Abstract
Tubulins play crucial roles in cell division, intracellular traffic, and cell shape. Tubulin concentration is autoregulated by feedback control of mRNA degradation via an unknown mechanism. Here, we identified tetratricopeptide protein 5 (TTC5) as a tubulin-specific ribosome-associating factor that triggers co-translational degradation of tubulin mRNAs in response to excess soluble tubulin. Structural analysis revealed that TTC5 binds near the ribosome exit tunnel and engages the N-terminus of nascent tubulins. TTC5 mutants incapable of ribosome or nascent tubulin interaction abolished tubulin autoregulation and showed chromosome segregation defects during mitosis. Our findings show how a subset of mRNAs can be targeted for coordinated degradation by a specificity factor that recognizes the nascent polypeptides they encode.
Alpha and beta tubulins form obligate heterodimers (hereafter αβ-tubulin) that reversibly and dynamically polymerize into microtubules—cytoskeletal elements that regulate cell shape, drive mitosis, provide platforms for intracellular transport, and mediate cell movement (1). Microtubule dynamics, and the various processes that depend on it (2, 3), are strongly influenced by the concentration of soluble (i.e., non-polymerized) αβ-tubulin (4). When cells detect an increase in soluble αβ-tubulin concentration, they trigger degradation of tubulin mRNAs via a process termed tubulin autoregulation (5–7).
Autoregulation requires translation, indicating that ribosome-engaged tubulin mRNAs are selectively targeted for degradation (8, 9). Analysis of β-tubulin autoregulation in mammalian cells indicates a critical role for the first four residues (Met-Arg-Glu-Ile, or MREI) common to all β-tubulin isoforms (10, 11). Because autoregulation is prevented by physical occlusion of the MREI motif (12), a factor is thought to engage this sequence on nascent tubulin to initiate degradation of the mRNA being translated.
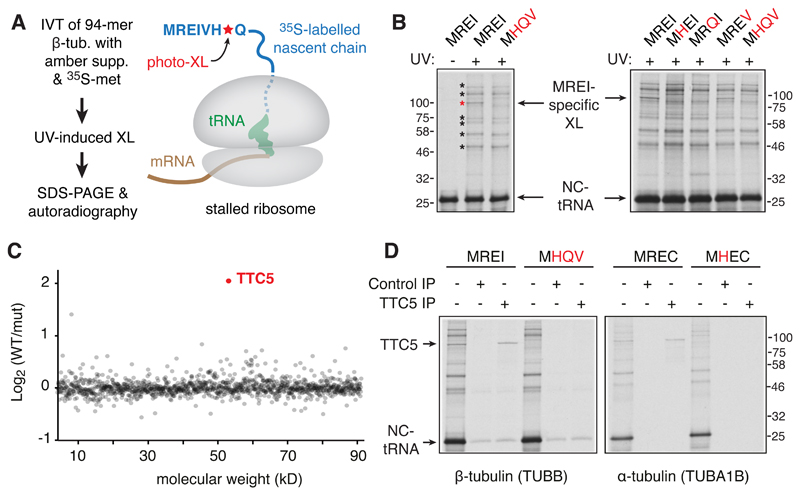
We used a site-specific photo-crosslinking strategy (Fig. 1A; fig. S1) to detect cytosolic factors that specifically recognize the N-terminal autoregulatory motif (MREI) of nascent β-tubulin early during its translation. A ribosome nascent chain complex (RNC) displaying the first 94 amino acids of 35S-methionine labelled human β-tubulin containing the UV-activated crosslinking amino acid p-benzoyl-l-phenylalanine (Bpa) was produced by in vitro translation in rabbit reticulocyte lysate. Irradiation of these RNCs with UV light generated nascent chain crosslinks to various proteins, only one of which was sensitive to mutation of residues 2, 3, and 4 of the MREI motif (Fig. 1B). This MREI-specific interaction partner was identified by quantitative mass spectrometry to be TTC5 (Fig. 1C), a highly conserved protein found widely across eukaryotes (fig. S2). TTC5 engaged the MREC motif at the N-terminus of nascent α-tubulin comparably to the MREI motif on β-tubulin (Fig. 1D), consistent with position 4 being less critical than positions 2 or 3 [Fig. 1B; (11)]. Thus, TTC5 is a nascent polypeptide binding protein specific for the N-termini of α- and β-tubulins.
Fig. 1. TTC5 interacts with the N-termini of nascent tubulins.
(A) Experimental strategy to detect interaction partners close to the N-terminus of nascent tubulin. The UV-activated crosslinking amino acid p-benzoyl-l-phenylalanine (Bpa) is introduced site-specifically at position 7 using amber suppression (see fig. S1). (B) Photo-crosslinking analysis of 35S-labelled 94-amino acid long ribosome-nascent chain complexes (RNCs) of human β-tubulin and mutants (indicated in red) in the N-terminal MREI motif. The positions of non-crosslinked tRNA-associated nascent chain (NC-tRNA) and a crosslinking partner specific to wild type tubulin (red asterisk) are indicated. Other nascent chain crosslinks agnostic to the MREI motif are indicated by black asterisks. (C) Quantitative mass spectrometry of proteins co-purified with wild type (WT) versus MHQV mutant (mut) β-tubulin RNCs plotted by molecular weight. (D) Photo-crosslinking and immunopreciptation (IP) analysis of 35S-labelled 94-amino acid long RNCs of α- or β-tubulin compared to the indicated mutants.
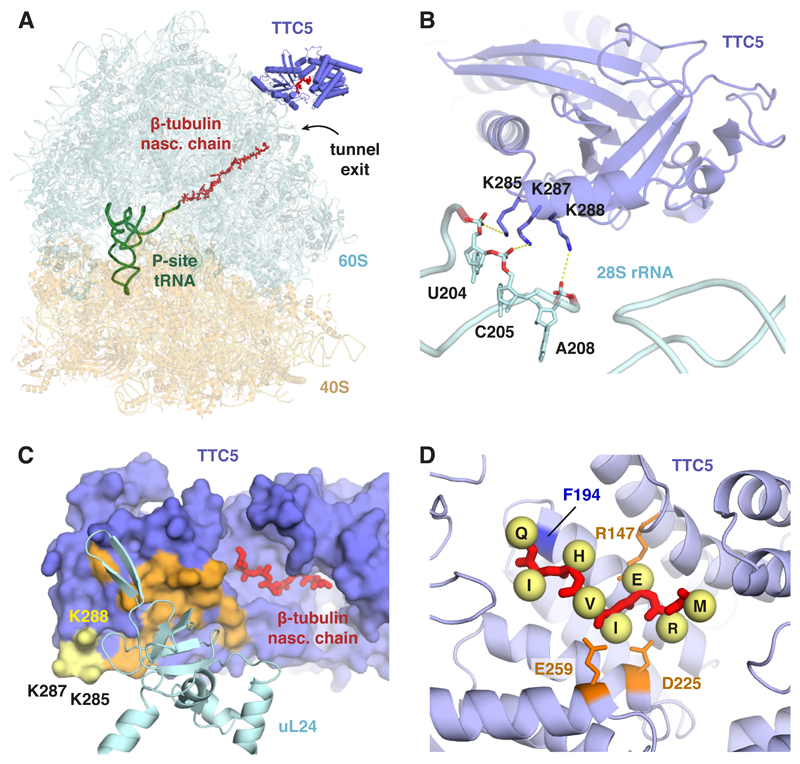
To understand how TTC5 engages its substrates on the ribosome, we purified nascent tubulin RNCs in complex with TTC5 (fig. S3) and determined its structure by single-particle electron cryomicroscopy (cryo-EM). The TTC5-RNC reconstruction (fig. S4, S5) showed the ribosome with a peptidyl-tRNA, nascent β-tubulin polypeptide within the ribosome exit tunnel, and TTC5 bound at the mouth of the tunnel (Fig. 2A). The heterodimeric nascent polypeptide associated complex (NAC) was observed at its previously established binding site (13, 14) opposite the exit tunnel from TTC5 (see fig. S4). NAC is not specific to tubulin RNCs (15, 16), does not contact TTC5 in the structure, and is not discussed further.
Fig. 2. Mechanism of ribosome-nascent chain engagement by TTC5.
(A) Overview of the cryo-EM derived structure of the complex between TTC5 and a ribosome containing the first 64 amino acids of β-tubulin. (B) Close-up view of the TTC5 interaction with 28S rRNA. Three conserved lysine residues in TTC5 within electrostatic interaction distance of the rRNA backbone are indicated. (C) The surface of TTC5 that interacts with ribosomal protein uL24 is indicated in orange. The 28S-interacting residues from panel B are shown in yellow. The N-terminal 8 amino acids of the β-tubulin nascent chain is shown in red within its binding groove of TTC5. (D) Close-up view of the N-terminal 8 amino acids of nascent β-tubulin (MREIVHIQ) within TTC5. Yellow spheres denote C-β atoms for the indicated side chains (not modelled). R147 is within salt-bridge distance of E3, and D225 and E259 are within salt-bridge distance of R2. F194 on the ‘floor’ of the binding groove, shown in Fig. 3A to crosslink with nascent β-tubulin, is indicated.
TTC5 was seen to make two contacts with the ribosome. The first contact involves three highly conserved lysine side chains in the oligonucleotide binding domain of TTC5 making electrostatic interactions with phosphates of the 28S rRNA backbone (Fig. 2B). The second contact involves ribosomal protein uL24 and buries ~ 500 Å2 of TTC5 adjacent to a deep groove formed by the tetratricopeptide repeat domain of TTC5 (Fig. 2C). The groove faces the mouth of the exit tunnel and contains cryo-EM density that we assigned to the first eight amino acids of β-tubulin (fig. S5), consistent with photo-crosslinking results (fig. S6).
The structural model allowed us to deduce likely interactions between the MREI motif and conserved side chains lining the TTC5 groove (Fig. 2D). Depending on its orientation, the R2 side chain of nascent tubulin is within salt bridge distance of E259 and D225 of TTC5. E3 in nascent tubulin would likely interact with R147 in TTC5. The side chain of I4 faces a moderately hydrophobic surface that could accommodate a cysteine (as in α-tubulins) or possibly other amino acids consistent with earlier mutagenesis (11). Collectively, the structure shows how TTC5 binds near the ribosome exit tunnel with its peptide binding groove positioned to engage nascent tubulins shortly after they emerge from the ribosome.
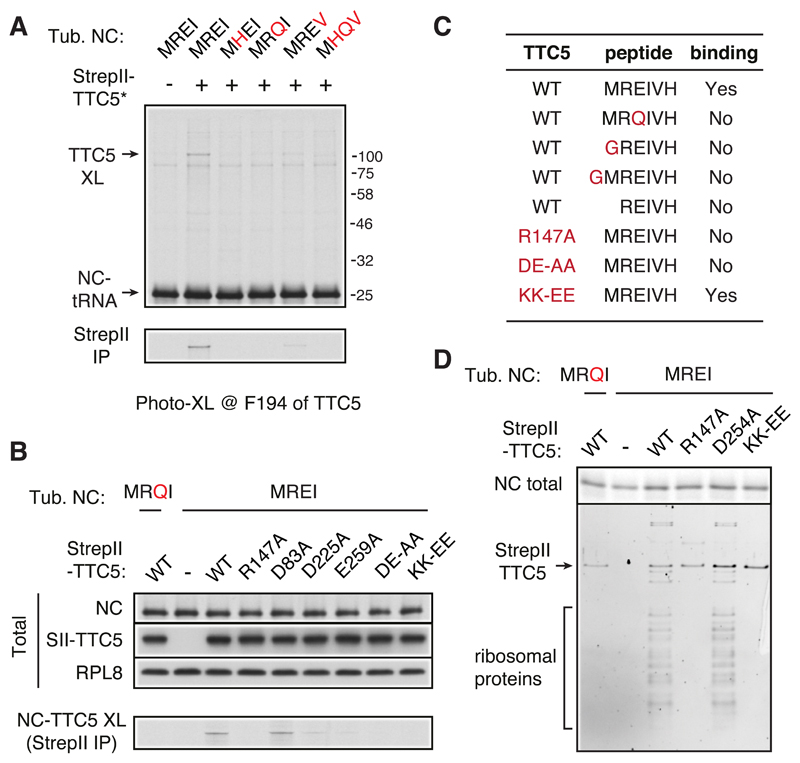
Recombinant TTC5 containing Bpa at position 194 in the ‘floor’ of the peptide binding groove (Fig. 2D) efficiently crosslinked with MREI-containing nascent chains, weakly crosslinked with MREV-containing nascent chains, and did not crosslink with any other mutants (Fig. 3A). Analysis of RNC crosslinking with various TTC5 mutants (Fig. 3B) validated R147, D225, and E259 as key residues within the groove that likely interact with R2 and E3 of nascent tubulin (see Fig. 2D). Binding assays with purified TTC5 and synthetic peptides (Fig. 3C; fig. S7) verified these findings and additionally showed that M1 is critical for TTC5 binding and must strictly be at the N-terminus. Thus, the structural analysis rationalizes all earlier β-tubulin mutagenesis studies on autoregulation requirement (11) and reveals the mechanistic basis of the exquisite specificity of autoregulation for α- and β-tubulins (5) that uniquely contain an MREI or MREC motif at the N-terminus (17).
Fig. 3. Avidity-based RNC binding imparts specificity to TTC5.
(A) Photo-crosslinking analysis of 35S-labelled 94-amino acid long ribosome-nascent chain complexes (RNCs) of human β-tubulin and N-terminal mutants (indicated in red) with recombinant StrepII-tagged TTC5 containing the photo-crosslinking residue Bpa at position F194. The nascent chain crosslink to TTC5 is indicated (TTC5-XL) and verified by pulldown via the StrepII tag (bottom panel). (B) Photo-crosslinking analysis using 35S-labelled 64-amino acid long ribosome-nascent chain complexes (RNCs) of human β-tubulin or the N-terminal MRQI mutant. Wild type or mutant recombinant StrepII-tagged TTC5 was included in the assay as indicated. The photo-crosslinking residue Bpa is at position 7 of the β-tubulin nascent chain. An aliquot of the total translation reaction was analyzed to verify equal levels of nascent chain (NC) synthesis by autoradiography and equal levels of recombinant TTC5 (SII-TTC5) by immunoblotting for the StrepII tag. The remainder was UV irradiated and TTC5 crosslinks were recovered via the StrepII tag and visualized by autoradiography (bottom panel). (C) Summary of interaction analysis between the indicated recombinant TTC5 proteins and the indicated synthetic peptides in a thermal shift denaturation assay (see fig. S7). (D) Wild type or mutant StrepII-tagged TTC5 was included during in vitro translation of wild type or mutant 64-mer β-tubulin RNCs as indicated. Equal translation of 35S-labelled nascent chain synthesis was verified (NC total). The remainder of each translation was affinity purified via the StrepII tag and analyzed by staining of total proteins (bottom panel).
Mutating the ribosome-interacting residues K285 and K287 of TTC5 to glutamic acid (KK-EE) completely abolished β-tubulin RNC binding in the crosslinking assay (Fig. 3B) despite unperturbed binding of TTC5 to synthetic tubulin autoregulatory peptide in a thermal shift assay (Fig. 3C; fig. S7). Affinity purification of recombinant TTC5 from in vitro translation reactions of nascent β-tubulin RNCs showed that no ribosomes were recovered with either TTC5(KK-EE) or the peptide binding mutant TTC5(R147A), in contrast to wild-type TTC5 (Fig. 3D). Thus, the avidity of bi-partite binding to the ribosome and nascent tubulin imparts high affinity and specificity to the TTC5-RNC interaction.
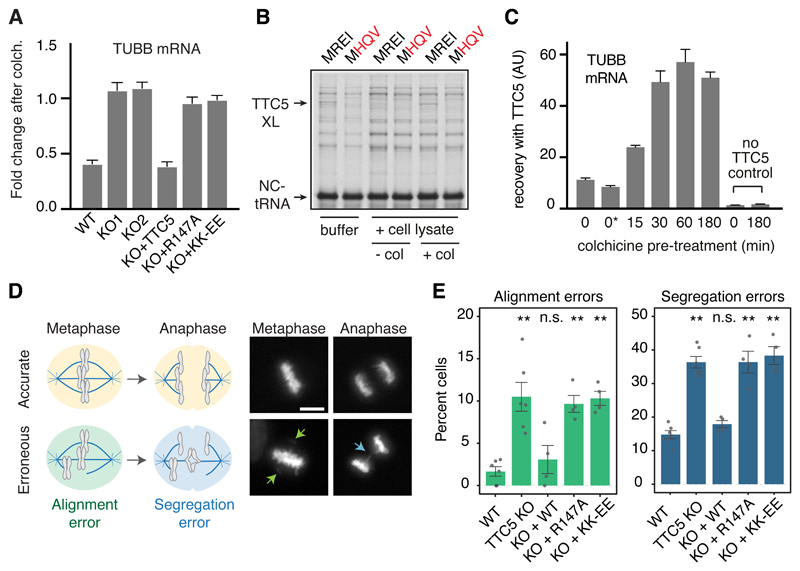
CRISPR-mediated disruption of TTC5 expression in multiple cell lines completely abolished the decay of α- and β-tubulin mRNAs in response to acute microtubule destabilization (Fig. 4A and fig. S8). Pulse labelling with 35S-methionine of wild type cells showed that of the major proteins visualised, tubulins were selectively reduced in their synthesis when cells are pre-treated with microtubule destabilizing agents (fig. S9). Selective reduction in tubulin protein synthesis was completely lost in TTC5 knockout cells, consistent with the failure to degrade tubulin mRNAs. Tubulin autoregulation, as judged by both mature- versus pre-mRNA levels (Fig. 4A) and rates of protein synthesis (fig. S9), could be restored to TTC5 knockout cells by re-expression of wild type TTC5 but not the peptide-binding mutant R147A or the ribosome-binding mutant KK-EE. Thus, TTC5 engagement of nascent tubulin at the ribosome is strictly required for tubulin mRNA degradation when cells initiate autoregulation. Access of TTC5 to the ribosome proved to be a regulated event.
Fig. 4. TTC5 is required for tubulin autoregulation and accurate mitosis.
(A) The indicated HEK293 cell lines were either left untreated or treated for 3 h with colchicine. The relative amounts of the indicated mRNAs or pre-mRNAs were quantified by RT-qPCR and normalized to a control ribosomal RNA. Plotted is the mean ± SD from three replicates. Similar results are seen in HeLa cells and with different microtubule destabilizing agents (fig. S8). (B) Pre-formed RNC-TTC5 complexes (see fig. S10) were mixed with buffer or cytosol from TTC5 knockout cells that had been pre-treated (+col) or not (-col) with colchicine for 3 hours. All samples were subjected to UV crosslinking to monitor the nascent chain interactions. The positions of the non-crosslinked tRNA linked nascent chain (NC-tRNA) and TTC5 crosslink (TTC5-XL) are indicated. (C) TTC5-knockout HEK293 cells were pretreated for the indicated times with colchicine and used to prepare lysates. One of the control samples included colchicine added after cell lysis (indicated as 0*). The products recovered by binding to recombinant TTC5 were analyzed for β-tubulin mRNA by quantitative RT-PCR. The relative recoveries are plotted (mean ± SD from three replicates). Similar results were seen for α-tubulin and when nocodazole was used instead of colchicine to trigger autoregulation (fig. S11). (D) Diagram (left) and examples (right) of accurate (top) and erroneous (bottom) chromosome alignment and segregation visualized with SirDNA dye during mitosis in HeLa cell lines (see fig. S13). Scale bar = 5 µm. (E) Quantification of errors in chromosome alignment and segregation in the indicated HeLa cell lines. Plotted is mean ± SEM from 4-6 independent biological replicates (dots) with 200-400 analyzed cells per replicate. Not significant (n.s.) and p<0.001 (**) in paired Student’s t-tests are indicated.
The TTC5-RNC complex was found to be disrupted by a cytosolic factor whose activity was lost when cells were pre-treated with colchicine to initiate autoregulation (Fig. 4B; fig. S10). Loss of this inhibitory activity under autoregulation conditions was accompanied by increased capacity of TTC5 to engage tubulin RNCs as measured by recovery of tubulin mRNAs (Fig. 4C; fig. S11). These results indicate that cells ordinarily contain an inhibitory factor that prevents TTC5 engagement of tubulin RNCs. This TTC5 inhibitor is inactivated when cells perceive excess αβ-tubulin, freeing TTC5 to engage tubulin RNCs and trigger tubulin mRNA degradation. TTC5 access to RNCs only during autoregulation explains why normally growing TTC5 knockout cells did not show notably elevated tubulin mRNA and protein (fig. S12). Because overexpressed TTC5 in the rescue cell lines did not trigger tubulin mRNA degradation until cells perceive excess αβ-tubulin (fig. S12), it seems the inhibitor is not easily saturated. Future work will identify the inhibitor and its mechanism of regulation.
Chromosome alignment and segregation during mitosis are sensitive to altered microtubule dynamics (18–21), motivating us to monitor these parameters in cells impaired in tubulin autoregulation (Fig. 4D). TTC5 knockout HeLa cells showed ~6.5-fold higher rate of chromosome alignment errors in metaphase (Fig. 4E, fig. S13), ~2.4-fold higher rate of segregation errors during anaphase (Fig. 4E, fig. S13), and a subtle but highly reproducible increase of mitotic duration (fig. S14). These phenotypes in TTC5 knockout cells were rescued by re-expression of wild type TTC5 but not the peptide binding or ribosome binding mutants of TTC5 (Fig. 4E, fig. S13, S14). Although the specific basis of mitotic defects in TTC5 knockout cells remains to be determined, we can ascribe the phenotypes to autoregulation, and not another TTC5 function (22–24), because the effects were not rescued by two unrelated point mutants of TTC5 that perturb autoregulation by different mechanisms.
TTC5 represents a highly selective and regulated ribosome-associating factor that only engages the ~2-3% of a cell’s ribosomes actively synthesizing α- and β-tubulins. By marking tubulin-translating ribosomes, TTC5 is ideally positioned to recruit yet unidentified downstream effectors to this site that trigger mRNA decay. More generally, the translating ribosome represents a platform from which to effect abundance control of key cellular proteins because translation initiation (25), elongation (26), polypeptide fate (27), and mRNA stability (28) can all be locally regulated from this site. Specificity for particular substrates would be imparted by recognition of the nascent protein emerging from the ribosome exit tunnel. Thus, cells may contain a family of substrate-specific ribosome-associating factors analogous to TTC5 that dynamically tune the abundance of key proteins such as histones (29) and chaperones (30). The methods and paradigm of co-translational abundance control established here should provide a framework for studying this mode of cellular regulation.
Supplementary Material
Acknowledgements
We thank V. Ramakrishnan for support and advice; S.-Y. Peak-Chew and M. Skehel for mass spectrometry analysis; J. Grimmett and T. Darling for advice, data storage and high-performance computing; P. Emsley for advice; the MRC Laboratory of Molecular Biology EM Facility for microscopy support, sample preparation and data collection; Brian Raught and Wade Harper for Flp-In TRex HeLa cells; the Nikon Imaging Center at Harvard Medical School for help with light microscopy.
Funding: This work was supported by the UK Medical Research Council (MC_UP_A022_1007 to R.S.H.), the US National Institutes of Health (NIH P50 107618 to T.J.M.), Harvard Medical School (S.S.) and Vallee Scholars Program (S.S.). Z.L. was supported by the Human Frontier Science Program postdoctoral fellowship. I.G. is a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG:2279-16). V.C. was supported by V. Ramakrishnan whose funding was from the MRC (MC_U105184332), the Wellcome Trust (WT096570 to V.R.), the Agouron Institute, and the Louis-Jeantet Foundation.
Footnotes
Author contributions: ZL discovered TTC5 and performed all biochemical analyses. IG and SS generated and validated HeLa cell lines. IG designed and performed phenotypic analysis of mitosis. VC performed structural analysis of the TTC5-ribosome complex. NP set up and characterized the in vitro photo-crosslinking system. SS, TJM, and RSH supervised different aspects of the project. RSH and ZL conceived the project, oversaw its implementation, and wrote the manuscript. All authors contributed to manuscript editing.
Competing interests: The authors declare no competing interests.
Data and material availability: The cryo-EM map has been deposited to the EMDB (EMD-10380) and atomic coordinates have been deposited to the Protein Data Bank (PDB 6T59). All other data are available in the manuscript or the supplementary material.
References
- 1.Borisy G, et al. Microtubules: 50 years on from the discovery of tubulin. Nat Rev Mol Cell Biol. 2016;17:322–328. doi: 10.1038/nrm.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desai A, Mitchison TJ. Microtubule Polymerization Dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 3.Brouhard GJ, Rice LM. Microtubule dynamics: an interplay of biochemistry and mechanics. Nat Rev Mol Cell Biol. 2018;19:451–463. doi: 10.1038/s41580-018-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker RA, et al. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasic I, Boswell SA, Mitchison TJ. Tubulin mRNA stability is sensitive to change in microtubule dynamics caused by multiple physiological and toxic cues. PLOS Biol. 2019;17:e3000225. doi: 10.1371/journal.pbio.3000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleveland DW, Lopata MA, Sherline P, Kirschner MW. Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell. 1981;25:537–46. doi: 10.1016/0092-8674(81)90072-6. [DOI] [PubMed] [Google Scholar]
- 7.Cleveland DW. Autoregulated instability of tubulin mRNAs: a novel eukaryotic regulatory mechanism. Trends Biochem Sci. 1988;13:339–43. doi: 10.1016/0968-0004(88)90103-x. [DOI] [PubMed] [Google Scholar]
- 8.Gay DA, Sisodia SS, Cleveland DW. Autoregulatory control of beta-tubulin mRNA stability is linked to translation elongation. Proc Natl Acad Sci U S A. 1989;86:5763–5767. doi: 10.1073/pnas.86.15.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pachter JS, Yen TJ, Cleveland DW. Autoregulation of tubulin expression is achieved through specific degradation of polysomal tubulin mRNAs. Cell. 1987;51:283–292. doi: 10.1016/0092-8674(87)90155-3. [DOI] [PubMed] [Google Scholar]
- 10.Yen TJ, Machlin PS, Cleveland DW. Autoregulated instability of β-tubulin mRNAs by recognition of the nascent amino terminus of βtubulin. Nature. 1988;334:580–585. doi: 10.1038/334580a0. [DOI] [PubMed] [Google Scholar]
- 11.Bachurski CJ, Theodorakis NG, Coulson RM, Cleveland DW. An amino-terminal tetrapeptide specifies cotranslational degradation of beta-tubulin but not alpha-tubulin mRNAs. Mol Cell Biol. 1994;14:4076–4086. doi: 10.1128/mcb.14.6.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theodorakis NG, Cleveland DW. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol Cell Biol. 1992;12:791–799. doi: 10.1128/mcb.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamerdinger M, et al. Early Scanning of Nascent Polypeptides inside the Ribosomal Tunnel by NAC. Mol Cell. 2019 doi: 10.1016/j.molcel.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 14.Wegrzyn RD, et al. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J Biol Chem. 2006;281:2847–57. doi: 10.1074/jbc.M511420200. [DOI] [PubMed] [Google Scholar]
- 15.Wiedmann B, Sakai H, Davis Ta, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- 16.Gamerdinger M, Hanebuth MA, Frickey T, Deuerling E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science (80-.) 2015;348:201–207. doi: 10.1126/science.aaa5335. [DOI] [PubMed] [Google Scholar]
- 17.T. UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699–2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vicente JJ, Wordeman L. The quantification and regulation of microtubule dynamics in the mitotic spindle. Curr Opin Cell Biol. 2019;60:36–43. doi: 10.1016/j.ceb.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petry S. Mechanisms of Mitotic Spindle Assembly. Annu Rev Biochem. 2016;85:659–683. doi: 10.1146/annurev-biochem-060815-014528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol. 2017;18:187–201. doi: 10.1038/nrm.2016.162. [DOI] [PubMed] [Google Scholar]
- 21.Kline-Smith SL, Walczak CE. Mitotic Spindle Assembly and Chromosome Segregation. Mol Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 22.Lynch JT, Somerville TDD, Spencer GJ, Huang X, Somervaille TCP. TTC5 is required to prevent apoptosis of acute myeloid leukemia stem cells. Cell Death Dis. 2013;4:e573–e573. doi: 10.1038/cddis.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demonacos C, Krstic-Demonacos M, La Thangue NB. A TPR motif cofactor contributes to p300 activity in the p53 response. Mol Cell. 2001;8:71–84. doi: 10.1016/s1097-2765(01)00277-5. [DOI] [PubMed] [Google Scholar]
- 24.Hu X, Mullins RD. LC3 and STRAP regulate actin filament assembly by JMY during autophagosome formation. J Cell Biol. 2019;218:251–266. doi: 10.1083/jcb.201802157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonenberg N, Hinnebusch AG. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat Struct Mol Biol. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marzluff WF, Koreski KP. Birth and Death of Histone mRNAs. Trends Genet. 2017;33:745–759. doi: 10.1016/j.tig.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Didomenico BJ, Bugaisky GE, Lindquist S. The heat shock response is self-regulated at both the transcriptional and posttranscriptional levels. Cell. 1982;31:593–603. doi: 10.1016/0092-8674(82)90315-4. [DOI] [PubMed] [Google Scholar]
- 31.Chin JW, et al. An Expanded Eukaryotic Genetic Code. Science (80-.) 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 32.Sakamoto K, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma A, Mariappan M, Appathurai S, Hegde RS. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol. 2010;619:339–63. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Q, Shao S. In vitro reconstitution of translational arrest pathways. Methods. 2018;137:20–36. doi: 10.1016/j.ymeth.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao S, et al. Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Cell. 2016;167:1229–1240.e15. doi: 10.1016/j.cell.2016.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams CJ, et al. The p53 cofactor Strap exhibits an unexpected TPR motif and oligonucleotide-binding (OB)-fold structure. Proc Natl Acad Sci. 2012;109:3778–3783. doi: 10.1073/pnas.1113731109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–53. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, et al. Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its αNAC subunit. Protein Cell. 2010;1:406–416. doi: 10.1007/s13238-010-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen VB, et al. MolProbity : all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.