SUMMARY
Dengue virus (DENV) can cause diseases ranging from dengue fever (DF) to more severe dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS). Whether antiviral T cells contribute to the protection against or pathogenesis of severe disease is not well defined. Here, we identified antigen-specific IL-10+IFN-γ+ double-positive (DP) CD4 T cells during acute DENV infection. While the transcriptomic signatures of DP cells partially overlapped with those of cytotoxic and type 1 regulatory CD4 T cells, the majority of them were non-cytotoxic/Tr1 and included IL21, IL22, CD109, and CCR1. Although we observed a higher frequency of DP cells in DHF, the transcriptomic profile of DP cells was similar in DF and DHF, suggesting that DHF is not associated with the altered phenotypic or functional attributes of DP cells. Overall, this study revealed a DENV-specific DP cell subset in patients with acute dengue disease and argues against altered DP cells as a determinant of DHF.
In Brief
Tian et al. identify and characterize antigen-specific IL-10+IFN-γ+ double-positive (DP) CD4 T cells in acute dengue patients. DP cells display similar transcriptomic profiles in mild DF and severe DHF, despite their increased frequency in DHF, suggesting that DHF is not associated with the altered phenotype or functionality of DP cells.
Graphical Abstract
INTRODUCTION
Dengue virus (DENV) is a serious public health problem, especially in tropical and subtropical areas, and infects up to ~390 million people annually (Bhatt et al., 2013). DENV infection is associated with a range of clinical manifestations, from asymptomatic to mild dengue fever (DF) to more severe and sometimes life-threatening dengue diseases, including dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). However, host immunological correlates of severe dengue disease, especially during the acute phase of DENV infection, have not been fully determined.
Both the pathological and protective effects of T cells have been reported during DENV infection (Ngono and Shresta, 2018; Rothman, 2011; Screaton et al., 2015; St John and Rathore, 2019; Tian et al., 2016c; Weiskopf and Sette, 2014). On the one hand, it has been reported that cross-reactive memory T cells that are specific for the primary infecting DENV serotype may expand and lead to immunopathology and ineffective viral clearance during a secondary heterologous infection (called original antigenic sin) (Halstead et al., 1983; Mongkolsapaya et al., 2003; Ngono and Shresta, 2018; Rothman, 2011; Screaton et al., 2015; St John and Rathore, 2019; Tian et al., 2016c; Weiskopf and Sette, 2014). On the other hand, accumulating evidence suggests that T cells may contribute to the control of DENV infection in both mice and humans (de Alwis et al., 2016; Elong Ngono et al., 2016; Grifoni et al., 2017; Prestwood et al., 2012; Tian et al., 2019; Weiskopf et al., 2013, 2015; Yauch et al., 2009, 2010; Zellweger et al., 2013, 2014, 2015; Zompi et al., 2012). We and others have previously shown that DENV-specific CD4 memory T cells can produce cytokines such as interferon g (IFN-γ), tumor necrosis factor α (TNF-α), and interleukin-2 (IL-2), which are usually associated with T helper type 1 (Th1) cells, following DENV infection and vaccination (Gwinn et al., 2003; Hatch et al., 2011; Lindow et al., 2012). Furthermore, human CD4 effector memory T cells re-expressing CD45RA (Temra cells) have been detected in healthy blood bank donors who have been infected with DENV multiple times and show an increased expression of numerous cytotoxic molecules, including CX3CR1, granzyme B, perforin, and CD107a (Weiskopf et al., 2015). Subsequent studies further revealed the transcriptomic profile and heterogeneity of CD4 Temra cells in apparently healthy cohorts and identified surface molecules such as GPR56 and CD244 that are uniquely expressed on cytotoxic CD4 Temra cells (Patil et al., 2018; Tian et al., 2017). However, the phenotype and transcriptomic profile of DENV-specific CD4 T cells during the acute phase of infection and their association with dengue disease severity have not been systematically defined.
In general, IL-10 is an immunosuppressive cytokine that has multifaceted functions in modulating T cell differentiation, memory formation, function, and exhaustion, as well as germinal center B cell responses (Cox et al., 2013; Laidlaw et al., 2015, 2017; Tian et al., 2016b; Xin et al., 2018). IL-10 can be produced by multiple cell types from both innate and adaptive immune systems, including dendritic cells (DCs), macrophages, B cells, and CD8 T cells, as well as various CD4 T cell subsets, including Th1 cells, Th2 cells, Foxp3+ regulatory T (Treg) cells, and Foxp3− type 1 regulatory T (Tr1) cells (Ouyang et al., 2011). Previous studies have reported that DENV infection can induce the production of IL-10 by monocytes, which may dampen anti-DENV immune responses and viral control (Adikari et al., 2016; Chareonsirisuthigul et al., 2007; Tsai et al., 2014; Ubol et al., 2010). In addition, the increased IL-10 level in the blood is associated with severe dengue disease (Butthep et al., 2012; Chen et al., 2006; Ferreira et al., 2015; Flores-Mendoza et al., 2017; Green et al., 1999; Malavige et al., 2013). However, the production of IL-10 by DENV-specific CD4 T cells and its association with dengue disease severity have not been fully elucidated.
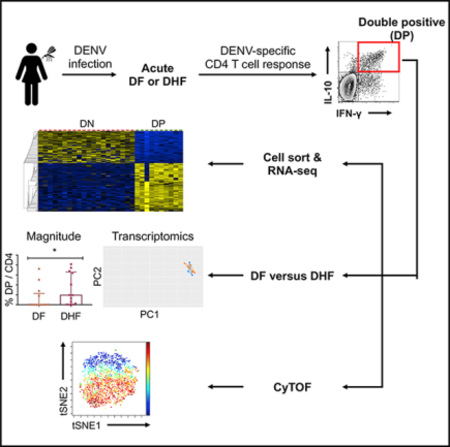
In this study, we set out to determine the production of IL-10 in conjunction with IFN-γ by DENV-specific CD4 T cells, their gene expression profiles, and their association with dengue disease severity. All of the patients with severe dengue who were included in this study had DHF but no other forms of severe dengue, including DSS and organ failure. Our data show that DENV-specific CD4 T cell response during the acute phase of infection consists of a prominent subset that co-produced IL-10 and IFN-γ. DENV-specific IL-10+IFN-γ+ double-positive (DP) cells were largely phenotypically homogeneous and exhibited non-cytotoxic/Tr1 gene signatures such as IL-21, IL-22, CD109, and CCR1. Although we detected more DENV-specific DP cells than DF during acute DHF, the phenotype of these cells was largely similar in acute DF and DHF samples. These results suggest that severe dengue disease is not associated with altered cytokine secretion or phenotype of this DP cell subset, arguing against an altered CD4 T cell response as being a determinant of DHF.
RESULTS
DENV-Specific IL-10+IFN-γ+ DP CD4 T Cells Are Prominent during Acute DENV Infection
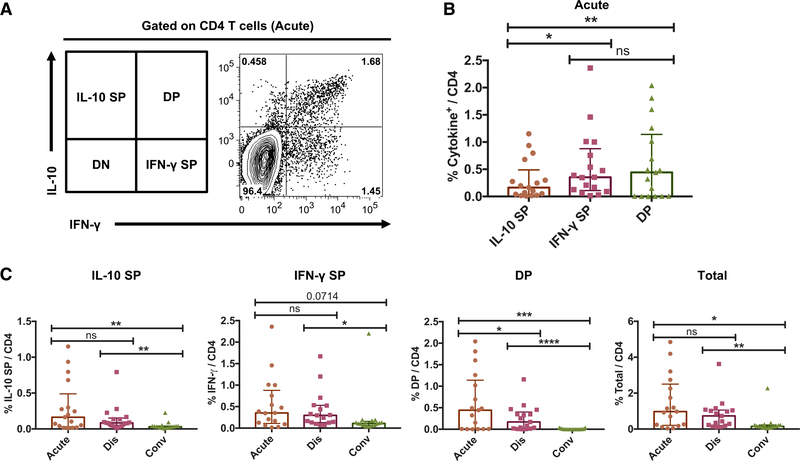
Previous studies have reported increased IL-10 production during acute DENV infection (Chareonsirisuthigul et al., 2007; Tsai et al., 2014; Ubol et al., 2010). However, whether DENV-specific CD4T cells may be responsible for IL-10 production was not addressed. We previously described a pool of 180 different CD4 T cell epitopes derived from DENV1–4 that allows broad coverage of anti-DENV CD4 T cell responses (Grifoni et al., 2017; Weiskopf et al., 2016). Hereafter, this epitope “megapool” is referred to as DENV MP. In the present study, peripheral blood mononuclear cells (PBMCs) from hospitalized DF or DHF Sri Lankan patients collected during the acute phase, discharge, and convalescent phase time points were stimulated ex vivo with the DENV MP, and the production of IFN-γ and IL-10 by DENV-specific CD4 T cells was assessed by cytokine secretion assay and flow cytometry analysis. As shown in Figure 1A for a representative donor, DENV-specific CD4 T cells from PBMCs collected during acute DENV infection produced IFN-γ and/or IL-10 in response to epitope stimulation. In addition to IL-10+IFN-γ− IL-10 single-positive (SP) and IL-10−IFN-γ+ IFN-γ SP cells, we observed a prominent IL-10+IFN-γ+ DP population (Figure 1A). When the responses of 17 different acute DENV infection samples were considered, the magnitudes of IFN-γ SP and DP responses were significantly higher than that of the IL-10 SP response, whereas no major difference was observed between the magnitudes of IFN-γ SP and DP responses (Figure 1B).
Figure 1. Identification of Human DENV-Specific IL-10+IFN-γ+ Double-Positive (DP) CD4 T Cells during the Acute Phase of Infection.
Human PBMCs isolated from DENV-infected patients at acute, discharge, and convalescent stages were stimulated with DENV MP, and DENV-specific CD4 T cells were identified by the production of IFN-γ and/or IL-10 using flow cytometry.
(A) Flow cytometry plot shows the production of IFN-γ and IL-10 by CD4 T cells during the acute phase.
(B) Bar graph shows the frequencies of DENV-specific IL-10 SP, IFN-γ SP, and DP cells among acute samples (n = 17).
(C) Bar graphs show the frequencies of IL-10 SP, IFN-γ SP, DP, and total cytokine-positive cells at acute, discharge (dis), and convalescent (conv) stages (n = 17).
Error bars show medians with interquartile ranges. Statistical significance was determined by two-tailed Wilcoxon test. ns, not significant, *p < 0.05, **p < 0.01, ***p<0.001,****p<0.0001.
We next assessed the kinetics of these DENV-specific CD4 T cell responses using longitudinal samples as a function of acute, discharge, and convalescent time points with a median of 4, 7, and 31 days from fever onset, respectively. As expected, the total DENV-specific CD4 T response (the sum of IL-10 SP, IFN-γ SP, and DP) diminished in discharge and convalescent samples (Figure 1C). This decrease was apparent for all subpopulations, as we observed a substantial decrease in the frequency of IL-10 SP and IFN-γ SP cells (Figure 1C). DENV-specific DP CD4 T cells were absent in the convalescent phase (Figure 1C). As a control, we examined the production of IL-10 and IFN-γ after stimulation with CD3/CD28 or DENV MP in healthy DENV seronegative donors from Sri Lanka and healthy control donors from non-dengue-endemic San Diego, California. Although IFN-γ SP cells were observed following CD3/CD28 stimulation, DP CD4 T cells were not detected in healthy donors (Figure S1, supporting the notion that this population is specific to acute dengue patients. These results reveal a prominent DENV-specific IL-10+IFN-γ+ DP CD4 T cell population during the acute phase, which progressively wanes upon discharge and in convalescence.
DP Cells Are Associated with a Prominent Transcriptome Profile
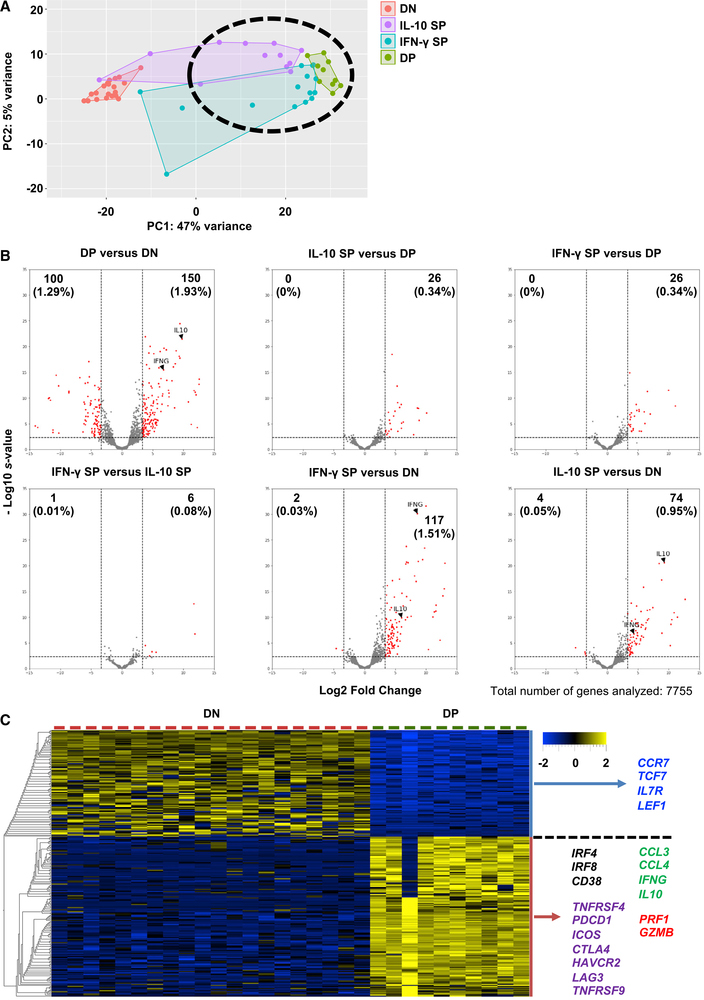
To further characterize the gene expression profiles of the DENV-specific CD4 T cell subsets defined by their production of IFN-γ and IL-10, we isolated DENV-specific IL-10 SP, IFN-γ SP, and DP cells by standard cytometry sorting and performed bulk RNA sequencing (RNA-seq) on these sorted subsets. As a control, we also performed RNA-seq on sorted IL-10− IFN-γ− double-negative (DN) CD4 T cells from the same antigen-stimulated cultures. Principal-component analysis (PCA) was used to visualize the global gene expression patterns of the four CD4 T cell populations. DN and DP cells formed two distinct clusters that were largely separated by principal component 1 (PC1) (Figure 2A). IL-10 SP and IFN-γ SP cells, while more heterogeneous, still mostly clustered closely to DP cells (Figure 2A).
Figure 2. Gene Expression Profiles of DENV-Specific CD4 T Cell Subsets.
(A) PCA analysis of gene expression data of DN (n = 20), IL-10 SP (n = 12), IFN-γ SP (n = 15), and DP cells (n = 10). N indicates the number of samples.
(B) Volcano plots show log2 fold changes versus −log10 s values for the comparisons between DP and DN, IL-10 SP and DP, IFN-γ SP and DP, IFN-γ SP and IL-10 SP, IFN-γ SP and DN, and IL-10 SP and DN. Genes with a log2 fold change of >3.3 or <−3.3 and an s value of <0.005 were considered significant and are shown in red. Numbers indicate the number of significant DE genes, and the numbers in parentheses indicate the percentages of significant DE genes (a total of 7,755 genes were analyzed).
(C) Heatmap shows the expression values after variance stabilizing transformation of the top 250 DE genes between DP and DN cells. Colored genes were associated with naive/memory T cells (blue), activation (black), cytokines and chemokines (green), co-stimulation/inhibition (purple), cytotoxicity (red).
We next analyzed the pattern of genes differentially expressed (DE) between the DP versus DN subsets. A large number of significantly DE genes was identified, and the top 250 DE genes were associated with a rather stringent cutoff threshold of absolute log2 fold change of 3.3, corresponding to ~10-fold. As expected, DP cells upregulated both IFNG and IL10 genes (Figure 2B). Next, pairwise analyses were performed to identify DE genes between the four CD4 T cell subsets, namely IL-10 SP versus DP, IFN-γ SP versus DP, IFN-γ SP versus IL-10 SP, IFN-γ SP versus DN, and IL-10 SP versus DN. Using the same log2 fold change of 3.3 threshold, only 26 DE genes were identified when comparing IL-10 SP versus DP and IFN-γ SP versus DP (Figure 2B). In addition, only 7 DE genes were observed between IL-10 SP and IFN-γ SP cells (Figure 2B), suggesting that the gene expression profiles of IL-10 SP and IFN-γ SP cells resemble that of DP cells, which is also consistent with the PCA analysis shown in Figure 2A. Fewer DE genes were identified by comparing IFN-γ SP versus DN and IL-10 SP versus DN (a total of 119 and 78, respectively, as compared to the 250 DE genes between DP and DN cells using the same cutoff; Figure 2B). IL10 transcripts were upregulated in IFN-γ SP cells and IFNG transcripts were upregulated in IL-10 SP cells (Figure 2B), supporting the notion that these two SP subsets may resemble DP cells but with a less prominent pattern of DE genes. These data also suggest that there is a gradient of differentiation states between these cell populations. These data show that DP cells are associated with a prominent gene expression profile and that IL-10 SP and IFN-γ SP cells may exhibit similar transcriptomic profiles, albeit less pronounced. Thus, we focused on DP cells for the rest of this study.
When the top 250 DE genes in DENV-specific DP cells (Table S1) were examined in more detail, we found that DP cells down-regulated several genes normally expressed by naive and memory T cells such as CCR7, TCF7, IL7R, and LEF1, which are also upregulated in memory precursors in an Foxo1-dependent manner (Hess Michelini et al., 2013). In contrast, these cells upregulated the expression of genes associated with T cell activation (IRF4, IRF8, CD38), cytotoxicity (PRF1, GZMB), cytokines (IFNG, IL10), chemokines (CCL3, CCL4), and co-stimulation and inhibition (TNFRSF4, PDCD1, ICOS, CTLA4, HAVCR2, LAG3, TNFRSF9) (Figure 2C). These data reveal a rich and complex gene signature associated with DP cells.
DENV-Specific DP Cells Partially Resemble Cytotoxic CD4 Temra Cells and Tr1-like Cells
Previous studies have described gene signatures associated with human cytotoxic CD4 Temra cells such as granzyme B, perforin, CCL4, CX3CR1, GPR56, T-bet, and CD107a (Patil et al., 2018; Tian et al., 2016c, 2017; Weiskopf et al., 2015), defined in healthy blood donors from hyperendemic areas (and thus corresponding to donors in the convalescent phase well past acute DENV infection). As shown in Figure S2A, DENV-specific DP cells associated with acute infection upregulated the expression of GZMB (encodes granzyme B), PRF1 (encodes perforin), and CCL4. However, DP cells did not significantly upregulate the expression of CX3CR1 and LAMP1 (encodes CD107a) (Figure S2A), and GPR56 was filtered out before DE analysis due to low expression (data now shown). Thus, DENV-specific DP cells express only a portion of the markers that are associated with cytotoxic CD4 Temra cells.
Tr1-like cells lack the expression of the transcription factor Foxp3 and are associated with the expression of multiple activation, inhibitory, and effector molecules such as CTLA4, ICOS, LAG-3, TIM-3, TIGIT, and PD-1, CD49b, and transforming growth factor β (TGF-β) (Roncarolo et al., 2018; White and Wraith, 2016; Zeng et al., 2015). Similar to Tr1-like cells, DP cells showed an elevated expression of IFNG, IL10, LAG3, HAVCR2 (encodes Tim-3), TIGIT, PDCD1 (encodes PD-1), ICOS, CTLA4, and TNFRSF18 (encodes GITR) (Figure S2B). However, they did not significantly upregulate ITGA2 (encodes CD49b), HLA-DRA, and TGFB1 (Figure S2B). These data suggest that DENV-specific DP cells express several markers associated with cytotoxic CD4 Temra cells and/or Tr1-like cells and may contain Tr1-like cells.
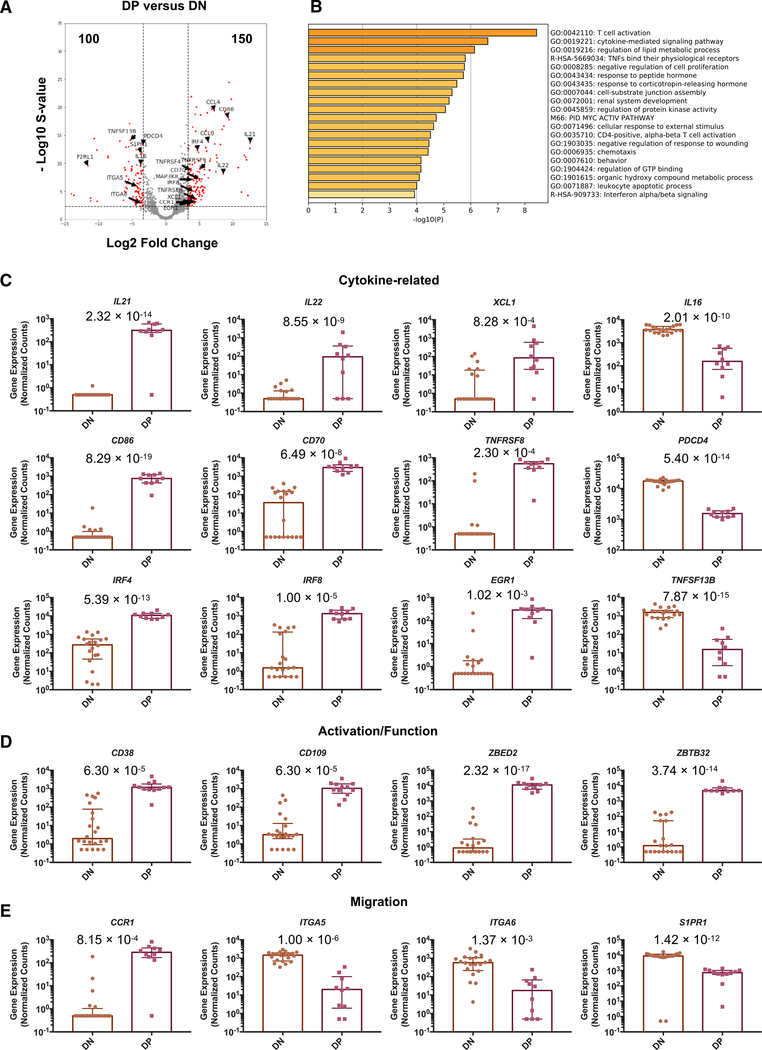
Non-cytotoxic/Tr1 DE Genes in DP Cells Are Associated with Cytokine Production, Activation, Function, and Migration
The vast majority (224, ~90%) of the top 250 DE genes did not overlap with the signature genes of cytotoxic CD4 Temra cells or Tr1-like cells, and we referred to these non-overlapping genes as “non-cytotoxic/Tr1” DE genes of DP cells in this study. DENV-specific DP cells upregulated genes such as IL21, IL22, CD86, CCL4, and CCL5, while downregulating genes such as F2RL1, TNFSF13B, S1PR1, and PDCD4 (Figure 3A). Pathway and process enrichment analysis revealed that these non-cytotoxic/ Tr1 DE genes are associated with pathways such as T cell activation, cytokine-mediated signaling pathway, and regulation of the lipid metabolic process (Figure 3B). DENV-specific DP cells upregulated numerous cytokine-related genes, including IL21, IL22, XCL1, CD86, CD70, TNFRSF8 (encodes CD30), IRF4, IRF8, and EGR1, while downregulating IL16, PDCD4, and TNFSF13B (Figure 3C). DENV-specific DP cells also showed an increased expression of T cell activation-associated genes, including CD38, CD109, ZBED2, and ZBTB32 (Figure 3D). In addition, DP cells upregulated the expression of CCR1, while downregulating the expression of ITGA5, ITGA5, and S1PR1, suggesting that DP cells may migrate to infected and/or inflammatory sites via migratory molecules such as CCR1 (Figure 3E). These data revealed the gene signatures of DP cells that are associated with cytokine activities, activation, function, and migration.
Figure 3. Non-cytotoxic/Tr1 DE Genes in DP Cells Are Associated with Cytokine Production, Activation, Function, and Migration.
(A) Volcano plot shows log2 fold changes versus −log10 s values for the comparisons between DP and DN with selected non-cytotoxic/Tr1 DE genes labeled. Significant DE genes are shown in red.
(B) Enrichment analysis results of the 224 non-cytotoxic/Tr1 DE genes.
(C–E) Bar graphs show the gene expression values in counts normalized by sequencing depth that is calculated by DESeq2 for (C) cytotoxicity-related,
(D) activation/function-related, and (E) migration-related genes in DN and DP cells. Error bars show medians with interquartile ranges. A pseudocount of 0.5 was added to allow for plotting in log scale. Statistical significance was determined by DESeq2 using the shrinkage estimator apeglm, and the computed s values were indicated in the graphs.
Expression of Transcription Regulators of CD4 T Cell Lineages
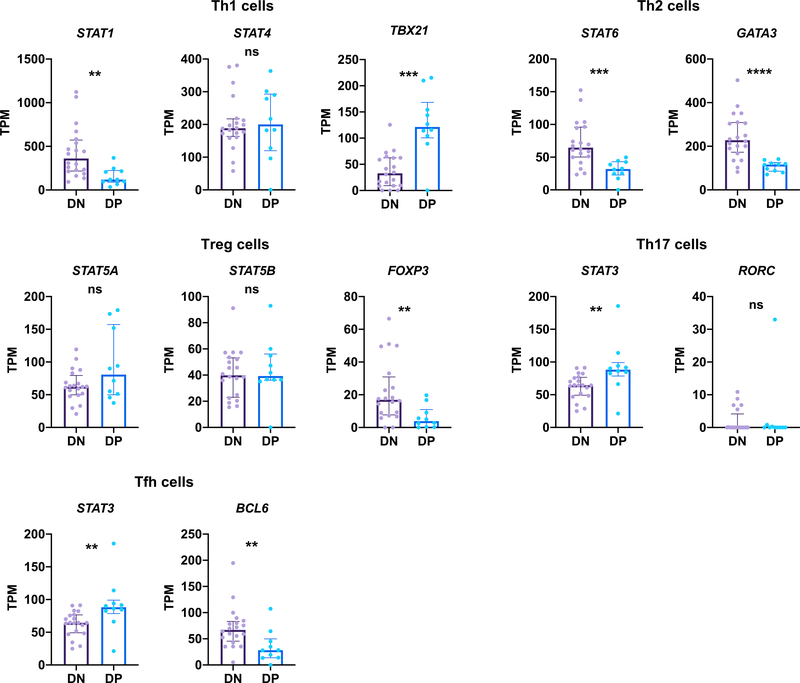
To further characterize the phenotype of DENV-specific DP cells, we assessed the gene expression of transcription factors that are crucial for the specification of several major CD4 T cell lineages, including Th1, Th2, Th17, Treg, and follicular T helper (Tfh) cells (Tripathi and Lahesmaa, 2014). In terms of early response signal transducer and activator of transcription (STAT) factors, DENV-specific DP cells had a lower expression of STAT1 and STAT6 but a higher expression of STAT3 than DN cells (Figure 4). STAT3 is involved in the differentiation of Th17 cells and Tfh cells (Tripathi and Lahesmaa, 2014); however, DP cells did not upregulate RORC and BCL6 (Figure 4), which are the master regulators of Th17 and Tfh cells, respectively (Tripathi and Lahesmaa, 2014). DP cells also had a lower expression of GATA3 and FOXP3 (Figure 4), suggesting that these cells were not Th2 or Treg cells. DP cells upregulated the expression of TBX21, which encodes T-bet that directs the differentiation of Th1 cells. Thus, these results indicate that DENV-specific DP cells may at least partially resemble Th1 cells.
Figure 4. Expression Levels of Transcription Regulators of Various CD4 T Cell Lineages by DENV-Specific DP Cells.
Bar graphs show gene expression values in transcripts per million (TPM) for various transcription factors in DN cells (n = 20) and DP cells (n = 10). Error bars show medians with interquartile ranges. Statistical significance was determined by two-tailed Mann-Whitney test. ns, not significant, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The Frequency but Not the Gene Expression Profile of DP Cells Differs between DHF and DF
We next investigated whether DP cells were associated with the DHF. The frequencies of IL-10 SP, IFN-γ SP, and total DENV-specific CD4 T cells were similar between DF and DHF during acute DENV infection, whereas the frequency of DP cells was significantly higher in acute DHF patients than in acute DF patients (Figure 5A). Nevertheless, similar to DN cells, DENV-specific DP CD4 T cells isolated from acute DF and DHF patients displayed very similar gene expression profiles and clustered closely to one another based on PCA (Figure 5B). Only 38 DE genes were identified when DP cells isolated from acute DF and DHF patients were directly compared (Figure 5C). We also compared IL-10 SP and IFN-γ SP cells between DF and DHF. Consistently, only 7 and 46 DE genes were detected between DF and DHF when IL-10 SP and IFN-γ SP cells were compared, respectively (Figure 5C). In addition, no significant difference was observed between DF and DHF in terms of the T cell receptor (TCR) repertoire diversity of DP cells (Figure 5D). We next examined the abundances of the top five productive TCR β chain (TRB, which accounts for most of the TCR repertoire variation; Hackl et al., 2016) clonotypes in each individual donor to investigate whether there were predominant TRB clonotypes. As shown in Figure 5E, the abundances of the top five productive clonotypes varied between donors. We further identified a few predominant TRB clonotypes such as CASSRGAGEVQETQYF, CASSRPSGGELFF, CAISEPLKGYEQYF, and CASSLGQGARVDGYTF, which constituted 39.6%, 45.5%, 33.8%, and 42.5% of the antigen-specific TRB repertoires in GS1430, GS1431, GS1458, and GS1470, respectively (Figure 5E; Table S2). Notably, none of these clonotypes were observed in two or more donors (Table S2). Future experiments are needed to investigate the antigen specificities of these predominant clonotypes. Although the magnitude of DENV-specific DP cell response may be higher in acute DHF samples, no major difference was observed in terms of the transcriptomic profile of DP cells, suggesting that severe dengue disease is not associated with the altered phenotype or function of DP cells.
Figure 5. The Frequency of DENV-Specific DP Cells Is Higher in Acute DHF Patients Than in DF Patients.
(A) Bar graphs show the percentages of IL-10 SP, IFN-γ SP, DP, and total cytokine-positive cells among acute DF (n = 13) and DHF (n = 13) samples.
(B) PCA of the gene expression data of DN cells (left panel) and DP cells (right panel) colored by DF and DHF (n = 10, 10, 3, and 7 for DN-DF, DN-DHF, DP-DF, and DP-DHF, respectively).
(C) Volcano plot shows log2 fold changes versus −log10 s values for the comparisons between DP-DHF and DP-DF (left panel), IL-10 SP-DHF (n = 7) and IL-10 SP-DF (n = 5) (center panel), and IFN-γ SP-DHF (n = 8) and IFN-γ SP-DF (n = 7) (right panel). Significant DE genes were shown in red.
(D) Dot plots show the observed diversity of the TCR α chain (TRA, left panel) and β chain (TRB, right panel) repertoires of DP cells in DF and DHF samples (n = 3 and 7 for DP-DF and DP-DHF, respectively).
(E) Bar graph shows the abundances of the top five TRB clonotypes in each donor.
Error bars show medians with interquartile ranges. Statistical significance was determined by two-tailed Mann-Whitney test. ns, not significant, *p < 0.05.
Validation of DE Genes in DP Cells and High-Dimensional Single-Cell Analysis by CyTOF
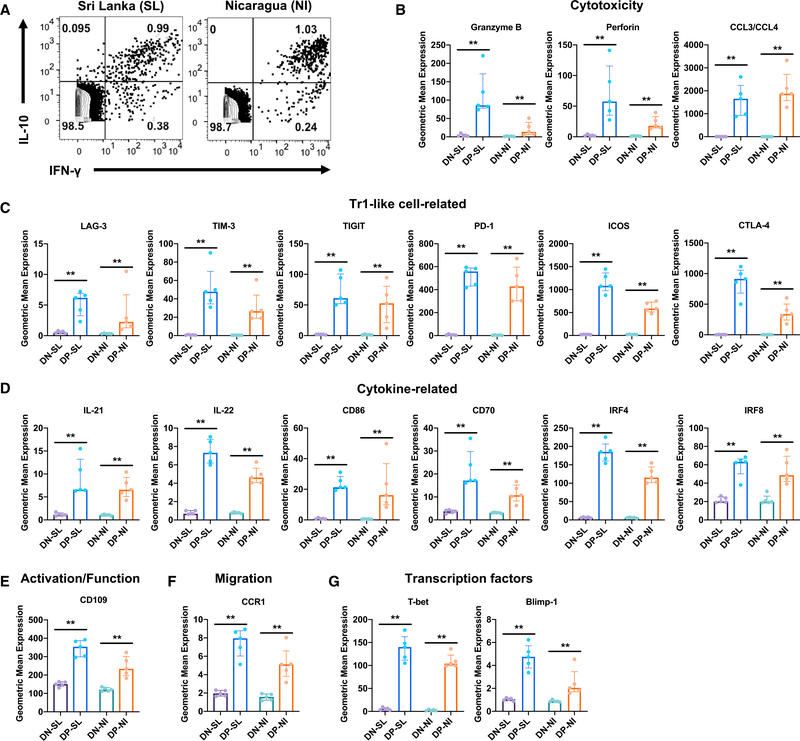
To validate the expression of DE genes in DENV-specific DP cells at the protein level, we performed cytometry by time-of-flight (CyTOF) analysis based upon selected DE genes for which antibodies were commercially available. We analyzed two independent sets of samples from two distinct geographical locations, namely Sri Lanka and Nicaragua. We focused our analysis on acute DHF samples, as DENV-specific DP cells were most abundant in these samples (Figures 1C and 5A). We detected the production of IFN-γ and IL-10 using intracellular staining (ICS) after ex vivo stimulation with the DENV MP and identified DENV-specific IL-10+IFN-γ+ DP cells in both Sri Lankan and Nicaraguan cohorts (Figure 6A), demonstrating that these cells may be a prominent antigen-specific CD4 T cell population that is commonly found in DHF patients at the acute stage, irrespective of geographic location. Furthermore, the CyTOF data validated the increased expression of several cytotoxic molecules, including granzyme B, perforin, and CCL3/CCL4 (the anti-CCL3 antibody used cross-reacts with CCL4) (Figure 6B), Tr1-like cell-related molecules, including LAG-3, TIM-3, TIGIT, PD-1, ICOS, and CTLA-4 (Figure 6C), as well as non-cytotoxic/Tr1 DE molecules, including IL-22, IL-22, CD86, CD70, IRF4, IRF8, CD109, and CCR1 (Figures 6D–6F) by DENV-specific DP cells at the protein level in the two cohorts from two different continents. As shown in Figure 6G, DENV-specific DP cells also had increased protein expression of the transcription factors T-bet and Blimp-1, which promote the production of IFN-γ and IL-10, respectively (Martins et al., 2006; Szabo et al., 2000).
Figure 6. Validation of DE Genes in DENV-Specific DP Cells at the Protein Level.
Human PBMCs isolated from Sri Lankan (SL, n = 5) and Nicaraguan (NI, n = 5) patients with acute DENV infection were stimulated with DENV MP, and the expression of various molecules was measured by CyTOF.
(A) Dot plots show the production of IFN-γ and IL-10 by CD4 T cells.
(B–G) Bar graphs show the protein expression in geometric mean values for molecules related to (B) cytotoxicity, (C) Tr1-like cells, (D) cytokines, (E) T cell activation/function, (F) migration, and (G) transcription factors in DN and DP cells.
Error bars show medians with interquartile ranges. Statistical significance was determined by two-tailed Mann-Whitney test. **p < 0.01.
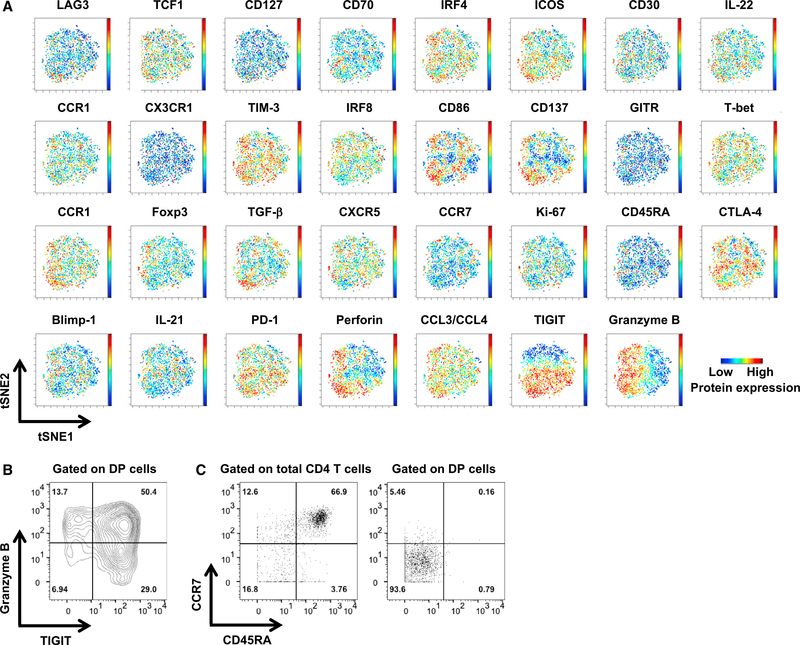
Next, we investigated potential heterogeneity within the DENV-specific DP cell population at a single-cell level. To this end, we measured the expression of a total of 31 proteins for each individual cell, and the resulting high-dimensional data were visualized in 2-dimensional maps generated by the visualization of stochastic neighbor embedding (viSNE) (Amir el et al., 2013). To obtain a sufficient number of cells for the viSNE analysis, we combined the data from both the Sri Lankan and Nicaraguan cohorts before running the viSNE analysis. As shown in Figure 7A, DENV-specific DP cells were arranged largely as one “island” in the viSNE maps. In addition, we did not observe distinctive expression patterns for the majority of the protein markers, except for TIGIT and granzyme B, which exhibited gradient expression among DENV-specific DP cells (Figure 7A). The cells with higher granzyme B expression also showed higher levels of perforin expression (Figure 7A). The gradients of TIGIT and granzyme B appeared to be orthogonal, which prompted us to “gate” the DP cells according to the expression of TIGIT and granzyme B. As shown in Figure 7B, ~50% of the DP cells co-express TIGIT and granzyme B. Furthermore, the majority of DENV-specific DP cells expressed low levels of CD45RA and CCR7, exhibiting a CD45RA−CCR7− effector or effector memory T (Tem) phenotype (Figures 7A and 7C), which was distinct from the Temra phenotype of cytotoxic CD4 T cells in DENV-exposed healthy individuals (Patil et al., 2018; Tian et al., 2017; Weiskopf et al., 2015). These data show that DENV-specific DP cells are largely similar and homogeneous for the majority of the markers tested, despite the fact that these markers were selected to maximize the potential detection of different subsets. Nevertheless, varied expression patterns were observed for two of the markers analyzed, TIGIT and granzyme B.
Figure 7. DENV-Specific DP Cells Are Largely Homogeneous, Except for the Expression of TIGIT and Granzyme B.
DP cells were merged from five Sri Lankan and five Nicaraguan donors.
(A) viSNE analysis was performed on DP cells based on the expression of 31 proteins. Plots show the expression of these 31 proteins in each individual cell.
(B) Contour plot shows the expression of TIGIT and granzyme B by DP cells. Numbers indicate the percentages of cells in each quadrant.
(C) Dot plots show the expression of CD45RA and CCR7 on total CD4 T cells (left panel) and DP cells (right panel). Numbers indicate the percentages of cells in each quadrant.
DISCUSSION
This study reveals several important characteristics of virus-specific CD4 T cells during the acute phase of DENV infection. First, a prominent DENV-specific IL-10+IFN-γ+ DP CD4 T cell population is detected in patients with acute DENV infection, which becomes undetectable at the convalescent stage. Second, DENV-specific DP cells have a different gene expression profile by comparison with DN cells, whereas DENV-specific IFN-γ+ SP and IL-10+ SP cells display similar gene expression patterns to DP cells. Albeit at a much lower level, DENV-specific IFN-γ+ SP cells upregulate IL10 gene expression. Conversely, DENV-specific IL-10+ SP cells upregulate IFNG gene expression, suggesting that these DENV-specific CD4 T cell populations may have a certain degree of plasticity and may be able to convert or transition into a different state. Third, although DENV-specific DP cells upregulate several cytotoxic CD4 T cell-associated and Tr1-like cell-associated molecules, the majority of DE genes such as IL21, IL22, CD109, and CCR1 in DP cells were non-cytotoxic/Tr1. Fourth, although the frequency of DENV-specific DP cells is higher in DHF patients than in DF patients, the transcriptomic profile and TCR feature of DENV-specific DP cells are similar between DF and DHF, suggesting that severe dengue disease is not associated with altered phenotypic or functional attributes of DP cells. Nevertheless, we could not rule out the possibility that the increased frequency of DP cells by itself may increase disease severity. Finally, we validated the expression of several DE genes of DENV-specific DP cells at the protein level using CyTOF in two independent cohorts from two continents. These results further showed that DENV-specific DP cells appear to be a largely homogeneous population except for the expression of TIGIT and granzyme B, which showed gradient expression among DP cells. Collectively, we identified an intriguing DENV-specific IL-10+IFN-γ+ DP CD4 T cell population during acute DENV infection and characterized their transcriptomic and proteomic profiles as well as their association with severe DENV disease.
Previous studies have reported an increased frequency of Treg cells during acute DENV infection (Jayaratne et al., 2018; Lühn et al., 2007). Since DP cells lacked the expression of the master transcription factor gene of Treg cells FOXP3 and STAT5A and STAT5B, our data indicate that they are distinct from Treg cells. We cannot rule out the possibility that the DP cell population may contain Tr1-like cells, as they share many molecular markers. It would be interesting to compare DP cells with Tr1-like cells from the same individuals. However, we were unable to distinguish them based on our CyTOF data at hand, supporting the notion that DP cells may contain Tr1-like cells.
IL-10+IFN-γ+ DP CD4 T cells have been previously reported in murine models of malaria, Leishmania major, Toxoplasma gondii, and chronic lymphocytic choriomeningitis virus (LCMV) infection (Anderson et al., 2007; Freitas do Rosário et al., 2012; Jankovic et al., 2007; Parish et al., 2014), and in humans with malaria, tuberculosis, and Borrelia burgdorferi infection (Boyle et al., 2017; Gerosa et al., 1999; Jagannathan et al., 2014, 2016; Pohl-Koppe et al., 1998; Walther et al., 2009). Malaria-specific DP cells are the most predominant in younger children who have a higher prior malaria incidence and have been recently exposed to malaria (Boyle et al., 2017; Jagannathan et al., 2014). In addition, children receiving antimalarial chemoprevention had reduced frequencies of DP cells (Jagannathan et al., 2016), suggesting that DP cells are not associated with protection in the context of malaria. In the present study, we extend these findings and report antigen-specific DP CD4 T cells during an acute viral infection. We show that DENV-specific DP cells mainly exhibit a CD45RA−CCR7− effector/Tem phenotype and upregulate both T-bet and Blimp-1 protein expression, which is consistent with previous findings on malaria-specific DP cells (Boyle et al., 2017; Jagannathan et al., 2014).
In addition to IL-10, DENV-specific DP cells showed an enhanced production of another member of the IL-10 family of cytokines—IL-22—which mainly acts on epithelial and stromal cells to mediate protection against various pathogens such as bacteria (Ouyang and O’Garra, 2019). IL-22 also has important functions in maintaining tissue homeostasis and facilitating tissue repair (Ouyang and O’Garra, 2019). Since severe dengue diseases are associated with tissue damage and plasma leakage (St John and Rathore, 2019), the IL-22 manufactured by DENV-specific DP cells may help to foster tissue regeneration. DENV-specific DP cells were also capable of producing IL-21, which exerts pleiotropic effects on the differentiation and function of both B and T cells (Spolski and Leonard, 2014; Tian et al., 2016a; Tian and Zajac, 2016). Notably, IL-21 plays important roles in promoting the generation of plasma cells and configuring germinal center (GC) and antibody responses by acting directly on GC B cells and indirectly on follicular CD4 T cell populations, including Tfh and follicular regulatory T (Tfr) cells (Spolski and Leonard, 2014; Tian and Zajac, 2016). This is of interest in DENV infection because DENV-specific antibodies not only can neutralize the virus but also may exacerbate dengue pathogenesis via mechanisms that include antibody-dependent enhancement (ADE), depending upon their titer, affinity, and avidity (St John and Rathore, 2019). IL-21 also promotes the synthesis of IL-22 in CD4 T cells (Yeste et al., 2014). Further studies will be necessary to define whether and how these cytokines secreted by DENV-specific DP cells modulate cellular and humoral immune responses against DENV and the development of dengue disease.
Previous studies have raised the concern that T cells may contribute to the pathogenesis of severe dengue disease, presumably via their production of proinflammatory cytokines, especially TNF (Rothman, 2011). However, we did not detect upregulated TNF gene expression in DENV-specific DP cells in this study. Although the frequency of DENV-specific DP cells was higher in DHF patients than in DF patients, the gene expression profile of these cells remained largely unchanged, suggesting that DENV-specific DP cells may play a minimal role in the development of DHF. It should be noted that DSS cases were not analyzed in this study. Since we focused on DENV-specific CD4 T cells in this study, we cannot rule out the possibility that CD8 T cells may be associated with immunopathology. Moreover, although the functional significance of antigen-specific DP CD4 T cells and their role in protection and/or immunopathology after DENV infection warrants further investigation, one may speculate that DENV-specific CD4 T cells may upregulate IL-10 production as a compensatory mechanism to calm the excessive inflammation in DHF patients. It has been postulated that Th1 cells can upregulate the production of IL-10 to restrict inflammation at the expense of antigen persistence and that the relative production of IFN-γ and IL-10 regulates the balance between antigen clearance and immunopathology (O’Garra and Vieira, 2007).
In summary, our results reveal an intriguing DENV-specific CD4 T cell subset that is capable of producing both the pro-inflammatory cytokine IFN-γ and the anti-inflammatory cytokine IL-10 during the acute phase of DENV infection. Our findings further demonstrate that the transcriptomic profile of these DP cells was similar in acute DF and DHF patients, arguing against the notion that altered CD4 T cell phenotype or function may be a determinant of DHF.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yuan Tian (ytian@lji.org). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study approval
Ethical approval to carry out this work is maintained through the La Jolla Institute for Immunology Institutional Review Board (protocols VD-085 and VD-101), the Medical Faculty of the University of Colombo (which served as a National Institutes of Health-approved institutional review board for Genetech and Kotelawala Defense University, protocols EC.15.002 and EC.15.095), the Nicaragua Ministry of Health (protocol CIRE-01/10/06–13.Ver14), and UC Berkeley Institutional Review Board (protocol 2010–06-1649). All clinical investigations have been conducted according to the principles expressed in the Declaration of Helsinki. All participants provided written informed consent prior to participation in the study.
Human PBMC samples
Sri Lanka acute donors
All samples have been collected in the North Colombo Teaching Hospital, Ragama in Gampaha District, Sri Lanka and the National Institute of Infectious Diseases, Gothatuwa, Angoda, Sri Lanka. These donations are derived from small blood volumes (5–10 ml) from anonymous patients at the time of diagnosis/admission (median day 4 from fever onset; acute fever sample), at discharge from the hospital (median day 7 from fever onset), and after recovery from the disease (median day 31 from fever onset; convalescent sample) as outlined in Table S3. One group comprises patients that were diagnosed as uncomplicated dengue fever (DF) cases with no sign of bleeding or pleural effusion, while the other group was classified as dengue hemorrhagic fever (DHF).
Nicaragua acute donors
Samples have been collected in The Hospital Infantil Manuel de Jesús Rivera (HIMJR) in the capital city of Managua. Acute fever samples have been collected on day 7 after fever onset. DHF was defined as fever with hemorrhagic manifestations, thrombocytopenia, and hemoconcentration or other signs of plasma leakage. The details of the donors used were listed in Table S3.
Sri Lanka healthy control donors
Blood donations from five healthy adults from this dengue-endemic area were obtained from healthy adult blood donors from the National Blood Center, Ministry of Health, Colombo, Sri Lanka in an anonymous fashion as previously described (Weiskopf et al., 2013). Donors were of both sexes and between 18 and 60 years of age. Seronegativity was confirmed by DENV specific IgG ELISAs and flow cytometry-based neutralization assays as previously described (Kraus et al., 2007). The details of the donors used were listed in Table S3.
San Diego healthy control donors
Blood donations from five healthy adults from this non-dengue-endemic area were obtained by the Clinical Studies Core at the La Jolla Institute for Immunology (LJI) in San Diego. The details of the donors used were listed in Table S3.
METHOD DETAILS
IFN-γ/IL-10 capture assay and cell sorting for RNA sequencing
Human PBMCs were rested overnight at 37°C in RPMI 1640 medium (catalog# RP-21, Omega Scientific, Inc.) supplemented with 5% human serum (catalog# 100–512, Gemini Bio-Products), 2 mM L-alanyl-L-glutamine (GlutaMAX-I, catalog# 35050061, Thermo Fisher Scientific), 100 U/ml penicillin, and 100 μg/ml streptomycin (catalog# 400–109, Gemini Bio-Products) and subsequently stimulated with DENV MP (1 μg/ml for individual peptides) or Dynabeads Human T-Activator CD3/CD28 for T Cell Expansion and Activation (catalog# 11132D, Thermo Fisher Scientific) for 3 h at 37°C. IFN-γ- and IL-10-producing cells were labeled using IFN-γ Secretion Assay - Detection Kit (PE), human (catalog# 130–054-202, Miltenyi Biotech) and IL-10 Secretion Assay – Detection Kit (APC), human (catalog# 130–090-761, Miltenyi Biotech), respectively, according to the manufacturer’s instructions. Subsequently, PBMCs were stained with anti-human CD3, CD4, CD8, CD14, CD19, CD45RA, and CCR7 (see Table S4 for antibody details). CD4 IL-10−IFN-γ− DN, IL-10+IFN-γ− IL-10 SP and IL-10−IFN-γ+ IFN-γ SP, and IL-10+IFN-γ+ DP cells were sorted using a FACSAria II cell sorter. Data were analyzed using FlowJo.
Microscaled RNA sequencing
For each condition, 100 cells were directly collected at 4°C in 8 μL of lysis buffer composed by 0.2% Triton X-100, 2 U/μl of recombinant RNase inhibitor (Takara), 5 mM dNTP mix (Life Technologies) in 0.2 mL PCR tube (Maxymum recovery, Axygen). Right after sort, tubes were vortexed at medium speed, spun, and stored at −80°C until the completion of the whole set of samples. Microscaled RNA-Seq library preparation was performed in technical duplicates for each biological condition. Briefly, 4 μL of each sample was amplified following the Smart-seq2 protocol (Picelli et al., 2014; Rosales et al., 2018). mRNA was captured using poly-dT oligos and directly reverse-transcribed into full-length cDNA using the described template-switching oligo (Picelli et al., 2014; Rosales et al., 2018). cDNA was amplified by PCR for 19 cycles and purified using AMPure XP magnetic bead (0.9:1 (vol:vol) ratio, Beckman Coulter). From this step, for each sample, 1 ng of cDNA was used to prepare dual-index barcoded standard NextEra XT libraries (NextEra XT DNA library prep kit and index kits; Illumina). Both whole-transcriptome amplification and sequencing library preparations were performed in a 96-well format to reduce assay-to-assay variability. Quality control steps were included to determine the optimal number of PCR preamplification cycles, and library fragment size. Samples that failed quality controls were eliminated from downstream steps. Library that passed strict quality controls were pooled at equimolar concentration, loaded and sequenced on the Illumina Sequencing platform, HiSeq2500 (Illumina). Technical libraries were sequenced and technical duplicate datasets merged in order to obtain more than 15 million 50-bp single-end reads (HiSeq Rapid Run Cluster and SBS Kit V2; Illumina) mapping uniquely to human transcriptome reference (GRCh38.gencode.v27B), generating a total of about 1.2 billion mapped reads.
RNA-sequencing analysis
The single-end reads that passed Illumina filters were filtered for reads aligning to tRNA, rRNA, adaptor sequences, and spike-in controls. The reads were then aligned to GRCh38 reference genome and Gencode 27 annotations using TopHat v1.4.1 (Trapnell et al., 2009). DUST scores were calculated with PRINSEQ Lite v0.20.3 (Schmieder and Edwards, 2011) and low-complexity reads (DUST > 4) were removed from the BAM files. The alignment results were parsed via the SAMtools (Li et al., 2009) to generate SAM files. Read counts to each genomic feature were obtained with the htseq-count program (Anders et al., 2015) using the “union” option. After removing absent features (zero counts in all samples), the raw counts were then imported to DESeq2 to identify differentially expressed genes between groups (Love et al., 2014). The shrinkage estimator apeglm was used to generate the shrunken log2 fold changes and compute s-values (Zhu et al., 2019). s-values provide the estimated rate of false sign among genes with equal or smaller s-value and are analogous to q-values (Stephens, 2017). Genes with an s-value of < 0.005 and a shrunken log2 fold change of > 1 or < −1 were considered significantly differentially expressed between groups. Enrichment analysis was performed using Metascape (Zhou et al., 2019).
CyTOF
PBMCs were stimulated with DENV MP (1 μg/ml for individual peptides) in the presence of brefeldin A (GolgiPlug, BD Biosciences, catalog# 51–2301KZ) for 6 h. Cells were then stained with the viability marker Cisplatin (194Pt, Fluidigm, catalog# 201194) followed by a surface antibody cocktail. Subsequently, cells were fixed in PBS with 2% paraformaldehyde overnight at 4°C. The following day, cells were stained with an intracellular/intranuclear antibody cocktail after fixation and permeabilization using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience, catalog# 00–5523-00). Before sample acquisition, cellular DNA was labeled with Cell-ID Intercalator-Ir (Fluidigm, catalog# 201192B). Samples were then acquired using a Helios mass cytometer (Fluidigm) with the wide-bore injector (Lee et al., 2019). Antibodies used in CyTOF were listed in Table S5. Data were analyzed using FlowJo and viSNE analysis of CyTOF data was conducted using Cytobank (Kotecha et al., 2010).
TCR analysis
MiXCR (Bolotin et al., 2015, 2017) was used to extract TCR beta-chain CDR3 repertoires from RNA-seq data of sorted CD4 T cell subsets. Subsequent TCR beta-chain diversity analysis was performed using VDJtools (Shugay et al., 2015).
QUANTIFICATION AND STATISTICAL ANALYSIS
Two-tailed Mann-Whitney (unpaired) or Wilcoxon (paired) test was used to determine statistical significance between groups using the Prism software (GraphPad, La Jolla, CA). Statistical details of experiments can be found in the figure legends. Statistical details of differential gene expression analysis can be found in the Method Details.
DATA AND CODE AVAILABILITY
The accession number for the RNA-sequencing data reported in this paper is Gene Expression Omnibus (GEO): GSE132367 and ImmPort: SDY 888. The accession number for the CyTOF data reported in this paper is ImmPort: SDY 888.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3, Alexa Fluor 700 | BioLegend | Cat#317340; Clone OKT3; RRID: AB_2563408 |
| CD4, APC eFluor 780 | eBioscience | Cat#47–0049–42; Clone RPA-T4; RRID: AB_1272044 |
| CD8, Brilliant Violet 650 | BioLegend | Cat#301042; Clone RPA-T8; RRID: AB_2563505 |
| CD14, V500 | BD Biosciences | Cat#561391; Clone M5E2; RRID: AB_10611856 |
| CD19, V500 | BD Biosciences | Cat#561121; Clone HIB19; RRID: AB_10562391 |
| CD45RA, eFluor 450 | eBioscience | Cat#48–0458–42; Clone HI100; RRID: AB_1272059 |
| CCR7, PerCP Cy5.5 | BioLegend | Cat#353220; Clone G043H7; RRID: AB_10916121 |
| CD14, 89Y | Biolegend | Cat#301802; Clone M5E2; RRID: AB_314184 |
| CD19, 89Y | Biolegend | Cat#302247; Clone HIB19; RRID: AB_2562815 |
| CD3, 115In | Biolegend | Cat#300402; Clone UCHT1; RRID: AB_314056 |
| CD223/LAG3, 141Pr | Biolegend | Cat#369302; Clone 11C3C65; RRID: AB_2616876 |
| TCF7, 142Nd | Biolegend | Cat#655202; Clone 7F11A10; RRID: AB_2562103 |
| IL-7R (CD127), 143Nd | Fluidigm | Cat#3143012B; Clone A019D5; RRID: AB_2810240 |
| CD70, 144Nd | Biolegend | Cat#355102; Clone 113–16; RRID: AB_2561429 |
| CD4, 145Nd | Fluidigm | Cat#3145001B; Clone RPA-T4; RRID: AB_2661789 |
| CD8a, 146Nd | Fluidigm | Cat#3146001B; Clone RPA-T8; RRID: AB_2687641 |
| IRF4, 147Sm | Biolegend | Cat#646402; Clone IRF4.3E4; RRID: AB_2280462 |
| CD278/ICOS, 148Nd | Fluidigm | Cat#3148019B; Clone C398.4A; RRID: AB_2756435 |
| CD30/TNFRSF8, 149Sm | R&D Systems | Cat#MAB229; Clone 81337; RRID: AB_2207591 |
| IL-22, 150Nd | Fluidigm | Cat#3150007B; Clone 22URTI; RRID: AB_2810972 |
| CCR1, 151Eu | R&D Systems | Cat#MAB145–100; Clone 53504; RRID: AB_2074021 |
| CX3CR1, 152Sm | Biolegend | Cat#341602; Clone 2A9–1; RRID: AB_1595422 |
| TIGIT, 153Eu | Fluidigm | Cat#3153019B; Clone MBSA43; RRID: AB_2756419 |
| HAVCR2 (TIM-3/CD366), 154Sm | Fluidigm | Cat#3154010B; Clone F38–2E2 |
| IRF8, 155Gd | Biolegend | Cat#656502; Clone 7G11A45; RRID: AB_2562395 |
| CD86, 156Gd | Fluidigm | Cat#3156008B; Clone IT2.2; RRID: AB_2661798 |
| CD137, 158Gd | Fluidigm | Cat#3158013B; Clone 4B4–1 |
| CD357/GITR, 159Tb | Fluidigm | Cat#3159020B; Clone 621 |
| T-bet, 160Gd | Fluidigm | Cat#3160010B; Clone 4B10; RRID: AB_2810251 |
| CD109, 161Dy | R&D Systems | Cat#MAB4385; Clone 496920; RRID: AB_10645622 |
| Foxp3, 162Dy | Fluidigm | Cat#3162024A; Clone 259D/C7 |
| TGFbeta, 163Dy | Fluidigm | Cat#3163010B; Clone TW4–6H10 |
| CXCR5 (CD185), 164Dy | Fluidigm | Cat#3164016B; Clone 51505; RRID: AB_2687858 |
| IFNg, 165Ho | Fluidigm | Cat#3165002B; Clone B27 |
| IL-10, 166Er | Fluidigm | Cat#3166008B; Clone JES3–9D7 |
| CCR7 (CD197), 167Er | Fluidigm | Cat#3167009A; Clone G043H7 |
| Ki-67, 168Er | Fluidigim | Cat#3168001B; Clone Ki-67; RRID: AB_2810856 |
| CD45RA, 169Tm | Fluidigim | Cat#3169008B; Clone HI100 |
| CD152 (CTLA-4), 170Er | Fluidigm | Cat#3170005B; Clone 14D3 |
| BLIMP1, 171Yb | Novus | Cat#NB600–235; Clone 3H2-E8; RRID: AB_10595225 |
| IL-21, 172Yb | Fluidigm | Cat#3172011B; Clone 3A3-N2; RRID: AB_2810975 |
| Granzyme B, 173Yb | Fluidigm | Cat#3173006B; Clone GB11; RRID: AB_2811095 |
| CD279 (PD-1), 174Yb | Fluidigm | Cat#3174020B; Clone EH12.2H7 |
| Perforin, 175Lu | Fluidigm | Cat#3175004B; Clone B-D48 |
| CCL3/CCL4, 176Yb | R&D Systems | Cat#MAB2701–100; Clone 93342; RRID: AB_2259652 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RPMI 1640 medium | Omega Scientific, Inc. | Cat#RP-21 |
| Human serum | Gemini Bio-Products | Cat#100–512 |
| GlutaMAX-I | Thermo Fisher Scientific | Cat#35050061 |
| Penicillin and streptomycin | Gemini Bio-Products | Cat#400–109 |
| DENV MP | Grifoni et al., 2017; Weiskopf et al., 2016 | N/A |
| Brefeldin A (GolgiPlug) | BD Biosciences | Cat#51–2301KZ |
| Cisplatin-194Pt | Fluidigm | Cat#201194 |
| Intercalator-Ir | Fluidigm | Cat#201192B |
| Foxp3/Transcription Factor Staining Buffer Set | eBioscience | Cat#00–5523–00 |
| Critical Commercial Assays | ||
| Dynabeads Human T-Activator CD3/CD28 for T Cell Expansion and Activation | Thermo Fisher Scientific | Cat#11132D |
| IFN-γ Secretion Assay – Detection Kit (PE), human | Miltenyi Biotech | Cat# 130–054–202 |
| IL-10 Secretion Assay – Detection Kit (APC), human | Miltenyi Biotech | Cat# 130–090–761 |
| Deposited Data | ||
| RNA-seq data | This paper | GEO: GSE132367; ImmPort: SDY 888 |
| CyTOF data | This paper | ImmPort: SDY 888 |
| Software and Algorithms | ||
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/ |
| GraphPad Prism 8 | GraphPad software | https://www.graphpad.com/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Metascape | Zhou et al., 2019 | http://metascape.org/gp/index.html#/main/step1 |
| Cytobank | Kotecha et al., 2010 | https://www.cytobank.org/ |
| MiXCR | Bolotin et al., 2015, 2017 | https://mixcr.readthedocs.io/en/master/ |
| VDJtools | Shugay et al., 2015 | https://vdjtools-doc.readthedocs.io/en/master/ |
Highlights.
DENV-specific IL-10+IFN-γ+ DP CD4 T cells are prominent during acute disease
Most DP cell DE genes are non-cytotoxic/Tr1 and include IL21, IL22, CD109, and CCR1
DP cells have similar gene expression in DF and DHF, despite higher frequency in DHF
Disease severity is not associated with altered DP cell phenotype or functionality
ACKNOWLEDGMENTS
We thank the La Jolla Institute for Immunology Flow Cytometry, Next-Generation Sequencing, and Bioinformatics core facilities for services, as well as the members of the Peters and Sette laboratories for their help. The FACSAria II cell sorter and the CyTOF mass cytometer were acquired through the Shared Instrumentation Grant (SIG) Program S10 RR027366 and S10 OD018499, respectively. The Illumina HiSeq 2500 Sequencer was purchased through NIH S10 OD016262. This work was performed as a project of the Human Immunology Project Consortium (HIPC) and supported by National Institute of Allergy and Infectious Diseases grants U19 AI118626 and P01 AI106695 and NIH contracts HHSN272200900042C and HHSN27220140045C. L.M.D.-O.-P.was the recipient of a postdoctoral fellowship from the Brazilian National Council for Scientific and Technological Development (CNPq 232745/2014-5). J.M. was supported by PhD student fellowships from the Departamento Administrativo de Ciencia, Tecnología e Innovación (COLCIENCIAS) and Pontificia Universidad Javeriana (Convocatoria 727 Doctorados Nacionales). Y.T. was supported through the American Association of Immunologists Intersect Fellowship Program for Computational Scientists and Immunologists. The female icon and mosquito icon used in the graphical abstract were made by Freepik and Kiranshastry, respectively, from https://www.flaticon.com.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.098.
REFERENCES
- Adikari TN, Gomes L, Wickramasinghe N, Salimi M, Wijesiriwardana N, Kamaladasa A, Shyamali NL, Ogg GS, and Malavige GN (2016). Dengue NS1 antigen contributes to disease severity by inducing interleukin (IL)-10 by monocytes. Clin. Exp. Immunol 184, 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, and Pe’er D (2013). viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol 31, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CF, Oukka M, Kuchroo VJ, and Sacks D (2007). CD4(+)CD25 (−)Foxp3(−) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J. Exp. Med. 204, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. (2013). The global distribution and burden of dengue. Nature 496, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381. [DOI] [PubMed] [Google Scholar]
- Bolotin DA, Poslavsky S, Davydov AN, Frenkel FE, Fanchi L, Zolotareva OI, Hemmers S, Putintseva EV, Obraztsova AS, Shugay M, et al. (2017). Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol 35, 908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle MJ, Jagannathan P, Bowen K, McIntyre TI, Vance HM, Farrington LA, Schwartz A, Nankya F, Naluwu K, Wamala S, et al. (2017). The Development of Plasmodium falciparum-Specific IL10 CD4 T Cells and Protection from Malaria in Children in an Area of High Malaria Transmission. Front. Immunol 8, 1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butthep P, Chunhakan S, Yoksan S, Tangnararatchakit K, and Chuansumrit A (2012). Alteration of cytokines and chemokines during febrile episodes associated with endothelial cell damage and plasma leakage in dengue hemorrhagic fever. Pediatr. Infect. Dis. J 31, e232–e238. [DOI] [PubMed] [Google Scholar]
- Chareonsirisuthigul T, Kalayanarooj S, and Ubol S (2007). Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J. Gen. Virol 88, 365–375. [DOI] [PubMed] [Google Scholar]
- Chen LC, Lei HY, Liu CC, Shiesh SC, Chen SH, Liu HS, Lin YS, Wang ST, Shyu HW, and Yeh TM (2006). Correlation of serum levels of macrophage migration inhibitory factor with disease severity and clinical outcome in dengue patients. Am. J. Trop. Med. Hyg 74, 142–147. [PubMed] [Google Scholar]
- Cox MA, Kahan SM, and Zajac AJ (2013). Anti-viral CD8 T cells and the cytokines that they love. Virology 435, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Bangs DJ, Angelo MA, Cerpas C, Fernando A, Sidney J, Peters B, Gresh L, Balmaseda A, de Silva AD, et al. (2016). Immunodominant Dengue Virus-Specific CD8+ T Cell Responses Are Associated with a Memory PD-1+ Phenotype. J. Virol 90, 4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elong Ngono A, Chen HW, Tang WW, Joo Y, King K, Weiskopf D, Sidney J, Sette A, and Shresta S (2016). Protective Role of Cross-Reactive CD8 T Cells Against Dengue Virus Infection. EBioMedicine 13, 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RA, de Oliveira SA, Gandini M, Ferreira Lda.C., Correa G, Abiraude FM, Reid MM, Cruz OG, and Kubelka CF (2015). Circulating cytokines and chemokines associated with plasma leakage and hepatic dysfunction in Brazilian children with dengue fever. Acta Trop. 149, 138–147. [DOI] [PubMed] [Google Scholar]
- Flores-Mendoza LK, Estrada-Jiménez T, Sederño-Monge V, Moreno M, Manjarrez MDC, González-Ochoa G, Millán-Pérez Peña L, and Reyes-Leyva J (2017). IL-10 and socs3 Are Predictive Biomarkers of Dengue Hemorrhagic Fever. Mediators Inflamm. 2017, 5197592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas do Rosário AP, Lamb T, Spence P, Stephens R, Lang A, Roers A, Muller W, O’Garra A, and Langhorne J (2012). IL-27 promotes IL-10 production by effector Th1 CD4+ T cells: a critical mechanism for protection from severe immunopathology during malaria infection. J. Immunol 188, 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerosa F, Nisii C, Righetti S, Micciolo R, Marchesini M, Cazzadori A, and Trinchieri G (1999). CD4(+) T cell clones producing both interferon-gamma and interleukin-10 predominate in bronchoalveolar lavages of active pulmonary tuberculosis patients. Clin. Immunol 92, 224–234. [DOI] [PubMed] [Google Scholar]
- Green S, Vaughn DW, Kalayanarooj S, Nimmannitya S, Suntayakorn S, Nisalak A, Rothman AL, and Ennis FA (1999). Elevated plasma interleukin-10 levels in acute dengue correlate with disease severity. J. Med. Virol 59, 329–334. [PubMed] [Google Scholar]
- Grifoni A, Angelo MA, Lopez B, O’Rourke PH, Sidney J, Cerpas C, Balmaseda A, Silveira CGT, Maestri A, Costa PR, et al. (2017). Global Assessment of Dengue Virus-Specific CD4+ T Cell Responses in Dengue-Endemic Areas. Front. Immunol 8, 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn W, Sun W, Innis BL, Caudill J, and King AD (2003). Serotype-specific T(H)1 responses in recipients of two doses of candidate live-attenuated dengue virus vaccines. Am. J. Trop. Med. Hyg 69 (6, Suppl), 39–47. [DOI] [PubMed] [Google Scholar]
- Hackl H, Charoentong P, Finotello F, and Trajanoski Z (2016). Computational genomics tools for dissecting tumour-immune cell interactions. Nat. Rev. Genet 17, 441–458. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Rojanasuphot S, and Sangkawibha N (1983). Original antigenic sin in dengue. Am. J. Trop. Med. Hyg 32, 154–156. [DOI] [PubMed] [Google Scholar]
- Hatch S, Endy TP, Thomas S, Mathew A, Potts J, Pazoles P, Libraty DH, Gibbons R, and Rothman AL (2011). Intracellular cytokine production by dengue virus-specific T cells correlates with subclinical secondary infection. J. Infect. Dis 203, 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess Michelini R, Doedens AL, Goldrath AW, and Hedrick SM (2013). Differentiation of CD8 memory T cells depends on Foxo1. J. Exp. Med 210, 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan P, Eccles-James I, Bowen K, Nankya F, Auma A, Wamala S, Ebusu C, Muhindo MK, Arinaitwe E, Briggs J, et al. (2014). IFNγ/IL-10 co-producing cells dominate the CD4 response to malaria in highly exposed children. PLoS Pathog. 10, e1003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan P, Bowen K, Nankya F, McIntyre TI, Auma A, Wamala S, Sikyomu E, Naluwu K, Nalubega M, Boyle MJ, et al. (2016). Effective Antimalarial Chemoprevention in Childhood Enhances the Quality of CD4+ T Cells and Limits Their Production of Immunoregulatory Interleukin 10. J. Infect. Dis 214, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Feng CG, Goldszmid RS, Collazo CM, Wilson M, Wynn TA, Kamanaka M, Flavell RA, and Sher A (2007). Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J. Exp. Med 204, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaratne HE, Wijeratne D, Fernando S, Kamaladasa A, Gomes L, Wijewickrama A, Ogg GS, and Malavige GN (2018). Regulatory T-cells in acute dengue viral infection. Immunology 154, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha N, Krutzik PO, and Irish JM (2010). Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom Chapter 10, Unit10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus AA, Messer W, Haymore LB, and de Silva AM (2007). Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J. Clin. Microbiol 45, 3777–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Cui W, Amezquita RA, Gray SM, Guan T, Lu Y, Kobayashi Y, Flavell RA, Kleinstein SH, Craft J, and Kaech SM (2015). Production of IL-10 by CD4(+) regulatory T cells during the resolution of infection promotes the maturation of memory CD8(+) T cells. Nat. Immunol 16, 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Lu Y, Amezquita RA, Weinstein JS, Vander Heiden JA, Gupta NT, Kleinstein SH, Kaech SM, and Craft J (2017). Interleukin-10 from CD4+ follicular regulatory T cells promotes the germinal center response. Sci. Immunol 2, eaan4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Kelly G, Bradford S, Davila M, Guo XV, Amir E-D, Thrash EM, Solga MD, Lannigan J, Sellers B, et al. (2019). A Modified Injector and Sample Acquisition Protocol Can Improve Data Quality and Reduce Inter-Instrument Variability of the Helios Mass Cytometer. Cytometry A 95, 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow JC, Borochoff-Porte N, Durbin AP, Whitehead SS, Fimlaid KA, Bunn JY, and Kirkpatrick BD (2012). Primary vaccination with low dose live dengue 1 virus generates a proinflammatory, multifunctional T cell response in humans. PLoS Negl. Trop. Dis 6, e1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lühn K, Simmons CP, Moran E, Dung NT, Chau TN, Quyen NT, Thao TT, Van Ngoc T, Dung NM, Wills B, et al. (2007). Increased frequencies of CD4+ CD25(high) regulatory T cells in acute dengue infection. J. Exp. Med 204, 979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavige GN, Gomes L, Alles L, Chang T, Salimi M, Fernando S, Nanayakkara KD, Jayaratne S, and Ogg GS (2013). Serum IL-10 as a marker of severe dengue infection. BMC Infect. Dis 13, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, and Calame K (2006). Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol 7, 457–465. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, et al. (2003). Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med 9, 921–927. [DOI] [PubMed] [Google Scholar]
- Ngono AE, and Shresta S (2018). Immune Response to Dengue and Zika. Annu. Rev. Immunol 36, 279–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Garra A, and Vieira P (2007). T(H)1 cells control themselves by producing interleukin-10. Nat. Rev. Immunol 7, 425–428. [DOI] [PubMed] [Google Scholar]
- Ouyang W, and O’Garra A (2019). IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity 50, 871–891. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, and Hymowitz SG (2011). Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol 29, 71–109. [DOI] [PubMed] [Google Scholar]
- Parish IA, Marshall HD, Staron MM, Lang PA, Brüstle A, Chen JH, Cui W, Tsui YC, Perry C, Laidlaw BJ, et al. (2014). Chronic viral infection promotes sustained Th1-derived immunoregulatory IL-10 via BLIMP-1. J. Clin. Invest 124, 3455–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O’Rourke P, de Silva AD, Harris E, Peters B, Seumois G, Weiskopf D, et al. (2018). Precursors of human CD4+ cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol 3, eaan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Pohl-Koppe A, Balashov KE, Steere AC, Logigian EL, and Hafler DA (1998). Identification of a T cell subset capable of both IFN-gamma and IL-10 secretion in patients with chronic Borrelia burgdorferi infection. J. Immunol 160, 1804–1810. [PubMed] [Google Scholar]
- Prestwood TR, Morar MM, Zellweger RM, Miller R, May MM, Yauch LE, Lada SM, and Shresta S (2012). Gamma interferon (IFN-γ) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-α/β receptor-deficient mice. J. Virol 86, 12561–12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarolo MG, Gregori S, Bacchetta R, Battaglia M, and Gagliani N (2018). The Biology of T Regulatory Type 1 Cells and Their Therapeutic Application in Immune-Mediated Diseases. Immunity 49, 1004–1019. [DOI] [PubMed] [Google Scholar]
- Rosales SL, Liang S, Engel I, Schmiedel BJ, Kronenberg M, Vijayanand P, and Seumois G (2018).A Sensitive and Integrated Approach to Profile Messenger RNA from Samples with Low Cell Numbers. Methods Mol. Biol 1799, 275–302. [DOI] [PubMed] [Google Scholar]
- Rothman AL (2011). Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol 11, 532–543. [DOI] [PubMed] [Google Scholar]
- Schmieder R, and Edwards R (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton G, Mongkolsapaya J, Yacoub S, and Roberts C (2015). New insights into the immunopathology and control of dengue virus infection. Nat. Rev. Immunol 15, 745–759. [DOI] [PubMed] [Google Scholar]
- Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, Pogorelyy MV, Nazarov VI, Zvyagin IV, Kirgizova VI, et al. (2015). VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput. Biol 77,e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolski R, and Leonard WJ (2014). Interleukin-21: a double-edged sword with therapeutic potential. Nat. Rev. Drug Discov 13, 379–395. [DOI] [PubMed] [Google Scholar]
- St John AL, and Rathore APS (2019). Adaptive immune responses to primary and secondary dengue virus infections. Nat. Rev. Immunol 19, 218–230. [DOI] [PubMed] [Google Scholar]
- Stephens M (2017). False discovery rates: a new deal. Biostatistics 18, 275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, and Glimcher LH (2000). A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669. [DOI] [PubMed] [Google Scholar]
- Tian Y, and Zajac AJ (2016). IL-21 and T Cell Differentiation: Consider the Context. Trends Immunol. 37, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Cox MA, Kahan SM, Ingram JT, Bakshi RK, and Zajac AJ (2016a).A Context-Dependent Role for IL-21 in Modulating the Differentiation, Distribution, and Abundance of Effector and Memory CD8 T Cell Subsets. J. Immunol. 196, 2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Mollo SB, Harrington LE, and Zajac AJ (2016b). IL-10 Regulates Memory T Cell Development and the Balance between Th1 and Follicular Th Cell Responses during an Acute Viral Infection. J. Immunol. 197, 1308–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Sette A, and Weiskopf D (2016c). Cytotoxic CD4 T Cells: Differentiation, Function, and Application to Dengue Virus Infection. Front. Immunol 7, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Babor M, Lane J, Schulten V, Patil VS, Seumois G, Rosales SL, Fu Z, Picarda G, Burel J, et al. (2017). Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat. Commun 8, 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Babor M, Lane J, Seumois G, Liang S, Goonawardhana NDS, De Silva AD, Phillips EJ, Mallal SA, da Silva Antunes R, et al. (2019). Dengue-specific CD8+ T cell subsets display specialized transcriptomic and TCR profiles. J. Clin. Invest 130, 1727–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi SK, and Lahesmaa R (2014). Transcriptional and epigenetic regulation of T-helper lineage specification. Immunol. Rev 261, 62–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TT, Chuang YJ, Lin YS, Chang CP, Wan SW, Lin SH, Chen CL, and Lin CF (2014). Antibody-dependent enhancement infection facilitates dengue virus-regulated signaling of IL-10 production in monocytes. PLoS Negl. Trop. Dis 8, e3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubol S, Phuklia W, Kalayanarooj S, and Modhiran N (2010). Mechanisms of immune evasion induced by a complex of dengue virus and preexisting enhancing antibodies. J. Infect. Dis 201, 923–935. [DOI] [PubMed] [Google Scholar]
- Walther M, Jeffries D, Finney OC, Njie M, Ebonyi A, Deininger S, Lawrence E, Ngwa-Amambua A, Jayasooriya S, Cheeseman IH, et al. (2009). Distinct roles for FOXP3 and FOXP3 CD4 T cells in regulating cellular immunity to uncomplicated and severe Plasmodium falciparum malaria. PLoS Pathog. 5, e1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, and Sette A (2014). T-cell immunity to infection with dengue virus in humans. Front. Immunol 5, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, Broadwater A, Kolla RV, De Silva AD, de Silva AM, et al. (2013). Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA 110, E2046–E2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, Crotty S, Peters B, and Sette A (2015). Dengue virus infection elicits highly polarized CX3CR1 + cytotoxic CD4+ T cells associated with protective immunity. Proc. Natl. Acad. Sci. USA 112, E4256–E4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Angelo MA, Grifoni A, O’Rourke PH, Sidney J, Paul S, De Silva AD, Phillips E, Mallal S, Premawansa S, et al. (2016). HLA-DRB1 Alleles Are Associated With Different Magnitudes of Dengue Virus-Specific CD4+ T-Cell Responses. J. Infect. Dis 214, 1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AM, and Wraith DC (2016). Tr1-Like T Cells-An Enigmatic Regulatory T Cell Lineage. Front. Immunol 7, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin G, Zander R, Schauder DM, Chen Y, Weinstein JS, Drobyski WR, Tarakanova V, Craft J, and Cui W (2018). Single-cell RNA sequencing unveils an IL-10-producing helper subset that sustains humoral immunity during persistent infection. Nat. Commun 9, 5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, and Shresta S (2009). A protective role for dengue virus-specific CD8+ T cells. J. Immunol 182, 4865–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, Sette A, and Shresta S (2010). CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J. Immunol 185, 5405–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeste A, Mascanfroni ID, Nadeau M, Burns EJ, Tukpah AM, Santiago A, Wu C, Patel B, Kumar D, and Quintana FJ (2014). IL-21 induces IL-22 production in CD4+ T cells. Nat. Commun 5, 3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Miller R, Eddy WE, White LJ, Johnston RE, and Shresta S (2013). Role of humoral versus cellular responses induced by a protective dengue vaccine candidate. PLoS Pathog. 9, e1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Eddy WE, Tang WW, Miller R, and Shresta S (2014). CD8+ T cells prevent antigen-induced antibody-dependent enhancement of dengue disease in mice. J. Immunol 193, 4117–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger RM, Tang WW, Eddy WE, King K, Sanchez MC, and Shresta S (2015). CD8+ T Cells Can Mediate Short-Term Protection against Heterotypic Dengue Virus Reinfection in Mice. J. Virol 89, 6494–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Zhang R, Jin B, and Chen L (2015). Type 1 regulatory T cells: a new mechanism of peripheral immune tolerance. Cell. Mol. Immunol 12, 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Ibrahim JG, and Love MI (2019). Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zompi S, Santich BH, Beatty PR, and Harris E (2012). Protection from secondary dengue virus infection in a mouse model reveals the role of serotype cross-reactive B and T cells. J. Immunol 188, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-sequencing data reported in this paper is Gene Expression Omnibus (GEO): GSE132367 and ImmPort: SDY 888. The accession number for the CyTOF data reported in this paper is ImmPort: SDY 888.