Abstract
Analysis of anticancer immunity aids in assessing the prognosis of patients with breast cancer. From 250 operated breast cancers, we focused on serum levels of C‐C motif chemokine ligand 5 (CCL5), which is involved in cancer immune reactions. Serum levels of CCL5 were measured using a cytometric bead‐based immunoassay kit and CCL5 expression in cancer cells was determined using immunohistochemical staining. In addition, mRNA in cancer and stromal cells was analyzed by microdissection and comparison with the public dataset. Disease‐free survival (DFS) of patients with high CCL5 levels (cut‐off, 13.87 ng/mL; n = 192) was significantly better than those with low CCL5 levels (n = 58; hazard ratio, 0.20; 95% confidence interval, 0.10‐0.39; P < .0001). An improved overall survival was observed in patients with high CCL5 levels compared to those with low CCL5 levels (P = .024). On the contrary, high immunohistochemical expression of CCL5 in cancer cells was significantly associated with decreased DFS. As serum CCL5 levels did not correlate with CCL5 expression in cancer cells and the relative expression of mRNA CCL5 was elevated in stromal cells in relation to cancer cells, serum CCL5 might be derived not from cancer cells, but from stromal cells. Expression of CCL5 in serum, but not in cancer cells, might contribute to improved patient prognosis mediating through not only immune reaction, but through other mechanisms. Determination of circulating CCL5 levels could be useful for predicting patient prognosis.
Keywords: breast cancer, CCL5, cytokine, immunity, prognosis
We found that high levels of serum C‐C motif chemokine ligand 5 (CCL5), a chemokine produced by immune cells, were significantly associated with improved prognosis of patients with operated breast cancer. As the relative expression of mRNA CCL5 was elevated in stromal cells, serum CCL5 seems to be derived not from cancer cells, but from stromal cells. CCL5 might contribute to improved patient prognosis mediating through not only immune reaction, but through other mechanisms.

Abbreviations
- CCL5
RANTES, C‐C motif chemokine ligand 5
- CI
confidence interval
- DC
dendritic cell
- DFS
disease‐free survival
- ER
estrogen receptor
- G protein
guanine nucleotide‐binding protein
- GNA
G protein subunit alpha
- GNB
G protein subunit beta
- GNG
G protein subunit gamma
- GNGT
GNG transducing
- GRK
G protein coupled receptor kinase
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- IL
interleukin
- MAX
MYC associated factor X
- MDSC
myeloid‐derived suppressor cell
- MYC
myelocytomatosis
- NK
natural killer
- NLR
neutrophil‐to‐lymphocyte ratio
- OS
overall survival
- RFS
recurrence‐free survival
- SDC4
syndecan‐4
- TIL
tumor‐infiltrating lymphocyte
- TN
triple‐negative
- Treg
regulatory T cell
1. INTRODUCTION
Recently, the involvement of immune reactions became well established in the initiation and progression of breast cancers.1, 2, 3 In particular, positive associations between high levels of TILs and favorable prognoses have been repeatedly confirmed by many studies.4, 5, 6 In a metaanalysis consisting of 22 964 patients, the DFS and OS of patients with TN breast cancers with high numbers of TILs were significantly better than those with few TILs (for DFS: HR, 0.82; 95% CI, 0.76‐0.88; for OS: HR, 0.79; 95% CI, 0.71‐0.87).6 In addition to TILs, the NLR in the blood has been reported to be a prognostic indicator of early breast cancers.7, 8 Similar to TILs, patients with low levels of NLR, which indicate relatively more lymphocytes than neutrophils, are associated with a significantly better prognosis than patients with high NLRs (for DFS: HR, 1.47; 95% CI, 1.06‐2.05; P = .025; for OS: HR, 2.21; 95% CI, 1.99‐4.11; P = .001).7
These data strongly indicate immunity against breast cancers is critical in terms of prognosis during the course of treatment of patients. However, the application of these biomarkers has been limited. Although elevated TIL numbers represent a T cell immune reaction, it is not known whether the function of these T cells is induced or suppressed by several factors, such as Tregs, MDSCs, tumor‐associated macrophages, or unfavorable cancer microenvironment.9, 10 Furthermore, the prognostic significance of the NLR is still controversial.11 Interestingly, positive associations between a high NLR and increased levels of circulating cytokines, including IL‐6, IL‐8, IL‐2Rα, hepatocyte growth factor, macrophage‐colony stimulating factor, and vascular epidermal growth factor, have been reported in metastatic colorectal cancers.12 As these inflammatory cytokines might be linked with anticancer immune responses, the NLR is speculated to be associated with patient prognosis by mediating through these cytokines.
C‐C motif chemokine ligand 5 is a chemokine produced by T lymphocytes, platelets, endothelial cells, macrophages, monocytes, NK cells, and DCs.13, 14, 15, 16 In addition, CCL5 secretion from mesenchymal stem cells and breast cancer cells are reported.17, 18, 19 It was found that CCL5 promotes breast cancer metastasis in a paracrine fashion, including enhancement of breast cancer motility, invasion, and metastatic ability.17 In addition, CCL5 promotes breast cancer metastasis by maintaining the immunosuppressive ability of MDSCs.20 Furthermore, a positive association between CCL5 and an aggressive phenotype or metastatic potential has been reported for breast cancer.21, 22 Therefore, CCL5 promotion of breast cancer progression and metastasis has been the focus of many studies 23 .
Supporting these in vitro and in vivo studies, high levels of CCL5 in patients with gastric cancer are associated with poor prognoses.24 Based on quantum‐dot‐based molecular imaging, high expression of CCL5 is significantly associated with poor DFS (P = .001) in patients with luminal B and HER2‐negative breast cancers.25 However, contrary to these studies, high CCL5 gene expression is significantly associated with good RFS in patients with TN breast cancer (HR, 0.39; 95% CI, 0.22‐0.71; P = .0012).26 Therefore, the prognostic significance of CCL5 in early breast cancers remains unclear.
In the present study, we examined serum CCL5 levels in terms of prognostic significance in patients with early breast cancer focusing on breast cancer subtypes. In addition, immunohistochemical staining of CCL5 in cancer cells and the relationship between serum CCL5 levels and TILs were evaluated.
2. MATERIALS AND METHODS
2.1. Patient recruitment
Patients with breast cancer operated on at the Hyogo College of Medicine Hospital between July 2009 and November 2016 were consecutively recruited for this retrospective study. During this period, 792 female patients were pathologically diagnosed with primary invasive breast carcinoma. We obtained informed consent for this study from 695 patients. Of these patients, 250 participants with enough volume of serum samples and that were available for this study were recruited. In addition, serum samples from noninvasive breast cancers (n = 29) and metastatic breast cancers (n = 49) were also analyzed. This study was approved by the ethics committee of the Hyogo College of Medicine (No. 106) and was in accordance with the Declaration of Helsinki. Written informed consent was obtained from each patient. Data on individual participants are unavailable because the Institute's ethics committee did not permit their publication.
2.2. Adjuvant treatments and patient follow‐up
Chemotherapies were given preoperatively (n = 12), postoperatively (n = 56), and both (n = 3). These included anthracycline‐containing (n = 5), taxane‐based (n = 24), sequential use of anthracycline and taxanes, (n = 38), and other (n = 4) chemotherapy regimens. A total of 184 patients were treated with endocrine therapies, including luteinizing hormone‐releasing hormone plus tamoxifen (n = 37), tamoxifen (n = 18), and aromatase inhibitors (n = 115). Endocrine therapies were switched from luteinizing hormone‐releasing hormone plus tamoxifen to aromatase inhibitors (n = 5) or luteinizing hormone‐releasing hormone plus aromatase inhibitors (n = 3), tamoxifen to aromatase inhibitors (n = 1), aromatase inhibitors to tamoxifen (n = 2) or luteinizing hormone‐releasing hormone plus tamoxifen (n = 1), and others (n = 2). Trastuzumab was used in 19 of 30 HER2‐positive cases.
The vast majority of the patients visited the hospital postoperatively every 3 to 6 months for 3 years and then every 6 to 12 months thereafter. The median follow‐up time was 45.7 months (range, 0.6‐101.2 months). During follow‐up, 29 patients had recurrence that was locoregional and in the lymph nodes (n = 10), ipsilateral or contralateral breast (n = 2), bone (n = 11), lungs (n = 6), liver (n = 2), or pleura (n = 2). Disease‐free survival was defined as the time from the operation to the first recurrence at any site, contralateral breast cancer, or death for any reason.
2.3. Measurements of serum CCL5
Blood samples were obtained on the same day of the operation and just before the start of the operation. For the 15 patients treated with preoperative chemotherapies, blood samples were collected before the start of these treatments. Serum levels of CCL5 were measured using a cytometric bead‐based immunoassay kit according to the manufacturer's protocol (Human RANTES Flex Set; BD Biosciences). Briefly, capture bead populations with distinct fluorescence intensities and coated with RANTES‐specific capture Ab were mixed together in equal volumes, where 50 μL of each sample and 50 μL of phycoerythrin‐conjugated detection Ab was added to 50 μL of a mixed‐bead population. This mixture was incubated for 3 hours at room temperature in the dark to form sandwich complexes. The beads were then washed with wash buffer and analyzed with a BD LSRFortessa X‐20 cell analyzer and FCAP Array software (BD Biosciences).
2.4. Immunohistochemical staining of CCL5, CD8, and FOXP3
From 250 patients, 160 breast cancer tissues were obtained for immunohistochemical staining of CCL5, in which adequate cancer cells were included. In addition, we were able to evaluate CD8 and FOXP3 expression levels in 156 and 143 samples, respectively. These samples were obtained during operation or biopsy prior to preoperative therapies. Formalin‐fixed, paraffin‐embedded tumor tissues were cut and deparaffinized, then antigen was retrieved for 8 minutes using a Cell Conditioning Solution (CC1; Ventana Medical Systems). Primary Ab (anti‐RANTES Ab, ab9679, rabbit polyclonal antibody; Abcam) was diluted to 1:100. We used an anti‐CD8 primary Ab (CONFIRM anti‐CD8 SP57 rabbit mAb; Roche Diagnostics) without dilution and 236A/E7 Ab (ab20034, mouse mAb; Abcam) against FOXP3 diluted to 1:500. The procedure was carried out in an automated immunostainer Ventana BenchMark ULTRA using the I‐VIEW DAB universal kit (Roche Diagnostics) for CCL5 and CD8, and in automated immunostainer BOND‐MAX using the Bond PolymerRefine kit (Leica Microsystems) for FOXP3. We used human skeletal muscle cells and normal breast epithelial cells as positive and negative controls, respectively, for CCL5. Positive controls for CD8 and FOXP3 were lymph node and tonsil cells, respectively, and normal breast epithelial cells were used as negative control. We evaluated cytoplasmic staining of cancer cells for CCL5, membrane staining of lymphocytes for CD8, and nuclear staining of lymphocytes for FOXP3. More than 500 cancer cells were counted in the areas of the stained lesions and the proportion of positive cells was calculated for CCL5. As for CD8 and FOXP3, positive cells were counted at a ×400 field and average counts of 4 fields were obtained.
We used the median value of 20% CCL5‐positive cells as a cut‐off value for DFS and divided breast cancers into CCL5‐high (>20%, n = 71) and CCL5‐low (≤ 20%, n = 89) groups. In order to compare cellular CCL5 levels with those in serum, cells were divided into 4 groups: 0% cancer‐positive cells and 3 groups with equal numbers of patients for 1% or more positive cancer cells (1%‐49%, n = 33; 50%‐69%, n = 29; 70% or more, n = 25).
2.5. Evaluation of TILs
Tumor‐infiltrating lymphocytes in breast cancer biopsy specimens treated with preoperative therapy (n = 13) and surgical specimens not exposed to preoperative therapy (n = 196) were measured according to a method described in a previous study.27 Briefly, lymphocytes and plasma cells within the tumor border were counted as TILs in a medium‐power field (×100) in the hot spot area. We classified TIL scores as low (less than 50%) or high (50% or more).
2.6. Public database
We calculated RFS and OS according to CCL5 gene expression levels provided by gene chip data using a specific probe (Affymetrix ID, 1555759_a_at) through the Kaplan‐Meier Plotter database (http://kmplot.com/analysis/).28 In this dataset, a total number of 1764 breast cancers was included and we used the median as a cut‐off value. Spearman's correlations between the mRNA levels of CCL5 and of other genes were determined using cBioPortal (http://www.cbioportal.org/)29, 30 for 1084 invasive breast carcinoma samples (The Cancer Genome Atlas, PanCancer Atlas). Network analysis of CCL5 was done using the public database on cBioPortal (http://www.cbioportal.org/). CCL5 expression in microdissected breast tumor cells and normal breast ductal cells was analyzed using microarray data (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE38959).31 In order to compare CCL5 mRNA expression levels between laser capture microdissected cancer cells and stromal cells, we used public dataset, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31192, including breast cancers from pregnant and nonpregnant women (CCL5 probe no. 1555759_a_at).32
2.7. Statistical analysis
The relationships between the clinicopathological characteristics and serum CCL5 levels were compared using Fisher's exact test or the Wilcoxon rank‐sum test. Differences in DFS in different subgroups were calculated using Kaplan‐Meier plots and log‐rank tests. A Cox proportional hazards model was used to obtain HRs and 95% CIs for univariable and multivariable analyses of clinical factors and CCL5 levels for DFS. Serum levels of CCL5 were compared among subgroups according to TIL levels or breast cancer types (noninvasive, invasive, or metastatic) using the Wilcoxon rank‐sum test. Statistical significance was set at P < .05 except for multiple CCL5 comparisons adjusted with the Bonferroni correction, for which significance was set at P < .0083 or P < .017. All statistical calculations were undertaken using JMP Pro 13 (SAS Institute).
3. RESULTS
3.1. Determination of serum CCL5 in noninvasive, invasive, and metastatic breast cancers and determination of the cut‐off value for DFS
The median value of CCL5 serum levels in invasive cancer of 32.90 ng/mL (range, 1.75‐646.64 ng/mL) was similar to the results obtained in noninvasive cancers (median, 31.39; range, 7.47‐102.68; P = .909), but significantly lower than those from metastatic cancers (median, 62.96; range, 5.60‐169.44; P < .0001, Figure S1). We determined the cut‐off value of CCL5 in invasive breast cancers to be 13.87 ng/mL for DFS based on receiver operating characteristic curves calculated as 0.676 using the Youden index for area under the curve as shown in Figure S2 (P = .0069). Using this cut‐off, we divided patients into CCL5‐high (n = 192) and ‐low (n = 58) (Table 1).
Table 1.
Clinicopathological characteristics of breast cancers according to C‐C motif chemokine ligand 5 (CCL5) levels
| Characteristics |
CCL5‐highb (n = 192) |
CCL5‐lowb (n = 58) |
P‐valuec |
|---|---|---|---|
| Age, years; median (range) | 58 (28‐86) | 68 (34‐90) | .0009 |
| Menopausal status | |||
| Pre‐ | 67 (83.8)a | 13 (16.3) | .0786 |
| Post‐ | 124 (73.4) | 45 (26.6) | |
| Otherd | 1 (100.0) | 0 (0.0) | |
| Tumor size, cm | |||
| ≤2 | 141 (80.6) | 34 (19.4) | .0495 |
| >2 | 51 (68.9) | 23 (31.1) | |
| Unknown | 0 (0.0) | 1 (100.0) | |
| Lymph node metastasis | |||
| Negative | 146 (79.8) | 37 (20.2) | .0825 |
| Positive | 43 (68.3) | 20 (31.8) | |
| Not examined | 3 (75.0) | 1 (25.0) | |
| No. of lymph node metastases | |||
| 0 | 146 (79.8) | 37 (20.2) | .349 |
| 1‐3 | 24 (66.7) | 12 (33.3) | |
| 4‐9 | 11 (68.8) | 5 (31.3) | |
| 10 and more | 8 (72.7) | 3 (27.3) | |
| Not examined | 3 (75.0) | 1 (25.0) | |
| Tumor grade | |||
| 1 | 115 (77.2) | 34 (22.8) | .7435 |
| 2 + 3 | 65 (79.3) | 17 (20.7) | |
| Unknown | 12 (63.2) | 7 (36.8) | |
| Estrogen receptor | |||
| Positive | 162 (78.6) | 44 (21.4) | .1676 |
| Negative | 30 (68.2) | 14 (31.8) | |
| HER2 status | |||
| Negative | 175 (79.6) | 45 (20.5) | .0178 |
| Positive | 17 (58.6) | 12 (41.4) | |
| Unknown | 0 (0.0) | 1 (100.0) | |
| Ki67 expression levele | |||
| Low | 102 (78.5) | 28 (21.5) | 1.0000 |
| High | 87 (78.4) | 24 (21.6) | |
| Unknown | 3 (33.3) | 6 (66.7) | |
| Chemotherapy treatment | |||
| No | 134 (75.3) | 44 (24.7) | .5068 |
| Yes | 57 (80.3) | 14 (19.7) | |
| Unknown | 1 (100.0) | 0 (0.0) | |
Abbreviation: HER2, human epidermal growth factor receptor 2.
Data are shown as n (%) unless otherwise indicated.
High: ≥ 13.87 ng/mL, low: < 13.87 ng/mL.
Fischer's exact test (unknown cases were excluded.).
Male breast cancer patient
Low: < 20%, high: ≥ 20%.
3.2. Relationship between clinicopathological characteristics and serum CCL5 levels
Clinicopathological factors in CCL5‐high and ‐low cohorts were compared in Table 1. Patients in CCL5‐high cancers were significantly younger than those in the CCL5‐low group (median age, 58 vs 68 years; P = .0009) and CCL5‐high breast cancers significantly frequently had a small tumor size (P = .0495) and HER2‐negative status (P = .0178). There were no significant differences for other factors between CCL5‐high and ‐low breast cancers.
3.3. Disease‐free survival of patients according to serum CCL5 levels
The DFS of CCL5‐high patients was significantly better than for those that were CCL5‐low (4‐year DFS, 0.93 and 0.61, respectively; HR, 0.20; 95% CI, 0.10‐0.39; P < .0001; Figure 1). Similarly, there was a significant difference in the OS of CCL5‐high and ‐low patients (P = .024; Figure 1). In the subgroup analysis, CCL5‐high patients consistently had a better DFS than CCL5‐low patients, irrespective of subgroup (Figure S3).
Figure 1.
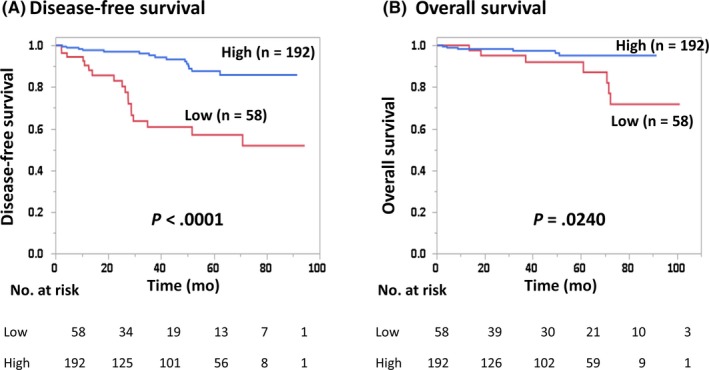
Disease‐free survival (A) and overall survival (B) of breast cancer patients with high and low serum C‐C motif chemokine ligand 5 (CCL5) levels
3.4. Univariable and multivariable analyses of prognostic factors of DFS, including CCL5 levels
Univariable analysis showed that menopausal status (P = .0380), tumor size (P = .0234), lymph node metastasis (P = .0001), tumor grade (P = .0024), Ki67 expression levels (P = .0017), CCL5 levels (P < .0001), and immunohistochemical staining of CCL5 (P = .0246) were significant prognostic factors for DFS (Table 2). We confirmed that serum CCL5 levels (HR, 0.28; 95% CI, 0.11‐0.70; P = .0068) was independent and significant prognostic factors for DFS as it was significant in the multivariable analysis (Table 2).
Table 2.
Univariable and multivariable analyses of disease‐free survival among breast cancer patients
| n | Univariable analysis HR (95% CI) | P‐value | Multivariable analysis HR (95% CI) | P‐value | |
|---|---|---|---|---|---|
| Menopausal status | |||||
| Pre‐ | 80 | 1.00 | .0380 | 1.00 | .0835 |
| Post‐ | 169 | 2.37 (1.05‐6.34) | 2.59 (0.89‐9.48) | ||
| Tumor size, cm | |||||
| ≤2 | 175 | 1.00 | .0234 | 1.00 | .7561 |
| >2 | 74 | 2.29 (1.12‐4.56) | 1.17 (0.41‐3.13) | ||
| Lymph node metastasis | |||||
| Negative | 183 | 1.00 | .0001 | 1.00 | .4570 |
| Positive | 63 | 4.11 (2.02‐8.48) | 1.47 (0.53‐4.12) | ||
| Tumor grade | |||||
| 1 | 149 | 1.00 | .0024 | 1.00 | .7291 |
| 2 + 3 | 82 | 2.97 (1.48‐6.08) | 1.21 (0.42‐3.86) | ||
| Estrogen receptor status | |||||
| Positive | 206 | 1.00 | .1581 | ||
| Negative | 44 | 1.78 (0.78‐3.71) | |||
| HER2 status | |||||
| Negative | 220 | 1.00 | .1148 | ||
| Positive | 29 | 2.06 (0.82‐4.51) | |||
| Ki67 expression levela | |||||
| Low | 130 | 1.00 | .0017 | 1.00 | .1067 |
| High | 111 | 3.10 (1.51‐6.82) | 2.76 (0.80‐9.71) | ||
| CCL5 levelb | |||||
| Low | 58 | 1.00 | <.0001 | 1.00 | .0068 |
| High | 192 | 0.20 (0.10‐0.39) | 0.28 (0.11‐0.70) | ||
| Immunohistochemical staining of CCL5c | |||||
| Low | 89 | 1.00 | .0246 | 1.00 | .1741 |
| High | 71 | 2.68 (1.14‐6.44) | 1.89 (0.75‐4.88) | ||
| Chemotherapy treatment | |||||
| No | 178 | 1.00 | .1924 | ||
| Yes | 71 | 1.62 (0.77‐3.26) | |||
Abbreviations: CCL5, C‐C motif chemokine ligand 5; CI, confidence interval; HER2, human epidermal growth factor receptor 2; HR, hazard ratio.
Low, <20%; high, ≥20%.
High, ≥13.87 ng/mL; low, <13.87 ng/mL.
Low: ≤20%, high >20%.
3.5. Disease‐free and overall survival of patients according to immunohistochemical expression of CCL5 levels in cancer cells
Positive staining of cytoplasm in invasive cancer cells by immune staining was evaluated as shown in Figure S4. Disease‐free survival of patients with CCL5‐high breast cancers (n = 71) was significantly worse than those with CCL5‐low breast cancers (n = 89) (P = .0194) (Figure 2). There was no significant difference in OS between CCL5‐high and ‐low patients (P = .1441) (Figure 2).
Figure 2.
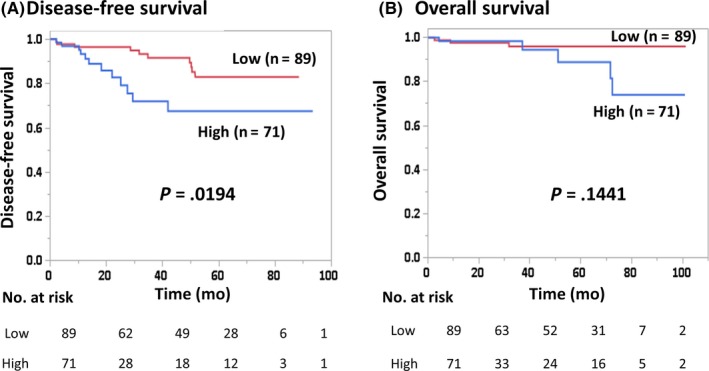
Disease‐free survival (A) and overall survival (B) of breast cancer patients according to the proportion of positive immunohistochemical staining of C‐C motif chemokine ligand 5 (CCL5) (high, >20%; low, ≤20%)
3.6. Correlations between serum CCL5 and immunohistochemical expression of CCL5 or TIL levels
There was no significant difference between serum levels of CCL5 and immunohistochemical staining of CCL5 in cancer cells separated by the 20% cut‐off value (P = .502; Figure 3). According to the further division of CCL5 immunohistochemical staining into 4 groups based on expression levels (0%, 1%‐49%, 50%‐69%, and 70% or higher), serum CCL5 levels in immunohistochemical CCL5‐positive groups (1% and above) were not significantly different when compared with the CCL5‐negative (0%) group (Figure 3).
Figure 3.
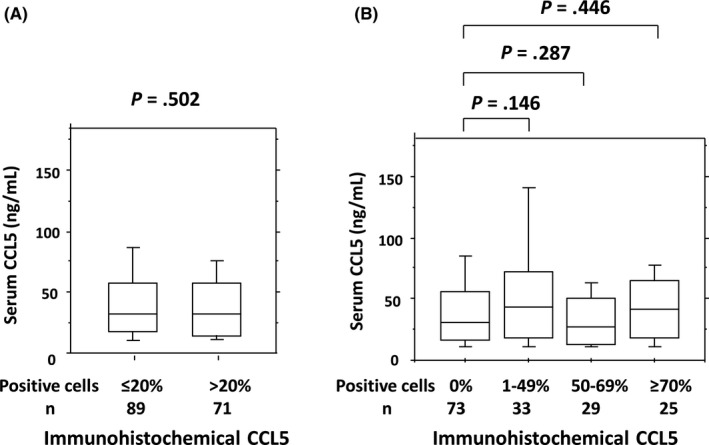
Correlation between serum C‐C motif chemokine ligand 5 (CCL5) levels and immunohistochemical staining of CCL5 in cancer cells. Positive cells were divided into (A) ≤20% (n = 89) or >20% (n = 71), and (B) 0% (n = 73), 1%‐49% (n = 33), 50%‐69% (n = 29), or ≥70% (n = 25)
Serum levels of CCL5 and immunohistochemical staining of CCL5 in cancer cells did not differ between TIL‐high and ‐low patients when all patients were considered (P = .257 or P = .427, Table S1). However, the frequency of patients with high serum levels of CCL5 was significantly higher in the TIL‐high group exclusively in the TN subtype. Furthermore, the CD8+ cells, FOXP3+ cells, or the FOXP3/CD8 ratio did not significantly associate with serum CCL5 levels. Both CD8+ cells and FOXP3+ cells were marginally increased in CCL5+ cancers (P = .0787 and P = .0769, respectively), but the FOXP3/CD8 ratio did not differ between CCL5+ and CCL5− cancers (P = .1625).
3.7. Associations of CCL5 gene expression levels with outcome and gene networks related to CCL5 in the public dataset
Using the Kaplan‐Meier Plotter public mRNA expression dataset, the prognostic significance of CCL5 mRNA expression was analyzed. Both the RFS and OS of patients with high CCL5 gene expression were significantly better than in patients with low CCL5 gene expression divided by the median value (Figure 4). Next, network analysis of CCL5 was carried out using the cBioPortal software. As shown in Figure 5, GNA11, GNB2, GNB4, GNG13, GNGT1, GRK6, SDC4, MYC, and MAX were found to be directly connected with CCL5. In addition, Spearman's correlations and P‐values of correlated genes are listed in Table S2. GRK6, GNB4, MAX, MYC, and GNGT1 were significantly and positively associated with CCL5. On the contrary, GNGA11, GNG13, and SDC4 were significantly connected to CCL5 in a negative manner.
Figure 4.
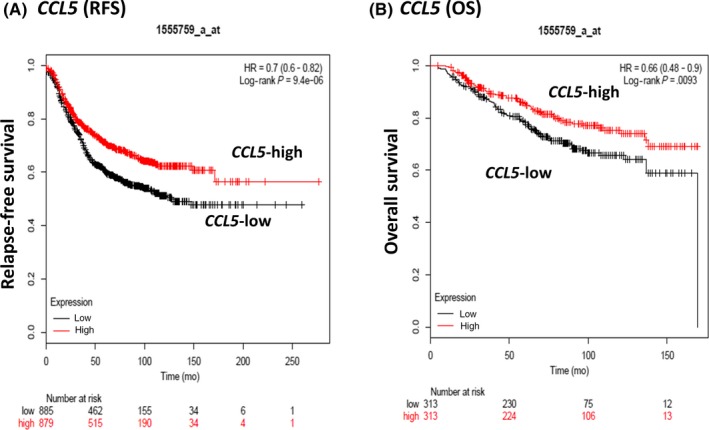
Relapse‐free survival (RFS) (A) and overall survival (OS) (B) of breast cancer patients according to CCL5 gene expression levels in the public dataset. Cut‐off value was set to the median value
Figure 5.
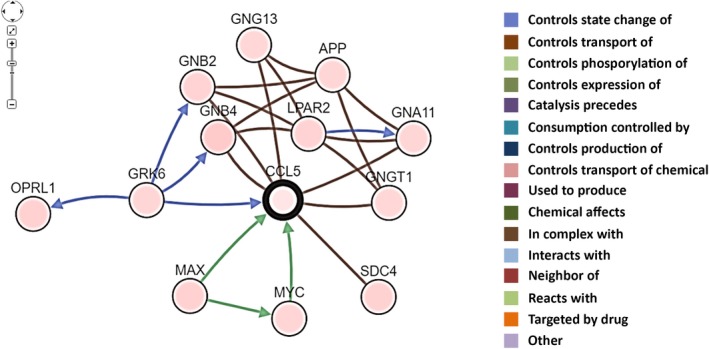
Schematic representation of network genes correlated with CCL5 in the cBioPortal (http://www.cbioportal.org/)
3.8. CCL5 mRNA expression levels in cancer and stromal cells
The mRNA expression levels of CCL5 were determined in normal breast tissue and in breast cancer tissues separated by laser capture microdissection considering 30 TN breast cancers in the public dataset. The relative expression levels of CCL5 were not significantly different between normal breast and breast cancer tissues (P = .079, Figure S5A). Next, we evaluated CCL5 expression levels in breast cancer tissues that were separated using the microdissection method into stromal cells and cancer cells regarding the public dataset including breast cancers from pregnant and nonpregnant patients. Expression levels of CCL5 in stromal cells were significantly higher than those in cancer cells (P = .0064) (Figure S5B).
4. DISCUSSION
In the present study, we found that the DFS of patients with high levels of CCL5 at baseline was significantly better than in those with low CCL5. This significant association between CCL5 and prognosis seems to be consistent irrespective of subtypes. It has been reported that β‐catenin induction results in an aggressive CCL5‐mediated phenotype in breast cancer that displays increased cell invasion and spheroid formation in breast cancer cells.33 Similarly, cooperative induction of CCL5 and IL‐6 induces an aggressive phenotype in breast cancer cells.22 Based on these studies, CCL5 has a direct role in the progression and metastasis of breast cancers. In line with these results, a poor patient prognosis is associated with high expression of CCL5 in breast cancers as determined by quantitative protein analysis.25 We identified a worse prognosis of patients with high expression of CCL5 in cancer cells, as determined by immunohistochemical staining. Contrary to these results, patients with high serum levels of CCL5 had a significantly improved DFS. The latter result was further supported by previous studies26, 33 as well as by the results obtained from the analysis of the public dataset presented in this study.
As mentioned above, it is speculated that CCL5 expressed in breast cancer cells might contribute to progression and enhance metastatic potential. However, Jayasinghe et al18 reported that tumor‐derived CCL5 had no impact on breast cancer growth rate or metastatic ability. In addition to cancer cells, CCL5 is derived from mesenchymal stem cells17 and peritumoral adipose tissue.34 As CCL5 is a chemokine, several functions associated with the immune reaction generated from cells other than cancer cells might also be involved in cancer progression and, thus, influence poor prognosis. It has been reported that CCL5 plays an important role mediating the migration of several sets of immune cells, including T lymphocytes, monocytes, eosinophils, NK cells, and macrophages.35, 36 In ER‐positive breast cancer, CCL5 increases infiltration of tumor‐associated macrophages.37 Furthermore, CCL5 leads to polarization of CD4+ cells into an immunosuppressive Th2 phenotype, which promotes metastasis in luminal breast cancer.38
In contrast to these immunosuppressive effects, CCL5 also increases migration and recruitment of lymphocytes into tumors. Upregulation of CCL5 induction as a result of microRNA‐21 inhibition promotes lymphocyte migration in MCF‐7 breast cancer cells.39 Considering that infiltration of CD8+ effector T cells into inflamed melanoma tissues depends on chemokines, including CCL5,40 CCL5 could play a role in immune‐mediated tumor regression. Furthermore, intratumoral injection of interferon‐beta induces the expression of CCL5 in melanoma and leads to the recruitment of CD8+ T cells.41 Recently, Araujo et al26 reported that high expression of CCL5 is associated with recruitment of CD8+ T cells, including expressions of CD8A and CD8B, activated CD4+ T cells, activated NK cells, and M1 macrophages. These results suggest the antitumor effects of CCL5 were induced by T cell‐mediated immunity. In line with this speculation, a positive correlation between CCL5 and TIL count was observed (P = .003).26 Interestingly, TIL count, but not CCL5 expression, was a significant and independent factor for distant RFS (HR, 0.336; 95% CI, 0.150‐0.753; P = .008) in a multivariable analysis, although both factors were significant in a univariable analysis.26
A positive association between CCL5 and TILs was consistent in the TN subtype but not in other subtypes in our study. The mechanism for these discrepant results depending on subtypes is currently unknown. According to the TIL gene signature, factors related to good prognosis, including CD8+ T cells, B cells, M1 macrophages, and DCs, were mainly involved in ER‐negative breast cancers.42 On the contrary, an abundance of molecules linked with poor prognosis, such as Tregs, MDSCs, and neutrophils, are related to ER‐positive breast cancers. The differences in the immune microenvironment of TN and other subtypes might partly explain discrepant association results between CCL5 and TILs. However, as the prognostic significance of serum CCL5 levels was obtained in this study regardless of the cancer subtype, CCL5 might be associated with prognosis, at least partly, due to the mediation of functions other than immunity against breast cancers.
On the basis of network analysis, we determined a positive link between CCL5 and GNB4, GNGT1, GRK6, Myc, and MAX (Figure 5; Table S2). In addition, we have encountered a negative association between CCL5 and GNA11, GNG13, and SDC4. Umar et al reported that the prognosis of patients with high GNB4 levels was significantly better than that of those with low levels in breast cancers treated with tamoxifen.43 In a mouse model, GRK6 deletion upregulated tumor‐infiltrating polymorphonuclear leukocyte and microvessel density, resulting in tumor progression and metastasis.44 In addition to the oncogenic function of Max in cooperation with Myc, MAX and the mitotic arrest deficient (Mad) network have been shown to antagonize Myc function through the mediation of proliferation, differentiation, and apoptosis.45 According to the Kaplan‐Meier Plotter database, high levels of Max, but not of Myc, are significantly associated with a better prognosis, which is consistent with CCL5 (data not shown), thus Max might contribute to favorable breast cancer prognosis. Additionally, SDC4 has been reported to promote transforming growth factor‐β1‐induced epithelial to mesenchymal transition in lung adenocarcinoma cells46 and the upregulation of GNA11 in metastatic cancers when compared with primary TN breast cancers.47 As the mRNA expression of SDC4 and GNA11 is negatively associated with CCL5, these studies could be in line with a good prognosis of CCL5‐high breast cancers mediated through low SDC4 and GNA11. This, CCL5 could contribute for the progression of breast cancers in cooperation with these molecules. The precise mechanisms underlying the prognostic significance of CCL5 remain to be determined, although multifunctional effects, including immune reactions and other functions mentioned above, are speculated to be involved.
Both expression of CCL5 in cancer cells and serum levels of CCL5 were linked with prognosis, but mediated through different mechanisms; based on multivariable analysis, expression of CCL5 in serum but not in cancer cells was found to be a significant and independent prognostic factor (Table 2). As serum levels of CCL5 in invasive breast cancers were lower than those in metastatic breast cancers (Figure S1), we speculate that CCL5 serum levels are not due to nonspecific inflammation, but to a cancer‐specific reaction. As shown in Figure 3, there was no significant association between the immunohistochemical expression of CCL5 in cancer cells and serum CCL5 levels. Even in breast cancer cells with no CCL5 expression, the serum CCL5 levels were similar to those in CCL5‐positive cancers. Furthermore, according to the samples analyzed by microdissection, mRNA expression levels of CCL5 in stromal cells were significantly higher when compared to those in cancer cells (Figure S5). Considering these results, we believe the great majority of serum CCL5 was derived not from cancer cells, but from stromal cells. The mRNA expression levels from the public database shown in Figure 4 might include both cancer and stromal cells; a positive association between CCL5 and prognosis was consistent with serum CCL5, but not with the immunohistochemical expression of CCL5 in cancer cells. Accordingly, evaluation of total CCL5 levels in the blood, including production by both cancer cells and other tissues, appears to be superior to examination of expression in cancer cells. To the best of our knowledge, this is the first study to reveal the prognostic significance of serum levels of CCL5 in breast cancers. The limitation of this study is that we were unable to determine a direct association between serum CCL5 and expression in stromal cells because most of the paraffin‐embedded tumor samples contained mainly cancer cells and inadequate stromal cells, especially in surrounding areas, and detailed analysis in stromal cells is infeasible. In addition, the functional mechanisms of CCL5 in favorable prognosis remain unsolved. These issues need to be addressed in future studies evaluating a larger sample population.
We showed that high serum CCL5 levels were significantly associated with improved prognosis of patients with early breast cancer. CCL5 could be associated with prognosis through multiple effects involving immune reactions and other factors. These data might be useful for predicting patient prognosis in clinical practice.
DISCLOSURE
Yasuo Miyoshi received lecture fee and research funds from Chugai, AstraZeneca, Eli Lilly, MSD, Kyowa‐Kirin, Taiho, Pfizer, and Eisai. Other authors declare no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
We thank Editage for English language editing.
Fujimoto Y, Inoue N, Morimoto K, et al. Significant association between high serum CCL5 levels and better disease‐free survival of patients with early breast cancer. Cancer Sci. 2020;111:209–218. 10.1111/cas.14234
Funding information
This study was supported by a Grant‐in‐Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (No. 15K10077) to M. Imamura.
REFERENCES
- 1. Burkholder B, Huang R‐Y, Burgess R, et al. Tumor‐induced perturbations of cytokines and immune cell networks. Biochim Biophys Acta. 2014;1845:182‐201. [DOI] [PubMed] [Google Scholar]
- 2. Ostrand‐Rosenberg S. Immune surveillance: a balance between pro‐ and anti‐tumor immunity. Curr Opin Genet Dev. 2008;18:11‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang X, Shapiro DJ. The immune system and inflammation in breast cancer. Mol Cell Endocrinol. 2014;382:673‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luen SJ, Savas P, Fox SB, Salgado R, Loi S. Tumor‐infiltrating lymphocytes and the emerging role of immunotherapy in breast cancer. Pathology. 2017;49:141‐155. [DOI] [PubMed] [Google Scholar]
- 5. Denkert C, von Minckwitz G, Darb‐Esfahani S, et al. Tumour‐infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40‐50. [DOI] [PubMed] [Google Scholar]
- 6. Mao Y, Zu Z, Chen X, et al. The prognostic value of tumor‐infiltrating lymphocytes in breast cancer: a systematic review and meta‐analysis. PLoS One. 2016;11(4):e0152500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei B, Yao M, Xing C, et al. The neutrophil lymphocyte ratio is associated with breast cancer prognosis: an updated systematic review and meta‐analysis. Onco Targets Ther. 2016;9:5567‐5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ethier JL, Desautels D, Templeton A, Shah PS, Amir E. Prognostic role of neutrophil‐to‐lymphocyte ratio in breast cancer: a systematic review and meta‐analysis. Breast Cancer Res. 2017;19:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490‐500. [DOI] [PubMed] [Google Scholar]
- 10. Theresa LW. The role of regulatory T cells in cancer immunology. Immunotargets Ther. 2015;4:159‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suppan C, Bjelic‐Radisic V, Garde ML, et al. Neutrophil/Lymphocyte ratio has no predictive or prognostic value in breast cancer patients undergoing preoperative systemic therapy. BMC Cancer. 2015;15:1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen ZY, Raghav K, Lieu CH, et al. Cytokine profile and prognostic significance of high neutrophil‐lymphocyte ratio in colorectal cancer. Br J Cancer. 2015;112:1088‐1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahn Y‐T, Huang B, McPherson L, Clayberger C, Krensky AM. Dynamic interplay of transcriptional machinery and chromatin regulates “late” expression of the chemokine RANTES in T lymphocytes. Mol Cell Biol. 2007;27:253‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nelson PJ, Kim HT, Manning WC, Goralski TJ, Krensky AM. Genomic organization and transcriptional regulation of the RANTES chemokine gene. J Immunol. 1993;151:2601‐2612. [PubMed] [Google Scholar]
- 15. Devergne O, Marfaing‐Koka A, Schall TJ, et al. Production of the RANTES chemokine in delayed‐type hypersensitivity reactions: involvement of macrophages and endothelial cells. J Exp Med. 1994;179:1689‐1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Antczak AJ, Vieth JA, Singh N, Worth RG. Internalization of IgG‐coated targets results in activation and secretion of soluble CD40 ligand and RANTES by human platelets. Clin Vaccine Immunol. 2011;18:210‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557‐563. [DOI] [PubMed] [Google Scholar]
- 18. Jayasinghe MM, Golden JM, Nair P, O'Donnell CM, Werner MT, Kurt RA. Tumor‐derived CCL5 does not contribute to breast cancer progression. Breast Cancer Res Treat. 2008;111:511‐521. [DOI] [PubMed] [Google Scholar]
- 19. Velasco‐Velázquez M, Pestell RG. The CCL5/CCR5 axis promotes metastasis in basal breast cancer. Oncoimmunology. 2013;2:e23660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y, Ly D, Kim HJ, et al. A novel role of hematopoietic CCL5 in promoting triple‐negative mammary tumor progression by regulating generation of myeloid‐derived suppressor cells. Cell Res. 2013;23:394‐408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sax MJ, Gasch C, Athota VR, et al. Cancer cell CCL5 mediates bone marrow independent angiogenesis in breast cancer. Oncotarget. 2016;7:85437‐85449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gallo M, Frezzetti D, Roma C, et al. RANTES and IL‐6 cooperate in inducing a more aggressive phenotype in breast cancer cells. Oncotarget. 2018;9:17543‐17553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khalid A, Wolfram J, Ferrari I, Mu C, Mai J, Yang Z. Recent advances in discovering the role of CCL5 in metastatic breast cancer. Mini Rev Med Chem. 2015;15:1063‐1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang T, Wei Y, Tian L, et al. C‐C motif chemokine ligand 5 (CCL5) levels in gastric cancer patient sera predict occult peritoneal metastasis and a poorer prognosis. Int J Surg. 2016;32:136‐142. [DOI] [PubMed] [Google Scholar]
- 25. Zhu YY, Chen C, Li JJ, Sun SR. The prognostic value of quantitative analysis of CCL5 and collagen IV in luminal B (HER2‐) subtype breast cancer by quantum‐dot‐based molecular imaging. Int J Nanomedicine. 2018;13:3795‐3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Araujo JM, Gomez AC, Aguilar A, et al. Effect of CCL5 expression in the recruitment of immune cells in triple negative breast cancer. Sci Rep. 2018;8:4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Watanabe T, Hida AI, Inoue N, et al. Abundant tumor infiltrating lymphocytes after primary systemic chemotherapy predicts poor prognosis in estrogen receptor‐positive/HER2‐negative breast cancers. Breast Cancer Res Treat. 2018;168(1):135‐145. [DOI] [PubMed] [Google Scholar]
- 28. Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725‐731. [DOI] [PubMed] [Google Scholar]
- 29. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Komatsu M, Yoshimaru T, Matsuo T, et al. Molecular features of triple negative breast cancer cells by genome‐wide gene expression profiling analysis. Int J Oncol. 2013;42:478‐506. [DOI] [PubMed] [Google Scholar]
- 32. Harvell DM, Kim J, O'Brien J, et al. Genomic signatures of pregnancy‐associated breast cancer epithelia and stroma and their regulation by estrogens and progesterone. Horm Cancer. 2013;4:140‐153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yasuhara R, Irie T, Suzuki K, et al. The β‐catenin signaling pathway induces aggressive potential in breast cancer by up‐regulating the chemokine CCL5. Exp Cell Res. 2015;338:22‐31. [DOI] [PubMed] [Google Scholar]
- 34. D'Esposito V, Liquoro D, Ambrosio MR, et al. Adipose microenvironment promotes triple negative breast cancer cell invasiveness and dissemination by producing CCL5. Oncotarget. 2016;7:24495‐24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Romagnani S. Cytokines and chemoattractants in allergic inflammation. Mol Immunol. 2002;38:881‐885. [DOI] [PubMed] [Google Scholar]
- 36. Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Svensson S, Abrahamsson A, Rodriguez GV, et al. CCL2 and CCL5 are novel therapeutic targets for estrogen‐dependent breast cancer. Clin Cancer Res. 2015;21:3794‐3805. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Q, Qin J, Zhong L, et al. CCL5‐mediated Th2 immune polarization promotes metastasis in luminal breast cancer. Cancer Res. 2015;75:4312‐4321. [DOI] [PubMed] [Google Scholar]
- 39. Wang Z, Han J, Cui Y, Zhou X, Fan K. miRNA‐21 inhibition enhances RANTES and IP‐10 release in MCF‐7 via PIAS3 and STAT3 signalling and causes increased lymphocyte migration. Biochem Biophys Res Comm. 2013;439:384‐389. [DOI] [PubMed] [Google Scholar]
- 40. Harlin H, Meng Y, Peterson AC, et al. Chemokine expression in melanoma metastases associated with CD8+ T‐cell recruitment. Cancer Res. 2009;69:3077‐3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Uehara J, Ohkuri T, Kosaka A, et al. Intratumoral injection of IFN‐β induces chemokine production in melanoma and augments the therapeutic efficacy of anti‐PD‐L1 mAb. Biochem Biophys Res Comm. 2017;490:521‐527. [DOI] [PubMed] [Google Scholar]
- 42. Hammerl D, Smid M, Timmermans AM, Sleijfer S, Martens JWM, Debets R. Breast cancer genomics and immuno‐oncological markers to guide immune therapies. Semin Cancer Biol. 2018;52:178‐188. [DOI] [PubMed] [Google Scholar]
- 43. Umar A, Kang H, Timmermans AM, et al. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol Cell Proteomics. 2009;8:1278‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raghuwanshi SK, Smith N, Rivers EJ, et al. G protein‐coupled receptor kinase 6 deficiency promotes angiogenesis, tumor progression, and metastasis. J Immunol. 2013;190:5329‐5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rottmann S, Lüscher B. The Mad side of the Max network: antagonizing the function of Myc and more. Curr Top Microbiol Immunol. 2006;302:63‐122. [DOI] [PubMed] [Google Scholar]
- 46. Toba‐Ichihashi Y, Yamaoka T, Ohmori T, Ohba M. Up‐regulation of Syndecan‐4 contributes to TGF‐β1‐induced epithelial to mesenchymal transition in lung adenocarcinoma A549 cells. Biochem Biophys Rep. 2015;5:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin FM, Yost SE, Wen W, et al. Differential gene expression and AKT targeting in triple negative breast cancer. Oncotarget. 2019;10:4356‐4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


