Abstract
Background
Double homeobox A pseudogene 8 (DUXAP8) has been identified as a key regulator at the posttranscriptional level in various types of cancers. However, whether DUXAP8 has a role in hepatocellular carcinoma (HCC) progression remains to be determined. Here, we aimed to investigate the potential clinical value of DUXAP8 as a pan-cancer marker, and its role in HCC development through an integrated analysis strategy and in vitro experimental validation.
Methods
Comprehensive analysis was performed using data mined from public databases to evaluate the expression patterns and clinical value of DUXAP8 in human pan-cancers. Bioinformatics analysis was performed to investigate the potential biological functions of DUXAP8 in HCC based on TCGA database. Real-time qPCR analysis was used to examine the expression levels of DUXAP8 in HCC tissue samples and cell lines. DUXAP8-siRNA was used to silence DUXAP8 in the Hep-G2 cell line to examine the role of DUXAP8 in HCC cell proliferation and invasion.
Results
DUXAP8 was significantly upregulated in various types of human cancers and could serve as a potential pan-cancer diagnostic and prognostic biomarker. Bioinformatics analysis suggested that DUXAP8 might be involved in the regulation of the biological processes of HCC cell cycle, cell division and cell proliferation. Additionally, downregulation of DUXAP8 inhibited HCC cell proliferation and invasion in vitro.
Conclusion
This study revealed that DUXAP8 may serve as a potential pan-cancer prognostic and diagnostic marker in humans. In addition, DUXAP8 promoted HCC cell proliferation and invasion, suggesting that it may represent a novel therapeutic target for HCC.
Keywords: DUXAP8, pan-cancer marker, hepatocellular carcinoma
Introduction
Despite the rapid advances in medical science in recent decades, cancer still represents the most significant threat to human health.1–3 Malignant tumors remain one of the leading causes of death worldwide, mainly due to the late diagnosis of most patients who are frequently not diagnosed until advanced stages as well as to the lack of effective therapy.4 Accordingly, the identification of novel cancer biomarkers and the understanding of their involvement in the underlying molecular mechanisms of tumorigenesis and cancer progression are urgent and essential for the development of more effective diagnostic methods and therapeutic strategies.
Pseudogenes have long been considered to be non-functional transcriptional relics of the genome of various organisms.5 Approximately 15,000 pseudogenes, including unprocessed and processed pseudogenes, have been identified in the human genome according to the latest annotation of the ENCODE project.6 The functional role of pseudogenes remains poorly understood and several studies have suggested that pseudogenes could play key roles in cancer progression by acting as competing endogenous RNAs (ceRNAs) in recent years.7,8
Double homeobox A pseudogene 8 (DUXAP8) is a member of the DUXA homeobox gene family. In 2017, Sun et al first determined by bioinformatics analysis that DUXAP8 was overexpressed in lung cancer tissues and then demonstrated that DUXAP8 promoted cell invasion and proliferation through epigenetic silencing of RHOB and EGR1 in non-small cell lung cancer.9 Recently, pseudogene DUXAP8 has been shown to be upregulated in multiple cancers and plays critical regulatory roles in cancer progression. For instance, DUXAP8 was shown to regulate cell growth by epigenetic silencing of PLEKHO1 expression in gastric cancer.10 In addition, knockdown of DUXAP8 inhibited cell proliferation and invasion in esophageal squamous cell cancer.11 Moreover, expression of DUXAP8 promoted pancreatic carcinoma cell growth by epigenetic silencing of KLF2 and CDKN1A.12 Also, high levels of DUXAP8 was associated with poor prognosis in patients with lung,9 gastric,10 esophageal,11 pancreatic,12 glioma,13 and bladder14 cancers. However, the degree and reliability of the prognostic value of DUXAP8 in human cancers has not been thoroughly evaluated due to the limited studies and small sample size. Furthermore, the expression pattern and biological function of DUXAP8 in HCC remain unclear.
In this study, due to the limited published studies on DUXAP8, RNA sequencing (RNA-Seq) count matrix and matched clinical data extracted from the Cancer Genome Atlas (TCGA) database were used to evaluate the expression of DUXAP8 and its clinical significance in human cancers. Additionally, we verified the DUXAP8 expression levels in 66 HCC samples and corresponding adjacent normal liver tissue samples by polymerase chain reaction (PCR) analysis and explored its biological function in HCC through in vitro cellular function assays.
Materials and Methods
Data Acquisition
Data from TCGA database (https://cancergenome.nih.gov/), RNA-Seq count matrices of human pan-cancers were extracted using the R software (https://www.r-project.org/). Datasets that lacked sufficient samples or survival information were excluded. Gene symbols corresponding to each Ensembl ID were acquired based on the gene transfer format file (Homo_sapiens.GRCh38.96.chr) using the R software. In addition, studies concerning DUXAP8 in the PubMed, EMBASE and Web of Science databases were comprehensively evaluated in this study.
DUXAP8 Expression and Its Clinical Value in Human Cancers
Expression levels of DUXAP8 in various human cancers were acquired from TCGA database to assess the expression patterns of DUXAP8 in pan-cancers. The area under the curve (AUC), specificity, and sensitivity of each dataset were acquired by receiver operating characteristic (ROC) curve analysis. Then, the pooled AUC was calculated by summary ROC (SROC) curve analysis to evaluate the diagnostic value of DUXAP8 in pan-cancers. The Deeks’ funnel plot asymmetrical test was performed to assess the potential publication bias in the SROC analysis.
The hazard ratio (HR) with 95% confidence interval (CI) was calculated to evaluate the association of DUXAP8 expression with overall survival (OS) in each study, and the pooled HR was calculated by meta-analysis to assess the prognostic importance of DUXAP8 in pan-cancers. Potential publication bias was assessed using the funnel plots with Begg’s test. High DUXAP8 expression was considered to be related to poor prognosis if the pooled HR>1 and the 95% CI did not overlap 1.
Tissue Specimens and HCC Cells
A total of 66 pairs of HCC-tissue and adjacent liver-tissue samples were collected from the Department of General Surgery at Beijing Tongren Hospital between 2011 and 2017. No patients received other treatments before sample collection. Written informed consent, which was in accordance with the Declaration of Helsinki, was obtained from each patient. The protocol of this study was approved by the institutional review board of Beijing Tongren Hospital, Capital Medical University.
HCC cell lines (BEL-7404, HEP-G2, BEL-7402, SMMC-7721, HUH-7, and HEP-3B) and the normal human liver cell line (LO-2) were obtained from the Cell Bank of Shanghai Institute of Biochemistry & Cell Biology (Shanghai, China). All the cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum.
Real-Time Quantitative PCR (RT-qPCR) Analysis
Total RNA was extracted from the tissue samples using Trizol (Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA). A reverse-transcription kit was used to synthesize cDNA. The real-time PCR analysis was performed using a real-time PCR system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Gene expression level was determined by comparing Ct (2−ΔΔCT) values.15
Cell Transfection
Three siRNA sequences (si-DUXAP8-1#: 5ʹ- AAGATAAAGGTGGTTTCCACAAGAA-3ʹ; si-DUXAP8-2#: 5ʹ-GGAACTTCCCAAACCTCCATGATTT-3ʹ; si-DUXAP8-3#: 5ʹ-CAGCATACTTCAAATTCACAGCAAA-3ʹ) were designed according to the cDNA sequence of DUXAP8 obtained from GeneBank. HCC cells were transfected with siRNA using lipofectamine (Invitrogen) according to the manufacturer’s instructions. In addition, a negative control sequence (si-NC) was synthesized for use as the negative control group.
Cell Viability and Colony Formation Assays
Cells from si-NC and si-DUXAP8 groups were seeded into 96-well plates. Cells viability was accessed at 24, 48, 72, and 96 h using the Cell Counting Kit 8 (CCK-8) assays according to the manufacturer’s instructions (Dojindo Laboratories, Kumamoto, Japan). For the colony formation assay, the colonies containing more than 50 cells were counted two weeks after seeding the cells into 6-well plates.
Transwell Assay and Wound Healing assay
Transwell chambers with Matrigel were used to evaluate cell invasion ability. Briefly, cell suspensions of the si-DUXAP8 and si-NC groups were added to the upper Transwell chambers containing serum-free medium. The corresponding lower chambers containing 10% FBS medium served as a chemo-attractant. Cells that migrated onto the lower surface of the membrane after 24 h incubation were fixed with formaldehyde and stained with hematoxylin for 30 min. Cells which migrated to the lower compartment of the chamber were counted using a microscope in 5 randomly selected fields. For the wound healing assay, cells from the 2 groups were seeded into 6-well plates. A scratch wound was made in each well by scratching the cell layer once confluence was reached. Cells were cultured in medium after washing away the detached cells with PBS. Photos were captured at 0, 8 and 24 h to determine the number of cells migrating into the scratch wound.
Statistical Analysis
Kaplan-Meier survival analysis and ROC curve analysis were performed using the GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, USA). Survival data was extracted from the Kaplan–Meier curve using the Engauge Digitizer software if the published article did not provide them, and then the HR was calculated using Tierney’s method.16 Meta-analysis and SROC curve analysis were conducted using the Stata software (College Station, TX, USA). All experiments in the current study were conducted in triplicate, and the results are represented as the mean ± SD. Comparison among different groups was performed by one-way analysis of variance (ANOVA) test or Student’s t-test, and P<0.05 indicates statistical significance.
Results
DUXAP8 Was Overexpressed and Had Excellent Clinical Value in Human Cancers
A total of 28 studies including 8301 patients were enrolled in this study. Among them, 17 TCGA datasets contained the expression data of DUXAP8 in sufficient tumor and normal samples. HRs with 95% CI for OS of cancer patients were calculated based on DUXAP8 expression and survival information extracted from 22 TCGA datasets. Six published articles detected OS comparisons between patients with high and low expression of DUXAP8. Detailed information of the collected studies is shown in Tables S1 and S2.
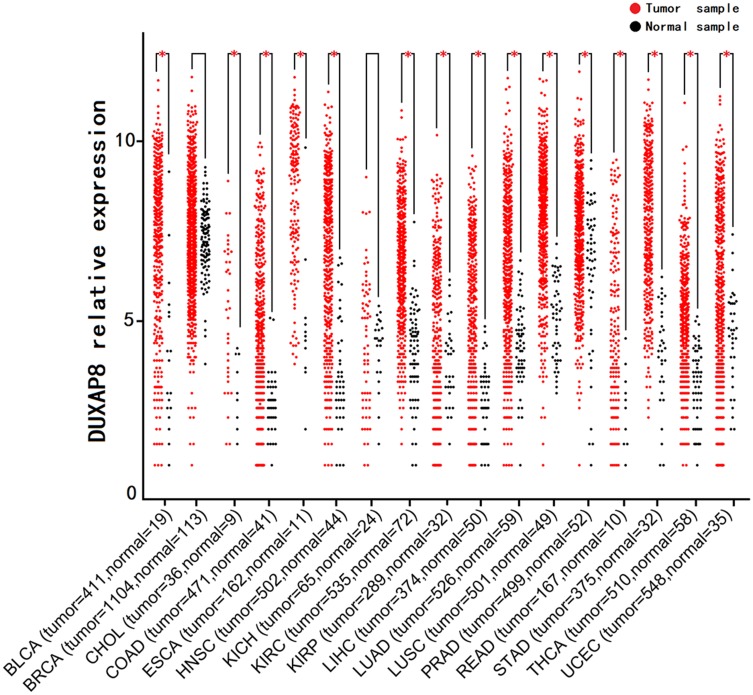
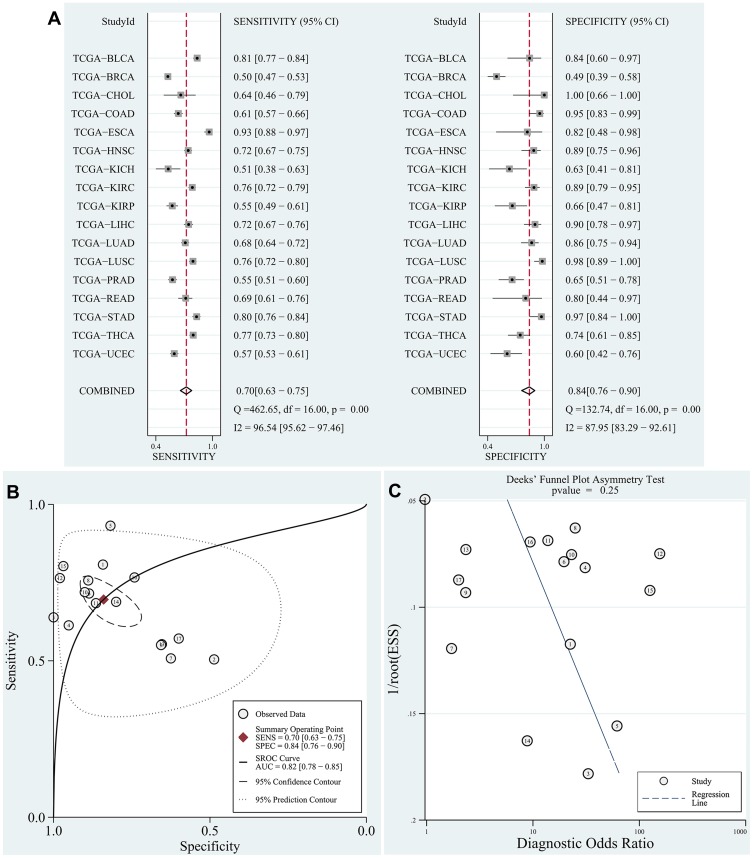
The data of DUXAP8 expression in human cancer -tissue samples and normal tissue samples from patients with various types of cancers were extracted from each RNA-Seq count matrix and then log2-transformed. DUXAP8 was significantly overexpressed in 15 types of human cancers, as shown in Figure 1. The AUC was obtained by ROC curve analysis for each TCGA dataset (Table S1) to determine the diagnostic value of DUXAP8 in each type of cancer. Then, the pooled specificity, sensitivity, and AUC were obtained by SROC curve analysis. As shown in Figure 2A and B, the pooled AUC was 0.82 [0.78–0.85], pooled sensitivity was 0.70 [0.63–0.75], and the pooled specificity was 0.84 [0.76–0.90]. The above results indicated that DUXAP8 had an excellent diagnostic value in pan-cancers. The results of Deeks’ funnel plot asymmetry test indicated that no significant publication bias existed for the studies in this SROC analysis (P =0.25) (Figure 2C).
Figure 1.
DUXAP8 expression levels in pan-cancers according to TCGA database (*P<0.05).
Abbreviation: TCGA, The Cancer Genome Atlas.
Figure 2.
SROC curve analysis according to TCGA database.
Notes: (A) Pooled sensitivity and pooled specificity were calculated to assess the diagnostic value of DUXAP8 in pan-cancers. (B) The results of pooled AUC (0.82 [0.78–0.85]) indicated that DUXAP8 has a good diagnostic value in pan-cancers. (C) No publication bias existed in the SROC analysis based on Deeks’ funnel plot asymmetrical test (P=0.25).
Abbreviations: SROC, summary receiver operating characteristic curve; TCGA, the cancer genome atlas; AUC, area under the curve.
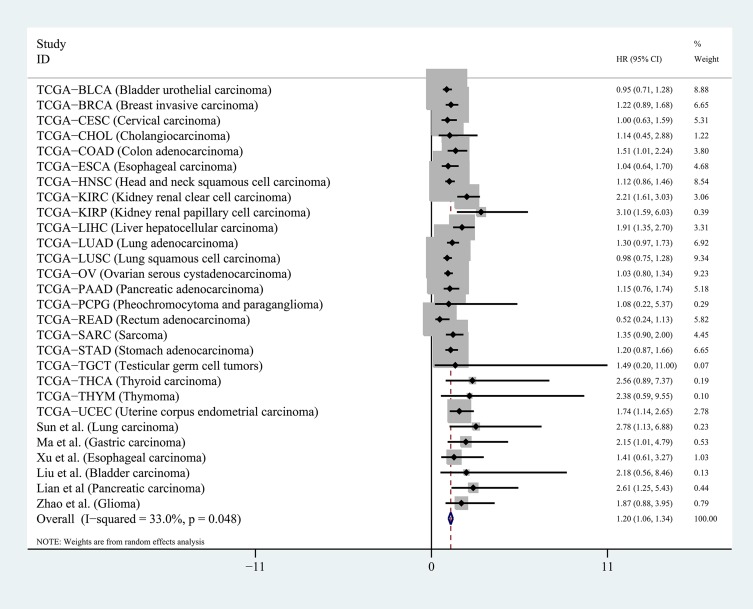
Next, we calculated the HR of each TCGA dataset based on the DUXAP8 expression data and matched OS information of the patients. For the published articles that were included in this study, the HR was obtained using the survival data excerpted from the Kaplan–Meier curves. The pooled HR obtained by meta-analysis suggested that DUXAP8 had an excellent prognostic value in human cancers (pooled HR=1.20[1.06–1.34], P<0.01, random), as shown in Figure 3. No significant publication bias existed for the studies in this meta-analysis according to the Begg’s funnel plot test (P =0.18). The results of this meta-analysis were reliable according to the sensitivity analysis, which indicated that all the datasets were within the 95% CI of the combined analysis.
Figure 3.
Pooled HR was calculated to assess the prognostic value of DUXAP8 in pan-cancers.
Note: Meta-analysis was performed to obtain the pooled HR.
Abbreviation: HR, hazard ratio.
The above results suggested that DUXAP8 was upregulated in human tumor tissues and could be used as a prognostic and diagnostic marker in human cancers.
Correlation Between DUXAP8 Expression and Clinicopathological Characteristics in HCC Patients
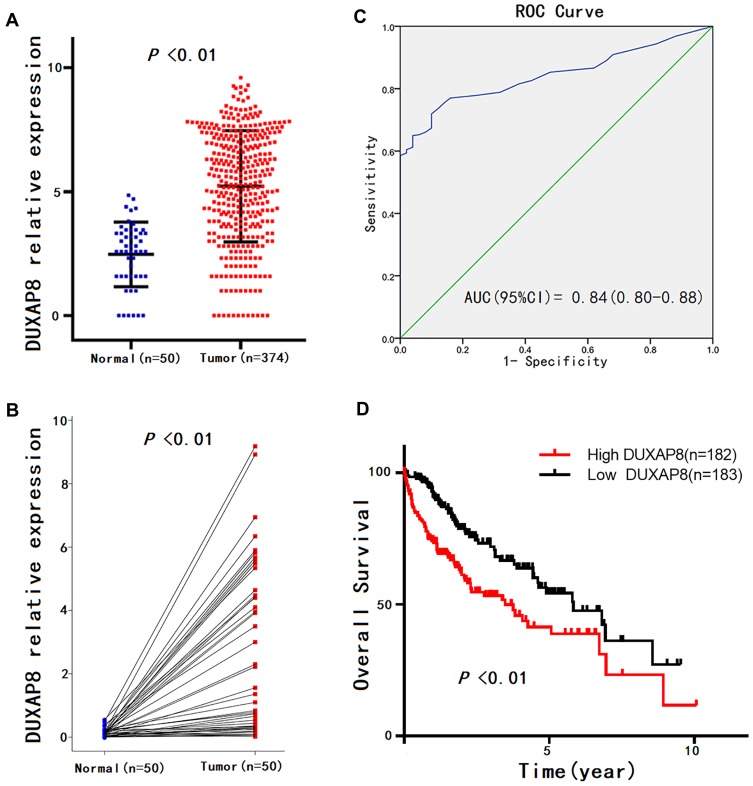
Given that the expression pattern and clinical significance of DUXAP8 in HCC were not yet characterized, we investigated the correlation between the DUXAP8 expression level of HCC tissue and the clinicopathologic characteristics of HCC patients based on the information extracted from TCGA database. The results revealed that DUXAP8 was significantly overexpressed in HCC tissue (Figure 4A and B), and DUXAP8 had excellent diagnostic value in HCC patients, according to the ROC curve analysis (AUC=0.84[0.80–0.88]) (Figure 4C). In addition, a high DUXAP8 expression level indicated poor OS in HCC patients (Figure 4D). Moreover, DUXAP8 was significantly overexpressed in patients over 60 years of age (Age≥ 60, 5.61 ± 0.15 vs. Age< 60, 4.73 ± 0.18; P< 0.01), with higher pathological stage (Stage III+IV, 5.67 ± 0.23 vs. StageI+II, 4.97 ± 0.14; P= 0.04), and with vascular invasion (Positive, 5.47 ± 0.23 vs. Negative 4.78 ± 0.15; P= 0.03), as shown in Table 1.
Figure 4.
DUXAP8 expression patterns in HCC according to TCGA database.
Notes: (A) DUXAP8 expression levels in HCC tissues compared with normal liver tissues, according to unpaired student’s t-test. (B) DUXAP8 expression level in HCC tissue compared with adjacent liver tissue from the same patient according to paired student’s t-test. (C) Significance of DUXAP8 in HCC according to the ROC curve analysis. (D) Overall survival (OS) plots of DUXAP8.
Abbreviations: HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas; ROC, receiver operating characteristic curve.
Table 1.
Correlation Between DUXAP8 Expression and Clinicopathological Characteristics in TCGA
| Characteristics | n | DUXAP8 Expression in TCGA Database | ||
|---|---|---|---|---|
| M ± SD | P-values | |||
| Tissues | HCC tissues | 371 | 5.22 ± 0.12 | <0.01* |
| Adjacent liver tissues | 50 | 2.47 ± 0.18 | ||
| Gender | Male | 250 | 5.27 ± 0.14 | 0.44 |
| Female | 121 | 5.08 ± 0.21 | ||
| Age | ≥60 | 202 | 5.61 ± 0.15 | <0.01* |
| <60 | 169 | 4.73 ± 0.18 | ||
| Tumor grade | G1+G2 | 232 | 5.11 ± 0.15 | 0.28 |
| G3+G4 | 134 | 5.37 ± 0.20 | ||
| Pathologic stage | I–II | 257 | 4.97 ± 0.14 | 0.04* |
| III–IV | 90 | 5.67 ± 0.23 | ||
| Vascular invasion | Negative | 206 | 4.78 ± 0.15 | 0.03* |
| Positive | 109 | 5.47 ± 0.23 | ||
| Recurrence | Yes | 95 | 4.97± 0.24 | 0.83 |
| No | 174 | 5.03± 0.16 | ||
Note: *P < 0.05.
Abbreviation: TCGA, the Cancer Genome Atlas.
Identification of DUXAP8 Related Protein-Coding Genes in HCC
To investigate the potential molecular regulatory mechanisms of DUXAP8 in HCC tumorigenesis, the protein-coding genes whose expression level was closely correlated with that of DUXAP8 were identified. Two-sided Pearson correlation coefficient analysis and the z-test were performed using the R software based on the gene expression data extracted from TCGA. |Pearson correlations| >0.50 and z-test p<0.001 were used as cutoff criteria. Ultimately, a total of 177 protein-coding genes were identified as DUXAP8-related genes (Table 2).
Table 2.
DUXAP8-Related Protein-Coding Genes in HCC Based on TCGA Database
| Pearson correlation>0.50 | MKRN3; FAM133A; COX7B2; COL24A1; CARD18; MAGEA1; MAGEA3; MAGEC2; SSX1; CNTNAP4; MAGEA6; CSMD1; DCAF4L2; CTAG2; SLCO6A1; DCAF8L2; GABRA3; PLCB1; PPP4R3C;VCX; MAGEC1; C5orf58; SPDL1; SHCBP1; NAA11; LIN28B; EME1; TRIP13; SLC44A5; RAD54L; TGIF2LX; ZC2HC1A; ZNF883; CIP2A; CDCA8; DDIAS; ZNF257; SYT1; MCM10;FSTL5; PKMYT1; GPR158; CC2D1B; NPFFR2; CEP131; TMEM206; CDC7; P3H1; DEPDC1; IBSP; RFPL4B; KIF4A; RNF220; KIF2C; GTSF1; RHEBL1; ZBTB17; NCAPH; ZNF681;ERCC6L; FADS3; PLK4; CCDC137; SGO2; LYPD8; TRAIP; POLQ; ZFP69B; ANKRD27; CSTF2; ZNF716; PDIA2; RAD51; NUP93; CDC45; ORC6; EXO1; GPSM2; C18orf54; GNGT1;BUB1; G6PD; PGS1; TRIM45; CDC20; CENPI; AGPAT4; TMEM241; CENPE; CSAG1; FANCD2; ZSCAN20; KPNA2; CCNA2; ANLN; MAGEA12; NEIL3; STIL; STX1A; ZNF99; C5orf34;TTF2; CENPU; SMARCD1; KCNK9; E2F2; DBF4B; RBM19; ABCC5; CLSPN; PIGU; HMMR; TEDC2; NRAS; CENPA; EIF2B5; BUB1B; IGF2BP3; CEP55; MAD2L1; GLMN; CDCA5;DIAPH3; NUF2; DDX53; TTK; STMN1; FANCA; C1orf52; SPATS2; SLC36A1; ZNF43; DLGAP5; TOP2A; CWC27; MELK; ITGB3BP; MED8; PLK1; ORC1; DPH2; NCAPG; SLC38A6;TPGS2; MTFR2; RUVBL1; ZIC2; MARS; SOHLH2; MDGA2; MAGEB6; WRAP73; CKAP2L; CENPK; CCDC88A; FANCB; KIF23; XRCC2; PDE6A; NPC1; ADAMTS20; RRM2; TUBG1; ZNF85;UTP6; ECT2; SGO1; STARD9; PRC1; KIF18B; ZNF730; CDKN2A; KIF18A; FAAP24; DCTN2; DCAF8L1; NDC1 |
| Pearson correlation<-0.50 | None |
Abbreviations: HCC, hepatocellular carcinoma; TCGA, the cancer genome atlas.
Gene-Annotation and Pathway Enrichment Analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and gene ontology (GO) annotation enrichment analyses were performed using the DAVID online tool (https://david.ncifcrf.gov/) to identify the potential signaling pathways and biological processes involved in HCC tumorigenesis and progression in which DUXAP8 participates.
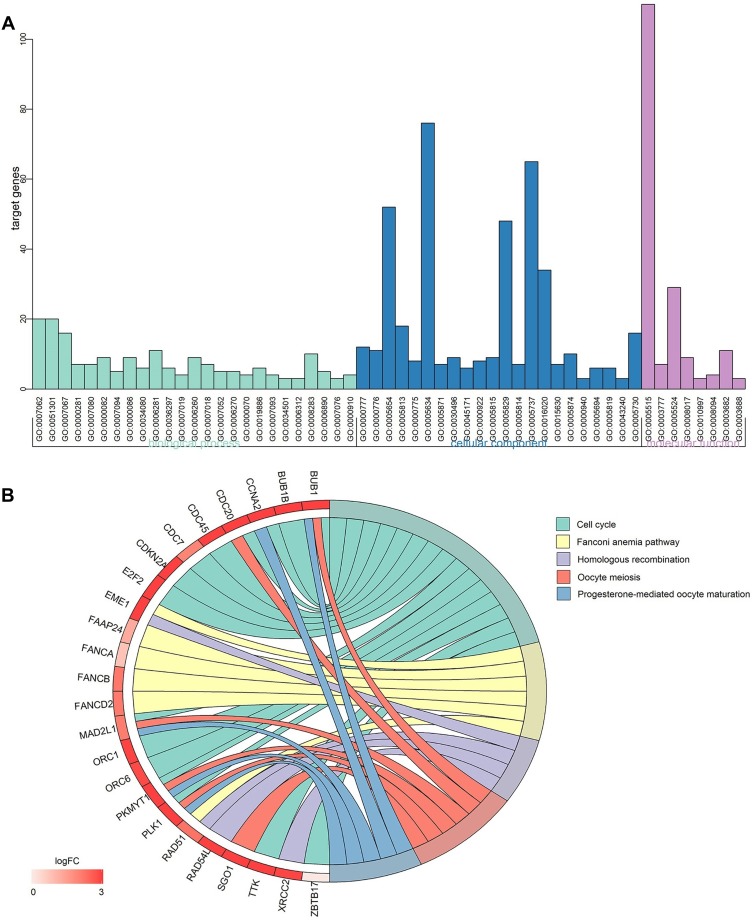
As shown in Figure 5A, regarding the GO enrichment analysis, there were 25 significant biological process annotations (P<0.01), which included G1/S and transition of mitotic cell cycle, cell division, G2/M transition of mitotic cell cycle, cell proliferation, etc. The GO enrichment analysis also revealed that there were 22 significant cellular component terms (P<0.01), which were correlated with condensed chromosome kinetochore, kinetochore, nucleoplasm, centrosome, etc. The GO analysis also revealed 8 significant molecular function terms (P<0.01), associated with protein binding, microtubule motor activity, ATP binding, DNA-dependent ATPase activity, etc. The detailed data are listed in Table 3.
Figure 5.
Gene-annotation and pathway enrichment analysis.
Notes: (A) GO enrichment analysis; (B) KEGG pathway enrichment analysis.
Abbreviations: GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 3.
The GO Analysis of DUXAP8-Related Protein-Coding Genes in HCC Based on TCGA Database
| GO ID | Term | Count | P-value |
|---|---|---|---|
| Biological process | |||
| GO:0007062 | Sister chromatid cohesion | 20 | 5.91×10−20 |
| GO:0051301 | Cell division | 20 | 4.13×10−10 |
| GO:0007067 | Mitotic nuclear division | 16 | 7.53×10−9 |
| GO:0000281 | Mitotic cytokinesis | 7 | 1.99×10−7 |
| GO:0007080 | Mitotic metaphase plate congression | 7 | 9.19×10−7 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 9 | 4.00×10−6 |
| GO:0007094 | Mitotic spindle assembly checkpoint | 5 | 2.79×10−5 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 9 | 3.50×10−5 |
| GO:0034080 | CENP-A containing nucleosome assembly | 6 | 4.15×10−5 |
| GO:0006281 | DNA repair | 11 | 5.63×10−5 |
| GO:0036297 | Interstrand cross-link repair | 6 | 7.87×10−5 |
| GO:0007019 | Microtubule depolymerization | 4 | 8.33×10−5 |
| GO:0006260 | DNA replication | 9 | 8.39×10−5 |
| GO:0007018 | Microtubule-based movement | 7 | 9.25×10−5 |
| GO:0007052 | Mitotic spindle organization | 5 | 1.47×10−4 |
| GO:0006270 | DNA replication initiation | 5 | 1.90×10−4 |
| GO:0000070 | Mitotic sister chromatid segregation | 4 | 0.001445018 |
| GO:0019886 | Antigen processing and presentation of exogenous peptide antigen via MHC class II | 6 | 0.001486498 |
| GO:0007093 | Mitotic cell cycle checkpoint | 4 | 0.00297531 |
| GO:0034501 | Protein localization to kinetochore | 3 | 0.003492615 |
| GO:0006312 | Mitotic recombination | 3 | 0.005947021 |
| GO:0008283 | Cell proliferation | 10 | 0.006165774 |
| GO:0006890 | Retrograde vesicle-mediated transport, Golgi to ER | 5 | 0.006535823 |
| GO:0007076 | Mitotic chromosome condensation | 3 | 0.007911301 |
| GO:0000910 | Cytokinesis | 4 | 0.009338818 |
| Cellular component | |||
| GO:0000777 | Condensed chromosome kinetochore | 12 | 2.49×10−10 |
| GO:0000776 | Kinetochore | 11 | 2.12×10−9 |
| GO:0005654 | Nucleoplasm | 52 | 7.95×10−8 |
| GO:0005813 | Centrosome | 18 | 1.99×10−7 |
| GO:0000775 | Chromosome, centromeric region | 8 | 6.18×10−7 |
| GO:0005634 | Nucleus | 76 | 2.42×10−6 |
| GO:0005871 | Kinesin complex | 7 | 6.57×10−6 |
| GO:0030496 | Midbody | 9 | 1.76×10−5 |
| GO:0045171 | Intercellular bridge | 6 | 3.92×10−5 |
| GO:0000922 | Spindle pole | 8 | 4.77×10−5 |
| GO:0005815 | Microtubule organizing center | 9 | 5.96×10−5 |
| GO:0005829 | Cytosol | 48 | 3.06×10−4 |
| GO:0005814 | Centriole | 7 | 4.70×10−4 |
| GO:0005737 | Cytoplasm | 65 | 0.001064211 |
| GO:0016020 | Membrane | 34 | 0.00122264 |
| GO:0015630 | Microtubule cytoskeleton | 7 | 0.001284983 |
| GO:0005874 | Microtubule | 10 | 0.001665627 |
| GO:0000940 | Condensed chromosome outer kinetochore | 3 | 0.002046274 |
| GO:0005694 | Chromosome | 6 | 0.002273943 |
| GO:0005819 | Spindle | 6 | 0.004189711 |
| GO:0043240 | Fanconi anaemia nuclear complex | 3 | 0.007372095 |
| GO:0005730 | Nucleolus | 16 | 0.008077134 |
| Molecular function | |||
| GO:0005515 | Protein binding | 110 | 2.48×10−6 |
| GO:0003777 | Microtubule motor activity | 7 | 9.34×10−5 |
| GO:0005524 | ATP binding | 29 | 2.12×10−4 |
| GO:0008017 | Microtubule binding | 9 | 6.83×10−4 |
| GO:0010997 | Anaphase-promoting complex binding | 3 | 0.001226464 |
| GO:0008094 | DNA-dependent ATPase activity | 4 | 0.003097754 |
| GO:0003682 | Chromatin binding | 11 | 0.003321884 |
| GO:0003688 | DNA replication origin binding | 3 | 0.00436336 |
Abbreviations: TCGA, the cancer genome atlas; GO, gene ontology; HCC, hepatocellular carcinoma.
Regarding the KEGG pathway enrichment analysis, there were 5 signaling pathways that were statistically significant (P<0.01), including cell cycle, Fanconi anemia pathway, homologous recombination, oocyte meiosis and progesterone-mediated oocyte maturation (Figure 5B).
DUXAP8 Was Overexpressed in HCC Tissue and Cell Lines
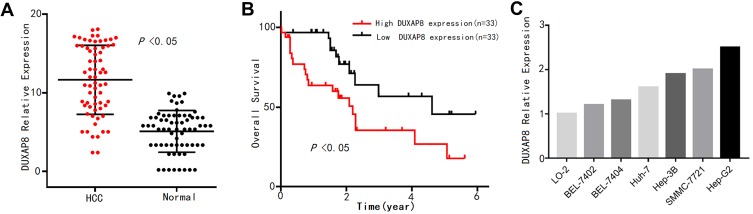
To verify the expression pattern of DUXAP8 in HCC, qPCR analysis was conducted to determine the expression levels of the 66 pairs of HCC tissue samples. As shown in Figure 6A, DUXAP8 was significantly overexpressed in HCC tissue, according to the paired Student’s t-test. Furthermore, high DUXAP8 expression was significantly associated with poor OS in HCC patients, according to the Kaplan–Meier curve analysis (Figure 6B), which was consistent with the result of the comprehensive analysis based on TCGA database.
Figure 6.
DUXAP8 expression pattern in HCC was verified by qPCR analysis.
Notes: (A) DUXAP8 expression levels in 66 pairs of HCC tissue and adjacent normal liver tissue samples. (B) Overall survival (OS) plots of DUXAP8 based on the 66 HCC patients’ clinical information. (C) DUXAP8 expression levels in normal liver cells and 6 HCC cell lines.
Abbreviation: HCC, hepatocellular carcinoma.
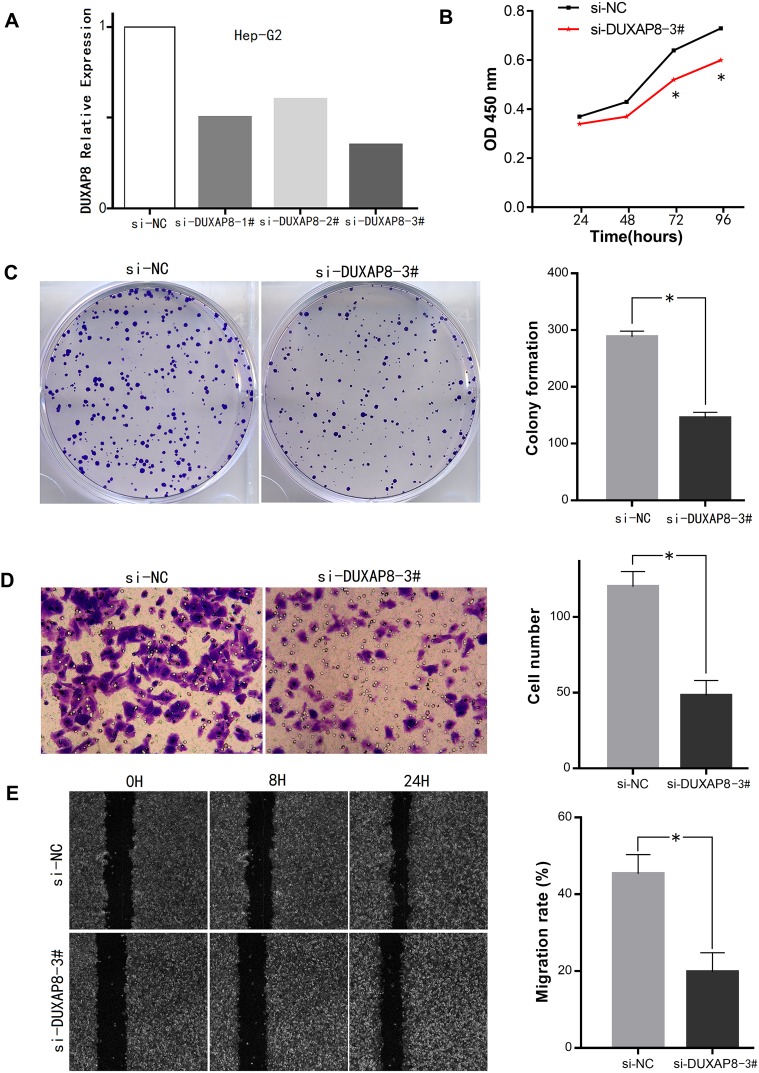
In addition, the expression levels of DUXAP8 in the 6 HCC cell lines were higher than that in the normal liver LO-2 cells (Figure 6C). The transfection of the Hep-G2 cells with three siRNAs using a lentiviral vector system revealed that the siRNA-DUXAP8-3# (5ʹ-CAGCATACTTCAAATTCACAGCAAA-3ʹ) had the highest interference efficiency according to qPCR analysis. (Figure 7A), thus it was used in subsequent experiments.
Figure 7.
Effects of DUXAP8 on HCC cell proliferation and invasion.
Notes: (A) Interference efficiency of the 3 si-RNAs in Hep-G2 cells. (B, C) Downregulation of DUXAP8 inhibited HCC cell proliferation and viability according to the CCK-8 assay and colony formation assay, respectively. (D, E) Invasion and migration ability of HCC cells was decreased after DUXAP8 knockdown according to the Transwell assay and wound healing assay (*P<0.05).
Abbreviations: HCC, hepatocellular carcinoma; CCK-8, Cell Counting Kit-8.
Effects of DUXAP8 Expression on HCC Cells Proliferation and Invasion
Cell viability in the DUXAP8 knockdown group was significantly inhibited compared with that in the si-NC group according to the growth curves obtained with the CCK-8 assay (Figure 7B). Additionally, according to the colony formation assay, HCC cells in the si-DUXAP8 group produced fewer colonies than those in the negative control group (Figure 7C).
The results of the Transwell assay suggested that downregulation of DUXAP8 inhibited the invasion ability of HCC cells (Figure 7D). The wound healing assay also revealed that the migration ability of HCC cells in the si-DUXAP8 group was reduced compared with that in the si-NC group (Figure 7E).
The results of the in vitro cellular function assays described above suggested that overexpression of DUXAP8 in HCC promoted cell proliferation and invasion.
Discussion
As a result of the development of next-generation sequencing technology, the integrated analysis through data mining, bioinformatics analysis, and meta-analysis has become one of the most powerful methods in cancer research.17 For example, Zhang et al found that the lncRNA MIR22HG could act as a cancer suppressor in HCC based on data extracted from TCGA portal.18 In addition, Ge et al showed that LINC00996 is significantly decreased in colorectal cancer tissues and the depletion of LINC00996 is associated with a poor outcome in colorectal cancer patients according to a meta-analysis using data extracted from the Gene Expression Omnibus (GEO) and TCGA databases.19
In our previous research, we used an integrated analysis approach based on the microarray data from the GEO and TCGA databases and found that DUXAP10, which is another member of the DUXA homeobox gene family, could act as a potential marker in pan-cancers.20 Additionally, we also showed that DUXAP10 could regulate cell growth by activating p-AKT in HCC.21 After comprehensively searching the studies on DUXAP8, we only found a few published articles in electronic databases. Therefore, we decided to mine the expression data of DUXAP8 from public databases, including TCGA and GEO.
Although, DUXAP8 has been identified in previous studies as an aberrantly expressed gene in lung cancer and gastric cancer by bioinformatics analysis based on GEO datasets,9,10 we could not find any data on DUXAP8 expression in human cancers from the GEO database, which might be due to the update of probe annotation files. Nevertheless, we successfully extracted the expression information of DUXAP8 from TCGA database. Comprehensive analysis including data mining, OS analysis, ROC curve analysis, SROC curve analysis, and meta-analysis were performed in this study, and the results revealed that DUXAP8 was overexpressed in human cancers tissue samples and had both excellent diagnostic and prognostic values in human cancers.
We also found that the DUXAP8 expression level in HCC tissue was significantly upregulated compared with that in adjacent normal liver tissue (log2FC>1 and P<0.01) based on the data extracted from TCGA. Also, high DUXAP8 expression was highly related to poor OS in HCC (P<0.01), and had excellent diagnostic value (AUC=0.84, 95% CI 0.80–0.88) in HCC according to the ROC curve analysis. Moreover, high DUXAP8 levels were detected in HCC patients with vascular invasion or high pathologic stage according to our analysis. However, to date, no study has been reported on the roles of DUXAP8 in HCC tumorigenesis and progression. Accordingly, we identified the DUXAP8 related protein-coding genes to investigate the potential biological process and signaling pathway in which DUXAP8 is involved. The results of the GO and KEGG pathway enrichment analysis suggested that DUXAP8-related genes were most significantly involved in the biological processes of cell division, cell cycle, etc. Also, according to the above results, we decided to experimentally examine the roles of DUXAP8 in HCC. The results of the qPCR analysis and in vitro cell function assays showed that DUXAP8 was upregulated in HCC tissue, and its downregulation inhibited HCC cell proliferation and invasion.
This study had a number of limitations that should be discussed. First, no clinicopathological characteristics were closely related with the DUXAP8 expression levels in the 66 HCC tissue samples that were collected in this study, which might be explained by the limited sample size. Second, the signaling pathways through which DUXAP8 might exert its regulatory effect on HCC progression were not explored in the current study. Thus, undoubtedly, this issue needs to be addressed through further investigation and verification.
Conclusions
To summarize, this study revealed that DUXAP8 was overexpressed in pan-cancers and could serve as a potential prognostic and diagnostic marker for cancer patients. In addition, we verified the DUXAP8 expression pattern in HCC and showed that DUXAP8 regulated HCC cell growth. To the best of our knowledge, this study is the first investigation of the regulatory roles of DUXAP8 in HCC and may contribute to the development of novel diagnosis markers and therapeutic approach for HCC.
Acknowledgments
This work was supported by Leader of the First Hospital of Shanxi Medical University (YD1607), Scientific Research Project Plan of Shanxi Provincial Health Planning Commission (2014029), Project Letter of Fostering Team for Precision Medical Key Innovation in the First Hospital of Shanxi Medical University (YT1603), and Shanxi Medical University Doctor’s Startup Fund Project (XD1802).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Goding Sauer A, Ortiz AP, et al. Cancer statistics for hispanics/ latinos, 2018. CA Cancer J Clin. 2018;68(6):425–445. doi: 10.3322/caac.21494 [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 5.Proudfoot N. Pseudogenes. Nature. 1980;286(5776):840–841. doi: 10.1038/286840a0 [DOI] [PubMed] [Google Scholar]
- 6.Frankish A, Harrow J. GENCODE pseudogenes. Methods Mol Biol. 2014;1167:129–155. doi: 10.1007/978-1-4939-0835-6_10 [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Guo ZY, Zhang R, et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis. 2013;34(8):1773–1781. doi: 10.1093/carcin/bgt139 [DOI] [PubMed] [Google Scholar]
- 8.An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med. 2017;21(1):185–192. doi: 10.1111/jcmm.12952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun M, Nie FQ, Zang C, et al. The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol Ther. 2017;25(3):739–751. doi: 10.1016/j.ymthe.2016.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Ma HW, Xie M, Sun M, et al. The pseudogene derived long noncoding RNA DUXAP8 promotes gastric cancer cell proliferation and migration via epigenetically silencing PLEKHO1 expression. Oncotarget. 2017;8(32):52211–52224. doi: 10.18632/oncotarget.11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu LJ, Yu XJ, Wei B, et al. Long non-coding RNA DUXAP8 regulates proliferation and invasion of esophageal squamous cell cancer. Eur Rev Med Pharmacol Sci. 2018;22(9):2646–2652. doi: 10.26355/eurrev_201805_14959 [DOI] [PubMed] [Google Scholar]
- 12.Lian Y, Yang J, Lian Y, Xiao C, Hu X, Xu H. DUXAP8, a pseudogene derived lncRNA, promotes growth of pancreatic carcinoma cells by epigenetically silencing CDKN1A and KLF2. Cancer Commun (Lond). 2018;38(1):64. doi: 10.1186/s40880-018-0333-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X, Hao S, Wang M, Xing D, Wang C. Knockdown of pseudogene DUXAP8 expression in glioma suppresses tumor cell proliferation. Oncol Lett. 2019;17(3):3511–3516. doi: 10.3892/ol.2019.9994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin MG, Hong YK, Zhang Y, Lin BB, He XJ. Mechanism of lncRNA DUXAP8 in promoting proliferation of bladder cancer cells by regulating PTEN. Eur Rev Med Pharmacol Sci. 2018;22(11):3370–3377. doi: 10.26355/eurrev_201806_15158 [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 16.Ge H, Liang C, Ren S, Yue C, Wu J. Prognostic value of DcR3 in solid tumors: a meta-analysis. Clin Chim Acta. 2018;481:126–131. doi: 10.1016/j.cca.2018.02.038 [DOI] [PubMed] [Google Scholar]
- 17.Yue C, Ren Y, Ge H, et al. Comprehensive analysis of potential prognostic genes for the construction of a competing endogenous RNA regulatory network in hepatocellular carcinoma. Onco Targets Ther. 2019;12:561–576. doi: 10.2147/OTT.S188913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang DY, Zou XJ, Cao CH, et al. Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for hepatocellular carcinoma. Theranostics. 2018;8(14):3751–3765. doi: 10.7150/thno.22493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge H, Yan Y, Wu D, Huang Y, Tian F. Potential role of LINC00996 in colorectal cancer: a study based on data mining and bioinformatics. Onco Targets Ther. 2018;11:4845–4855. doi: 10.2147/OTT.S173225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue C, Ren Y, Ge H, et al. Pseudogene DUXAP10 can be used as a diagnostic and prognostic biomarker in human cancers. J Cell Physiol. 2019;234(12):23685–23694. doi: 10.1002/jcp.28937 [DOI] [PubMed] [Google Scholar]
- 21.Yue C, Liang C, Ge H, et al. Pseudogene DUXAP10 acts as a diagnostic and prognostic marker and promotes cell proliferation by activating PI3K/AKT pathway in hepatocellular carcinoma. Onco Targets Ther. 2019;12:4555–4566. doi: 10.2147/OTT.S210623 [DOI] [PMC free article] [PubMed] [Google Scholar]