITGA11 in tumor cells itself. Westcott et al reported ITGA11 as a key invasion‐promoting gene for breast cancer cells in collective invasion.31 Although we found no change in proliferation rate in cells overexpressing ITGA11, overexpression of ITGA11 in lung cancer cells led to higher migration and invasion capacity on the collagen matrix, similar to other integrins.25, 26 In tumor cells, ITGA11 seems to mainly regulate cell migration and invasion rather than cell proliferation or tumor growth.
In our study, no significant relationship was found between ITGA11 expression and vascular or lymphatic invasion in clinical samples, despite increased cell motility in in vitro results. Intravasation of cancer cells is considered to be a complex process that involves cells invading the tumor ECM and crossing the endothelial junctions,32, 33 which require an interaction between cancer cells and endothelial cells.34, 35 Previous reports showed that tumor matrix metalloproteinase 1 (MMP‐1) activated endothelial proteinase‐activated receptor 1 (PAR1) to facilitate vascular intravasation by regulating endothelial permeability.36 ITGA11 expression in cancer cells is likely not the only factor that is necessary to invade the vascular or lymphatic system.
The present study has several limitations. Samples in this study were collected from patients with resected NSCLC, with most patients having earlier, stage I or II, disease. In addition, the expression level of ITGA11 and recurrence‐free survival were retrospectively analyzed. We need further in vivo studies to confirm our results.
In conclusion, high expression of ITGA11 in NSCLC was associated with higher stage and postoperative recurrence. Our findings in human lung cancer cell lines suggest that ITGA11 plays a significant role in cancer migration and invasion, which may lead to a higher recurrence rate.
DISCLOSURE
The authors declare no conflicts of interest for this article.
Supporting information
ACKNOWLEDGMENTS
This study was supported by a Grant‐in‐Aid for Young Scientists (B) (JSPS KAKENHI No. 15K20917 and 18K15266), a Grant‐in‐Aid for Scientific Research (C) (JSPS KAKENHI No. 16K09574) and a Grant‐in‐Aid for Scientific Research (A) (JSPS KAKENHI No. 16H02653) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Ando T, Kage H, Matsumoto Y, et al. Integrin α11 in non–small cell lung cancer is associated with tumor progression and postoperative recurrence. Cancer Sci. 2020;111:200–208. 10.1111/cas.14257
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367‐1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Howlader NNAM, Krapcho M, Miller, D, et al. SEER Cancer Statistics Review, 1975–2016. Bethesda, MD: National Cancer Institute; 2019. [Google Scholar]
- 5. Borghaei H, Paz‐Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non–small‐cell lung cancer. N Engl J Med. 2015;373:1627‐1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR‐mutated advanced non–small‐cell lung cancer. N Engl J Med. 2018;378:113‐125. [DOI] [PubMed] [Google Scholar]
- 7. Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901‐3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3:a004994‐a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673‐687. [DOI] [PubMed] [Google Scholar]
- 10. Sheppard D. Epithelial integrins. BioEssays. 1996;18:655‐660. [DOI] [PubMed] [Google Scholar]
- 11. Chapman HA, Li X, Alexander JP, et al. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121:2855‐2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adachi M, Taki T, Higashiyama M, Kohno N, Inufusa H, Miyake M. Significance of integrin alpha5 gene expression as a prognostic factor in node‐negative non–small cell lung cancer. Clin Cancer Res. 2000;6:96‐101. [PubMed] [Google Scholar]
- 13. Roman J, Ritzenthaler JD, Roser‐Page S, Sun X, Han S. alpha5beta1‐integrin expression is essential for tumor progression in experimental lung cancer. Am J Respir Cell Mol Biol. 2010;43:684‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan P, Zhu H, Yin L, et al. Integrin alphavbeta6 promotes lung cancer proliferation and metastasis through upregulation of IL‐8‐mediated MAPK/ERK signaling. Transl Oncol. 2018;11:619‐627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Popova SN, Rodriguez‐Sanchez B, Liden A, Betsholtz C, Van Den Bos T, Gullberg D. The mesenchymal alpha11beta1 integrin attenuates PDGF‐BB‐stimulated chemotaxis of embryonic fibroblasts on collagens. Dev Biol. 2004;270:427‐442. [DOI] [PubMed] [Google Scholar]
- 16. Talior‐Volodarsky I, Connelly KA, Arora PD, Gullberg D, McCulloch CA. alpha11 integrin stimulates myofibroblast differentiation in diabetic cardiomyopathy. Cardiovasc Res. 2012;96:265‐275. [DOI] [PubMed] [Google Scholar]
- 17. Bansal R, Nakagawa S, Yazdani S, et al. Integrin alpha 11 in the regulation of the myofibroblast phenotype: implications for fibrotic diseases. Exp Mol Med. 2017;49:e396‐e396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu N, Carracedo S, Ranta J, Heuchel R, Soininen R, Gullberg D. The human alpha11 integrin promoter drives fibroblast‐restricted expression in vivo and is regulated by TGF‐beta1 in a Smad‐ and Sp1‐dependent manner. Matrix Biol. 2010;29:166‐176. [DOI] [PubMed] [Google Scholar]
- 19. Carracedo S, Lu N, Popova SN, Jonsson R, Eckes B, Gullberg D. The fibroblast integrin alpha11beta1 is induced in a mechanosensitive manner involving activin A and regulates myofibroblast differentiation. J Biol Chem. 2010;285:10434‐10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial‐mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ke XS, Qu Y, Goldfinger N, et al. Epithelial to mesenchymal transition of a primary prostate cell line with switches of cell adhesion modules but without malignant transformation. PLoS ONE. 2008;3:e3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C. The T‐box transcription factor Brachyury promotes epithelial‐mesenchymal transition in human tumor cells. J Clin Invest. 2010;120:533‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Navab R, Strumpf D, To C, et al. Integrin alpha11beta1 regulates cancer stromal stiffness and promotes tumorigenicity and metastasis in non–small cell lung cancer. Oncogene. 2016;35:1899‐1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li A, Zhou T, Guo L, Si J. Collagen type I regulates beta‐catenin tyrosine phosphorylation and nuclear translocation to promote migration and proliferation of gastric carcinoma cells. Oncol Rep. 2010;23:1247‐1255. [DOI] [PubMed] [Google Scholar]
- 26. Grzesiak JJ, Bouvet M. The alpha2beta1 integrin mediates the malignant phenotype on type I collagen in pancreatic cancer cell lines. Br J Cancer. 2006;94:1311‐1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vuoristo M, Vihinen P, Vlaykova T, Nylund C, Heino J, Pyrhonen S. Increased gene expression levels of collagen receptor integrins are associated with decreased survival parameters in patients with advanced melanoma. Melanoma Res. 2007;17:215‐223. [DOI] [PubMed] [Google Scholar]
- 28. Tiger CF, Fougerousse F, Grundstrom G, Velling T, Gullberg D. alpha11beta1 integrin is a receptor for interstitial collagens involved in cell migration and collagen reorganization on mesenchymal nonmuscle cells. Dev Biol. 2001;237:116‐129. [DOI] [PubMed] [Google Scholar]
- 29. Schnittert J, Bansal R, Mardhian DF, van Baarlen J, Ostman A, Prakash J. Integrin alpha11 in pancreatic stellate cells regulates tumor stroma interaction in pancreatic cancer. FASEB J. 2019;33:6609‐6621. [DOI] [PubMed] [Google Scholar]
- 30. Primac I, Maquoi E, Blacher S, et al. Stromal integrin alpha11 regulates PDGFR‐beta signaling and promotes breast cancer progression. J Clin Invest. 2019;130:4609‐4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Westcott JM, Prechtl AM, Maine EA, et al. An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. J Clin Invest. 2015;125:1927‐1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chiang SP, Cabrera RM, Segall JE. Tumor cell intravasation. Am J Physiol Cell Physiol. 2016;311:C1‐C14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han W, Chen S, Yuan W, et al. Oriented collagen fibers direct tumor cell intravasation. Proc Natl Acad Sci USA. 2016;113:11208‐11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zervantonakis IK, Hughes‐Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Three‐dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc Natl Acad Sci USA. 2012;109:13515‐13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reymond N, d’Agua BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13:858‐870. [DOI] [PubMed] [Google Scholar]
- 36. Juncker‐Jensen A, Deryugina EI, Rimann I, et al. Tumor MMP‐1 activates endothelial PAR1 to facilitate vascular intravasation and metastatic dissemination. Cancer Res. 2013;73:4196‐4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


