Abstract
Background
Vaginal and seminal microbiomes have gained increasing interest for their involvement in reproductive health and fertility. However, their role in reproductive outcome is not fully understood yet. In this study, we aimed to correlate the vaginal and the seminal microbiome of 23 couples with idiopathic infertility to the clinical pregnancy rate after intrauterine insemination (IUI).
Methods
Vaginal swabs and seminal fluids were collected on the day of IUI procedure and analyzed through polymerase chain reaction amplification of variable regions 3 and 4 (V3–V4) of 16S ribosomal ribonucleic acid genes and Illumina MiSeq sequencing. The taxonomic data were then correlated to IUI success.
Results
Idiopathic infertile women showed a different average composition of vaginal microbiome compared with control sequences, whereas for seminal counterpart no relevant differences were observed. Furthermore, among idiopathic infertile women, different patterns of Lactobacillus species dominations were observed, with a predominance either of Lactobacillus crispatus, a marker of a healthy vaginal ecosystem, or of Lactobacillus iners and Lactobacillus gasseri, associated with a more dysbiosis-prone environment. More important, considering all investigated variables, vaginal L crispatus domination was the only factor strongly associated to IUI success (P = .0002).
Conclusions
Our results strengthen the potential role of L crispatus in promoting a favorable environment for pregnancy and suggest that microbiome characterization could be useful, together with standard clinical and laboratory assessments, in the pre-IUI evaluation of infertile couples.
Keywords: idiopathic infertility, intrauterine insemination (IUI) success, seminal microbiome, vaginal lactobacilli, vaginal microbiome
The vaginal microbiota plays a pivotal role in maintaining the physiological homeostasis of the environment and protects from the colonization by opportunistic pathogens [1]. A healthy vaginal ecosystem is characterized by a predominance of Lactobacillus species, mainly Lactobacillus crispatus, Lactobacillus iners, Lactobacillus gasseri, and Lactobacillus jensenii [2], which actively contribute to lower vaginal pH (<4.5) through lactic acid production. However, Lactobacillus species differ in their ability to promote the stability of vaginal microflora: community states dominated by L crispatus are associated with lower pH values (<4.0), whereas L gasseri and L iners are associated with higher pH values (>4.0) and with a less stable vaginal microflora [3]. Vaginal dysbiosis can result in bacterial vaginosis (BV), the most common vaginal syndrome of reproductive-age women. Besides representing a risk factor for acquiring sexually transmitted infections [4, 5], BV is known to be among the causes of adverse pregnancy outcome [6] and pelvic inflammatory disease [7].
Differently from the vaginal microbiota, semen is characterized by a polymicrobial flora with high species richness and variability [8]. Alterations in seminal microbiota have been related to reduced quality of semen [9] and to genitourinary infections, potentially responsible for up to 15% of cases of male infertility [8]. A recent study described the correlation between the differential abundance in semen of specific bacterial genera, such as Prevotella, Staphylococcus, and Lactobacillus, with abnormal sperm motility and morphology [10].
According to the World Health Organization (WHO), infertility is defined as a couple’s failure to achieve pregnancy after 1 year of regular and unprotected intercourses [11]. Up to 30% of infertile couples are diagnosed with idiopathic or unexplained infertility, a condition characterized by the absence of a definable cause after evaluation of tubal patency, ovulatory function, and semen analysis [12]. Commonly, in young women, intrauterine insemination (IUI) is proposed as a first-line fertility treatment for idiopathic infertility.
Recent evidences suggest a possible association between an alteration of vaginal and seminal microbiota and infertility [9, 13, 14]. Evidence from several studies shows a differential vaginal microbiota composition in infertile patients compared with healthy and fertile women [14–16]. An abnormal vaginal microbiota has been associated with a poor reproductive outcome in patients undergoing in vitro fertilization [17], as well as with early spontaneous abortion in patients undergoing in vitro fertilization (IVF) treatment [18]. However, the relationship of genital microbiome composition and reproductive outcomes is not fully ascertained yet. In this study, we aimed to characterize vaginal and seminal microbiome in 23 couples with idiopathic infertility undergoing IUI treatment, correlating it to the clinical pregnancy rate after IUI.
METHODS
Patients Recruitment and Sample Collection
This observational prospective study included 25 consecutive couples with primary idiopathic infertility undergoing their first IUI treatment at Centro Scienze della Natalità of Ospedale San Raffaele, Milan, Italy, from May 2015 to November 2017. Of these, we recorded 2 drop outs for independent reasons obtaining a final sample size of n = 23 couples. All participants were of Caucasian ethnicity and with reproductive age (33 ± 3 years for women, 34 ± 4 years for men). A comprehensive physiological and medical history has been collected for both members of every couple. Female diagnostic work-up has included several clinical variables such as measured body mass index (BMI), smoking, along with a basal (day 3–5 of the menstrual cycle) hormonal profile assessment (follicle-stimulating hormone [FSH], luteinizing hormone, thyroid-stimulating hormone, anti-Müllerian hormone, and prolactin), and a single assessment of progesterone (midluteal phase). All women underwent a hysterosonosalpingogram to assess tubal patency. At least 2 different semen analyses were performed. Exclusion criteria included the presence of female/male hormonal diseases, such as polycystic ovary syndrome, endometriosis, tubal pathology, and/or oligoasthenoteratozoospermia, according to WHO 2010 criteria [11].
Controlled ovarian stimulation was performed using a daily low dose (50–75 international units [IU]) of recombinant-FSH (Gonal-F; Merck, Milano, Italy). When a maximum of 2 follicles of diameter >16 mm were observed, trigger of ovulation was induced with 5000 IU of human chorionic gonadotropin (hCG) (Gonasi; IBSA, Lodi, Italy), and IUI was scheduled after 36 hours.
The study was approved by the Institutional Ethical Committee, and an informed consent (OSR/MICROBIOMA 10/12/2014 version 1.0) was obtained from all subjects before sampling. Vaginal samples were collected under direct visualization from the posterior fornix using a BBL CultureSwab MaxV Liquid Amies swab (Becton, Dickinson and Company, Oxford, UK). Semen specimens were collected by masturbation after 2–4 days of sexual abstinence and stored in sterile microcentrifuge tubes. Both vaginal and seminal samples were collected on the day of IUI procedure and immediately frozen and stored at −80°C until further processing for the study.
For sample size calculation, 22 couples were considered to achieve a test power of 0.75 and with an effect size of 1.53 and significance level of 0.05. Based on our previous clinical experience, we considered an overall sample of 25 patients to take into account a drop probability of 0.14. At the end, only 2 drop outs were recorded.
Sample Processing and Illumina MiSeq Sequencing
Vaginal swabs were resuspended in transport buffer (Amies Liquid Medium) by vortexing. Processing of semen samples was performed on total ejaculates. Semen samples were processed using a density gradient (Sperm Gradient Kit; Cook Medical, Limerick, Irleand) following the manufacturer’s protocol. Deoxyribonucleic acid (DNA) extraction was performed from vaginal and seminal samples with QIAamp BiOstic Bacteremia DNA kit (QIAGEN, Milano, Italy) following the manufacturer’s protocol. The concentration of extracted DNA was measured using a Qubit double-stranded DNA (dsDNA) HS Assay Kit (Qubit) on a Qubit 2.0 Fluorometer (Qubit; Life Technologies Corporation, Carlsbad, CA). Variable regions 3 and 4 (V3–V4) of 16S ribosomal ribonucleic acid (rRNA) genes were polymerase chain reaction (PCR) amplified as previously reported [19]. Amplicons were purified with Agencourt AMPure XP PCR Purification Kit (Beckman Coulter, Indianapolis, IN), and the size of the amplicon library was assessed on the Agilent Bioanalyzer 2100 using DNA High Sensitivity kit (Agilent Technologies, Santa Clara, CA). The amplicon library was diluted to 4 nM and pooled after 16S Metagenomic Sequencing Library Preparation protocol (Illumina, San Diego, CA). Fifteen percent of PhiX Control library (v3; Illumina) was combined with the pooled library. The libraries were sequenced on the MiSeq Illumina platform with a 300-base pairs (bp) paired-end read protocol. Data quality trimming and reads demultiplexing were performed on the MiSeq instrument.
For bacterial DNA quantification, a real-time PCR was performed with an ABI Prism 7000 Sequence Detection System thermalcycler (Applied Biosystem) using TaqMan Gene Expression Master Mix and TaqMan Gene Expression probe for pan-bacterial detection of 16S rRNA (Ba04230899_s1; Thermo Fisher Scientific). To create the standard curve, we did 10-fold dilution series starting from a known load of Escherichia coli Dh5α. We used the regression line derived from the standard curve to determine the copy number of the unknown samples, and we normalized the number of 16S copies to the total ng of loaded DNA.
Sequence Analysis
Sequence reads processing was performed using QIIME (Quantitative Insights Into Microbial Ecology, version 1.9.0) [20]. Paired-end reads were assembled with join_paired_ends.py. Operational taxonomic units (OTUs) were defined at 97% similarity using open-reference OTU picking [21]. Taxonomic classifications were assigned to each sequence using Ribosomal Database Project (RDP) Naive Bayesian Classifier v.2.2 [22] trained on the Greengenes database. For taxonomic analysis, bacterial taxa with a relative abundance <1% were excluded. Rarefaction analysis was performed with alpha_rarefaction.py for the Shannon index to assess alpha diversity, and paired t test was used to compare the mean Shannon indices between groups.
For descriptive purposes, we compared our taxonomic results with sequences related to healthy subjects. For the vaginal microbiome analysis, sequences from the Vaginal 16S rDNA Reference Database deposited at NCBI’s Sequence Read Archive (BioProject ID: PRJNA46877) were used as control [23]. For the seminal microbiome, we used sequences from healthy male subjects that we analyzed in a previous study (NM, RP, EP, MCl, MCa [2016] unpublished data). Clinical parameters of control male subjects are reported in Supplementary Table S2. For both vaginal and seminal control sequences, the metagenomics analysis was performed as described above.
Statistical Analysis
Statistical analysis was conducted with R software version 3.5.0 (http://www.r-project.org) and SPSS version 25. Hierarchical clustering was performed in R with hclust function to group women according to relative abundance of Lactobacillus species. Independent-samples Mann-Whitney U test was performed to correlate anamnestic and clinical parameters of both partners with IUI pregnancy outcome. To compare taxonomic data with clinical variables, we performed Mann-Whitney U test with exact P values computed by permutation methods. To compare the microbial composition profile observed in the clusters of interest, we performed a multivariate ADONIS analysis considering age, BMI, and sperm count as possible confounding factors. Exact P values were calculated by permutation analysis and subsequently corrected for multiple comparisons with false discovery rate (FDR). Kruskal-Wallis test was used to compare Shannon diversity indexes for the investigated groups.
RESULTS
A total of 8.945.927 sequences were obtained from all vaginal (23) and seminal (23) samples, with a mean length (±standard deviation [SD]) of 464 (± 5.3) bp after joining of paired-end reads. Only taxa featuring >1% relative abundance were considered in the taxonomic analysis. No significant differences were observed in overall bacterial load within vaginal and seminal samples (Supplementary Figure S1 and Supplementary Table S1).
Both vaginal and seminal microbiome data were compared with control sequences. For the vaginal samples, sequences from healthy women were downloaded from the Vaginal 16S rDNA Reference Database [23]; whereas for seminal samples, sequence reads from healthy male subjects, already present in our dataset from a previous unpublished study, were used as control. The clinical parameters of control male subjects are reported in Supplementary Table S2.
Microbiome data were correlated to several anamnestic and clinical parameters. All taxonomic data, patients’ baseline characteristics, and ovulation induction parameters were then correlated to the clinical pregnancy rate after IUI (Figure 1). Clinical characteristics and laboratory values of the cohort of the infertile couples are reported in Table 1.
Figure 1.
Study design. BMI, body mass index; IUI, intrauterine insemination.
Table 1.
Demographics and Characteristics of the Patientsa
| IUI Success (n = 5) | No IUI Success (n = 18) | ||||
|---|---|---|---|---|---|
| Female Partners | Mean | SD | Mean | SD | P Value |
| Age (years) | 31.60 | 3.45 | 33.39 | 3.45 | .11 |
| Weight (kg) | 55.00 | 10.27 | 58.72 | 10.27 | .54 |
| BMI | 20.64 | 3.43 | 21.51 | 3.43 | .36 |
| FSH (mIU/mL) | 6.02 | 1.65 | 6.72 | 1.65 | .76 |
| LH (mIU/mL) | 4.98 | 2.13 | 6.42 | 2.13 | .41 |
| PRL (mIU/mL) | 14.18 | 100.10 | 51.66 | 100.10 | .68 |
| TSH (mIU/mL) | 1.65 | 0.91 | 1.98 | 0.91 | .65 |
| AMH (ng/mL) | 5.52 | 1.42 | 2.67 | 1.42 | .06 |
| Follicle number | 1.40 | 0.55 | 1.22 | 0.55 | .81 |
| E2 (pg/mL) | 294.75 | 145.87 | 293.53 | 145.87 | .73 |
| Endometrial thickness (mm) | 7.86 | 2.06 | 7.91 | 2.06 | .64 |
| Male Partners | |||||
| Age (years) | 38.80 | 5.81 | 34.50 | 4.09 | .20 |
| Semen volume (mL) | 2.10 | 1.14 | 3.09 | 1.17 | .18 |
| Sperm preconcentration (106/mL) | 67.00 | 35.99 | 33.38 | 17.88 | .05 |
| Sperm premotility (%progressive) | 43.00 | 10.37 | 47.50 | 15.28 | .39 |
| Sperm postconcentration (106/mL) | 48.00 | 31.14 | 42.00 | 31.50 | .60 |
| Sperm postmotility (%progressive) | 61.00 | 8.94 | 59.67 | 14.20 | .87 |
Abbreviations: AMH, anti-Müllerian hormone; BMI, body mass index; E2, estradiol; FSH, follicle-stimulating hormone; IUI, intrauterine insemination; LH, luteinizing hormone; post, after semen capacitation (used for IUI treatment); pre, before semen capacitation; PRL, prolactin; SD, standard deviation; TSH, thyroid-stimulating hormone.
aCorrelation of anamnestic, clinical and laboratory features of female and male partners with IUI pregnancy outcome.
Vaginal and Seminal Microbiome Composition
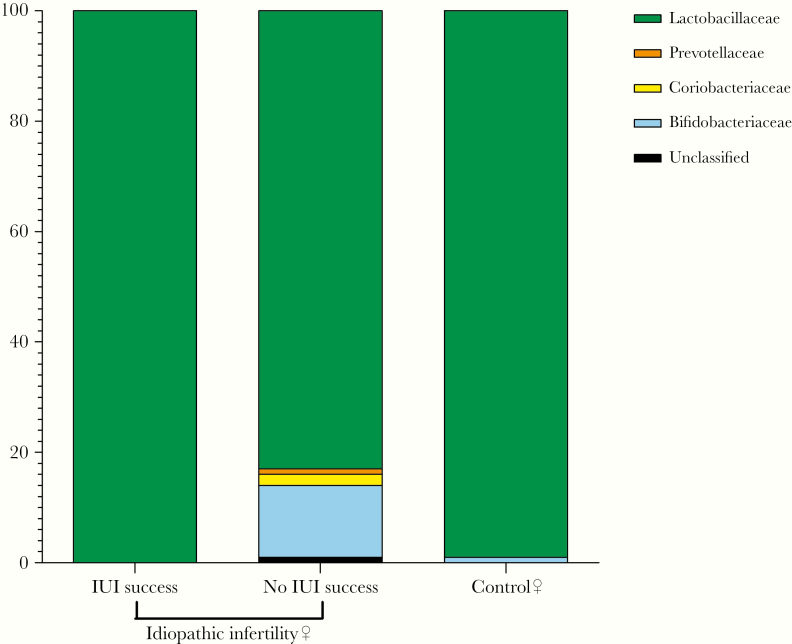
Vaginal microbiome of women with idiopathic infertility showed a different composition compared with that of control sequences (Figure 2), with an increase in the diversity of taxa and a reduction in Lactobacillaceae together with an increase in Bifidobacteriaceae. The same changes in microbial composition were observed at the genus level, with a marked reduction in Lactobacillus and an increase in Bifidobacterium among idiopathic infertile women. Although potentially relevant under a biological point of view, these differences do not reach the statistical significance.
Figure 2.
Vaginal microbiome taxonomic profile at family level. Comparison between women with idiopathic infertility, stratified by intrauterine insemination (IUI) outcome, and controls.
As shown in Supplementary Figure S2, only 3 women featured a relevant increase in Bifidobacteriaceae, showing a vaginal microbiome pattern similar to that observed during vaginal dysbiosis. To investigate this point, we performed the analysis at the species level for Bifidobacterium genus, and we identified Bifidobacterium breve and Gardnerella vaginalis as the most abundant species in these women (Supplementary Table S3).
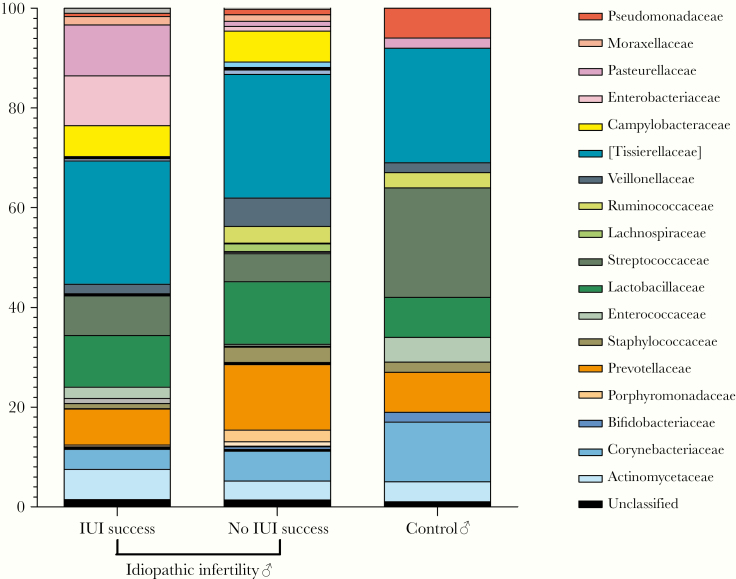
For the male counterpart, no significant differences were observed between the seminal microbiota of men with idiopathic infertility and controls (Figure 3). In both groups, the most abundant taxa at family level were [Tissierellaceae], followed by Lactobacillaceae, Streptococcaceae, Prevotellaceae, and Corynebacteriaceae (Figure 3). For both female and male partners, no significant correlations were detected between the microbiome data and any of the recorded clinical variables.
Figure 3.
Seminal microbiome taxonomic profile at family level. Comparison between men with idiopathic infertility, stratified by intrauterine insemination (IUI) outcome, and controls.
Correlation of Vaginal and Seminal Microbiome to Intrauterine Insemination Outcome
Five of the 23 women included in our study had a successful pregnancy after the IUI procedure. None of the investigated clinical variables correlated with IUI pregnancy outcome (Table 1). Only sperm concentration, before sperm separation, was higher in the male partners of the couples experiencing IUI success, with a borderline statistical significance (Table 1).
It is interesting to note that the vaginal microbial composition showed a different pattern between the 2 groups based on IUI outcome (Figure 2). The clinical pregnancy was associated with a more evident Lactobacillus spp domination, comparable with that observed in controls (Figure 2); IUI failure was associated with a reduced relative amount of Lactobacillaceae, ranging from 100% to 83% ± 34% (mean ± SD), and an increase in Bifidobacteriaceae, from 0% to 12% ± 22%. In line, the analysis of the alpha diversity (within-sample diversity) revealed a significant difference between IUI success and IUI failure, with a lower Shannon index in women experiencing IUI success (0.8 ± 0.9 vs 1.5 ± 1.1; P = .003) (Supplementary Figure S3A).
On the contrary, no relevant differences were observed in seminal microbiome in male partners of couples with different IUI outcome (Figure 3 and Supplementary S4). In line, the Shannon index result was comparable between the 2 groups of idiopathic infertile men, although it was significantly higher than controls (4.8 ± 0.5 for IUI success and 4.3 ± 1.1 for IUI failure versus 3.1 ± 1.3 for controls, P = .004) (Supplementary Figure S3B).
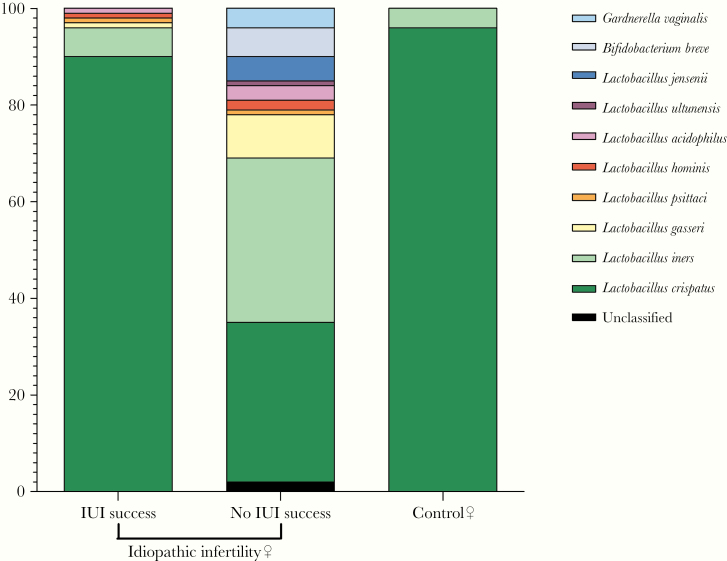
At the lowest taxonomic level, a different pattern in relative abundance of Lactobacillus species was observed between women with different IUI outcome (Figure 4) (Supplementary Table S3). Despite most (19 of 23) of the women within our cohort showing a clear Lactobacillus domination (relative abundance ≥95%) in their vaginal microbiome, several differences emerged at the species level. In particular, the 5 women experiencing IUI success showed a >85% (relative abundance ranging from 87% to 91%) domination by L crispatus (Supplementary Table S3), the species mostly associated with a physiological vaginal environment. In contrast, among the 18 women experiencing IUI failure, 6 featured L crispatus domination, even if at low level (from 50% to 82%); in 8 subjects, non-L crispatus species were observed, with L iners domination in 6 women (from 53% to 92%) and L gasseri in 2 cases (from 55% to 68%), including 1 with only 74% relative amount of lactobacilli detected. A codomination by 3 different Lactobacillus species were observed in 1 case (L jensenii 34%, L crispatus 31%, L acidophilus 29%). Finally, in 2 cases, no clear Lactobacillus spp domination was observed (from 1% to 30%) (Supplementary Table S3).
Figure 4.
Vaginal microbiome taxonomic profile at species level. Comparison between women with idiopathic infertility, stratified by intrauterine insemination (IUI) outcome, and controls.
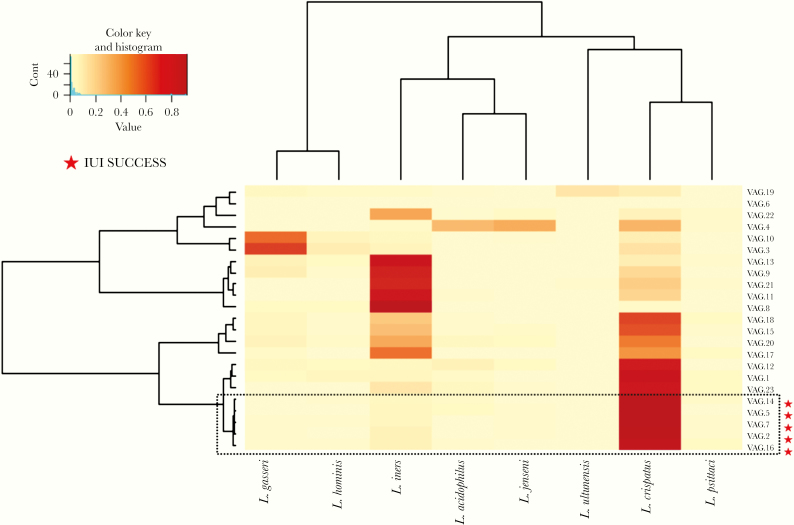
To further investigate the observed differences, women were clustered according to the relative abundance of discrete Lactobacillus species observed in their vaginal microbiome. After this approach, a discrete cluster identified the group of 5 women with a favorable IUI outcome (Figure 5), significantly separating it from the remnant population. In this analysis, we considered age, BMI, and sperm count as possible confounding factors (multivariate ADONIS analysis, R2 = 0.21588, P = .000996). The analysis also showed that L crispatus was the species mostly differentiating the vaginal microbiome between the 2 cohorts of women with discrete IUI outcome (FDR adjusted P = .0002) (Supplementary Table S4). The same analysis was performed for seminal microbiome, revealing no significant differences in the relative abundance of Lactobacillus species among the cohort of idiopathic infertile men (Supplementary Figure S5, Supplementary Tables S5 and S6).
Figure 5.
Heatmap of the relative abundance of Lactobacillus species found in the vaginal microbiome of 23 women with idiopathic infertility. Color key is indicated in the upper left corner. The dashed-black box identifies the cluster relative to intrauterine insemination (IUI) success group (P = .0117).
Discussion
The absence of a definable cause makes idiopathic infertility extremely frustrating for couples trying to conceive, thus highlighting the need of dedicated studies in this specific field. The role played by genital microbial community on reproductive outcome is not fully understood yet, despite recent evidence suggesting an association between an altered vaginal and seminal microbiome composition and infertility [9, 13, 14]. To the best of our knowledge, this is the first study evaluating both the vaginal and the seminal microbiome of couples with idiopathic infertility and considering the IUI success as the primary outcome. Only 1 previous study investigated the mutual interactions of the vaginal and seminal microbiome after sexual intercourse in couples with infertility of different etiologies, revealing the complementarity of the partners’ genital microbiome [24]. However, it did not evaluate the association with fertility [24].
Several studies previously investigated the correlation of the seminal microbiome with sperm quality, evidencing the role of even subclinical infections in cases of male infertility. For example, a study on men of infertile couples showed that an abundance in seminal lactobacilli was associated with healthy semen, whereas the presence of proinflammatory Gram-negative genera, such as Pseudomonas spp and Prevotella spp, was associated with low-quality semen [9]. Similar findings were not evident in our idiopathic infertile male cohort probably because, by definition of idiopathic infertility, all their seminal parameters fell within the range of normal semen quality.
Moreover, in our study, no significant microbiome differences were observed between the male partners of couples with positive and negative IUI outcomes. The only non-microbiological factor that somehow emerged, reaching the limit of statistical significance, was the higher sperm concentration observed, before sperm isolation, in the male partners of couples with positive IUI outcome. However, no significant differences were observed after semen capacitation, thus dramatically decreasing the clinical importance of this finding.
On the contrary, more interesting data emerged from the analysis of the female partners of our cohort. First of all, a more dysbiotic vaginal community was evident in our patients, when comparing their profiles with control sequences available online. Indeed, they featured a microbial pattern somehow similar to that observed in women affected by BV [14], even if in the absence of any overt BV-related symptoms. A possible interpretation of this finding is that idiopathic infertility in women may be associated with a condition of subclinical vaginal dysbiosis, consisting either in an overall reduction of lactobacilli or, as more commonly observed in our cohort, in a domination of Lactobacillus species associated with a less physiological environment.
This interpretation is further supported by the finding that the level of vaginal dysbiosis somehow correlated to IUI outcome. As a matter of fact, in our cohort, infertile women with a successful outcome showed a more physiological vaginal microbiome, whereas women who did not achieve a successful IUI showed an altered microbial composition with a significantly higher alpha diversity. It is known that several microorganisms colonizing the vaginal tract can cross the cervical barrier causing gynecologic and reproductive complications, such as pelvic inflammatory disease, preterm delivery, and miscarriage. Although the uterine cavity has been traditionally considered a sterile environment [25], recent findings reported the existence of an endometrial microbiota [26, 27]. Moreover, the presence of non-Lactobacillus bacteria in the endometrium has already been associated with adverse reproductive outcomes and implantation failure [27]. Likewise, Wittemer et al [28] reported that the presence of endocervical microorganisms (such as Candida albicans, Ureaplasma urealyticum, G vaginalis) may interfere with embryonic implantation process, thus affecting IVF outcomes. In our cohort, we found a proportion of vaginal G vaginalis in 2 of the idiopathic infertile women. Indeed, these women featured an altered vaginal microbiota, which could be the reason for IUI failure. Therefore, because the stability and homeostasis of the vaginal microbiome is crucial for reproductive health, the alterations in vaginal microbiome observed in this study may be somehow involved in the etiology of idiopathic infertility. Bacterial metabolites and/or compounds may cause an inflammatory response in the endometrium that could interfere with embryo implantation after the IUI procedure.
Given the differences of vaginal Lactobacillus spp domination, a species-level analysis of microbiome datasets is essential to a deep understanding of the vaginal microbial composition. In our study, despite that the majority of women showed a totality of lactobacilli in their vaginal microbiota, several differences emerged among the dominant species, evidencing a strong association between L crispatus vaginal domination and IUI success. Lactobacillus crispatus is considered as a marker of a healthy vaginal ecosystem. Vaginal communities dominated by L crispatus are characterized by lower pH values (<4.5) [2] and are more stable; therefore, they are less often associated with transitions to a BV state [29]. Lactobacillus species associated with higher pH values, such as L iners and L gasseri, are more conducive to the development of BV. A perturbation of vaginal microbiota, including BV, has been significantly associated with a reduction in pregnancy rate after IVF [17, 30]. Furthermore, recent findings enlighten the possible beneficial role of Lactobacilli in assisted reproductive technology. A vaginal microbiota dominated by Lactobacillus spp (>90%) has been associated with higher implantation and pregnancy rates, which is considered to be reproductive success [27]. Thus, our findings strengthen the potential role of L crispatus in promoting a favorable environment for pregnancy.
We are aware that the sample size is the main limitation of our study, especially considering the lack of differences observed in the male cohort. Moreover, the evaluation of vaginal microbiota in infertile women on a larger cohort could also be strengthened by the use of more precise bioinformatics pipelines based on the detection of amplicon sequence variants [31]. However, we think that the exclusion of other known causes of infertility strengthens what we observed in the vaginal microbiome of our female cohort. Thereof, we suggest that vaginal microbiome characterization could be useful for women with idiopathic infertility, especially considering that all of the enrolled couples underwent their first IUI cycle. The modulation of vaginal microbiota could improve the likelihood of either spontaneous or IUI-induced pregnancy. Probiotics may be an option, but their clinical effectiveness is still debated depending on several issues, including the selected strain, dosing, and delivery route (oral or topical). For example, 1 strain in particular, L crispatus CTV-05, has demonstrated vaginal colonization efficiency in women lacking endogenous L crispatus [32], as well as safety and tolerability in a Phase 2 trial in women with BV [33].
Conclusions
However, other approaches may deserve further attention, somehow following what was already performed in other settings [34, 35]. A previous study showed that vaginal fluids from the mothers could be used to colonize the upper respiratory tract of cesarean-delivered newborns with more physiological bacteria [36]. In a recent study, vaginal microbiome transplantation from healthy donors has been successfully tested for the first time as a treatment for intractable and recurrent BV [37]. Likewise, our results could pave the way to interventional studies in the pre-IUI modulation of vaginal microbiota. Instead of administering single probiotic strains, fluids from fertile women with L crispatus-dominated microbiota could be transferred to women with vaginal dysbiosis to restore a healthier vaginal flora and a more “pregnancy-friendly” environment.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We thank Massimo Alfano, Irene Locatelli, and Filippo Pederzoli for quantitative polymerase chain reaction analysis.
Financial support. This work was funded by the Italian Ministry of Health (5X1000, Ospedale San Raffaele).
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Lewis FM, Bernstein KT, Aral SO. Vaginal microbiome and its relationship to behavior, sexual health, and sexually transmitted diseases. Obstet Gynecol 2017; 129:643–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011; 108(Suppl 1):4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verstraelen H, Verhelst R, Claeys G, et al. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol 2009; 9:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wiesenfeld HC, Hillier SL, Krohn MA, et al. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis 2003; 36:663–8. [DOI] [PubMed] [Google Scholar]
- 5. Cherpes TL, Meyn LA, Krohn MA, et al. Association between acquisition of herpes simplex virus type 2 in women and bacterial vaginosis. Clin Infect Dis 2003; 37:319–25. [DOI] [PubMed] [Google Scholar]
- 6. Donati L, Di Vico A, Nucci M, et al. Vaginal microbial flora and outcome of pregnancy. Arch Gynecol Obstet 2010; 281:589–600. [DOI] [PubMed] [Google Scholar]
- 7. Ness RB, Kip KE, Hillier SL, et al. A cluster analysis of bacterial vaginosis-associated microflora and pelvic inflammatory disease. Am J Epidemiol 2005; 162:585–90. [DOI] [PubMed] [Google Scholar]
- 8. Hou D, Zhou X, Zhong X, et al. Microbiota of the seminal fluid from healthy and infertile men. Fertil Steril 2013; 100:1261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weng SL, Chiu CM, Lin FM, et al. Bacterial communities in semen from men of infertile couples: metagenomic sequencing reveals relationships of seminal microbiota to semen quality. PLoS One 2014; 9:e110152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baud D, Pattaroni C, Vulliemoz N, et al. Sperm microbiota and its impact on semen parameters. Front Microbiol 2019; 10:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cooper TG, Noonan E, von Eckardstein S, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update 2010; 16:231–45. [DOI] [PubMed] [Google Scholar]
- 12. Gunn DD, Bates GW. Evidence-based approach to unexplained infertility: a systematic review. Fertil Steril 2016; 105:1566–74.e1. [DOI] [PubMed] [Google Scholar]
- 13. van Oostrum N, De Sutter P, Meys J, Verstraelen H. Risks associated with bacterial vaginosis in infertility patients: a systematic review and meta-analysis. Hum Reprod 2013; 28:1809–15. [DOI] [PubMed] [Google Scholar]
- 14. Campisciano G, Florian F, D’Eustacchio A, et al. Subclinical alteration of the cervical-vaginal microbiome in women with idiopathic infertility. J Cell Physiol 2017; 232:1681–8. [DOI] [PubMed] [Google Scholar]
- 15. Sirota I, Zarek SM, Segars JH. Potential influence of the microbiome on infertility and assisted reproductive technology. Semin Reprod Med 2014; 32:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wee BA, Thomas M, Sweeney EL, et al. A retrospective pilot study to determine whether the reproductive tract microbiota differs between women with a history of infertility and fertile women. Aust N Z J Obstet Gynaecol 2018; 58:341–8. [DOI] [PubMed] [Google Scholar]
- 17. Haahr T, Jensen JS, Thomsen L, et al. Abnormal vaginal microbiota may be associated with poor reproductive outcomes: a prospective study in IVF patients. Hum Reprod 2016; 31:795–803. [DOI] [PubMed] [Google Scholar]
- 18. Haahr T, Zacho J, Bräuner M, et al. Reproductive outcome of patients undergoing in vitro fertilisation treatment and diagnosed with bacterial vaginosis or abnormal vaginal microbiota: a systematic PRISMA review and meta-analysis. BJOG 2019; 126:200–7. [DOI] [PubMed] [Google Scholar]
- 19. Fadrosh DW, Ma B, Gajer P, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014; 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rideout JR, He Y, Navas-Molina JA, et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2014; 2:e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fettweis JM, Serrano MG, Sheth NU, et al. ; Vaginal Microbiome Consortium (additional members) Species-level classification of the vaginal microbiome. BMC Genomics 2012; 13(Suppl 8):S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mändar R, Punab M, Borovkova N, et al. Complementary seminovaginal microbiome in couples. Res Microbiol 2015; 166:440–7. [DOI] [PubMed] [Google Scholar]
- 25. Romero R, Espinoza J, Mazor M. Can endometrial infection/inflammation explain implantation failure, spontaneous abortion, and preterm birth after in vitro fertilization? Fertil Steril 2004; 82:799–804. [DOI] [PubMed] [Google Scholar]
- 26. Mitchell CM, Haick A, Nkwopara E, et al. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol 2015; 212:611.e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moreno I, Codoñer FM, Vilella F, et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol 2016; 215:684–703. [DOI] [PubMed] [Google Scholar]
- 28. Wittemer C, Bettahar-Lebugle K, Ohl J, et al. Abnormal bacterial colonisation of the vagina and implantation during assisted reproduction. Gynecol Obstet Fertil 2004; 32:135–9. [DOI] [PubMed] [Google Scholar]
- 29. Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 2012; 4:132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hyman RW, Herndon CN, Jiang H, et al. The dynamics of the vaginal microbiome during infertility therapy with in vitro fertilization-embryo transfer. J Assist Reprod Genet 2012; 29:105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 2017; 11:2639–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Antonio MA, Meyn LA, Murray PJ, et al. Vaginal colonization by probiotic Lactobacillus crispatus CTV-05 is decreased by sexual activity and endogenous Lactobacilli. J Infect Dis 2009; 199:1506–13. [DOI] [PubMed] [Google Scholar]
- 33. Hemmerling A, Harrison W, Schroeder A, et al. Phase 2a study assessing colonization efficiency, safety, and acceptability of Lactobacillus crispatus CTV-05 in women with bacterial vaginosis. Sex Transm Dis 2010; 37:745–50. [DOI] [PubMed] [Google Scholar]
- 34. Hocquart M, Lagier JC, Cassir N, et al. Early fecal microbiota transplantation improves survival in severe Clostridium difficile infections. Clin Infect Dis 2018; 66:645–50. [DOI] [PubMed] [Google Scholar]
- 35. van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407–15. [DOI] [PubMed] [Google Scholar]
- 36. Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 2016; 22:250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lev-Sagie A, Goldman-Wohl D, Cohen Y, et al. Vaginal microbiome transplantation in women with intractable bacterial vaginosis. Nat Med 2019; 25:1500–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.