Abstract
Long noncoding RNAs (lncRNAs) have recently been considered as central regulators in diverse biological processes controlling tumorigenesis. Small nucleolar RNA host gene 16 (SNHG16) is an important tumor-associated lncRNA mainly involved in tumorigenesis and progression by competing with endogenous RNA (ceRNA) which sponges tumor-suppressive microRNA (miRNA), and by its recruitment mechanism. SNHG16 is overexpressed in tumor tissues and cell lines of different kinds of cancers, and its presence is associated with a poor clinical prognosis. Reviewing all publications about SNHG16 revealed that it plays a key role in the different hallmarks that define human cancer, including promoting proliferation, activating migration and invasion, inhibiting apoptosis, affecting lipid metabolism and chemoresistance. This review highlights the role that the aberrant expression of SNHG16 plays in the development and progression of cancer, and suggests that SNHG16 may function as a potential biomarker and therapeutic target for human cancers.
Keywords: long noncoding RNA, biomarker, cancer, SNHG16
Plain Language Summary
Studies have shown that the long non-coding RNA SNHG16 plays a functional role in various human cancers and is closely associated with tumor growth, metastasis and poor prognoses.
SNHG16 is upregulated in hepatocellular carcinoma, osteoblastoma, lung cancer, colorectal cancer, glioma, ovarian cancer, bladder cancer, breast cancer, gastric cancer, retinoblastoma, cervical cancer, and thyroid cancer.
SNHG16 is overexpressed in several types of cancer and is implicated in multiple different hallmarks that define human cancer.
In order to have a deeper comprehension of this novel biomarker for prognosis and therapy in cancers, more studies are need for further analysis.
Introduction
Cancer is a group of malignant diseases in which cell growth, migration, and invasion are uncontrolled. The high mortality and disability rates associated with cancer are a cause of distress and burden on patients and society as a whole.1 Surgical treatment, chemotherapy, and radiation therapy are currently the main treatments for cancer. However, the therapeutic effects of these primary treatments remain limited.2 Therefore, early diagnostic markers and new treatments are urgently needed.
About 70–90% of the human genome is transcribed into RNA, but only 2% of the total RNA is translated into protein. The remaining RNA that is not translated into protein is called noncoding RNA (ncRNA).3 An ncRNA that is 200 nucleotides in length or shorter is categorized as a short ncRNA, whereas an ncRNA longer than 200 nucleotides is considered a long ncRNA (lncRNA).4 It was previously thought lncRNA did not play a role during transcription.5 However, studies have shown that lncRNAs can regulate gene expression at the epigenetic, transcriptional, and post-transcriptional levels. A number of lncRNAs are involved in biological processes, including genomic imprinting, histone modification, chromatin remodeling, and cell cycle regulation.6–8 Furthermore, studies have found a clear correlation between lncRNAs and tumorigenesis.9 Differences in lncRNA expression in tumor tissues and adjacent tissues, as well as in cancer cell lines and corresponding normal cells, have also been reported in various types of cancer. This differential expression may promote angiogenesis and induce metastasis by affecting the cell cycle and cytological behavior of tumors, thus promoting tumor development.10,11
Small nucleolar RNA host gene 16 (SNHG16), which is encoded by a 7571‐bp region at chromosome 17q25.1, has recently been recognized as a cancer-related lncRNA.12 The SNHG16 was initially discovered in neuroblastoma.13 The dysregulation of SNHG16 has been detected in various types of cancer, including colorectal cancer (CRC),14 hepatocellular carcinoma (HCC),15 osteosarcoma,16 and glioma.17 Recent studies have demonstrated that SNHG16 expression is upregulated in multiple types of tumors. Furthermore, downregulation of SNHG16 in cancer cells inhibits cell proliferation, invasion, and migration; induces apoptosis; and results in decreased N-cadherin and increased E-cadherin,15,18–23 suggesting that SNHG16 also acts as an oncogenic lncRNA in cancer.
Understanding the role of SNHG16 may provide a new perspective to study the mechanisms of cancer development. This paper reviews current studies on the expression, function, mechanism, and clinical significance on tumor development of SNHG16 and highlights its impact on the hallmarks of cancer.
Mechanisms of SNHG16 in Human Cancers
Further evidence shows that SNHG16 promotes tumor development by acting as a competitive endogenous RNA (ceRNA).24,25 The ceRNA hypothesis states that various RNA species, including mRNAs encoding proteins and RNAs without protein-encoding capabilities (eg, pseudogenes, lncRNA, and circular RNA), share miRNA binding sites, allowing for competitive binding to common miRNAs with pro- or anti-cancer effects. Thus, the inhibition of miRNAs on other targets can be alleviated. In this manner, lncRNAs can have tumor-promoting or -inhibiting abilities. The ceRNA model is a very important biological pathway and, through this pathway, lncRNAs can participate in the entire process of tumorigenesis.26 It is evident from previous literature that SNHG16 acts as a ceRNA to regulate mRNA by sponging corresponding miRNAs in multiple cancers.17,27 Argonaute-crosslinking and immunoprecipitation (AGO-CLIP) analysis led to the hypothesis that SNHG16 heavily binds to AGO. It has 27 AGO/miRNA target sites along its length and may act as a ceRNA for miRNA in cancer.14 In addition to the ceRNA mechanism, SNHG16 can also play a role in promoting cancer through other mechanisms. p21, a cell cycle-dependent protein kinase inhibitor belonging to the kinase inhibitory protein/cyclin-dependent kinase (CDK) -interacting protein(KIP/CIP) family, can form a cell cycle G1 checkpoint with p53. If DNA damage is not repaired, the cell cannot progress through the cell cycle, which reduces the replication and accumulation of damaged DNA, thus reducing the risk of oncogenesis.28 SNHG16 can directly inhibit the expression of p21 by enriching enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), thereby promoting proliferation and inhibiting apoptosis in bladder cancer cells.29 The function of SNHG16 in various biological processes is conferred primarily by binding with miRNAs as sponges or by interacting with proteins, including those active in cell proliferation, migration, invasion, and apoptosis.
Overexpression of SNHG16 and Clinical Significance in Cancer
It has been shown that SNHG16 expression is significantly upregulated in tumor tissues and cell lines, such as hepatocellular carcinoma (HCC), lung cancer (LC), colorectal cancer (CRC) and glioma.15,17,30,31 In addition, overexpression of SNHG16 indicates poor prognosis and is usually correlated with tumor size, lymph node metastasis, tumor grade, disease-free survival, and overall survival (OS). For example, to gain more in‐depth knowledge of tumor biology, nine co-expression modules were identified in the Cancer Genome Atlas database after screening and analysis. These modules identified four important prognosis‐related lncRNAs, including SNHG16, which may provide insights into the biological significance and clinical application of lncRNAs in breast cancer.32 The expression of SNHG16 can be upregulated by c-Myc, which is a key determinant gene in the pathogenesis of human malignancies, in oral squamous cell carcinoma.33 SNHG16 overexpression and its known clinical significance in human cancers are summarized in Tables 1 and 2.
Table 1.
Functional Characterization of SNHG16 in Various Cancers
| Cancer Type | Expression | Numbers of Cases | Mainly Assessed Cell Lines | Related Genes | Phenotypes Affected | Role | References |
|---|---|---|---|---|---|---|---|
| Non-small cell cancer | Up | 66 | A549 | miR-146a, MUC5AC, PCNA, MMP2, MMP9 | Proliferation, migration, invasion, tumorigenesis | Oncogenic | [32] |
| Hepatocellular carcinoma | Up | 40 | HepG2, SMMC7721 | miR-195 | Proliferation, migration, invasion, tumorigenesis | Oncogenic | [29] |
| Up | 50 | Hep-3B, Sk-hep-1 | miR-186, ROCK1 | Proliferation, migration, invasion | Oncogenic | [14] | |
| Up | 30 | HepG2 | Has-miR-17-5p, p62, mTOR, NF-κB | Proliferation, migration, invasion, cell cycle, apoptosis | Oncogenic | [26] | |
| Up | 34 | Huh7, HepG2 | miR-302a-3p, FGF19 | Proliferation | Oncogenic | [28] | |
| Up | 88 | SMMC7721 | miR-4500, STAT3, E-cadherin, N-cadherin | Proliferation, migration, invasion, EMT | Oncogenic | [27] | |
| Up | 103 | HepG2/SOR | miR-140-5p | Sorafenib resistance | Oncogenic | [25,30] | |
| Cervical cancer | Up | 78 | SiHa | miR-216-5p, ZEB1, E-cadherin, N-cadherin | Proliferation, migration, EMT | Oncogenic | [34,35] |
| Osteosarcoma | Up | 30 | SAOS2 | miR-16, ATG4B | Proliferation, migration, invasion | Oncogenic | [41] |
| Up | 96 | U2OS, SAOS2 | miR-98-5p, ZEB1, E2F5, STAT3 | Proliferation, migration, invasion, cell cycle, apoptosis | Oncogenic | [38] | |
| Up | 38 | U2OS, MG-63 | miR-1301, BCL-9 | Proliferation, migration, invasion | Oncogenic | [40] | |
| Up | 25 | U2OS, Hfob1.19, SaOS2 | miR-340, Caspase 3, caspase 7 | Viability, migration, invasion, apoptosis | Oncogenic | [15] | |
| Up | 20 | MG-63, U2OS | miR-205, ZEB1, caspase3, PARP | Proliferation | Oncogenic | [39] | |
| Papillary thyroid cancer | Up | 48 | IHH-4, TPC-1 | miR-497, BPNA, YAP1 | Proliferation, migration, invasion, apoptosis | Oncogenic | [42] |
| Glioma | Up | 40 | U251, LN229 | miR-373, PI3K, AKT, EGFR, MMP2, MMP9 | Proliferation, migration, invasion, apoptosis | Oncogenic | [48] |
| Up | 40 | LN229 | P21, cyclinD1, cyclinB1 | Proliferation, apoptosis, cell cycle | Oncogenic | [49] | |
| Up | 48 | U251, H4 | miR-4518, PRMT5, Bcl, Bax, PI3K, AKT, caspase3 | Proliferation, apoptosis | Oncogenic | [50] | |
| Colorectal cancer | Up | 56 | SW480, L0V0 | miR-200a-3p | Proliferation, migration, invasion, tumorigenesis | Oncogenic | [45] |
| Up | 281 | HCT116, SW480 | c-Myc | Viability, apoptosis, migration | Oncogenic | [13] | |
| Ovarian cancer | Up | 103 | SKOV3, H08910 | AKT, PAKT, MMP9 | Proliferation, migration, invasion | Oncogenic | [53] |
| Bladder cancer | Up | 275 | J82 | miR-17-5p, caspase3, E-cadherin, N-cadherin, TIMP3 | Viability, apoptosis, EMT, proliferation | Oncogenic | [35] |
| Up | 19 | T24 | Bcl, Bax, capase3, E-cadherin, N-cadherin, Wnt1, c-myc, P2Tkip1 | Proliferation, viability, migration, invasion, apoptosis | Oncogenic | [34] | |
| Up | 46 | T24, BIU67 | P21 | Proliferation, migration, invasion, apoptosis, cell cycle | Oncogenic | [56] | |
| Breast cancer | Up | 30 | MDA-MB-231, MCF-7 | miR-98, E2F5 | Migration | Oncogenic | [62] |
| Retinoblastoma | Up | 30 | SORB-50, Y-79 | miR-140-5p | Proliferation, colony formation | Oncogenic | [48] |
| Gastric cancer | Up | 54 | BGC-823, SGC-7901 | miR-628, NRP1 | Apoptosis | Oncogenic | [68] |
| Up | 122 | MGC-803 | JAK2, p-STAT3, miR-135a | Proliferation, migration, invasion, apoptosis | Oncogenic | [66] | |
| Pancreatic cancer | Up | 46 | ASPC-1 | miR-218-5p, HMGB1 | Migration, invasion | Oncogenic | [41] |
| Lymphoma | Up | 48 | OCL-LY7 | miR-497-5p, P1M1 | Proliferation, migration, invasion, apoptosis, cell cycle | Oncogenic | [50] |
Table 2.
Clinical Significance of SNHG16 in Diverse Cancers
| Cancer Type | Overexpression of SNHG16 and Clinical Features | References |
|---|---|---|
| Non-small cell cancer | Tumor size, TNM stage, lymph node metastasis, shorter DFS and OS | [32] |
| Hepatocellular carcinoma | TNM stage, metastasis | [29] |
| Tumor size, TNM stage, ALT (U/L) | [14] | |
| Shorter DFS and OS, multiple tumors, macro vascular invasion | [26] | |
| Shorter DFS and OS | [28] | |
| Lymph node involvement, TNM stage | [27] | |
| Tumor size, TNM stage, vascular invasion | [25] | |
| Tumor size, TNM stage, metastasis, portal vein tumor thrombus (PVTT) | [30] | |
| Cervical cancer | Tumor size, FIGO stage, lymph node metastasis, poor differentiation | [34,35] |
| Osteosarcoma | Tumor size, TNM stage, metastasis, shorter OS | [39] |
| Tumor size, TNM stage, distance, metastasis, shorter OS | [41] | |
| Papillary thyroid cancer | TNM stage, lymph nodes, | [42] |
| Glioma | Tumor size, WHO stage, shorter OS and PFS, | [51] |
| Colorectal cancer | Metastasis, lymph node metastasis | [13] |
| Ovarian cancer | Clinical stage, distance metastasis, shorter OS | [53] |
| Bladder cancer | Historical grade, tumor stage, lymph node metastasis | [35] |
| Metastasis, lymph nodes, pathological stage | [34] | |
| Gastric cancer | Lymph node metastasis, invasive depth, TNM stage | [67] |
| Pancreatic cancer | Tumor size, TNM stage, lymph node metastasis, differentiation | [41] |
More recently, SNHG16 has been studied most frequently in HCC. Several studies have found that the expression of SNHG16 is significantly increased in HCC tissues compared to adjacent noncancerous tissues. The expression of SNHG16 is also remarkably upregulated in HCC cell lines.15,18–23 Further analysis showed that high expression of SNHG16 is associated with high tumor grade, multiple tumors, macrovascular invasion, large tumor size, and lymph node status, as well as poor disease-free survival and shorter overall survival.15,19–21,23 In HCC patients, multivariate analysis identified SNHG16 overexpression as a significant independent predictor of poor prognosis.20,23
Moreover, there are other studies of SNHG16 and related clinical effects in non-small cell lung cancer (NSCLC), osteosarcoma, glioma, bladder cancer (BC), ovarian cancer (OC), pancreatic cancer (PC), and papillary thyroid cancer (PTC). SNHG16 expression is significantly upregulated in NSCLC tissues and cell lines.30 Reduced SNHG16 is correlated with poor prognosis and factors associated with tumor progression, including larger tumor size, advanced pathological stage, lymph node metastasis, and shorter OS, revealing that SNHG16 may serve as an independent predictor of poor prognosis in NSCLC.30 SNHG16 was also overexpressed in human osteosarcoma tissues and cell lines when compared to matched normal tissues and cell lines, respectively.16,36–39 Osteosarcoma patients with high SNHG16 expression are more likely to have shorter survival, larger tumors, more advanced TNM stage, metastasis, and poorer prognosis.37,39 Zhou et al revealed that high expression of SNHG16 was associated with poor clinical pathological parameters and shorter OS and progression-free survival, which indicated that it can act as an independent prognostic factor of glioma.40 Peng et al revealed that SNHG16 was positively correlated with miR-17-5p. High expression of SNHG16 and miR-17-5p was related to unfavorable clinicopathological factors, such as higher historical grade and more advanced tumor stage in bladder cancer. Yang et al reported that SNHG16 expression was significantly increased in human ovarian cancer (OC) as compared to normal ovarian tissue. In addition, higher SNHG16 expression was significantly associated with higher clinical grade, tumor size, and lymph node and distant metastasis in OC patients. Liu et al revealed that SNHG16 was upregulated in PC tissues and cell lines. Moreover, increased expression of SNHG16 was correlated with poor differentiation, advanced TNM stage, lymph node metastasis, and shorter survival.41 SNHG16 is upregulated in both papillary thyroid tissues and cell lines, which indicates advanced TNM stage and lymph node metastasis.42
In addition to sequencing in tumor tissues, lncRNA levels can also be measured in exosomes. Exosomes, which are microvesicles (70–120 nm) derived from endosomes secreted by many cell types, can participate in intercellular communication by transferring intracellular substances (such as proteins, lipids, and nucleic acids, including lncRNA).43 Certain lncRNAs within exosomes secreted by tumor cells have been described as candidate biomarkers to predict prognosis.25 Zhang et al evaluated the serum exosomal expression of SNHG16 in bladder cancer patients and found that SNHG16 expression was correlated with TNM stage and invasion and predicted poor prognosis.44 As a noninvasive marker of cancer prognosis, the measurement of lncRNA in exosomes is of great significance. Therefore, further studies are needed, including larger clinical samples, multicenter studies, and functional analysis.
These findings indicate that SNHG16 may serve as a potential biomarker for human cancer diagnosis, prognosis, and effective treatment.
Implication of SNHG16 in the Hallmarks of Cancer
Promoting Proliferation
Proliferation is considered to be the most basic feature of cancer cells.45 In normal cells, the regulation of proliferation is strictly controlled. The production and signal transmission of growth-promoting factors must be fully controlled, and checkpoints must be set up at each node of cell division to ensure tissue homeostasis (cell number, tissue structure, and function).46 In cancer cells, this balance is disrupted. SNHG16 has been shown to sustain proliferation of cancer cells in different types of cancers.
Many related mechanisms have been reported. SNHG16 is negatively correlated with known miRNAs that act as tumor suppressors. In HCC, Xie et al revealed that knockdown of SNHG16 inhibited cell proliferation of HCC cells by inhibiting the expression of proliferation protein Ki67 and miR195 acted as a direct target of SNHG16.22 Li et al found that the SNHG16/miR-302a-3p/fibroblast growth factor 19 (FGF19) axis was an important pathway in promoting proliferation of HCC cells.21 Lin et al found that SNHG16 modulates signal transducer and activator of transcription 3 (STAT3) expression by competitively binding miR-4500.20 In addition, Knockdown of SNHG16 hinders osteosarcoma cell proliferation. However, in osteosarcoma, mechanistic investigations revealed that SNHG16 could act as a sponge for miR-98-5p, miR-1301, miR-16, miR-340, and miR-205.16,36–39 Inhibition of miR-98-5p and miR205 increases zinc finger E-box binding homeobox 1 (ZEB1), E2F transcription factor 5 (E2F5), and STAT3 expression to promote tumorigenesis.36,37 Li et al reported that SNHG16 was revealed to bind with miR-200a-3p, leading to the proliferation of CRC cells.31 Zhou et al found that SNHG16 regulated proliferation and invasion of glioma cells through the miR-373/epidermal growth factor receptor (EGFR) axis by the phosphatidylinositol 3-kinase (PI3K) -protein kinase B (AKT) pathway.47 Xu et al explored the molecular mechanism and revealed that SNHG16 acts as a miR-140-5p sponge in RB carcinogenesis. In addition, suppressing SNHG16 can slow tumor growth in vivo.48
Moreover, SNHG16 can also promote cancer cell proliferation by activating cell cycle progression through G1/S transition. Knockdown of SNHG16 in osteosarcoma results in the inhibition of cell proliferation in vitro by the induction of a G0/G1 arrest.49 Zhu et al revealed that silencing of SNHG16 inhibited proliferation, migration, and invasion, in addition to causing cell cycle arrest. SHNG16 acts by repressing miR-497-5p, blocking its repression of interacting with Moloney murine leukaemia virus 1 in diffuse large B-cell lymphoma (DLBCL).50 SNHG16 can also stimulate cell cycle progression through repressing p21 by recruiting enhancer of zeste homolog 2 (EZH2) to its promoter region. p21 belongs to a family of cyclin-dependent kinases (CDK) inhibitors and has been reported to be able to block cell cycle progression at the G0/G1 checkpoint.51 p21 can therefore act as a tumor suppressor in bladder cancer.29
These results show the significance of SNHG16 in the molecular etiology of proliferation of cancer cells and suggest the potential application of SNHG16 as an essential diagnostic marker.
Activating Migration and Invasion
Metastasis is the main cause of cancer treatment failure and death. Its molecular mechanism is complex, involving multistep, multistage, multigene changes.2 At the initial stage of cancer formation, epithelial cells are transformed into cells with a mesenchymal phenotype through specific signaling pathways.52 The main characteristics of this transformation are decreased expression of cell adhesion molecules (such as E-cadherin), vimentin, rather than cytokeratin, as the main cytoskeletal protein, and the cells taking on the morphological characteristics of mesenchymal cells.53 This phenotypic plasticity, named epithelial-to-mesenchymal transition (EMT), causes epithelial cells to lose cell polarity, lose the connection with basement membrane and other epithelial phenotypes, and gain higher migration and invasion ability.52
Several studies have shown that the overexpression of SNHG16 can promote EMT and migration, thus promoting the invasion of cancer cells. Lin et al showed that inhibition of SNHG16 in HCC reverses EMT and represses cell migration and invasion in vitro.20 Peng et al reported metalloproteinases 3(TIMP3) can be downregulated by elevated SNHG16 and miR-17-5p, promoting proliferation, migration, invasion, and EMT in bladder cancer cells.54 Previous studies have shown that activating the Wnt/β-catenin pathway affected cell metastasis and induced EMT in cancers. Feng et al found that suppressing SNHG16 can inhibit proliferation, migration, invasion, and EMT and promote apoptosis in bladder cancer cells. They further stated that SNHG16 could sponge miR-98 and target STAT3 through the Wnt/β-catenin pathway to act as an oncogene in bladder cancer.55 And SNHG16 acts as a ceRNA in colorectal cancer cells to suppress miR-200a-3p and increase the expression of ZEB1 and, thereby, promote the expression of EMT-related genes, such as cadherins and vimentin, and promote cell migration and invasion. Similarly, Zhu et al revealed that silencing SNHG16 inhibits cell migration and invasion, and EMT in vitro, indicating that SNHG16 is involved in the tumorigenesis of cervical cancer (CC). Although the oncogenic function of SNHG16 in CC is not completely known, it is known to act in a miR-216-5p-dependent manner and positively regulate the miR-216-5p target ZEB1 in CC cells.34,35
In addition, SNHG16 can also regulate migration and invasion by sponging miRNA through transcriptional activation of mRNA. Chen et al reported SNHG16 might function as a ceRNA for miR-186 to regulate rho-associated coiled-coil-containing protein kinase 1 (ROCK1) expression, which plays important roles in regulating cell polarity and migration in HCC.15 Su et al have found that SNHG16 directly bind to miR-340 to promote OS cells migration and invasion.16 Additionally, Wang et al found that SNHG16 promotes B Cell Lymphoma 9 (BCL9) expression by sponging miR-1301 to facilitate the migration and invasion of osteosarcoma cells.38 Cai et al revealed that SNHG16 competitively binding miR-98 with E2F5 may promote breast cancer metastasis in an miR-98-dependent manner.56 Interestingly, in this model, SNHG16 transcript levels were significantly positively associated with E2F5 mRNA levels, indicating an activation loop of that metastasis process.56 A similar situation was posited in another study. Wang et al found that knockdown of SNHG16 suppressed gastric cancer cell migration and invasion in vitro. Further, mechanistic investigation revealed that SNHG16 could mediate Janus kinase 2 (JAK2) and STAT3 expression as a ceRNA of miR-135a. Additionally, STAT3 can affect the expression of SNHG16.57 Another study found that miR-628 is regulated by SNHG16 and its target Neuropilin-1 (NRP1) can promote tumor metastasis.58 SNHG16 was shown to promote cell invasion and migration in pancreatic cancer cells via mediating high mobility group box 1 (HMGB1) (a known oncogene in PC) expression through sponging miR-218-5p.41 Wen et al found that knockdown of SNHG16 inhibits migration and invasion of TPC-1 cells via regulating miR-497.42 Interestingly, Vahid found a correlation between SNHG16 expression and the vitamin D receptor (VDR), which supports the hypothesis of an interactive network between SNHG16, VDR, and miR-98 in the context of cancer.59 SNHG16 is expected to become a novel molecular marker for diagnosis, prognosis, and treatment of BC.
Moreover, matrix metalloproteinases (MMPs) are highly expressed in tumor tissues. MMPs cause degradation of protein components in the extracellular matrix and increased tumor invasion and metastatic risk. Xie et al found SNHG16 significant increase in expression of MMP-2 and MMP-9 through a novel SNHG16-miR-195 axis in HCC cells. SNHG16 was shown to induce OC cell migration and invasion through the upregulation of P-AKT and MMP-9, indicating its potential in promoting tumor progression by regulating the PI3K/AKT pathway and MMPs.60
Inhibiting Apoptosis
Apoptosis is a natural, multistep process that plays an important role in development and homeostasis.61 Cancer cells exhibit enhanced tolerance to both environmental and genomic stresses, resulting in resistance to apoptosis and tumor progression.62 The B-cell lymphoma protein 2 (Bcl-2) family proteins, PI3K-AKt pathway, and nuclear factor κB (NF-ΚB) signaling are vital regulators of apoptosis.63,64 It has been shown that overexpressed SNHG16 directly interacted with miR-4518 and promoted protein arginine methyltransferase 5 to inhibit apoptosis in glioma cells. Furthermore, downregulation of SNHG16 could lead to the repression of anti-apoptotic Bcl-2 family members and PI3K/AKt signaling.40 Additionally, Zhou et al revealed that SNHG16 suppressed the expression of p21, caspase 3, and caspase 9, while promoting cyclin D1 and cyclin B1 expression, to inhibit apoptosis in glioma cells.65 Silencing SNHG16 increases caspase 3/7 to promote apoptosis in osteosarcoma cells.16 NF-κB is a eukaryotic transcription factor that can cause abnormal expression of tumor necrosis factors and promote carcinogenesis and apoptosis. Zhang et al found that SNHG16 may act as an endogenous sponge of miR‐17‐5p to upregulate its target p62 and activate the mammalian target of rapamycin (mTOR) pathway to promote metabolism and proliferation in HCC cells, as well as activating PI3K/AKt and NF-κB signaling to resist apoptosis.19 Christensen et al reported SNHG16 overexpression is an early event in CRC, which is regulated by the Wnt pathway and c-Myc. They also found that knockdown of SNHG16 can reduce cell viability and induce apoptotic death.14 Another study found that miR-628 is regulated by SNHG16 and the expression of miR-628-3p is down in GC tissues and GC cell lines, and its target NRP1 can inhibit cell apoptosis.58
Affecting Lipid Metabolism
Lipid biosynthesis has been frequently observed in tumor tissues.66 Enhanced lipid synthesis is required for the metabolic reprograming of cancer cells and tumor development.66 Christensen et al found that SNHG16 is enriched in the cytoplasm of CRC cell lines and associated with ribosomes, which may lend a better understanding of its function and means of action.14 Overexpression of SNHG16 is positively correlated to the expression of Wnt-regulated transcription factors, including achaete-scute complex homolog 2 (ASCL2) and c-Myc. Moreover, knockdown of SNHG16 predominantly affects genes involved in lipid metabolism. Interestingly, these genes contain a common sequence motif, suggesting that a broad spectrum of lncRNA-miRNA targets may converge on a few co-target genes.14
Inhibiting Inflammation
There are few data available about the link between SNHG16 and cancer inflammation. NF-κB is a eukaryotic transcription factor that helps regulate the immune and inflammatory response, cell proliferation, apoptosis, tumor growth, and differentiation. NF-κB signal transduction can cause the abnormal expression of tumor necrosis factors (TNF) and promote tumorigenesis and apoptosis. Zhang et al found that SNHG16 may act as an endogenous sponge of miR‐17‐5p to upregulate its target p62 and activate the mTOR pathway to promote metabolism and proliferation in HCC cells, as well as activating the NF-κB pathway to aggravate inflammation.19
Chemoresistance and Vasculogenic Mimicry
There are evidences that SNHG16 is involved in drug resistance. Ye et al and Guo et al both found that knocking down SNHG16 increased sensitivity to sorafenib in HCC cells via acting as an endogenous sponge for miR-140-5p, which indicates that SNHG16 might be a promising therapeutic target to boost the effectiveness of chemotherapy for HCC patients.18,23 Osteosarcoma is a rare cancerous bone tumor characterized by high metastasis, rapid progression, and poor prognosis.67 In the past few decades, there has been great progress in the diagnosis and treatment of osteosarcoma, but the OS is still unsatisfactory.68 The molecular mechanisms underlying the development and progression of osteosarcoma remain unclear, and multidrug resistance severely limits the development and use of neoadjuvant chemotherapy.69 Therefore, it is particularly urgent to find feasible molecular therapeutic targets for osteosarcoma. Interestingly, increases in autophagy-related genes (such as ATG4B) via downregulation of miR-16 can reduce cell apoptosis by inducing cell autophagy, which leads to chemoresistance.39 This finding may signify novel therapeutic directions in osteosarcoma.
In addition, glioma is one of the most common and aggressive malignant primary brain tumors, with a 5-year survival rate of less than 10% and a median survival period of only 14 months.70 Glioma is poorly understood, and the dismal clinical outcome makes the study of molecular mechanisms in glioma urgent.71 The anti-angiogenic treatment of malignant glioma cells is an effective method to treat high-grade gliomas. However, the effect is limited in glioma due to the presence of vasculogenic mimicry (VM).72 Wang reported that knockdown of SNHG16 can promote the expression of miR-212-3p and inhibit VM to sensitize glioma to anti-angiogenic treatment, which provides deeper comprehension and a novel direction for the treatment of glioma.72 The results indicate that SNHG16 plays a key role in glioma and it can be a novel therapeutic target.
Conclusion and Future Perspectives
A growing number of studies have shown that dysregulated lncRNAs are potential oncogenes or tumor suppressors that play important roles in tumorigenesis and tumor progression. LncRNAs can be used as biological markers and therapeutic targets for tumors and have broad applications in the diagnosis and treatment of tumors. SNHG16 is a newly discovered lncRNA, and it has been shown to be upregulated in many tumor types and associated with tumor stage, lymph node metastasis, and tumor size, indicators of poor prognosis. SNHG16 binds to endogenous miRNAs, resulting in abnormal expression of downstream target genes or dysregulation of classical signaling pathways (Table 1, Figure 1). The studies discussed here highlight that the study of SNHG16 is of great significance for further study of the mechanism of tumor development and progression, and SNHG16 may be considered as a new diagnostic/prognostic biomarker and a therapeutic target for cancer. Although the role of SNHG16 in cancer has been studied, there are still many areas that need further research. For example, there are few studies on thyroid cancer, breast cancer and retinoblastoma, and the understanding of the molecular mechanisms by which it drive carcinogenesis is not deep enough, instead focusing on in vitro experiments. Further research should be focused on investigating the precise molecular regulatory mechanisms of SNHG16, and a larger cohort of tumor samples should also be included to facilitate the clinical application of SNHG16 as early as possible.
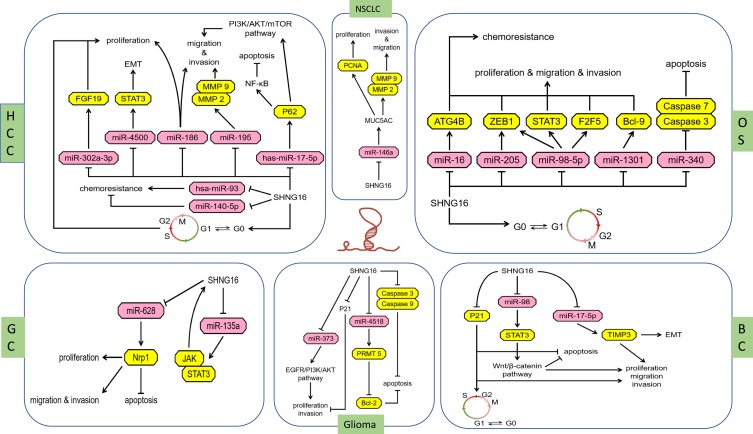
Figure 1.
The different regulatory mechanisms of SNHG16 in different human cancers.
Abbreviations: HCC, Hepatocellular carcinoma; NSCLC, non-small cell lung cancer; OS, Osteosarcoma; GC, gastric cancer; BC, bladder cancer.
Acknowledgment
All the authors thank the library of Beijing Tongren Hospital for assistance with literature search.
Funding Statement
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Abbreviations
BC, breast cancer; CC, cervical cancer; ceRNA, competitive endogenous RNA; CRC, colorectal cancer; DLBCL, diffuse large B-cell lymphoma; EMT, epithelial-mesenchymal transition; GC, gastric cancer; HCC, hepatocellular carcinoma; lncRNA, long noncoding RNA; MMP, matrix metalloproteinase; ncRNA, noncoding RNA; NSCLC, non-small cell lung cancer; OC, ovarian cancer; OS, overall survival; PC, pancreatic cancer; PI3K, phosphatidylinositol 3-kinase; PTC, papillary thyroid carcinoma; RB, retinoblastoma; SNHG16, small nucleolar RNA host gene 16; STAT3, Signal Transducers and Activators of Transcription 3; TNM, tumor, node, metastasis; VDR, vitamin D receptor; VM, vasculogenic mimicry; ZEB1, zinc finger E-box binding protein 1.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jones RM, Pattwell SS. Future considerations for pediatric cancer survivorship: translational perspectives from developmental neuroscience. Dev Cogn Neurosci. 2019;38:100657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peixoto A, Relvas-Santos M, Azevedo R, Santos LL, Ferreira JA. Protein glycosylation and tumor microenvironment alterations driving cancer hallmarks. Front Oncol. 2019;9(undefined):380. doi: 10.3389/fonc.2019.00380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi LL, Han F, Zhang XM, Li YY. LncRNA MT1JP acts as a tumor inhibitor via reciprocally regulating Wnt/β-Catenin pathway in retinoblastoma. Eur Rev Med Pharmacol Sci. 2018;22(13):4204–4214. doi: 10.26355/eurrev_201807_15414 [DOI] [PubMed] [Google Scholar]
- 5.Shang Y. LncRNA THOR acts as a retinoblastoma promoter through enhancing the combination of c-myc mRNA and IGF2BP1 protein. J Biomed Parmacother. 2018;106(undefined):1243–1249. doi: 10.1016/j.biopha.2018.07.052 [DOI] [PubMed] [Google Scholar]
- 6.Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18(1):206. doi: 10.1186/s13059-017-1348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricciuti B, Mencaroni C, Paglialunga L, et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33(2):18. doi: 10.1007/s12032-016-0731-2 [DOI] [PubMed] [Google Scholar]
- 8.Ellatif SKA, Gutschner T, Diederichs S. Long noncoding RNA function and expression in cancer. Noncoding RNA Res. 2012;4(2):45. [Google Scholar]
- 9.Dong P, Xiong Y, Yue J, et al. Exploring lncRNA-mediated regulatory networks in endometrial cancer cells and the tumor microenvironment: advances and challenges. Cancers. 2019;11(2):undefined. doi: 10.3390/cancers11020234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaforui-Fard S, Taheri M. Nuclear Enriched Abundant Transcript 1 (NEAT1): A long non-coding RNA with diverse functions in tumorigenesis. Biomed Pharmacotherapie. 2018;111(undefined):51–59. doi: 10.1016/j.biopha.2018.12.070 [DOI] [PubMed] [Google Scholar]
- 11.Weidle UH, Birzele F, Kollmorgen G, Rueger R. Long non-coding RNAs and their role in metastasis. Cancer Genom Proteom. 2017;14(3):143–160. doi: 10.21873/cgp.20027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin B, Liu Y, Li G, Xu Y, Cui W. The role of lncRNA SNHG16 in myocardial cell injury induced by acute myocardial infarction and the underlying functional regulation mechanism. Panminerva Med. 2019;undefined(undefined):undefined. [DOI] [PubMed] [Google Scholar]
- 13.Yu M, Ohira M, Li Y, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol. 2009;34(4):931–938. doi: 10.3892/ijo_00000219 [DOI] [PubMed] [Google Scholar]
- 14.Christensen LL, True K, Hamilton MP, et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10(8):1266–1282. doi: 10.1016/j.molonc.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Li M, Huang P. LncRNA SNHG16 promotes hepatocellular carcinoma proliferation, migration and invasion by regulating miR-186 expression. J Cancer. 2019;10(15):3571–3581. doi: 10.7150/jca.28428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su P, Mu S, Wang Z. Long noncoding RNA SNHG16 promotes osteosarcoma cells migration and invasion via sponging miRNA-340. DNA Cell Biol. 2019;38(2):170–175. doi: 10.1089/dna.2018.4424 [DOI] [PubMed] [Google Scholar]
- 17.Yang BY, Meng Q, Sun Y, Gao L, Yang JX. Long non-coding RNA SNHG16 contributes to glioma malignancy by competitively binding miR-20a-5p with E2F1. J Biol Regul Homeost Agents. 2018;32(2):251–261. [PubMed] [Google Scholar]
- 18.Ye J, Zhang R, Du X, Chai W, Zhou Q. Long noncoding RNA SNHG16 induces sorafenib resistance in hepatocellular carcinoma cells through sponging miR-140-5p. Onco Targets Ther. 2019;12(undefined):415–422. doi: 10.2147/OTT.S175176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong JH, Xiang X, Wang YY, et al. The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. J Cell Physiol. 2019;undefined(undefined):undefined. [DOI] [PubMed] [Google Scholar]
- 20.Lin Q, Zheng H, Xu J, Zhang F, Pan H. LncRNA SNHG16 aggravates tumorigenesis and development of hepatocellular carcinoma by sponging miR-4500 and targeting STAT3. J Cell Biochem. 2019;undefined(undefined):undefined. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Xu W, Song JS, Wu T, Wang WX. LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19 axis in hepatocellular carcinoma. Neoplasma. 2019;66(3):397–404. doi: 10.4149/neo_2018_180720N504 [DOI] [PubMed] [Google Scholar]
- 22.Xie X, Xu X, Sun C, Yu Z. Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp Cell Res. 2019;383(1):111501. doi: 10.1016/j.yexcr.2019.111501 [DOI] [PubMed] [Google Scholar]
- 23.Guo Z, Zhang J, Fan L, et al. Long noncoding RNA (lncRNA) small nucleolar RNA host gene 16 (SNHG16) predicts poor prognosis and sorafenib resistance in hepatocellular carcinoma. Med Sci Monit. 2019;25:2079–2086. doi: 10.12659/MSM.915541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grüll MP, Massé E. Mimicry, deception and competition: the life of competing endogenous RNAs. Interdiscip Rev RNA. 2019;10(3):e1525. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J, Li L, Han ZY, Wang ZX, Qin LX. Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am J Cancer Res. 2019;9(7):1367–1381. [PMC free article] [PubMed] [Google Scholar]
- 26.Lian Y, Xiong F, Yang L, et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37(1):253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao W, Fu H, Zhang S, Sun S, Liu Y. LncRNA SNHG16 drives proliferation, migration, and invasion of hemangioma endothelial cell through modulation of miR-520d-3p/STAT3 axis. Cancer Med. 2018;7(7):3311–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zohny SF, Al-Malki AL, Zamzami MA, Choudhry H. p21: its paradoxical effect in the regulation of breast cancer. Breast Cancer. 2019;26(2):131–137. [DOI] [PubMed] [Google Scholar]
- 29.Cao X, Xu J, Yue D. LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther. 2018;25(1):10–17. [DOI] [PubMed] [Google Scholar]
- 30.Han W, Du X, Liu M, Wang J, Sun L, Li Y. Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int J Biol Macromol. 2019;121:270–278. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Lu Y, Chen Y. Long non-coding RNA SNHG16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer via sponging miR-200a-3p. Biosci Rep. 2019;39(5):undefined. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Gao C, Liu C. Four lncRNAs associated with breast cancer prognosis identified by coexpression network analysis. J Cell Physiol. 2019;234(8):14019–14030. doi: 10.1002/jcp.28089 [DOI] [PubMed] [Google Scholar]
- 33.Li S, Zhang S, Chen J. c-Myc induced upregulation of long non-coding RNA SNHG16 enhances progression and carcinogenesis in oral squamous cell carcinoma. Cancer Gene Ther. 2019. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H, Zeng Y, Zhou CC, Ye W. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys. 2018;637:1–8. [DOI] [PubMed] [Google Scholar]
- 35.Aalijahan H, Ghorbian S. Long non-coding RNAs and cervical cancer. Exp Mol Pathol. 2019;106:7–16. [DOI] [PubMed] [Google Scholar]
- 36.Zhu C, Cheng D, Qiu X, Zhuang M, Liu Z. Long noncoding RNA SNHG16 promotes cell proliferation by sponging MicroRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell Physiol Biochem. 2018;51(1):429–440. [DOI] [PubMed] [Google Scholar]
- 37.Liao S, Xing S, Ma Y. LncRNA SNHG16 sponges miR-98-5p to regulate cellular processes in osteosarcoma. Cancer Chemother Pharmacol 2019;83(6):1065–1074. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Hu K, Chao Y, Wang L. LncRNA SNHG16 promotes proliferation, migration and invasion of osteosarcoma cells by targeting miR-1301/BCL9 axis. Biomed Pharmacother. 2019;114:108798. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Gu S, Li H, Wang J, Wei C, Liu Q. SNHG16 promotes osteosarcoma progression and enhances cisplatin resistance by sponging miR-16 to upregulate ATG4B expression. Biochem Biophys Res Commun. 2019;518(1):127–133. doi: 10.1016/j.bbrc.2019.08.019 [DOI] [PubMed] [Google Scholar]
- 40.Lu YF, Cai XL, Li ZZ. LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in glioma. Cell Physiol Biochem. 2018;45(5):1975–1985. [DOI] [PubMed] [Google Scholar]
- 41.Liu S, Zhang W, Liu K, Liu Y. LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p. Biomed Pharmacother. 2019;114:108862. [DOI] [PubMed] [Google Scholar]
- 42.Wen Q, Zhao L, Wang T, et al. LncRNA SNHG16 drives proliferation and invasion of papillary thyroid cancer through modulation of miR-497. Onco Targets Ther. 2019;12:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Nabet BY, Qiu Y, Shabason JE. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. 2017;170(2):352–366.e313. doi: 10.1016/j.cell.2017.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S, Du L, Wang L, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23(2):1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Liu L, Cai X, Yao Z, Huang J. Progress in the study of long noncoding RNA in tongue squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2019. [DOI] [PubMed] [Google Scholar]
- 46.Lecerf C, Le Bourhis X, Adriaenssens E. The long non-coding RNA H19: an active player with multiple facets to sustain the hallmarks of cancer. Cell Mol Life Sci. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2019. [DOI] [PubMed] [Google Scholar]
- 48.Xu C, Hu C, Wang Y, Liu S. Long noncoding RNA SNHG16 promotes human retinoblastoma progression via sponging miR-140-5p. Genomics. 2019;117:109153. [DOI] [PubMed] [Google Scholar]
- 49.Lian D, Amin B, Du D, Yan W. Enhanced expression of the long non-coding RNA SNHG16 contributes to gastric cancer progression and metastasis. Cancer Biomarkers. 2017;21(1):151–160. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Q, Li Y, Guo Y, et al. Long non-coding RNA SNHG16 promotes proliferation and inhibits apoptosis of diffuse large B-cell lymphoma cells by targeting miR-497-5p/PIM1 axis. J Cell Mol Med. 2019;23(11):7395–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9(6):400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehnman M, Chaabane W, Haglund F, Tsagkozis P. The tumor microenvironment of pediatric sarcoma: mesenchymal mechanisms regulating cell migration and metastasis. Curr Oncol Rep. 2019;21(10):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang XJ, Wang W, Hann SS. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie. 2019. [DOI] [PubMed] [Google Scholar]
- 54.Peng H, Li H. The encouraging role of long noncoding RNA small nuclear RNA host gene 16 in epithelial-mesenchymal transition of bladder cancer via directly acting on miR-17-5p/metalloproteinases 3 axis. Mol Carcinog. 2019;58(8):1465–1480. doi: 10.1002/mc.23028 [DOI] [PubMed] [Google Scholar]
- 55.Feng F, Chen A, Huang J, Xia Q, Chen Y, Jin X. Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/β-catenin pathway axis. J Cell Biochem. 2018;119(11):9408–9418. doi: 10.1002/jcb.27257 [DOI] [PubMed] [Google Scholar]
- 56.Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485(2):272–278. doi: 10.1016/j.bbrc.2017.02.094 [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Kan J, Han J, Zhang W, Bai L, Wu H. LncRNA SNHG16 functions as an oncogene by sponging MiR-135a and promotes JAK2/STAT3 signal pathway in gastric cancer. J Cancer. 2019;10(4):1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pang W, Zhai M, Wang Y, Li Z. Long noncoding RNA SNHG16 silencing inhibits the aggressiveness of gastric cancer via upregulation of microRNA-628-3p and consequent decrease of NRP1. Cancer Manag Res. 2019;11:7263–7277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Oskooei VK, Geranpayeh L, Omrani MD, Ghafouri-Fard S. Assessment of functional variants and expression of long noncoding RNAs in vitamin D receptor signaling in breast cancer. Cancer Manag Res. 2018;10:3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang XS, Wang GX, Luo L. Long non-coding RNA SNHG16 promotes cell growth and metastasis in ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(3):616–622. [DOI] [PubMed] [Google Scholar]
- 61.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407(6805):770–776. doi: 10.1038/35037710 [DOI] [PubMed] [Google Scholar]
- 62.Kerr JFR, Winterford CM, Harmon BVJ. Apoptosis. Its significance in cancer and cancer therapy. Cancer. 2015;73(8):2013–2026. doi: [DOI] [PubMed] [Google Scholar]
- 63.Wang CY, Mayo MW, Baldwin AS. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274(5288):784–787. doi: 10.1126/science.274.5288.784 [DOI] [PubMed] [Google Scholar]
- 64.Wang HG, Miyashita T, Takayama S, et al. Apoptosis regulation by interaction of Bcl-2 protein and Raf-1 kinase. Oncogene. 1994;9(9):2751–2756. [PubMed] [Google Scholar]
- 65.Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 exerts oncogenic functions in promoting proliferation of glioma through suppressing p21. Pathol Oncol Res. 2019. [DOI] [PubMed] [Google Scholar]
- 66.Lee YE, He HL, Shiue YL, et al. Li CF The prognostic impact of lipid biosynthesis-associated markers, HSD17B2 and HMGCS2, in rectal cancer treated with neoadjuvant concurrent chemoradiotherapy. Tumor Biol. 2015;36(10):7675–7683. doi: 10.1007/s13277-015-3503-2 [DOI] [PubMed] [Google Scholar]
- 67.Zhao G, Zhang L, Qian D, Sun Y, Liu W. miR-495-3p inhibits the cell proliferation, invasion and migration of osteosarcoma by targeting C1q/TNF-related protein 3. Onco Targets Ther. 2019;12:6133–6143. doi: 10.2147/OTT.S193937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Li J, Wang Y, Jing J, Li J. The roles of circular RNAs in osteosarcoma. Med Sci Monit. 2019;25:6378–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng ZQ, Lu RB, Xiao DM, Xiao ZM. Increased expression of the lncRNA BANCR and its prognostic significance in human osteosarcoma. Genet Mol Res. 2016. 15(1). [DOI] [PubMed] [Google Scholar]
- 70.Peng ZA, Lu RB, Xiao DM, Xiao ZM. RETRACTION of articles with plagiarism in common with other publications. Genet Mol Res GMR. 2016. 15(3). [DOI] [PubMed] [Google Scholar]
- 71.Tai PA, Liu YL, Wen YT, et al. The development and applications of a dual optical imaging system for studying glioma stem cells. Mol imaging. 2019;18:1536012119870899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang D, Zheng J, Liu X, et al. Knockdown of USF1 inhibits the vasculogenic mimicry of glioma cells via stimulating SNHG16/miR-212-3p and linc00667/miR-429 axis. Mol Ther Nucleic Acids. 2019;14:465–482. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]