Abstract
The role of long noncoding RNAs (lncRNAs) in the epithelial‐mesenchymal transition (EMT) in pancreatic ductal adenocarcinoma (PDAC) is unclear. Some lncRNAs can be transferred by extracellular vesicles (EVs) and have potential as biomarkers. Here, we identify an lncRNA that could serve as a biomarker for PDAC and show the functional roles of the lncRNA. Expression profiling of lncRNAs revealed that highly upregulated in liver cancer (HULC) was highly expressed, and induced, by transforming growth factor‐β in PDAC cells and their EVs. Knockdown of HULC decreased PDAC cell invasion and migration by inhibiting the EMT. Thus, HULC could be transferred by EVs, and promote EMT, invasion, and migration in recipient PDAC cells. To assess the roles of HULC, PDAC cell xenografts in nude mice were established. Knockdown of HULC in PDAC cells implanted in mice inhibited tumor growth. Moreover, microRNA‐133b suppressed PDAC cell invasion and migration by inhibiting the EMT through targeting HULC. Furthermore, serum samples were obtained from 20 PDAC and 22 intraductal papillary mucinous neoplasm (IPMN) patients, as well as 21 healthy individuals. Analysis of serum EV HULC expression by digital PCR showed that HULC expression was significantly increased in PDAC patients compared to healthy individuals or IPMN patients. Additionally, HULC showed good predictive performance for discriminating PDAC, suggesting that the analysis of EV‐encapsulated HULC would contribute to the diagnosis for human PDAC. Extracellular vesicle‐transported HULC promotes cell invasion and migration by inducing the EMT, and microRNA‐133b suppresses the EMT by targeting HULC. Extracellular vesicle‐encapsulated HULC could be a potential circulating biomarker for human PDAC.
Keywords: epithelial‐mesenchymal transition, exosome, liquid biopsy, long noncoding RNA, microRNA
Extracellular vesicle HULC can regulate cell invasion and migration through induction of epithelial‐mesenchymal transition. Extracellular vesicle HULC could compare favorably with CA19‐9 as a novel biomarker in liquid biopsy for human pancreatic ductal adenocarcinoma.
Abbreviations
- AUC
area under the curve
- CA19‐9
carbohydrate antigen 19‐9
- CEA
carcinoembryonic antigen
- CRC
colorectal cancer
- CREB
cAMP response element binding protein
- CRNDE‐h
colorectal neoplasia differentially expressed‐h
- EMT
epithelial‐mesenchymal transition
- EV
extracellular vesicle
- exRNA
extracellular RNA
- HCC
hepatocellular carcinoma
- HULC
highly upregulated in liver cancer
- IPMN
intraductal papillary mucinous neoplasm
- lncRNA
long noncoding RNA
- miR
microRNA
- miRNA
microRNA
- ncRNA
noncoding RNA
- qPCR
quantitative PCR
- PDAC
pancreatic ductal adenocarcinoma
- TGF‐β
transforming growth factor‐β
- TUG1
taurine upregulated gene 1
1. INTRODUCTION
Pancreatic ductal adenocarcinoma is the fourth leading cause of cancer‐related death in the United States. Pancreatic ductal adenocarcinoma is a highly invasive and metastatic malignancy; less than 20% of cases are localized and thus potentially curable.1, 2 The EMT, which involves fundamental changes in gene expression that disrupt epithelial polarity and establish a mesenchymal phenotype, can be induced by TGF‐β.3 The zinc finger transcription factor Snail is upregulated by TGF‐β and could be a key inducer of the EMT.4 In cancers, this process is involved in resistance to cell death, senescence, chemotherapy, and immunotherapy.5 Although the EMT can trigger invasion and metastasis of several cancers, including PDAC,6 the regulatory mechanisms by which EMT pathways are regulated in PDAC are unclear.
Long noncoding RNAs are defined as nonprotein‐coding RNAs more than 200 nucleotides in length. Long noncoding RNAs have been implicated in the regulation of gene expression and the pathogenesis of various diseases, including cancers. Some lncRNAs, such as H19, HULC, MALAT‐1, and HOTAIR, were reported to be highly expressed, and to function as oncogenic lncRNAs, in several cancers.7, 8 Other lncRNAs are being increasingly recognized to contribute to biological processes through diverse mechanisms in PDAC. HOTAIR was reported to be a negative prognostic factor, and to regulate cell proliferation, the cell cycle, and apoptosis in PDAC.9 MALAT‐1 decreased the sensitivity to chemotherapy and accelerated tumor angiogenesis by enhancing stem cell‐like phenotypes in PDAC.10 HOTTIP was also found to promote PDAC cell proliferation and invasion by regulating HOXA13.11 Although several other lncRNAs, such as H19, GAS5, and PVT1, are related to PDAC development, the role of lncRNAs in the EMT pathway in PDAC is not well understood.12, 13, 14 Moreover, the interrelationship between 2 classes of ncRNA, lncRNAs and miRNAs, reportedly contributes to the epigenetic regulation of gene expression in several diseases.15 Thus, there is a need to identify lncRNAs and miRNAs that regulate the EMT, and elucidate the underlying regulatory mechanisms, in human PDAC.
Extracellular vesicles, such as microvesicles and exosomes, are membranous vesicles released by most cells.16 Extracellular vesicles contain proteins, lipids, and RNAs from donor cells, can be taken up by, and transfer their contents to, recipient cells, and modulate recipient cell behavior.17, 18, 19, 20 Noncoding RNAs are also contained and carried by EVs. Although several miRNAs are transferred by EVs, the mechanisms of intercellular transfer of lncRNA in EVs are unclear. We have previously reported that lncRNAs, such as linc‐RoR and linc‐VLDLR, can be carried by EVs and modulate signaling pathways related to resistance to chemotherapy or hypoxic stress in recipient hepatocellular cancer cells.21, 22 Based on these findings, we hypothesized that intercellular signaling by EV lncRNAs mediates tumor cell invasion and metastasis by regulating the EMT pathway in human PDAC.
Although CA19‐9 and CEA are currently used as clinical biomarkers for human PDAC, they are insufficient for its diagnosis, particularly at an early stage.23 Recently, circulating tumor cells, cell‐free DNA, and EVs (including exosomes) have been evaluated in terms of liquid biopsy. Liquid biopsy constitutes a promising noninvasive alternative to traditional tissue biopsy for diagnosis and monitoring of the evolution and therapeutic response of a tumor.24 Although EV‐encapsulated small ncRNAs reportedly modulate cancer cell behavior and/or serve as biomarkers for cancers, there is very limited information regarding lncRNAs in this context.
In this study, we aimed to identify lncRNAs involved in the EMT pathway and investigate their functional roles during PDAC cell invasion and migration. Moreover, we identified miRNAs that negatively regulate lncRNAs and revealed the contribution of the interrelationship between lncRNAs and miRNAs to the regulatory mechanisms of PDAC development. Furthermore, we identified an lncRNA in serum EVs that could serve as a novel biomarker for human PDAC.
2. MATERIALS AND METHODS
2.1. Cell lines, culture, and reagents
The nonmalignant human pancreatic ductal epithelial cell line hTERT‐HPNE was obtained from ATCC and cultured as recommended by the supplier. The human pancreatic cancer cell lines Panc‐1, MiaPaCa‐2, and BxPC‐3 were obtained from ATCC, and KP‐3 and QGP‐1 were purchased from the JCRB Cell Bank. Panc‐1 cells were cultured in high‐glucose DMEM (Thermo Fisher Scientific) containing 10% FBS and 1% penicillin‐streptomycin (Thermo Fisher Scientific). MiaPaCa‐2 cells were cultured in high‐glucose DMEM containing 10% FBS, 2.5% horse serum, and 1% penicillin‐streptomycin. BxPC‐3, KP‐3, and QGP‐1 cells were cultured in RPMI‐1640 medium (Thermo Fisher Scientific) containing 10% FBS and 1% penicillin‐streptomycin. All cells were cultured at 37°C in an atmosphere containing 5% CO2. For all EV studies, EV‐depleted medium was prepared using exosome‐depleted FBS (Thermo Fisher Scientific). Transforming growth factor‐β was obtained from EMD Millipore. Cells were treated with 10 ng/mL TGF‐β for 72 hours to induce the EMT.
2.2. Human samples
Human studies were approved by the Asahikawa Medical University Institutional Review Board (protocol no. 15 084), and informed consent was obtained from all subjects. Serum samples from 20 PDAC patients, 22 branch duct‐type IPMN patients, and 21 healthy individuals seen at Asahikawa Medical University Hospital from 2016 to 2019 were included in this study. Details of the patients are summarized in Table 1.
Table 1.
Clinical and laboratory characteristics of the 63 study participants
| Sample | Sex | Age (y) | Diagnosis | Serum CA19‐9 (kU/L) | Serum CEA (kU/L) | TNM cancer stage |
|---|---|---|---|---|---|---|
| 1 | M | 75 | Healthy | 14 | 3.7 | |
| 2 | M | 64 | Healthy | 14 | 1.7 | |
| 3 | F | 65 | Healthy | 18 | 2 | |
| 4 | F | 81 | Healthy | 4 | 1.5 | |
| 5 | M | 48 | Healthy | 4 | 0.2 | |
| 6 | F | 69 | Healthy | 5 | 0.7 | |
| 7 | F | 70 | Healthy | N/A | N/A | |
| 8 | M | 67 | Healthy | 13 | 2.1 | |
| 9 | M | 80 | Healthy | 12 | 2.4 | |
| 10 | F | 74 | Healthy | 23 | 2.4 | |
| 11 | F | 69 | Healthy | 6 | 1.4 | |
| 12 | F | 65 | Healthy | 11 | 1.3 | |
| 13 | M | 68 | Healthy | 10 | 1.9 | |
| 14 | M | 74 | Healthy | 13 | 3.2 | |
| 15 | M | 82 | Healthy | 12 | 2.4 | |
| 16 | F | 44 | Healthy | 13 | 0.3 | |
| 17 | M | 82 | Healthy | 15 | 2.1 | |
| 18 | F | 80 | Healthy | 7 | 2.4 | |
| 19 | M | 80 | Healthy | 13 | 5.5 | |
| 20 | F | 77 | Healthy | 6 | 2.2 | |
| 21 | F | 59 | Healthy | 10 | 1 | |
| Median (IQR) | 72 (66.5) | 12.0 (6.8) | 2.1 (1.4) | |||
| 22 | F | 80 | IPMN | 1 | 4.4 | |
| 23 | F | 67 | IPMN | 5 | 2.9 | |
| 24 | F | 68 | IPMN | 8 | 1.6 | |
| 25 | M | 76 | IPMN | 1 | 4.3 | |
| 26 | M | 80 | IPMN | 5 | 0.4 | |
| 27 | M | 69 | IPMN | N/A | N/A | |
| 28 | M | 79 | IPMN | 1 | 2.6 | |
| 29 | M | 58 | IPMN | 8 | 5.5 | |
| 30 | F | 71 | IPMN | 2 | 4.5 | |
| 31 | M | 62 | IPMN | N/A | N/A | |
| 32 | F | 75 | IPMN | N/A | N/A | |
| 33 | F | 70 | IPMN | 5 | 2 | |
| 34 | F | 58 | IPMN | 71 | 2.7 | |
| 35 | F | 61 | IPMN | 1 | 2.8 | |
| 36 | F | 87 | IPMN | 20 | 6.9 | |
| 37 | F | 70 | IPMN | 7 | 2.6 | |
| 38 | F | 86 | IPMN | 13 | 4.1 | |
| 39 | F | 70 | IPMN | 9 | 0.8 | |
| 40 | F | 64 | IPMN | 10 | 5.3 | |
| 41 | F | 71 | IPMN | 9 | 2.4 | |
| 42 | M | 82 | IPMN | 10 | 2.3 | |
| 43 | M | 70 | IPMN | N/A | 3.4 | |
| Median (IQR) | 70 (66.3) | 7.5 (2.8) | 2.8 (2.4) | |||
| 44 | F | 74 | PDAC | 224 | 1.3 | T3N0M0 |
| 45 | M | 68 | PDAC | 5673 | 1.7 | T3N1M1 |
| 46 | M | 62 | PDAC | 31 809 | 196.4 | T4N1M1 |
| 47 | M | 62 | PDAC | 319 754 | 378.3 | T4N1M1 |
| 48 | F | 67 | PDAC | 9 | 0.9 | T3N0M0 |
| 49 | F | 86 | PDAC | 245.4 | 1.6 | T3N0M0 |
| 50 | F | 63 | PDAC | 96 | 4.4 | T4N1M0 |
| 51 | F | 86 | PDAC | 2 | 18.2 | T4N1M1 |
| 52 | F | 71 | PDAC | 138 | 1.8 | T4N1M0 |
| 53 | F | 86 | PDAC | 2768 | 1.8 | T4N1M1 |
| 54 | M | 65 | PDAC | 1423 | 29.9 | T3N1M1 |
| 55 | F | 78 | PDAC | 1924 | 8.4 | T3N1M1 |
| 56 | F | 65 | PDAC | 219 | 1.4 | T4N1M0 |
| 57 | F | 84 | PDAC | 66 | 1.5 | T3N1M0 |
| 58 | F | 68 | PDAC | 85 | 1.9 | T3N1M1 |
| 59 | F | 73 | PDAC | 1801 | 2.2 | T3N0M0 |
| 60 | M | 57 | PDAC | 2145 | 2.8 | T4N1M1 |
| 61 | M | 80 | PDAC | 14 | 2.8 | T3N1M1 |
| 62 | M | 74 | PDAC | 10 | 1.4 | T3N0M0 |
| 63 | M | 66 | PDAC | 223 713 | 3.7 | T4N1M1 |
| Median (IQR) | 69.5 (65) | 234.7 (80.3) | 2.1 (1.6) |
Median and interquartile range (IQR) of each group are indicated.
Abbreviations: CA19‐9, carbohydrate antigen 19‐9; CEA, carcinoembryonic antigen; F, female; Healthy, healthy individual; IPMN, intraductal papillary mucinous neoplasm; M, male; PDAC, pancreatic ductal adenocarcinoma.
Other materials and methods are described in Data S1.
3. RESULTS
3.1. High expression of HULC in PDAC cells and expression induced by TGF‐β
Transforming growth factor‐β induces the EMT pathway, which triggers invasion and metastasis in several cancers. To investigate whether TGF‐β induces the EMT in PDAC cells, we examined the expression of EMT‐related genes and morphological changes after incubation with 10 ng/mL TGF‐β. Epithelial‐mesenchymal transition is associated with the downregulation of epithelial markers and the upregulation of mesenchymal markers at the molecular level. We verified that the epithelial marker E‐cadherin was downregulated, and that the EMT‐inducible transcription factor Snail, and the mesenchymal markers N‐cadherin and vimentin were upregulated, in TGF‐β‐treated PDAC cells (Figure S1A). As the EMT is morphologically characterized by changes from an epithelial cell phenotype to a spindle‐like appearance, and functionally characterized by decreased cell adhesion, we also confirmed that PDAC cells became spindle shaped and grew in isolation after TGF‐β treatment (Figure S1B).
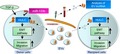
Next, to identify lncRNAs induced by TGF‐β in PDAC, we examined the expression profiles of 90 known lncRNAs in Panc‐1 PDAC cells after incubation with or without TGF‐β by RT‐qPCR. We identified several lncRNAs with greater than 0.5 log2 (fold change) alteration in expression in Panc‐1 cells treated with TGF‐β compared to the control (Figure 1A). We next analyzed the expression of lncRNAs in EVs derived from Panc‐1 cells after TGF‐β treatment. Compared to control EVs (derived from Panc‐1 cells incubated without TGF‐β), the expression of 21 lncRNAs was increased by more than 0.5 log2 (fold change) in EVs derived from Panc‐1 cells treated with TGF‐β (Figure 1B). These included 6 deregulated lncRNAs in TGF‐β‐treated Panc‐1 cells (Figure 1C). Among them, HULC was the only lncRNA whose expression was induced by TGF‐β in a panel of pancreatic cancer cells (Figure 1D) and altered by 1.7‐ to 4.7‐fold in several pancreatic cancer cells compared to values in hTERT‐HPNE normal pancreatic epithelial cells (Figure 1E). Thus, we focused on HULC as a candidate EMT‐related, as well as oncogenic, lncRNA.
Figure 1.
Long noncoding RNA (lncRNA) expression in pancreatic ductal adenocarcinoma (PDAC) cells is upregulated by transforming growth factor (TGF)‐β. A, LncRNA expression was assessed in Panc‐1 cells after incubation with 10 ng/µL TGF‐β. B, LncRNA expression in extracellular vesicles (EVs) released by Panc‐1 cells treated with TGF‐β. Each analysis was carried out on 3 independent samples for each lncRNA. Each bar represents the relative expression of an lncRNA. Changes in lncRNA expression of at least 2‐fold, in cells or EVs treated with TGF‐β compared to the controls, are shown. C, Venn diagram of the number of lncRNAs whose expression was increased in Panc‐1 cells or EVs treated with TGF‐β compared to the controls. D,E, RNA was extracted and RT‐quantitative PCR was undertaken. D, HULC expression was assessed in PDAC cells incubated with or without TGF‐β for 72 h. E, Basal expression level of HULC in a nonmalignant human pancreatic ductal epithelial cell line (hTERT‐HPNE) and PDAC cell lines. Expression of HULC was normalized to that of RNU6B. Bars are means ± SEM of 3 independent experiments. *P < .05
3.2. Knockdown of HULC attenuates cell invasion and migration by inhibiting the EMT in PDAC cells
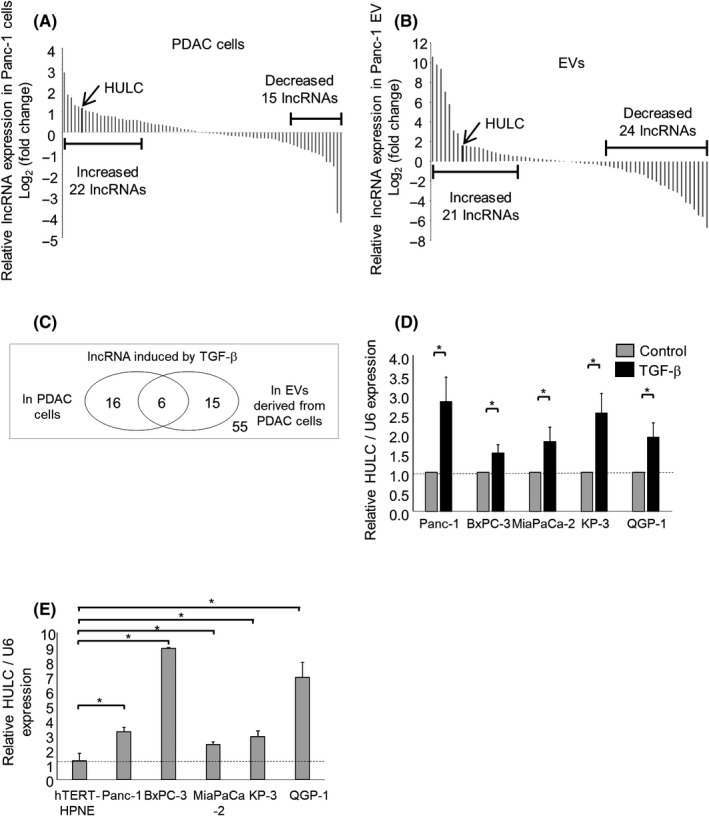
Having identified HULC as a TGF‐β‐regulated and candidate EMT‐related lncRNA, we next assessed its role in the EMT pathway. First, we confirmed the efficiency of HULC siRNAs and used them in further experiments. The mRNA level of the EMT‐inducible transcription factor Snail was reduced by HULC knockdown compared to the control. An siRNA to HULC also decreased the N‐cadherin and vimentin mRNA levels, whereas it increased that of E‐cadherin, in Panc‐1 and MiaPaCa‐2 cells (Figure 2A and Figure S2). Consistent with these changes, the N‐cadherin, vimentin, and Snail protein levels were decreased, and that of E‐cadherin was increased, by HULC knockdown (Figure 2B). Moreover, cells transfected with an siRNA to HULC showed significantly reduced proliferation, viability, invasion, and migration (Figure 2C‐F). Therefore, HULC is a TGF‐β‐inducible lncRNA that is aberrantly expressed in tumor cells and can modulate PDAC cell phenotypes in part by regulating the EMT pathway.
Figure 2.
Effect of HULC knockdown on the epithelial‐mesenchymal transition and phenotype of pancreatic ductal adenocarcinoma cells. Panc‐1 cells were transfected with an siRNA to HULC 1, HULC 2, or a control siRNA. A, After 48 h, RNA was isolated and quantitative PCR for HULC, E‐cadherin, N‐cadherin, vimentin, and Snail was carried out. B, After 72 h, protein was extracted and western blotting was carried out using Abs against E‐cadherin, N‐cadherin, vimentin, and Snail. Relative expression normalized to that of β‐actin is shown. C‐F, Panc‐1 cells were transfected with an siRNA to HULC 1, HULC 2, or a control siRNA. C,D, After 24, 48, 72, and 96 h, cell proliferation was examined by cell counting using Trypan blue (C) and cell viability was examined by MTS assay (D). E, After 24 h, cell invasion was assessed by Transwell assay under an inverted microscope. F, After 24 h, cell migration was assessed by wound healing assay. Bars are means ± SEM of 3 independent experiments. *P < .05. ref, reference; rel., relative
3.3. Tumor cell growth modulated by HULC suppressing EMT in vivo
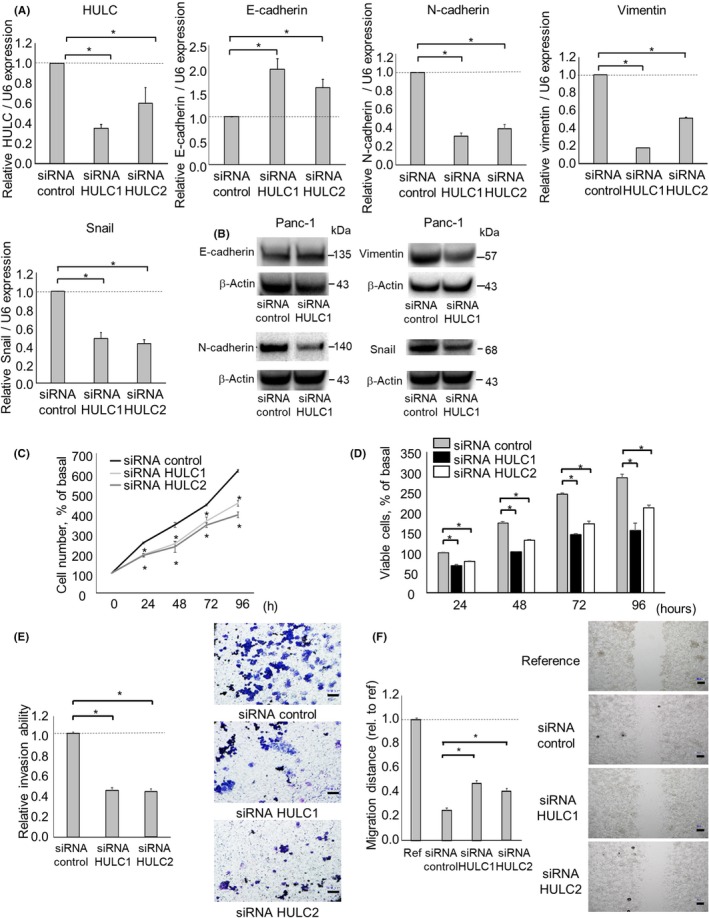
We next examined the effect of HULC knockdown on the growth of PDAC cell xenografts in vivo. The PDAC cells were s.c. injected following transfection with siRNA to HULC or a control siRNA. The PDAC cell growth and HULC expression were decreased in xenograft tumors transfected with an siRNA to HULC (Figure 3A,B). We also assessed whether HULC regulates the EMT pathway in xenograft tumors. Consistent with our observation that HULC knockdown suppressed the EMT of PDAC cells in vitro, N‐cadherin, vimentin, and Snail expression was decreased, and that of E‐cadherin was increased, in xenograft tumors (Figure 3C, D). These in vivo results are consistent with those obtained in vitro and confirm that HULC modulates tumor growth by inducing the EMT in PDAC cells.
Figure 3.
Effect of HULC knockdown in tumor xenografts in vivo. Xenograft tumors were established following ex vivo transfection of Panc‐1 cells with an siRNA to HULC or a control siRNA. A, Tumor volume was estimated at the indicated time points. Data are mean estimated tumor volumes from 3 separate xenografts. B, Tumors were excised at 4 weeks after implantation. Bars are means ± SEM xenograft tumor weights. Scale bar = 10 mm. C, RNA was isolated from xenograft tumors and quantitative PCR was carried out for HULC, E‐cadherin, N‐cadherin, vimentin, and Snail. D, Protein was isolated from xenograft tumors and western blotting was undertaken using Abs against E‐cadherin, N‐cadherin, vimentin, and Snail. Relative expression normalized to that of β‐actin is shown. Bars are means ± SEM of 3 independent experiments. *P < .05
3.4. Extracellular HULC transfer can promote PDAC cell invasion and migration
Extracellular vesicle contents are transferred from donor to recipient cells. We have reported that EV‐mediated transfer of lncRNAs can modulate the behavior of neighboring HCC cells.21, 22 Thus, we explored the ability of EV‐encapsulated HULC to regulate the EMT in recipient PDAC cells. We collected EVs from PDAC cells by ultracentrifugation and visualized their morphology by electron microscopy. The collected EVs were approximately 100 nm microstructures with a lipid bilayer membrane (Figure 4A). We next evaluated the effect of the transfer of EV HULC released from PDAC donor cells on the EMT pathway in recipient PDAC cells. Incubation with EVs derived from PDAC cells after transfection with control siRNA significantly increased HULC expression compared to incubations without EVs or with EVs derived from PDAC cells after transfection with siRNA HULC1. Moreover, E‐cadherin expression was decreased, and that of N‐cadherin, vimentin, and Snail was increased in recipient cells incubated with EVs isolated from PDAC cells after transfection with control siRNA (Figure 4B,C), suggesting that the promoting roles of EMT by EV derived from PDAC cells would be HULC‐mediated. Furthermore, incubation with EVs derived from PDAC cells significantly increased recipient cell proliferation, viability, invasion, and migration (Figure 4D‐G). These effects were further enhanced by EVs derived from PDAC cells treated with TGF‐β. These observations indicate that HULC could be transferred from donor to recipient cells by EVs, which could promote cancer cell invasion and migration by inducing the EMT pathway.
Figure 4.
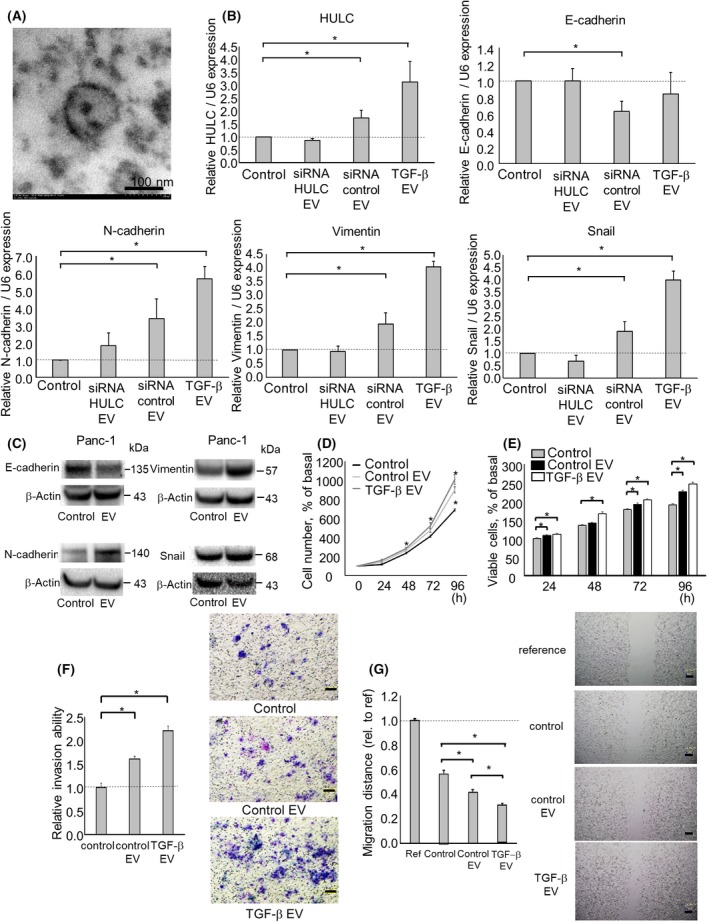
Intercellular HULC transfer by extracellular vesicles (EVs) during the epithelial‐mesenchymal transition and the phenotype of pancreatic ductal adenocarcinoma cells. Panc‐1 cells (1 × 106 per 10 cm dish) were cultured for 72 h, and EVs were isolated from the conditioned medium by ultracentrifugation. A, Transmission electron micrograph of EVs from Panc‐1 cells. A homogeneous population of particles was obtained. B‐G, EVs were isolated from Panc‐1 cells after transfection with an siRNA to HULC 1 or a control siRNA, or incubated with 10 ug/mL transforming growth factor (TGF‐β for 72 h. Then, EVs were added to recipient Panc‐1 cells. B, RNA was isolated from recipient cells and HULC, E‐cadherin, N‐cadherin, vimentin, and Snail expression was assayed by quantitative RT‐PCR. RNA expression was normalized to that of RNU6B and expressed relative to the value of the control. C, Protein was isolated from recipient Panc‐1 cells and the E‐cadherin, N‐cadherin, vimentin, and Snail levels were analyzed by western blotting. Protein levels were normalized to that of β‐actin. D,E, After 24, 48, 72, and 96 h, cell proliferation was examined by cell counting using Trypan blue (D) and cell viability was examined by MTS assay (E). F, After 24 h, cell invasion was assessed by Transwell assay under an inverted microscope. G, After 24 h, cell migration was assessed by wound healing assay. Bars are means ± SEM of 3 independent experiments. *P < .05. ref, reference; rel., relative
3.5. HULC is a direct target of miR‐133b
The interrelationship between 2 classes of ncRNAs, miRNAs and lncRNAs, reportedly contributes to epigenetic regulation in several cancers.25 Therefore, to reveal the regulatory mechanisms of HULC, we identified candidate miRNAs that target HULC and are downregulated by TGF‐β. We postulated that candidate tumor‐suppressing miRNAs would suppress EMT signaling by targeting HULC. A microarray analysis identified 187 miRNAs whose expression was decreased by less than 0.87‐fold in Panc‐1 cells treated with TGF‐β (Figure 5A,B). Of these, miR‐133b was predicted to target HULC by a bioinformatics analysis on miRNA.org (Figure 5C). Moreover, miR‐133b expression was reduced by TGF‐β in a panel of pancreatic cancer cells (Figure 5D). The posttranscriptional regulation of gene expression by miRNAs is the result of their complementary binding to the 3′‐UTR of their RNA targets.26 Bioinformatics analysis predicted that HULC has a 7‐nucleotide miR‐133b binding site. Therefore, to determine whether HULC is a direct target of miR‐133b, we cotransfected a miR‐133b mimic and a firefly luciferase reporter vector containing the target site of HULC into Panc‐1 and MiaPaCa‐2 cells. The HULC firefly luciferase activity was significantly decreased in miR‐133b‐overexpressing cells (Figure 5E), confirming the specificity of the interaction between miR‐133b and HULC. Based on these results, we focused on miR‐133b as a candidate EMT‐suppressing miRNA in further experiments.
Figure 5.
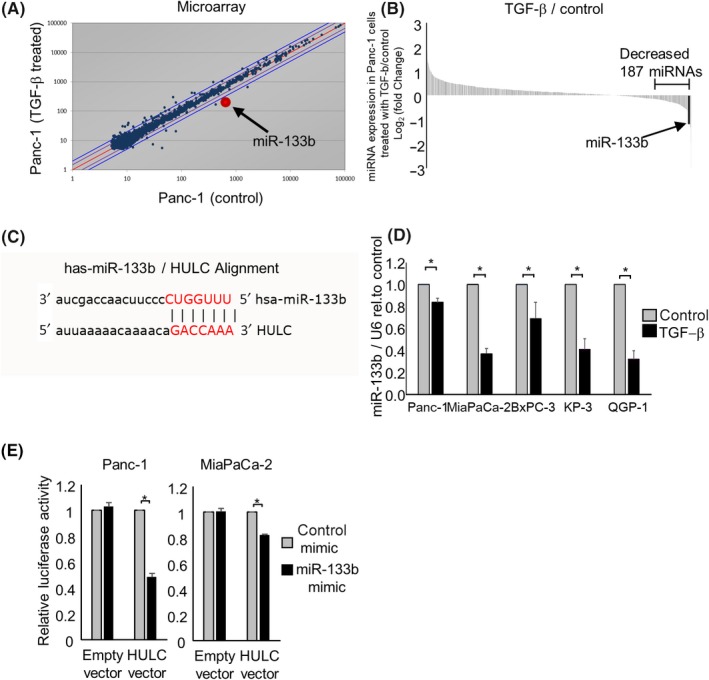
MicroRNAs (miRNAs) that regulate HULC expression in pancreatic ductal adenocarcinoma (PDAC) cells. Panc‐1 cells were treated with 0 (control) or 10 ng/mL transforming growth factor (TGF)‐β. After incubation for 72 h, RNA was isolated, and expression profiling of 2555 miRNAs was carried out. Expression of 1719 miRNAs was detected in Panc‐1 cells. A, Scatter plot of the microarray intensities of TGF‐β‐treated Panc‐1 cells against those of control cells. B, Waterfall plot of the 187 miRNAs whose expression was decreased <0.87‐fold in Panc‐1 cells treated with TGF‐β. C, MicroRNA (MiR)‐133b was predicted to target HULC by bioinformatics analysis using miRNA.org. D, RNA was extracted and miR‐133b expression was assessed by quantitative RT‐PCR in PDAC cells after incubation with 10 ng/mL TGF‐β for 72 h. MiR‐133b expression relative to the controls is shown. Gene expression was normalized to that of RNU6B. E, Panc‐1 and MiaPaCa‐2 cells were transfected with 12.5 nmol/L miR‐133b or control mimic, and after 24 h, were cotransfected with 2.0 µg HULC firefly luciferase reporter vector or empty vector and 0.1 μg Renilla luciferase reporter pRL‐SV40. After a further 24 h, relative firefly luciferase activity was measured and normalized to Renilla activity. Bars are means ± SEM of 3 independent experiments. *P < .05
3.6. MicroRNA‐133b suppresses cell invasion and migration by inhibiting EMT by targeting HULC
To assess whether miR‐133b suppresses cell invasion and migration by targeting HULC, we next investigated the functional contribution of miR‐133b to the EMT pathway. We used a miRNA mimic to overexpress miR‐133b and confirmed its effect on miR‐133b by qPCR. The miR‐133b mimic decreased HULC expression in PDAC cells. MicroRNA‐133b overexpression significantly reduced N‐cadherin, vimentin, and Snail expression, and increased that of E‐cadherin (Figure 6A,B). Also, overexpression of miR‐133b decreased cell proliferation, viability, invasion, and migration (Figure 6C–F).
Figure 6.
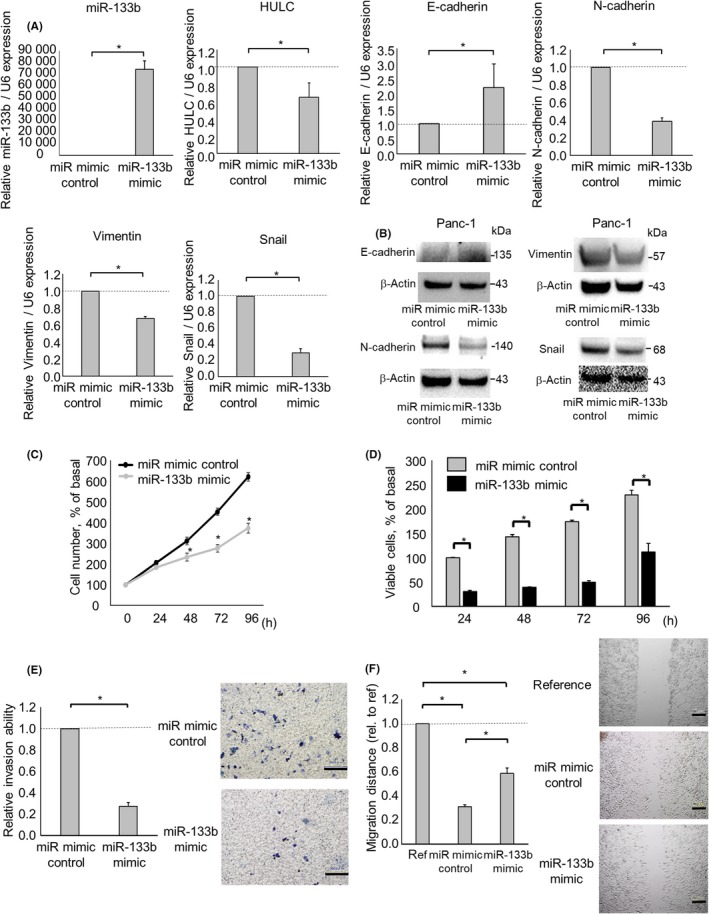
Effect of microRNA (miR)‐133b overexpression on the epithelial‐mesenchymal transition and phenotype of pancreatic ductal adenocarcinoma cells. Panc‐1 cells were transfected with 12.5 nmol/L miR‐133b or the control mimic. A, After 48 h, RNA was isolated and quantitative RT‐PCR for miR‐133b, HULC, E‐cadherin, N‐cadherin, vimentin, and Snail was carried out. B, After 72 h, protein was extracted, and western blotting was undertaken using Abs against E‐cadherin, N‐cadherin, vimentin, and Snail; protein levels normalized to that of β‐actin are shown. C,D, After 24, 48, 72, and 96 h, cell proliferation was examined by cell counting using Trypan blue (C) and cell viability was examined by MTS assay (D). E, After 24 h, cell invasion was assessed by Transwell assay under an inverted microscope. F, After 24 h, cell migration was assessed by wound healing assay. Bars are means ± SEM of 3 independent experiments. *P < .05. ref, reference; rel., relative
To further evaluate the impact of miR‐133b on the EMT, a miR‐133b inhibitor was used to attenuate miR‐133b activity in PDAC cells. We assessed the expression levels of EMT‐related genes in Panc‐1 cells after incubation with the miR‐133b inhibitor. Inhibition of miR‐133b significantly increased HULC, N‐cadherin, vimentin, and Snail expression, and decreased that of E‐cadherin (Figure S3A,B). We next analyzed tumor cell behavior after miR‐133b inhibition. The miR‐133b inhibitor significantly increased the proliferation, viability, invasion, and migration of PDAC cells (Figure S3C‐F). Therefore, miR‐133b suppressed PDAC cell invasion and migration by inhibiting the EMT through targeting HULC.
3.7. Extracellular vesicle‐encapsulated HULC derived from serum is a potential biomarker for human PDAC
Finally, we investigated the potential of EV HULC as a novel biomarker for human PDAC. Serum samples from 20 PDAC patients, 22 branch duct‐type IPMN patients, and 21 healthy individuals were collected. Extracellular vesicle RNA derived from serum was isolated and subjected to digital PCR analysis. The subjects’ data are shown in Table 1. The subjects comprised 6 stage II, 3 stage III, and 11 stage IV PDAC patients; no stage 0 or I PDAC patients were analyzed. Extracellular vesicle HULC expression was significantly increased in PDAC patients compared to healthy individuals. Extracellular vesicle HULC expression was also increased in PDAC patients compared to IPMN patients (Figure 7A,B). Moreover, analysis using only PDAC patient samples showed that there was no significant correlation between EV HULC expression and stage, N factor, M factor, or the other clinical factors analyzed (Figure 7C). A receiver operating characteristic curve was plotted to assess the diagnostic performance of EV HULC expression, CA19‐9, and CEA for PDAC. Extracellular vesicle HULC showed a high AUC for discriminating PDAC from healthy individuals and IPMN patients. If healthy individuals and IPMN patients were regarded as a non‐PDAC group, HULC showed good predictive performance, with an AUC of 0.92 for discriminating PDAC from non‐PDAC. The AUC values of CA19‐9 and CEA were 0.9 and 0.54, respectively, and there was no significant difference in the AUC between HULC and CA19‐9 (Figure 7D). These results suggest that HULC performs similarly to CA19‐9 as a potential biomarker for human PDAC, particularly for predicting the malignant potential of IPMN.
Figure 7.
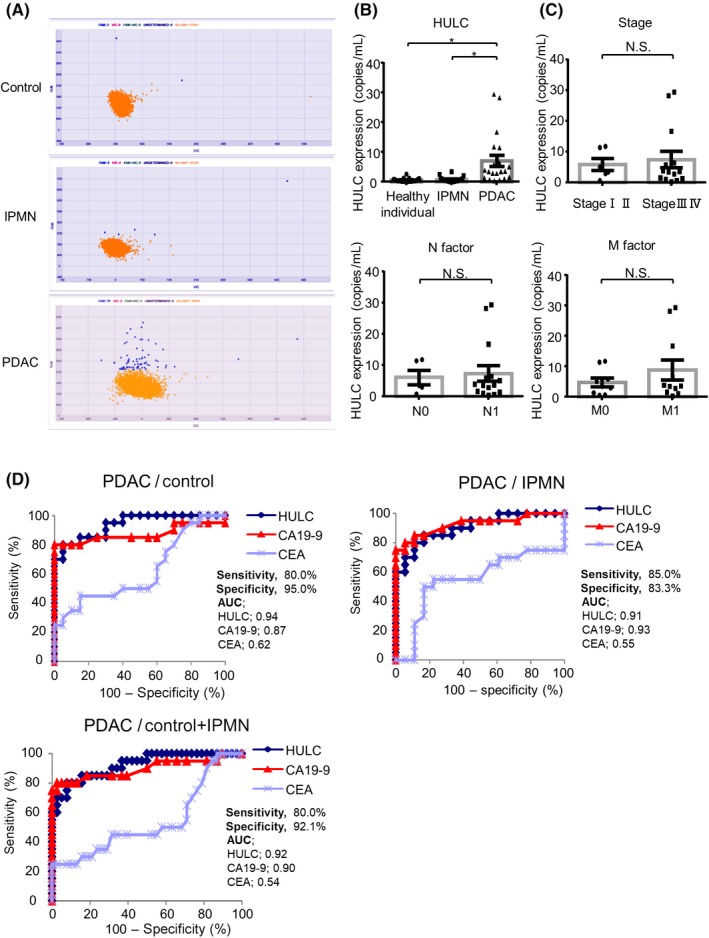
Evaluation of extracellular vesicle (EV) HULC as a biomarker for pancreatic ductal adenocarcinoma (PDAC). EV HULC expression was analyzed in serum from PDAC patients, intraductal papillary mucinous neoplasm (IPMN) patients, and healthy individuals. A, Blue dots, wells containing HULC; yellow dots, wells without HULC. B, Expression of HULC. C, Samples from PDAC patients were analyzed to evaluate the correlation between EV HULC expression and stage, N factor, and M factor. Bars are means ± SEM. *P < .05. D, Discriminatory ability of EV HULC expression, carbohydrate antigen 19‐9 (CA19‐9), and carcinoembryonic antigen (CEA) in PDAC patients compared to healthy individuals, IPMN patients, and both healthy individuals and IPMN patients. Sensitivity and specificity of HULC expression are presented as area under the curve (AUC) values. Cut‐off (probability) values of HULC, CA19‐9, and CEA are 0.43, 0.44, and 0.6 (PDAC patients vs healthy individuals), and 0.22, 0.39, and 0.26 (PDAC patients vs both healthy individuals and IPMN patients), respectively. N.S., not significant
4. DISCUSSION
The mechanisms by which tumor cells respond to TGF‐β and induce the EMT pathway are crucial in cancer invasion and metastasis, and in particular for tumors such as PDAC, which frequently show local invasion and metastasis at an early stage. Long noncoding RNA HULC was identified as an inducer of the EMT and was aberrantly expressed in PDAC in vitro and in vivo. HULC is enriched within EVs released by tumor cells after incubation with TGF‐β and can modulate cellular signaling and tumor invasion and migration in recipient cells. This lncRNA is the most upregulated gene in HCC.27 HULC copurifies with ribosomes in the cytoplasm, is involved in the regulation of cancer cell proliferation by downregulating p18, and is related to ATM/ATR‐ and p53‐dependent signaling.28 Also, HULC negatively regulates the expression of several miRNAs, including miR‐372. Downregulation of miR‐372 decreases translational repression of PRKACB, which induces phosphorylation of CREB. As phosphorylated CREB forms part of the RNA pol II transcriptional machinery, which activates HULC expression, HULC might act as an endogenous miRNA sponge to regulate miRNA activities in HCC cells.7, 29 Our studies revealed several important roles for this EMT‐inducible lncRNA, and identified the mechanism by which HULC coordinates cancer cell invasion and migration. Indeed, HULC has been shown to promote the EMT in HCC by upregulating ZEB1.30 Further studies of the roles of this lncRNA in the EMT and behavior of tumor cells using a metastatic tumor mouse model, or studies about the effects of HULC on the expression of TGF‐β receptors or secretion of TGF‐β, could lead to the discovery of therapeutic targets.
Long noncoding RNAs can epigenetically modulate gene expression by chromatin remodeling or controlling transcription, posttranscriptional mRNA processing, protein function or localization, and intercellular signaling.7 Although many lncRNAs contribute to the development of digestive cancers and other gastroenterological diseases, only a handful of lncRNAs are involved in the pathogenesis of PDAC. Of these, HOTAIR, H19, and TUG1 are related to PDAC cell invasion and metastasis.14 HOTAIR was reported to promote tumor cell invasion in part by regulating HOXA13. Lymph node metastasis in PDAC patients was correlated with HOXA13 expression in tumor tissue.11 TUG1 promotes cell migration and contributes to the EMT in PDAC cells.31 In this study, we found that HULC expression is induced by TGF‐β and promotes cell invasion and migration by regulating the EMT pathway in PDAC cells. Thus, HULC is a potential oncogenic gene in human PDAC, as in other gastroenterological cancers. Recently, the interrelationship between miRNAs and lncRNAs has been reported to contribute to the epigenetic regulation of gene expression in several diseases.15 HULC is a target of miR‐488. MicroRNA‐488 suppressed cell invasion by inhibiting the EMT pathway through targeting ADAM9, and attenuated cell proliferation by inhibiting HULC expression through sponging to HULC in HCC cells.32 Our study revealed that miR‐133b targets HULC directly and attenuates PDAC cell invasion and migration by inhibiting HULC expression. These results provide new insights into the miRNA‐lncRNA interaction and suggest potential strategies to inhibit invasion and metastasis in human PDAC. As a single miRNA can target multiple RNAs, further investigations, such as rescue studies by HULC overexpression, are required to fully understand the role of the miR‐133b‐HULC interaction in the regulation of the EMT.
Although most (but not all) exRNA is contained within EVs, which are selectively isolated within exRNA preparations, incubation of EVs obtained from PDAC cells transferred HULC and enhanced tumor cell viability, invasion, and metastasis by promoting the EMT, suggesting that extracellular HULC could be packaged within EVs.16, 17 Other factors in EVs, such as mRNAs, proteins, and ncRNAs, could affect cell phenotype or induce the EMT. However, expression profiling of lncRNAs within PDAC cell‐derived EVs identified HULC as one of the most highly enriched lncRNAs. Moreover, the HULC content of EVs was increased by TGF‐β treatment, and incubation with these EVs further increased HULC expression and induced the EMT pathway in recipient PDAC cells. Although further studies are needed to evaluate the role of HULC in PDAC development, our findings show that EV HULC promotes the EMT, as well as the invasion and migration, of PDAC cells.
Circulating nucleic acids, including mRNAs and ncRNAs, can be useful for liquid biopsy, which can provide diagnostic and prognostic information. Circulating EVs have potential for liquid biopsy because they can transport cargo, such as mRNAs, ncRNAs, and proteins.33, 34 There are few reports regarding liquid biopsy using circulating EV lncRNAs.35 For instance, CRNDE‐h is expressed in CRC tissue. The serum exosomal CRNDE‐h level was elevated and could serve as a diagnostic and prognostic biomarker for CRC.36 Long noncoding RNA H19 is highly expressed in HCC tissue, mainly in cholangiocytes. Cholangiocyte‐derived exosome‐mediated transfer of H19 promotes cholestatic injury in hepatocytes. Moreover, the serum exosomal H19 level gradually increased during liver injury in a mouse model.37 The potential of EV lncRNA as a biomarker for pancreatic cancer is unclear. In this study, EV HULC was highly expressed in the serum of PDAC patients; the AUC was comparable to that of CA19‐9. Moreover, EV HULC might enable detection of early PDAC in IPMN patients. Although the limitation of this study is the small number of samples, and further analysis of the usefulness of EV HULC for liquid biopsy using a greater number of samples is warranted, our data suggest that EV HULC has potential as a biomarker for early diagnosis of human PDAC.
We evaluated the role of EV lncRNA‐miRNA signaling in tumor invasion and migration in PDAC cells, and identified EV‐encapsulated lncRNA as a biomarker for PDAC (Figure 8). These findings provide mechanistic insights into cancer cell invasion and metastasis, identify novel therapeutic targets for cancer, and suggest the potential of EV HULC for early diagnosis of PDAC.
Figure 8.

Schematic overview of the roles of extracellular vesicle (EV) HULC as regulator of development and liquid biopsy for pancreatic ductal adenocarcinoma. EMT, epithelial‐mesenchymal transition; lncRNA, long noncoding RNA; miR, microRNA; TGF‐β, transforming growth factor‐β
DISCLOSURE
The authors have no conflict of interest to declare.
Supporting information
ACKNOWLEDGMENTS
This project was supported in part by Japan Society for the Promotion of Science KAKENHI Grant No. JP15K19303. We are grateful to Hiroki Bochimoto for assistance with transmission electron microscopy and Shinichi Chiba for help with the digital PCR analysis, and thank all members of our department for helpful discussions.
Takahashi K, Ota Y, Kogure T, et al. Circulating extracellular vesicle‐encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. 2020;111:98–111. 10.1111/cas.14232
Contributor Information
Kenji Takahashi, Email: t-kenji@asahikawa-med.ac.jp.
Tsuguhito Ota, Email: ota@asahikawa-med.ac.jp.
REFERENCES
- 1. Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605‐1617. [DOI] [PubMed] [Google Scholar]
- 2. Ma J, Siegel R, Jemal A. Pancreatic cancer death rates by race among US men and women, 1970–2009. J Natl Cancer Inst. 2013;105:1694‐1700. [DOI] [PubMed] [Google Scholar]
- 3. Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of disease: epithelial‐mesenchymal transition–does cellular plasticity fuel neoplastic progression? Nat Clin Pract Oncol. 2008;5:280‐290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M. Role of Ras signaling in the induction of snail by transforming growth factor‐beta. J Biol Chem. 2009;284:245‐253. [DOI] [PubMed] [Google Scholar]
- 5. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial‐mesenchymal transitions in development and disease. Cell. 2009;139:871‐890. [DOI] [PubMed] [Google Scholar]
- 6. Satoh K, Hamada S, Kimura K, et al. Up‐regulation of MSX2 enhances the malignant phenotype and is associated with twist 1 expression in human pancreatic cancer cells. Am J Pathol. 2008;172:926‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology. 2014;60:744‐753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta RA, Shah N, Wang KC, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro‐oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616‐1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiao F, Hu H, Han T, et al. Long noncoding RNA MALAT‐1 enhances stem cell‐like phenotypes in pancreatic cancer cells. Int J Mol Sci. 2015;16:6677‐6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Z, Zhao X, Zhou Y, et al. The long non‐coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu X, Fang Y, Wang Z, et al. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013;354:891‐896. [DOI] [PubMed] [Google Scholar]
- 13. Huang C, Yu W, Wang Q, et al. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106:143‐149. [PubMed] [Google Scholar]
- 14. Ma C, Nong K, Zhu H, et al. H19 promotes pancreatic cancer metastasis by derepressing let‐7's suppression on its target HMGA2‐mediated EMT. Tumour Biol. 2014;35:9163‐9169. [DOI] [PubMed] [Google Scholar]
- 15. Bian EB, Xiong ZG, Li J. New advances of lncRNAs in liver fibrosis, with specific focus on lncRNA‐miRNA interactions. J Cell Physiol. 2018. [DOI] [PubMed] [Google Scholar]
- 16. Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226‐1232. [DOI] [PubMed] [Google Scholar]
- 18. Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581‐593. [DOI] [PubMed] [Google Scholar]
- 19. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654‐659. [DOI] [PubMed] [Google Scholar]
- 20. Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle‐mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237‐1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia‐signaling pathways by extracellular linc‐RoR. J Cell Sci. 2014;127:1585‐1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of extracellular vesicle long noncoding RNA (linc‐VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res. 2014;12:1377‐1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu L, Xu H, Wang W, et al. A preoperative serum signature of CEA+/CA125+/CA19‐9 >/= 1000 U/mL indicates poor outcome to pancreatectomy for pancreatic cancer. Int J Cancer. 2015;136:2216‐2227. [DOI] [PubMed] [Google Scholar]
- 24. Yadav DK, Bai X, Yadav RK, et al. Liquid biopsy in pancreatic cancer: the beginning of a new era. Oncotarget. 2018;9:26900‐26933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiong DD, Li ZY, Liang L, et al. The LncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to Mir‐193a‐3p as a competitive endogenous RNA. Cell Physiol Biochem. 2018;48:905‐918. [DOI] [PubMed] [Google Scholar]
- 26. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118‐126. [DOI] [PubMed] [Google Scholar]
- 27. Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up‐regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330‐342. [DOI] [PubMed] [Google Scholar]
- 28. Du Y, Kong G, You X, et al. Elevation of highly up‐regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down‐regulating p18. J Biol Chem. 2012;287:26302‐26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J, Liu X, Wu H, et al. CREB up‐regulates long non‐coding RNA, HULC expression through interaction with microRNA‐372 in liver cancer. Nucleic Acids Res. 2010;38:5366‐5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li SP, Xu HX, Yu Y, et al. LncRNA HULC enhances epithelial‐mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR‐200a‐3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431‐42446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. The Lncrna‐TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and emt phenotype formation through sponging Mir‐382. Cell Physiol Biochem. 2017;42:2145‐2158. [DOI] [PubMed] [Google Scholar]
- 32. Hu D, Shen D, Zhang M, et al. MiR‐488 suppresses cell proliferation and invasion by targeting ADAM9 and lncRNA HULC in hepatocellular carcinoma. Am J Cancer Res. 2017;7:2070‐2080. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut. 2018. [DOI] [PubMed] [Google Scholar]
- 34. Matsuzaki J, Ochiya T. Extracellular microRNAs and oxidative stress in liver injury: a systematic mini review. J Clin Biochem Nutr. 2018;63:6‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang S, Li X. Recent advances in extracellular vesicles enriched with non‐coding RNAs related to cancers. Genes Dis. 2018;5:36‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE‐h as a novel serum‐based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551‐85563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li X, Liu R, Huang Z, et al. Cholangiocyte‐derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology. 2018;68:599‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


