Abstract
We have developed a reagentless optical biosensor for glutamine based on the Escherichia coli glutamine binding protein (GlnBP). Site-directed mutagenesis was performed to engineer single cysteine mutants which were covalently modified with environmentally sensitive sulfhydryl-reactive probes. The fluorescence emission of acrylodan and 2-(4′-(iodoacetamido)anilino)naphthalene-6-sulfonic acid (IAANS) attached to GlnBP mutant S179C was shown to decrease 65 and 35%, respectively, upon titration with increasing amounts of glutamine (0 to 6.4 μM; KDapp 160 nM). No significant changes in the fluorescence intensity were observed for the structurally similar amino acids glutamate, asparagine, and arginine. Time-resolved intensity decays showed a 2.4-fold decrease in mean lifetime for GlnBP S179C-acrylodan upon the addition of glutamine, indicating the possibility of a lifetime-based assay. Anisotropy decay measurements for GlnBP S179C-acrylodan showed a 13-ns rotational correlation time in the ligand-free state, whereas multiple correlation times were assigned in the glutamine-bound conformation. The decrease in fluorescence intensity of S179C-acrylodan was adapted to polarization sensing of glutamine. The engineered GlnBP is a first step toward the development of a nonenzymatic biosensor capable of determining glutamine concentrations in cell cultures.
In recent years, fluorescent sensors have been developed based upon the natural affinity and specificity between a protein and its substrate (1, 2). Site-directed mutagenesis has allowed alterations in amino acid sequences, resulting in changes in protein binding constants and insertion of new positions for reporter group labeling. Such amino acid changes have allowed the signal transduction of the binding event to be quantitated using a variety of physical and chemical techniques (3, 4). In particular, optical methods of detection using fluorescence energy transfer, polarization, and solvent sensitivity have been shown to offer high signal-to-noise ratios and the potential to construct simple and robust devices (5, 6). As a result, the use of such methods has allowed for the development of highly sensitive optical protein biosensors for a variety of analytes, including amino acids, sugars, and metabolic byproducts.
The Escherichia coli periplasmic space contains a diverse group of binding proteins whose main function is to present various biomolecules such as sugars, amino acids, peptides, and inorganic ions for transport into the cell (7). Although these proteins range in size and primary sequence, they have remarkably similar structures (8, 9). The three-dimensional structures are characterized by two globular domains connected by a hinge region which forms a cleft for substrate binding. X-ray crystallographic data consistently show that the binding of substrate induces large conformational changes in the tertiary structure of the protein. The changes in global structure of these proteins make them compelling targets to serve as the basis for new biosensor design.
Glutamine is an important energy and nitrogen source used in the culturing of eukaryotic cells (10, 11). Since the catabolism of glutamine leads to the buildup of ammonia which may become toxic to the growing culture and thus inhibit growth (12, 13), monitoring glutamine concentrations throughout the growth cycle is important. Current biosensors available for glutamine are based on immobilized glutaminase (4, 14). Complications in the determination of glutamine using this type of enzymatic biosensor arise due to the high degree of chemical similarity to glutamate as well as other amino acids. Consequently, additional proteins and electrodes must be added to take into account the interfering signals. In the present report, we introduce an optical assay for glutamine based upon the E. coli glutamine binding protein (GlnBP).1 Our design takes advantage of the high chemical specificity between GlnBP and the glutamine substrate. We have constructed single cysteine mutants of GlnBP to which environmentally sensitive fluorophores have been covalently attached, allowing for the determination of glutamine concentrations in solution. The engineered GlnBP is a first step toward a simple, optical biosensor for the determination of glutamine concentrations in cell cultures.
MATERIALS AND METHODS
Cloning
The gene encoding the periplasmic GlnBP was amplified from the E. coli genome using PCR. An upstream primer (5′-CTGCAGAGAAAAAAGGAAATGCTATGAA) including the Shine–Dalgarno sequence of GlnBP as well as the first three amino acid residues of the signal sequence was designed to introduce a PstI site. A downstream primer (5-AAGCTTTATTATTTCGGTTCAGTACCGA) covering the last five amino acids and the stop codon of GlnBP was used to introduce a HindIII site. The amplified 777 bp fragment was cloned into the PstI/HindIII site of the high copy number vector, pTZ18U (Bio-Rad, Hercules, CA). Transformation and subsequent expression of the resulting gene product was performed in E. coli strain HB101 (15). Single cysteine mutations were made using the Quick Change Mutagenesis kit from Stratagene (La Jolla, CA) as outlined by the manufacturer.
Purification
Purification of the wild-type and mutant binding proteins was performed as previously described (16). Release of the periplasmic proteins was achieved using either the chloroform shock method (17) or osmotic shock. The chloroform shock method was used to screen for E. coli expressing new mutants. Cells from 2-ml overnight cultures were pelleted and resuspended in approximately 20 μl of Luria broth (LB). Chloroform (20 μl) was added to the cells which were then vortexed briefly and allowed to sit for 10 min at room temperature. Phosphate buffer, pH 7.5 (80 μl; 20 mM), was added. Samples were vortexed briefly and spun for 20 min at 6000g. The supernatant containing the periplasmic proteins was treated with 1 μl fluoresceiniodoacetamide (20 mg/ml). The presence of a fluorescent band at 26 kDa on 15% Coomassie-stained SDS–PAGE denoted the expression of a single cysteine GlnBP mutant.
Osmotic shock was used for larger preparations of GlnBP. HB101 expressing GlnBP were grown overnight at 37°C in LB supplemented with 100 μg/ml ampicillin. Cells were harvested by centrifugation for 10 min at 6000g and resuspended in 20% sucrose, 30 mM Tris (pH 8.0), and 1 mM EDTA (200 ml per liter of cell culture). The suspension was shaken at room temperature for 30 min. Cells were collected by centrifugation (8000g, 10 min), and the periplasmic proteins were released by the addition of 5 ml ice-cold 10 mM MgSO4 per gram cells. Treated cells were shaken on ice for 10 min followed by centrifugation for 10 min at 6000g. Crude periplasmic preparations were equilibrated with phosphate buffer (pH 7.5) and applied to a DEAE–Sepharose column (Aldrich). Due to the high basicity of GlnBP, there was no adsorption at this pH, and thus, the protein appears in the flowthrough fractions. These fractions were pooled and concentrated using Amicon (Beverly, MA) Centriprep-10 concentrators. At this stage fluorophore labeling as described in the next section was performed to identify fractions containing single cysteine mutants of GlnBP. Yields were typically 10 mg/L. Pooled DEAE–Sepharose fractions were electrofocused using a Rotofor 60 ml focusing chamber cell from Bio-Rad containing 1% ampholytes with a pH gradient of 3 to 10. Phosphoric acid (0.1 M) was used at the anode and sodium hydroxide (0.1 M) at the cathode. The ampholytes were focused for 2 h or until the voltage leveled off at 10 W. Protein purity was checked using Coomassie-stained SDS–PAGE.
Fluorophore Coupling
Fluorophores were covalently attached to the engineered mutants via a single cysteine residue. 6-Acryloyl-2-dimethylaminonaphthalene (acrylodan), (((2-(iodoacetoxy)ethyl)methyl)amino)-7-nitrobenz-2-oxa-1,3-diazole (IANBD), 2-(4′-(iodoacetamido)anilino)naphthalene-6-sulfonic acid (IAANS), N-(4,6-dimethylamino-2-benzofuranyl)phenyl maleimide (DAFP), and tris-(2-carboxyethyl)phosphine, HCl (TCEP) were obtained from Molecular Probes (Eugene, OR) and used without further purification. A threefold excess of fluorophore in DMSO was added dropwise to a phosphate-buffered protein solution (pH 7.2) containing a slight excess of TCEP (Molecular Probes). Conjugation was allowed to occur for 2 h at room temperature or overnight at 4°C. Removal of unreacted dye was preformed by gel filtration over Sephadex G-25 (Sigma-Aldrich) using the same phosphate buffer. The extent of labeling was calculated from the protein concentration determined by the Pierce Coomassie Plus protein assay reagent (Rockford, IL) and the peak extinction coefficients for the individual fluorophores, assuming no change occurred upon protein labeling resulting in dye-to-protein ratios of 0.6 to 0.8. A minimum of three labeling preparations were performed for each experiment. Less than 3% variability in emission intensity changes was observed between preparations regardless of the size of the culture or the amount of protein labeled. The conjugates were stable for a period of approximately 5 months at 4°C as determined by glutamine binding assays.
Binding Assay
All measurements were carried out using 500 nM labeled GlnBP in 20 mM phosphate buffer (pH 7.2) unless otherwise stated. Steady-state fluorescence data were performed on a SLM 8000 spectrofluorometer. Apparent binding constants were calculated from titration data using the equation
| [1] |
where ΔF is the change in fluorescence emission, ΔFmax is the change in fluorescence in the presence of saturating amounts of glutamine, S is the concentration of glutamine, and KD is the apparent binding constant.
Time-Resolved Spectroscopy
Time-resolved frequency domain intensity and anisotropy decays were performed on a previously described GHz instrument (18). Excitation was accomplished using the frequency-doubled output of a cavity dumped pyridine-2 dye laser at 365 nm synchronously pumped by a mode-locked argon ion laser (514 nm). Acrylodan emission was monitored using a Corning 3–71 long pass filter. For intensity decay measurements, excitation and emission polarizers were set at 0 and 54°, respectively, to eliminate the intrinsic polarization of the system. DCS (4-(dimethylamino)-4′-cyanostilbene) in methanol with a lifetime of 0.46 ns was used as reference.
The intensity decay data were described by the multiexponential model,
| [2] |
where αi is the amplitude of each component of the decay, τi is the respective decay time, and ∑αi = 1.0 The amplitude average lifetime is given by The differential phase and modulated anisotropy data were fit to the multicorrelation model,
| [3] |
where r0j is the amplitude of the total anisotropy which decays with a correlation time θj. Analysis was performed using nonlinear least squares as previously described (19–21).
RESULTS
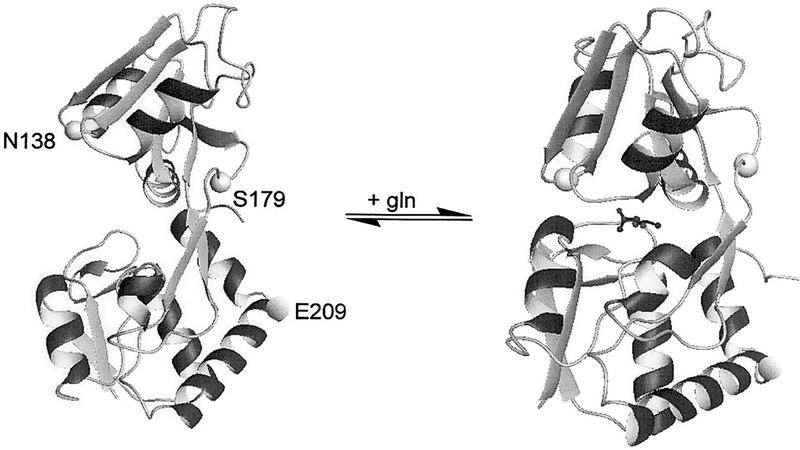
A number of E. coli periplasmic binding proteins have been utilized for the development of potential biosensors for phosphate (22–24), glucose (25, 26), and maltose (27). Due to the favorable sensitivity and specificity for glutamine, GlnBP was selected as the basis for the optical biosensor. The X-ray crystal structure of GlnBP has been determined in both the ligand-free and ligand-bound states (9, 28). Based upon the known structures of each conformation as well as the exposure of residues to water solvent, amino acid residues were selected for mutagenesis (Fig. 1). Residues S179 and E209 showed the greatest change in solvation and were therefore modified to a cysteine residue for labeling with environmentally sensitive fluorophores. In addition, a N138C mutation was made based upon previous data using other binding proteins which showed that fluorophores placed near the binding pocket displayed sensitivity to ligand binding (22, 26).
FIG. 1.
Structure of E. coli GlnBP with and without glutamine. The amino acids which were mutated to cysteine residues are indicated. Structures were prepared using the program MOLMOL (36).
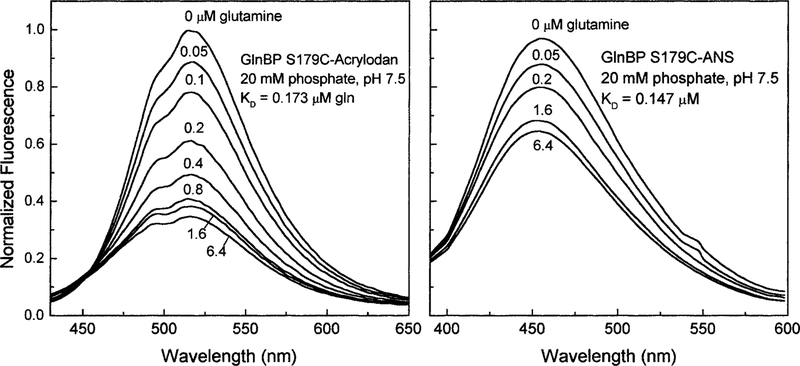
Each of the single cysteine mutants was covalently modified with four different environmentally sensitive fluorophores (Table 1). The largest changes in fluorescence emission were observed for S179C GlnBP. For this mutant, acrylodan and IAANS showed the most significant decreases in emission intensity of 65 and 35%, respectively (Fig. 2).
TABLE 1.
Response to Glutamine Using Mutants of E. coli Glutamine Binding Protein Covalently Modified with Solvent-Sensitive Fluorophores
| fluorophore | % change | ||
|---|---|---|---|
| N138C | S179C | E209C | |
| Acrylodan | 10.9 | −65 | nda |
| NBDamide | 8.4 | −2 | −3 |
| DAFP | 10 | −4.6 | nd |
| IAANS | −7 | −35 | 20 |
No detectable change.
FIG. 2.
The fluorescence intensity response of the GlnBP mutant S179C labeled with two different environmentally sensitive fluorophores, acrylodan (left) and 1,8-IAANS (right).
Because Ser179 is located away from the ligand binding site, it was predicted that there would be minimal effects on the interaction of glutamine with the binding protein. Using the emission data (Fig. 2) and Eq. [1], binding constants for glutamine were determined to be approximately 160 ± 42 nM. This result correlates well with a dissociation constant of 100 to 300 nM which was determined for the wild-type protein (16).
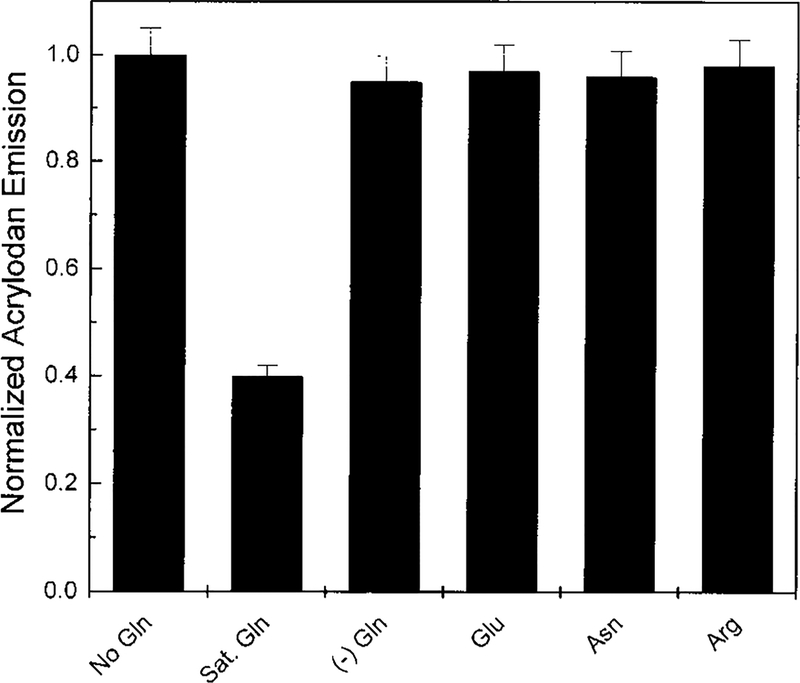
A major drawback to currently used glutamine biosensors, most of which are designed around glutaminase (4), is the interference from glutamate. Antiglutamate layers are frequently needed to correct for this potential problem which results in the increased complexity and cost of the biosensor (14, 29, 30). Therefore, it is necessary to consider the specificity of the GlnBP S179C-acrylodan system. No significant change in the acrylodan fluorescence is observed upon the addition of 20 μM glutamate or structurally similar amino acids, asparagine, and arginine (Fig. 3). These data agree with previous equilibrium dialysis experiments which showed that of the naturally occurring amino acids only glutamine is able to interact with the GlnBP (16). A detailed description of the interactions between glutamine and the GlnBP is given by the crystallographic data reported by Sun et al. (28). This group postulates that binding specificity of GlnBP for glutamine is conferred by His156 and Lys115 which form hydrogen bonds with Oϵ1 which is unique to the neutral amide side chain of glutamine.
FIG. 3.
Effect of structurally similar amino acids on the fluorescence emission of GlnBP S179C-acrylodan. (−) Gln shows the increase of the emission intensity of the system once glutamine has been removed.
Since the system under study is a binding protein, a second consideration is reversibility, which is an indicator of reusability of this type of sensor. The on-rate and off-rate of GlnBP are reported to be 9.8 × 107 M−1 s−1 and 16 s−1, respectively (16). As expected from the rapid off-rate of GlnBP, results showed that the GlnBP S179C-acrylodan system was freely reversible for glutamine (Fig. 3). This was determined by dilution of a concentrated sample of ligand saturated S179C-acrylodan and measurement of the change in fluorescence emission over time. The acrylodan emission returned to maximal level during the time it took to mix the sample. These data suggest the possibility for the real-time measurement of glutamine concentrations in solution.
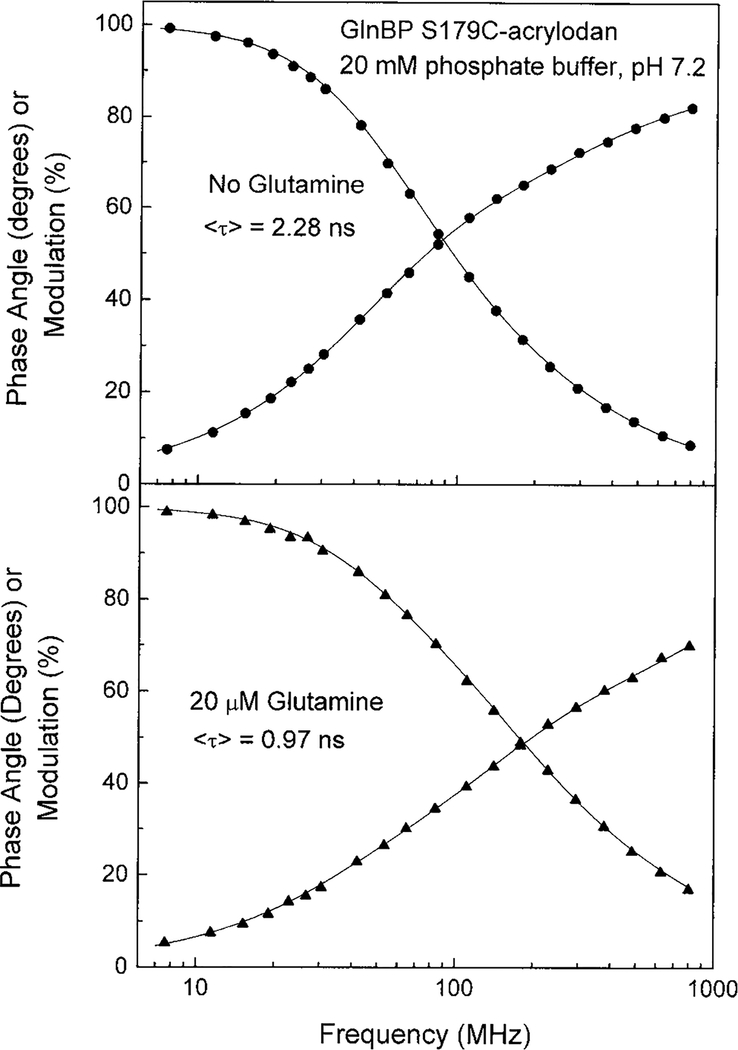
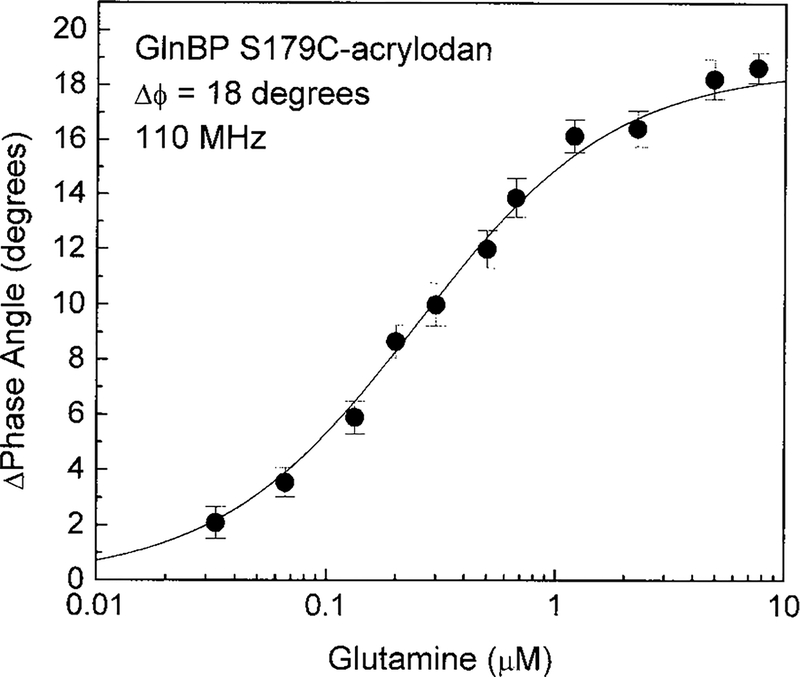
Advantages of lifetime-based sensing include insensitivity to probe concentration, photobleaching, excitation intensity, and emission path (31). Since there is no possibility for quantitative measurement of glutamine using ratiometric sensing of S179C-acrylodan emission alone, it was necessary to determine if a change in fluorescence lifetime accompanied the decrease in emission intensity of mutant S179C. Upon binding glutamine, the lifetime of S179C-acrylodan decreased from 2.28 to 0.97 ns (Fig. 4), which correlates well with the intensity changes observed (Fig. 2). The frequency at which the phase angle displayed the largest change was used to construct a plot of glutamine concentration versus phase angle of the acrylodyl probe (Fig. 5). There was a nonlinear response to glutamine with a maximum change in the phase angle at 110 MHz of 18°. Although ANS covalently attached to position 179 showed a 40% reduction in emission intensity (Table 1), no change in the fluorescence lifetime was observed (data not shown) which precludes the possibility of lifetime-based sensing using the ANS-labeled GlnBP.
FIG. 4.
Time-resolved intensity decays of the GlnBP S179C-acrylodan in the absence (top) and presence (bottom) of glutamine.
FIG. 5.
The phase angle measured at 110 MHz of GlnBP S179C-acrylodan as a function of glutamine concentration.
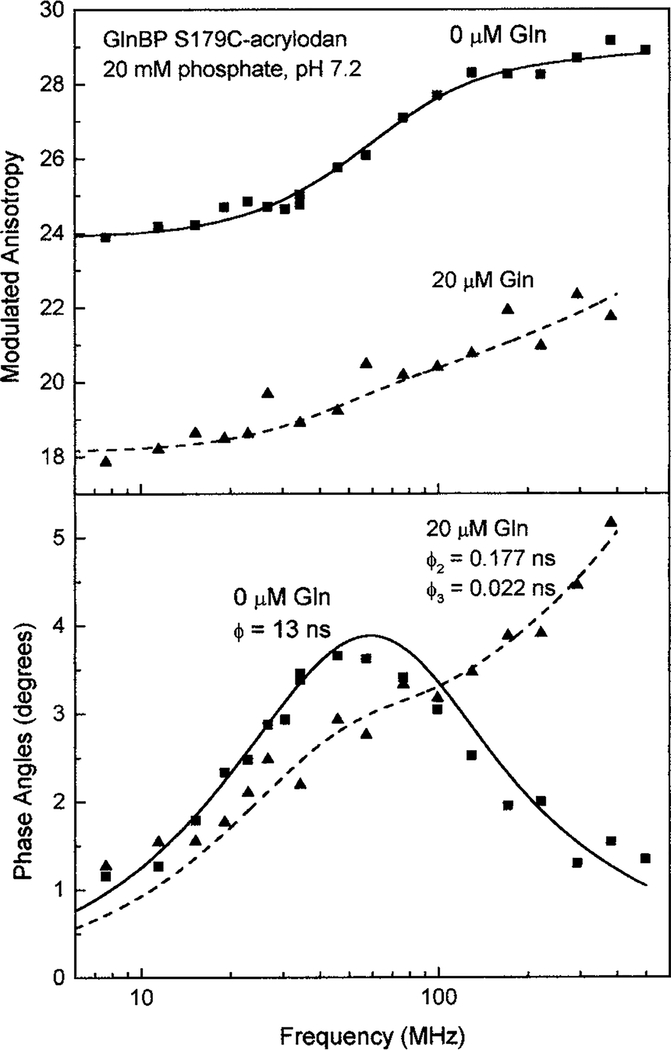
The anisotropy decay was measured in the presence and absence of glutamine to determine some characteristics of the microenvironment of acrylodan bound to position 179 (Fig. 6). Residue 179 was found to be localized at the interface between the large upper domain and hinge 2 of GlnBP (28). In the absence of glutamine, a single rotational correlation time of 13.1 ns for acrylodan emission was found. This suggested that the fluorophore was able to interact with the protein backbone around the modified region and thus displayed hindered rotation. The recovered correlation time was in good agreement with a predicted time of 11.7 ns for a hydrated 26-kDa protein, as calculated using the definition of rotational correlation time (32). Upon adding 20 μM glutamine to GlnBP S179C-acrylodan, two additional correlation times of 0.177 and 0.022 ns were resolved (Table 2). The longer correlation time most likely resulted from an increased mobility of the protein segment to which the acrylodan moiety was bound, while the shorter indicated free rotation of the probe following the conformational change. The GlnBP undergoes a 34.3° rotation around the ψ angle of Glu181. It is possible that the acrylodan was able to interact with nearby amino acid residues within hinge 2 (181–185) which following rotation were no longer available. Although the crystallographic data for GlnBP showed a more compact structure following the addition of glutamine, the anisotropy data suggested that the modified region of the protein may become less ordered after substrate binding.
FIG. 6.
Modulated anisotropy (top) and differential phase angles (bottom) of GlnBP S179C-acrylodan in the ligand free (circles) and bound (triangles) state.
TABLE 2.
Anisotropy Decay Data for GlnBP S179C-Acrylodan-Free and Saturated with Glutamine
| ϕi (ns) | r0i | ∑r0i | χθR | |
|---|---|---|---|---|
| No gln | 13.1 | 0.29 | 0.29 | 2.7 |
| 20 μM gln | 9.5 | 0.202 | ||
| 0.177 | 0.060 | |||
| 0.022 | 0.050 | 0.31 | 1.7 |
Note. The errors in the measurements were taken to be δϕ = 0.04 and δm = 0.007 for phase and modulation, respectively.
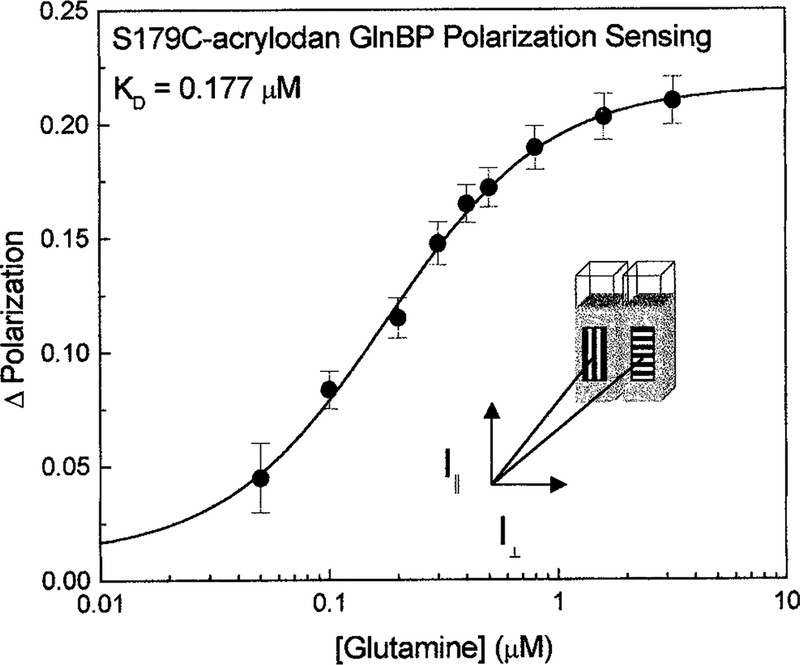
An additional characterization of the GlnBP S179C-acrylodan system was made using a new method recently been developed in our lab that allows for quantitative polarization measurements on any fluorophore displaying changes in emission intensity to be made (33–35). These measurements do not give the actual polarization of the samples, but rather code a sample and reference emission by orthogonally polarized light. Polarizing film oriented orthogonally to each other was placed in the emission path on the outside of two cuvettes. A second, rotatable polarizer was inserted downstream in the emission path (Fig. 7, inset). Measurement of the combined emission from the cuvettes housing identical solutions of GlnBP S179C-acrylodan was made, while glutamine was titrated into one of the cuvettes. By rotating the downstream polarizer between 0 and 90°, simple intensity data were transformed into a ratiometric measurement. An apparent increase of 220 mP with increasing glutamine concentrations was observed (Fig. 7). Since it is well known that polarizations values can be accurately determined to better than 0.005, this method offers the possibility of accurate and quantitative glutamine sensing using only the emission intensity.
FIG. 7.
Glutamine-dependent polarization sensing using S179C-acrylodan.
CONCLUSIONS
We report a first generation reagentless, optical assay for glutamine using the E. coli. GlnBP with a single cysteine mutation of serine179. To develop the assay system presented here into a working biosensor for the measurement of glutamine in cell cultures, several considerations must be taken into account. Glutamine concentrations in cell culture media ranges from 0.2 to 5 mM. Due to the nanomolar binding constant for glutamine, significant dilution of cell culture media would be necessary to make it possible to measure this level of glutamine using the current GlnBP system. However, due to the fast on- and off-rates for GlnBP, the assay is reversible, which opens the possibility to real-time measurements in the nanomolar range. In addition, the high degree of specificity with which GlnBP recognizes glutamine may remove the need for additional proteins. Hence, the complexity and cost of a glutamine biosensor based on GlnBP may be reduced. Although this sensor performs well in solution, many factors may affect the local environment of a fluorophore resulting in changes in the emission properties. Future research into the development of a glutamine biosensor using GlnBP will be directed toward other quenching strategies based on energy-transfer-type mechanisms.
ACKNOWLEDGMENTS
The authors thank Govind Rao and Leah Tolosa for many helpful discussions. J.D.D. thanks Mathew Foshrink and Tammy Halle from the Department of Medical and Research Technology for their contributions. This research was supported by a grant from the NIH National Center for Research Resources (RR-08119).
Footnotes
Abbreviations used: GlnBP, glutamine binding protein; acrylodan, 6-acryloyl-2-dimethylaminonaphthalene; IAANS, 2-(4′-(iodoacetamido)anilino)naphthalene-6-sulfonic acid; DAFP, N-(4,6-dimethylamino-2-benzofuranyl)phenyl maleimide; IANBD, (((2-(iodoacetoxy)ethyl)methyl)amino)-7-nitrobenz-2-oxa-1,3-diazole; TCEP, tris-(2-carboxyethyl)phosphine, HCl; LB, Luria broth.
REFERENCES
- 1.Coulet PR, and Bardeletti G (1991) Biochem. Soc. Trans 19, 1–4. [DOI] [PubMed] [Google Scholar]
- 2.Hellinga HW, and Marvin JS (1998) Trends Biotechnol. 16, 183–189. [DOI] [PubMed] [Google Scholar]
- 3.Ramsden JJ (1997) J. Mol. Recogn 10, 109–120. [DOI] [PubMed] [Google Scholar]
- 4.Sorochinskii VV, and Kurganov BI (1997) Appl. Biochem. Microbiol 33, 515–529. [Google Scholar]
- 5.Giuliano KA, Post PL, Hahn K, and Taylor DL (1995) Annu. Rev. Biophys. Biomol. Struct 24, 405–434. [DOI] [PubMed] [Google Scholar]
- 6.Giuliano KA, and Taylor DL (1998) Trends Biotechnol. 16, 135–140. [DOI] [PubMed] [Google Scholar]
- 7.Higgins CF (1992) Annu. Rev. Cell Biol 8, 67–113. [DOI] [PubMed] [Google Scholar]
- 8.Adams MD, and Oxender DL (1989) J. Biol. Chem 264, 15739–15742. [PubMed] [Google Scholar]
- 9.Hsiao CD, Sun UJ, Rose J, and Wang B-C (1996) J. Mol. Biol 262, 225–242. [DOI] [PubMed] [Google Scholar]
- 10.Thomas JN (1986) in Mammalian Cell Technology (Thilly WG, Ed.), pp. 109–130, Butterworths, Boston. [Google Scholar]
- 11.Zielke HR, Zielke CL, and Ozand PT (1984) Fed. Proc 43, 121–125. [PubMed] [Google Scholar]
- 12.Ozturk SS, and Palsson BO (1990) Biotechnol. Progr 6, 121–128. [DOI] [PubMed] [Google Scholar]
- 13.Oh GS, Izuishi T, Inoue T, W. S. H, and Yoshida TJ (1996) J. Ferment. Bioeng 81, 329–336. [Google Scholar]
- 14.Madaras MB, Spokane RB, Johnson JM, and Wookward JR (1997) Anal. Chem 69, 3674–3678. [DOI] [PubMed] [Google Scholar]
- 15.Brown TA (1991) in Molecular Biology Lab Fax, p. 3, Bios Scientific Publishers Limited. [Google Scholar]
- 16.Weiner JH, and Heppel LA (1971) J. Biol. Chem 246, 6933–6941. [Google Scholar]
- 17.Ames G, F-L., Prody C, and Kustu S (1984) J. Bacteriol 160, 1181–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laczko G, Gryczynski I, Gryczynski Z, Wiczk W, Malak H, and Lakowicz JR (1990) Rev. Sci. Instrum 61, 2331–2337. [Google Scholar]
- 19.Lakowicz JR, Gratton E, Laczko G, Cherek H, and Limkeman M (1984) Biophys. J 46, 463–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gratton E, Lakowicz JR, Maliwal B, Cherek H, Laczko G, and Limkeman M (1984) Biophys. J 46, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakowicz JR, Cherek H, Kusba J, Gryczynski I, and Johnoson ML (1993) J. Fluorescence 3, 103–116. [DOI] [PubMed] [Google Scholar]
- 22.Burne M, Hunter JL, Corrie JET, and Webb MR (1994) Biochemistry 33, 8262–8271. [DOI] [PubMed] [Google Scholar]
- 23.Barman T, Brune M, Lionne C, Piroddi N, Poggesi C, Stehle R, Tesi C, Travers F, and Webb MR (1998) Biophys. J 74, 3120–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundgren JS, Sallins LLE, Kaneva I, and Daunert S (1999) Anal. Chem 71, 589–595. [DOI] [PubMed] [Google Scholar]
- 25.Tolosa L, Gryczynski I, Eichhorn L, Dattelbaum JD, Castellano FN, Rao G, and Lakowicz JR (1999) Anal. Biochem 267, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marvin JS, and Hellinga HW (1998) J. Am. Chem. Soc 120, 7–11. [Google Scholar]
- 27.Sloan DJ, and Hellinga HW (1998) Protein Eng 11, 819–823. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y-J, Rose J, Wang B-C, and Hsiao C-D (1998) J. Mol. Biol 278, 219–229. [DOI] [PubMed] [Google Scholar]
- 29.White SF, Turner APF, Biltewski U, Bradley J, and Schmid RD (1995) Biosensors Bioelectronics 10, 543–551. [DOI] [PubMed] [Google Scholar]
- 30.Cattaneo MV, Luong JHT, and Mercille S (1992) Biosensors Bioelectronics 7, 329–334. [DOI] [PubMed] [Google Scholar]
- 31.Szmacinski H, and Lakowicz JR (1992) in Topics in Fluorescence Spectroscopy (Lakowicz JR, Ed.), Vol. 4, Plenum, New York. [Google Scholar]
- 32.Lakowicz JR (2000) Principles of Fluorescence Spectroscopy, Plenum, New York. [Google Scholar]
- 33.Gryczynski I, Gryczynski Z, and Lakowicz JR (1999) Anal. Chem 71, 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lakowicz JR, Gryczynski I, Gryczynski Z, Tolosa L, Dattelbaum JD, and Rao G (1999) Appl. Spectrosc 53, 1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakowicz JR, Gryczynski I, Gryczynski Z, and Dattelbaum JD (1999) Anal. Biochem 267, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koradi R, Billeter M, and Wuthrich K (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]