Abstract
Programmable nucleic acid nanoparticles (NANPs) provide controlled coordination of therapeutic nucleic acids (TNAs) and other biological functionalities. Beyond multivalence, recent reports demonstrate that NANP technology can also elicit a specific immune response, adding another layer of customizability to this innovative approach. While the delivery of nucleic acids remains a challenge, new carriers are introduced and tested continuously. Polymeric platforms have proven to be efficient in shielding nucleic acid cargos from nuclease degradation while promoting their delivery and intracellular release. Here, we venture beyond the delivery of conventional TNAs and combine the stable cationic poly-(lactide-co-glycolide)-graft-polyethylenimine with functionalized NANPs. Furthermore, we compare several representative NANPs to assess how their overall structures influence their delivery with the same carrier. An extensive study of various formulations both in vitro and in vivo reveals differences in their immunostimulatory activity, gene silencing efficiency, and biodistribution, with fibrous NANPs advancing for TNA delivery.
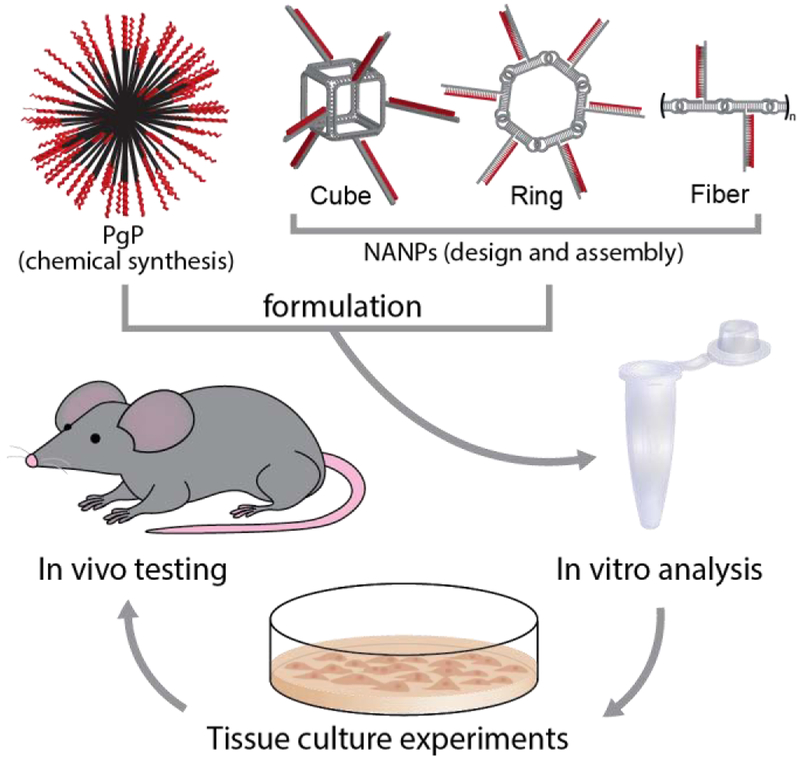
Graphical abstract
Functional nucleic acid nanoparticles (NANPs) are combined with the novel polymeric carrier for potent gene silencing and controllable immunostimulation. Optimization of the formulation is achieved and efficacy is demonstrated against several targets. Biodistribution, hemocompatibility, and immunotoxicity are assessed to identify how NANP structure determines therapeutic outcomes.
BACKGROUND
Therapeutic nucleic acids (TNAs) have garnered considerable attention as potential therapeutics for assorted diseases. RNA interference (RNAi) inducers, aptamers, and immunostimulatory nucleic acids have been studied for their ability to treat both endogenous and infectious diseases. The latest success of the first RNAi-based therapeutic agent (Patisiran) (1), FDA-approved in 2018, makes advances in the composition and delivery of TNAs timely and important (2-5). Recently, a new generation of nucleic acid-based nanoparticles (NANPs) functionalized with TNAs has been introduced. NANPs are composed of multiple nucleic acid strands programmed to self-assemble into defined 3D structures (6) with further possibilities for embedded functionalities (7-13). Various self-assembling NANPs have been generated to both increase stability and enzymatic resistance, as well as to coalesce multiple pharmaceuticals (14-20). The ability to finely control the size, shape, multivalencey, and therapeutic payload makes NANP technology an attractive option for biomedical applications. Despite advances, hurdles to the rapid translation of NANPs from benchtop to clinic include their poor resistance to enzymatic degradation in blood serum, their inability to cross biological membranes, and the potential for deleterious immune responses (21,22). Therefore, combining potent customizable therapeutic NANPs with stable, non-toxic, and non-immunogenic carriers would greatly increase the clinical potential of such agents for the treatment of a plethora of diseases; however, the effects of NANP size, shape, and composition on relative delivery efficiency with the same carrier have not yet been determined.
A number of effective TNA carriers composed of a wide variety of materials have been investigated to combat the difficulties associated with the use of NANPs and each possesses distinct features (23-26). In addition, each new formulation varies in terms of toxicity, biodistribution, accumulation, in vivo stability, and excretion. Furthermore, different formulations have shown varied success in crossing the blood brain barrier (BBB), a major challenge for the treatment of central nervous system (CNS) diseases. Finally, the drug loading capabilities of each formulation varies, allowing for various payload magnitudes to be delivered in a single multi-modal platform.
Cationic lipids, liposomes, and polymers, including polyethylenimine (PEI), poly (β-amino esters), and polyamidoamine (PAMAM) dendrimers, have been evaluated as non-viral vectors for pDNA and small RNA therapeutics (27-29). Among these carriers, branched PEI (bPEI; 25kDa), considered the gold standard for gene transfection, has exhibited the highest transfection efficiency among non-viral vectors in serum-free conditions due to its ability to form stable polyplexes with nucleic acids and its buffering capacity, which facilitates its endosomal escape via the proton sponge effect (30). However, the low transfection efficiency of bPEI in the presence of serum limits its potential for use as a vector in vivo.
In the present work, we employed the previously described cationic amphiphilic co-polymer, poly (lactide-co-glycolide)-graft-polyethylenimine (PgP) (31,32) as a carrier for a series of NANPs of different shapes and sizes that include three-dimensional cubic (14,15,33-35), planar ring-like (36-39), and fibrous (36,40) RNA self-assembling NANPs. PgP, a micelle-forming co-polymer composed of poly (lactic-co-glycolic acid) (PLGA) and bPEI, has previously been characterized and demonstrated to be an efficient carrier of siRNA and pDNA in vitro in various cell lines and in the rat normal spinal cord in vivo (32). PgP has also been demonstrated to deliver siRNA targeting RhoA to spinal cord lesion sites in a rat model of compression injury, achieving sustained RhoA gene expression knockdown for up to four weeks and supporting its efficacy as an in vivo TNA delivery system (41). Here, we investigated the effect of NANP size, shape, and composition on PgP/NANP polyplex stability, intracellular uptake, silencing efficiency, toxicity, immunostimulatory activity, hemocompatibility, and biodistribution, in vitro and/or in vivo. Scheme 1 illustrates the experimental design of the current work.
Scheme 1.
Experimental design of the current work.
METHODS
All methods and materials are detailed in supplementary information.
Synthesis of PgP and NANPs.
PgP was synthesized as previously described using PLGA (4kDa, 50:50, Durect Corporation Pelham, AL) and bPEI (MW 25 kDa, Sigma). NANPs were synthesized and assembled as previously described using T7 RNA polymerase in vitro run-off transcription followed by a one-pot assembly.
Nuclease protection assay of PgP/DNA duplex polyplexes.
To assess the ability of PgP to protect nucleic acids, a DSDNA carrying an Alexa Fluor 488 (Al488) (5’ sense) and an Iowa Black Quencher (3’ anti-sense) were conjugated with PgP at various concentrations and treated with DNase. The fluorescence resulting from the digestion of the DNA was measured every 30 seconds.
Physical characterization of NANPs.
Atomic force microscopy (AFM) was performed for each NANP on APS-modified mica using a MultiMode AFM Nanoscope IV system (Bruker Instruments, Santa Barbara, CA) in tapping mode. Non-denaturing polyacrylamide gel electrophoresis (Native-PAGE) was performed using 8% acrylamide, 37.5:1 acrylamide/bisacrylamide, run at 4° C. A heparin competition assay was performed to ensure structural integrity was maintained. For this assay, each NANP was bound to PgP at a 30:1 N/P ratio, then released by electrostatic competition with heparin, and run through Native-PAGE.
Fluorescent microscopy and cellular uptake.
Confocal microscopy imaged were taken using a LSM710 confocal microscope (Carl Zeiss, Germany) equipped with a 63X/1.4 magnification lens. To confirm uptake, PgP/DS RNA-Al546 was transfected into MDA-MB-231 cells and incubated for 6 hours, then fixed with 4% paraformaldehyde for 20 minutes at room temperature. To confirm endosomal co-localization, cells were treated with endosomal targeting Rab5 antibodies (Sana Cruz, Dallas, TX) and a secondary antibody labeled with Al647. To further confirm endosomal uptake, cells were transfected at both 4° C and 37° C with PgP/DS RNA-Al488 or PgP/DS DNA-Al488. Uptake of the fluorescent polyplexes was confirmed by both flow cytometry as well as fluorescent microscopy.
Specific gene silencing and cell viability.
Assessment of gene knockdown potential was confirmed for all NANPs complexed with PgP for two different genes, green fluorescent protein (GFP) and RhoA. For GFP, cubes, rings, and fibers were functionalized with DS RNAs targeting GFP. The PgP/NANP polyplexes were transfected into MDA-MB-231 cells expressing GFP. Knockdown was confirmed by both flow cytometry as well as fluorescent microscopy. Cell viability was confirmed using an MTS assay (Promega, WI). For gene silencing of RhoA, cubes, rings, and fibers were designed carrying DS RNAs against RhoA. B35 neuroblastoma cells were transfected with PgP/NANP polyplexes. The expression of RhoA was measured by real-time PCR. Furthermore, the cell viability of the B35 cells post transfection was measured using an MTT assay (Sigma-Aldrich).
Immunostimulation in vitro.
To quantify the immunostimulation from PgP/NANP polyplexes, the IL-6 and IFN-β expression was measured post-transfection in human microglia-like cell line (hμglia) via ELISA. The hμglia cells were generously provided from the laboratory of Dr. Jonathan Karn (Department of Molecular Biology and Microbiology, Case Western Reserve University). This cell line was generated from primary human microglia cells purchased from ScienCell and transformed with lentiviral vectors expressing SV40 and hTert antigens. These cells have been classified as microglia due to their microglia-like morphology, migratory and phagocytic activity, presence of the surface markers CD11b, TGFβR, and P2RY12, and characteristic microglial RNA expression profile. Furthermore, contribution from specific toll-like receptor and intracellular signaling pathways were QUANTIfied using HEK-Blue™ hTLR 3 and 7 cells, and THP1-Dual™ cells, respectively. HEK-Blue™ hTLR 3 and 7 cells are HEK cells which have been engineered to express a single Toll-like receptor. The Toll-like receptor leads downstream to secreted alkaline phosphatase (SEAP), which can be easily detected and QUANTIfied using reagent QUANTI-Blue™. THP1-Dual™ cells are monocytes engineered in a similar fashion to express SEAP when the NF-κB pathway is stimulated, or luciferase when the IRF pathway is stimulated. Both can be readily QUANTIfied using either QUANTI-Blue™ or QUANTI-Luc™, respectively. The HEK-Blue™ and THP1-Dual™ cells were used for specific identification of the molecular pathways involved in the immune-recognition of the PgP/NANP polyplexes.
Hemolysis assay in vitro.
For hemocompatability, PgP/NANP polyplexes were incubated with rat erythrocytes for 1 hour at 37° C. Following incubation, suspensions were centrifuged at 1000 x g for 10 minutes and absorbance of supernatant was measured at 540 nm using a Synergy HT plate reader (Biotek, Winooski, VT). PBS and Triton X-100 were used as controls for 0 and 100 % hemolysis. Hemolysis was QUANTIfied using the following formula:
where ASample, APBS, ATriton are the absorbance of the sample, PBS, and Triton-X, respectively. The erythrocytes pellet were reconstituted in PBS and the cells were were imaged using an inverted microscope (Zeiss Axiovert 200, Göttingen, Germany).
Biodistribution of PgP/NANP polyplexes after systemic injection.
The PgP/NANP polyplexes were administered to male CD-1 mice (7-8 weeks-old, Chares River Laboratories, MA) via tail vein injection. The mice were imaged 1, 2, 6, and 24 hours after injection by live animal fluorescence imaging system (IVIS Luminar XR, Caliper Life Sciences) under anesthesia with isoflurane gas. At 24 hours post-injection, the animals were euthanized by CO2 and their organs were harvested for ex vivo organ imaging. The percent of organ distribution was measured by the fluorescence of each organ.
Statistics.
Experimental results are presented as the mean ±SEM. Statistical significance was determined using one-way Anova using GraphPad Prism Software Version 7.
RESULTS
PgP/DNA stability, binding, and nuclease protection assay.
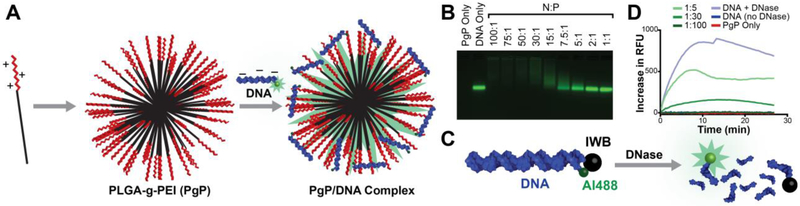
For cost efficiency, all initial in vitro optimization experiments were performed using Alexa 488-labeled DNAs rather than labeled RNA. To identify the formation of stable complexes of PgP and DNA duplexes (Fig. 1A), a fixed amount of fluorescently tagged DNA duplex was mixed with PgP at varying N/P ratios and run through a 2% (w/v) agarose gel. Duplexes not complexed with PgP migrated freely through the gels, while the mobility of duplexes electrostatically complexed with PgP was limited. Complete retardation was observed at an N:P ratio of 30:1 (Fig. 1B). Above this ratio, additional PgP did not demonstrate any difference in the gel. This demonstrated a visual representation of the PgP’s maximum loading capacity and confirmed attachment of PgP to nucleic acids. Next, we investigated the ability of the PgP to protect the nucleic acid cargo from enzymatic degradation. DNA duplexes tagged with Alexa 488 at the 5’-side and Iowa Black quencher at the corresponding 3’-side were treated with RQ1 DNase (Fig. 1C-D). In untreated duplexes, the close proximity of Iowa Black completely quenched the fluorescence of Alexa 488. However, upon DNase treatment, the degradation of duplexes and further spatial separation of fluorophore and quencher led to activation of the fluorescent signal. PgP was shown to successfully shield the attached nucleic acids from enzymatic activity for several hours, as evidenced by the minimal increase in fluorescence as compared to the rapidly increasing signal in that of free duplexes.
Figure 1.
Binding of DNA to PgP leads to protection from nuclease activity. (A) Schematic showing electrostatically-driven PgP binding to nucleic acids, (B) gel retardation of various PgP/DNA polyplexes prepared using various PgP/DNA ratios (N/P ratio), and (C-D) nuclease protection assay of PgP/DNA polyplexes after incubation with DNase I.
Intracellular uptake of PgP/RNA or PgP/DNA complexes.
To confirm intracellular uptake and localization, PgP complexed with Alexa 488-labeled DS RNAs (PgP/RNA-Al488) were added to breast cancer cells (MDA-MB-231). MDA-MB-231 cells are an immortalized triple negative breast cancer cell line used frequently in the study of cancer therapeutics. The cells proliferate quickly, and are frequently studied as transfection targets for nucleic acid therapeutics. Confocal microscopy showed that PgP/RNA-Al488 polyplexes were successfully internalized into the cells and distributed in the cytoplasm for extended periods of time (Fig. 2A). To confirm that the PgP was taken in via endocytosis, cells were again exposed to PgP/RNA-Al488 and co-localization with protein Rab5, which localizes around endosomes to promote trafficking (42), was demonstrated (Fig. 2B). The co-localization of the Alexa 488 (green) and the anti-Rab5 antibodies (red) demonstrates that the PgP/RNA-Al488 polyplexes were taken up and processed via an endosomal pathway. To further confirm this observation, MDA-MB-231 cells were exposed to PgP/RNA-Al488 or PgP/DNA-Al488 at either 37 °C or 4 °C (Fig. 2C). Endocytosis is a temperature-dependent process that has been previously demonstrated to be absent in cells under 4 °C conditions (43). Our flow cytometry and microscopy results show that cells exposed to PgP/DNA-Al488 or PgP/RNA-Al488 complexes at 4 °C showed a negligible increase in fluorescence, while those transfected with the same sample at 37 °C showed a marked increase in fluorescence (Fig 2C-D). These data therefore support our hypothesis that PgP/DNA and PgP/RNA polyplexes are taken into cells via endocytosis.
Figure 2.
Intracellular uptake of PgP/DS RNA polyplexes assessed in MDA-MB-231 human breast cancer cells. (A) Visualization of intracellular uptake of polyplexes by confocal microscope; (B) Co-localization of polyplexes with early endosomal marker Rab5 confirms an endocytic pathway of internalization; (C) Flow cytometry measuring the effect of temperature on intracellular uptake of polyplexes (**** denotes p < 0.0005); (D) Fluorescent microscopy and bright-field overlay of MDA-MB-231 human breast cancer cells transfected with either PgP/DS RNA-Al488 or PgP/DNA-Al488 at either 4 °C or 37 °C, 6 hours after transfection.
Characterization of PgP/NANP polyplexes.
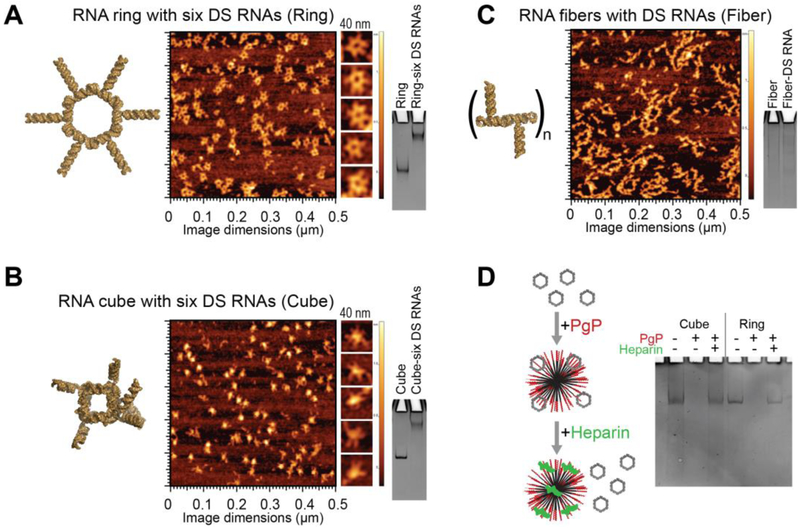
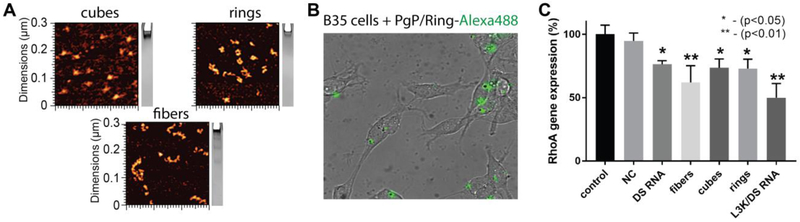
The NANPs included in this study were generated utilizing various design strategies and were formulated to assemble into robust constructs with distinct connectivity, shapes, and sizes. Three-dimensional hexameric RNA cubes assemble via intermolecular Watson-Crick base pairing, while planar hexameric RNA rings and linear RNA fibers both assemble via magnesium-dependent intramolecular Watson-Crick base pairing to facilitate intermolecular kissing loop interactions (120° ColE1-like for rings and 180° HIV-like for fibers). The cubic RNA structures are designed to form solely due to intermolecular forces, avoiding secondary structures within individual strands and forming a 3D cube. The rings and fibers are designed to fold via intramolecular interactions, exposing kissing loops for strand-strand interactions to form a planar ring. By extending the individual strands of each scaffold, cubes, rings, and fibers, can be functionalized with DS RNAs against any target and generated via one-pot assembly as previously described (15,21,37). The assemblies of resulting functional NANPs can then be confirmed by both gel electrophoresis (native-PAGE) and atomic force microscopy (AFM) as shown in Fig. 3A-C. An important aspect of this study was to determine the effect of functional NANP compositions on their delivery and efficacy. As such, it was essential to demonstrate that the NANPs stay intact following release from PgP. A heparin competition assay was performed to disrupt the PgP/NANP interaction and release the NANP following PgP binding. Gel electrophoresis demonstrates that the model NANPs remain intact following their release (Fig. 3D).
Figure 3.
Characterization of various functionalized NANPs by AFM and native-PAGE: (A) RNA rings functionalized with six Dicer substrate (DS) RNAs, (B) RNA cubes functionalized with six DS RNAs, and (C) DS functionalized RNA fibers. (D) Native-PAGE results confirm the integrity of NANPs upon heparin-assisted release from PgP polyplexes.
Gene silencing with PgP/NANP(GFP or RhoA) polyplexes.
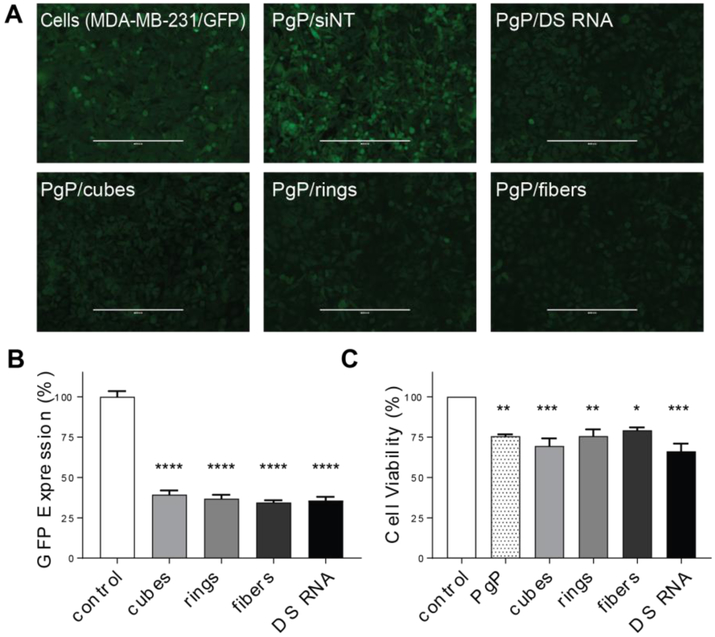
We have previously demonstrated that NANPs functionalized with DS RNA against GFP efficiently silence GFP expression when transfected into various cell lines using common transfection agents such as Lipofectamine 2000 (11,12,21,38,44). To confirm the efficacy of PgP as a delivery agent, we transfected the breast cancer cell line MDA-MB-231/GFP with the panel of PgP/NANP(GFP) polyplexes. The NANPs with multiple DS RNAs against GFP, as well as individual GFP DS RNA complexed with PgP, were shown to successfully silence the expression of GFP as demonstrated by fluorescent microscopy and flow cytometry (Fig. 4A-B), without significant changes in cell viability (Fig. 4C). All transfections were performed at concentrations normalized to the amounts of DS RNAs (50 nM). Despite their structural differences, no NANP was determined to be significantly better than the others. To further demonstrate therapeutic potential in central nervous system injury repair, NANPs decorated with DS RNAs targeting RhoA were tested in rat neuroblastoma (B35) cells. Previously, PgP complexed with siRNA against RhoA has been demonstrated to treat spinal cord injuries in rats (41). We sought to determine whether RhoA DS RNA-functionalized NANPs (Fig. 5A) have enhanced silencing capabilities versus conventional DS RNAs. PgP/NANPs(RhoA) were transfected into B35 cells and uptake was confirmed (Fig. 5B). We identified that fibers functionalized with RhoA DS RNAs induced greater knockdown than cube NANPs, ring NANPs, or standard RhoA siRNA duplexes (Fig. 5C). This enhanced knockdown occurred in the absence of significant changes in cell viability (data not shown).
Figure 4.
Specific gene silencing with PgP/NANP(GFP) polyplexes and cell viability assays tested against GFP expressing breast cancer cells (MDA-MB-231/GFP). Fluorescent microscopy (A) shows representative images of GFP knockdown by PgP/NANP(GFP) polyplexes. (B) GFP knockdown efficiency by cubes (PgP/cubes(GFP)), rings (PgP/rings(GFP)), and fibers (PgP/fibers(GFP)) are compared to free DS RNAs(GFP) and negative control (siNT), PgP only (0.1 mg/mL) used as an additional control. (N=3, **** denotes statistically significant vs. untreated cells with p<0.0005, # denotes statistical significance vs. cubes with p<0.05). (C) Cell viability by PgP/NANP(GFP) polyplexes (N=3, * denotes statistical significance with p<0.05, ** with p<0.005, *** with p<0.005).
Figure 5.
(A) AFM images of RhoA NANPs and (B,C) specific gene silencing with PgP/NANP polyplexes targeting RhoA in rat neuroblastoma cells (B35). In all experiments, cubes (PgP/cubes(RhoA)), rings (PgP/rings(RhoA)), fibers (PgP/fibers(RhoA)), and individual DS RNAs(RhoA) are compared. Negative control siRNA (siNT) and DS RNA transfected with Lipofectamine 3000 (L3K) are used as controls. Intracellular uptake of fluorescently labeled PgP/Ring-Al488 by confocal microscopy (A) and RhoA gene silencing assessed by RT PCR (B).
Hemocompatibility and immunotoxicity of PgP/NANP polyplexes.
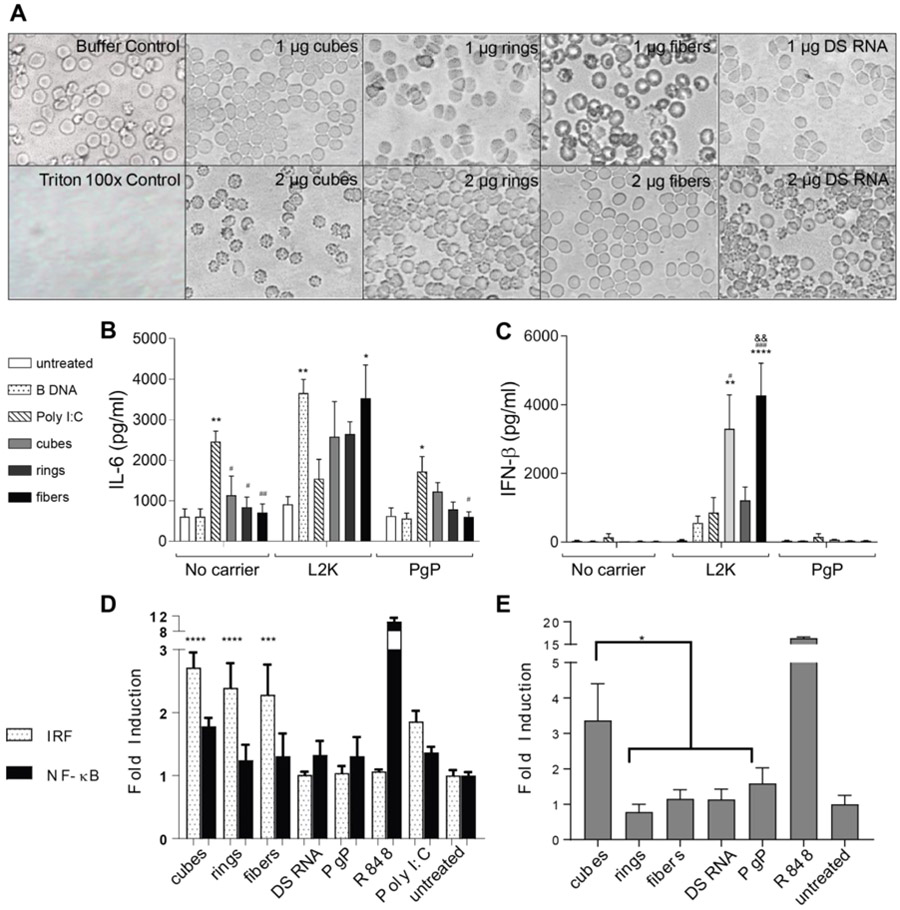
To assess the potential for detrimental toxicity that would limit the therapeutic potential of PgP/NANP polyplexes, we have assessed the hemocompatibility of these complexes by incubating them with rat erythrocytes and investigating their effects on blood cell morphology. We observed that erythrocytes treated with Triton X-100 showed complete hemolysis, while the morphology of erythrocytes treated with various PgP/NANP polyplexes were intact and hemolysis was not significantly different than that of erythrocytes treated with PBS (Supplementary Table S1). Figure 6A shows the representative images of erythrocytes treated with PBS, Triton X-100, and various PgP/NANP polyplexes.
Figure 6.
Blood compatibility and immunostimulation with PgP/NANP(GFP) polyplexes. In all experiments, cubes (PgP/cubes(GFP)), rings (PgP/rings(GFP)), fibers (PgP/fibers(GFP)), and individual DS RNAs(GFP) are compared. (A) Hemolysis assay was conducted using primary rat erythrocytes and demonstrated no lysis. (B-C) Immunostimulation of PgP/NANP(GFP) polyplexes measured via ELISA of inflammatory cytokines and type I interferons in hμglia cells. (D) IRF and NF-κB stimulation as measured by luciferase production and SEAP production, respectively, in THP1-Dual™ cells, and (E) TLR stimulation from polyplexes as measured by SEAP secretion from HEK-Blue™ hTLR7 cells. In B and C, statistical significance relevant to cells, poly I:C, and ring is denoted by *, #, and & respectively (*/# p<0.05, **/##/&& p<0.005, ### p<0.001, **** p<0.0001). (D) The role of IRF and NF-κB stimulation were measured using THP1-Dual™ cells. Statistical significance compared to cells denoted by *** with p<0.0005 and **** with p< 0.0001). (E) HEK-Blue™ hTLR7 cells were transfected with the various PgP/NANP complexes and the TLR stimulation was measured using QUANTI-Blue™ detection media (* denotes p<0.005).
Another important pharmaceutical consideration is immunostimulatory activity. Previously, NANPs have been shown to elicit the secretion of a number of pro-inflammatory cytokines and chemokines (6,21,45). In addition, we conducted a QUANTItative structure-activity relationship study using hμglia cells to identify physicochemical contributors to immunostimulatory activity (45). These studies, however, were limited to commercially available carriers such as L2K, which has no potential for clinical use. We therefore investigated the immunogenic properties of PgP/NANPs and determined whether structural differences influenced them. NANPs were delivered to hμglia cells using PgP or L2K and the release of the cytokines interleukin-6 (IL-6) and interferon-beta (IFN-β) was determined at 24 hours postexposure by specific capture ELISAs. Poly I:C and poly dA:dT, known inducers of immune responses, were used as positive controls in these studies. Delivery of NANPs with L2K induced the release of both IL-6 and IFN-β. Interestingly, cube and ring-shaped NANPs complexed with PgP induced the same release of IL-6 compared to a positive control, while fibers induced a significantly lower response (Fig. 6B), thus indicating that the structure of NANPs can influence the immunostimulatory properties of polyplexes. Additionally, we observed that the delivery of NANPs or positive controls complexed with PgP showed much lower IL-6 and IFN-β release than transfection using L2K. Only cubes delivered with PgP stimulated detectable release of IL-6 and IFN-β when compared to positive control, indicating a link between NANP structure and immunogenic properties consistent with previous observations (6). Importantly, these data suggest that delivery of NANPs with PgP may reduce potentially detrimental immunogenicity when compared to the conventional polycationic lipid-like carriers.
Furthermore, the specific contribution made by cellular signaling pathways and individual toll-like receptors (TLR) to NANP-induced responses was evaluated using HEK-Blue™ hTLR and THP1-Dual™ cells. Engineered THP1-Dual™ cells express either SEAP or secreted luciferase upon activation of either NF-κB or IRF pathways, respectively. Our results demonstrate that the majority of the response elicited by the PgP/NANP constructs occurs via the IRF pathway (Fig 6D). The HEK-Blue™ hTLR cell model expresses a single human TLR, which upon stimulation activates the production of SEAP for further QUANTIfiable detection. TLRs 3, 7, and 8 are responsible for RNA detection in endosomal compartments, with TLR3 recognizing dsRNA and TLRs 7 and 8 responsible for ssRNA detection. Therefore, we selected TLR3 and TLR7 receptor-expressing in vitro models for use in the present study. Our data demonstrate that the TLR7-mediated immune stimulation is mainly caused by the PgP/cube polyplexes (Fig 6E) while other NANPs demonstrated negligible stimulation. Additionally, TLR3 did not show any utilization in PgP/NANP recognition (Supporting Figure S1). Overall, these results are consistent with our studies on L2K-mediated NANP delivery in human PBMCs (6,46).
Biodistribution of PgP/NANP polyplexes after systemic in vivo administration.
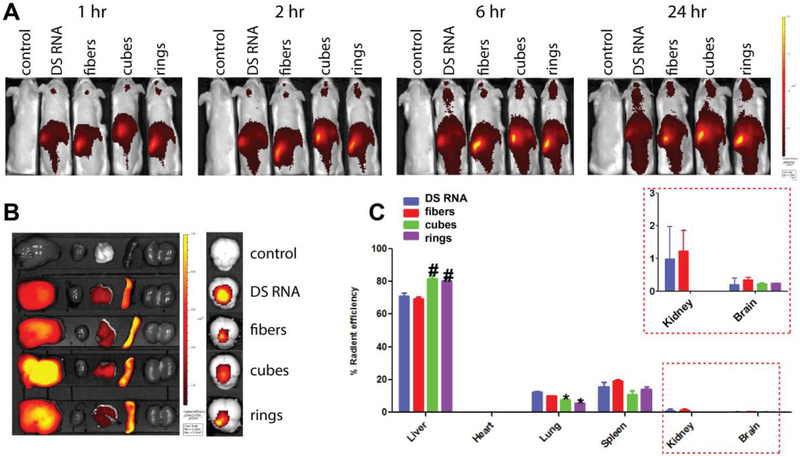
Biodistribution of various DiR-PgP/NANP polyplexes in CD-1 mice was monitored over 24 hours after intravenous injection using an IVIS live animal imaging system (Fig. 7). As shown in Figure 7A, the PgP/NANP polyplexes were distributed throughout the body after 24 hours. Interestingly, we observed strong signals in the head by in vivo animal imaging (Fig. 7A), suggesting that PgP/NANP polyplexes might be capable of crossing the BBB. However, only low-level signal expression was detectable in brain tissue ex vivo (Figure 7B, right), and the same analysis showed that the majority of the polyplexes were located in the liver, spleen, and lungs (Figure 6B, left). Figure 7C shows the percent organ biodistribution, with a rank order of distribution of liver (>70%), spleen, and then lung, with minor distribution to other organs. We observed that the percentage biodistribution of both DiR-PgP/cube(GFP) and DiR-PgP/ring(GFP) polyplexes to the liver and lung was significantly higher than DS RNA. Interestingly, we observed that all DiR-PgP/NANP polyplexes were detectable in brain tissue for up to 24 hours postadministration, but it should be noted that their percentage distribution to this site was less than 1%. Clearly, further studies will be required to confirm the BBB penetrance of PgP/NANP polyplexes.
Figure 7.
Biodistribution of various PgP/NANPs after systemic injection via tail vein in mice. The results were analyzed in vivo (A), and ex vivo (B-C). Significant difference (p<0.05) when compared to PgP/DS RNA for PgP/NANPs’ accumulation in lungs (*) and in livers (#) are shown.
DISCUSSION
A principle goal of this study was to determine how the shape and structure of functional NANPs affects the stability and efficacy of PgP/NANP polyplexes. The RNA nanoparticle structures were selected due to their differences in assembly and shape; the cubes, rings, and fibers differ in terms of dimensionality. The cubes are a globular-type structure (3D), whereas the rings are planar (2D), and the fibers form long chains of various lengths (1D). The fibers can be considered a more pliant structure, which could lead to increased binding to PgP due to ease of bending to conform to various surfaces. Such features could impact the delivery and therapeutic efficacy of each NANP. In order for duplexes, fibers, or rings to successfully bind to PgP, simple bending is needed for the entire structure to be electrostatically bound to the surface; however, due to their three dimensional globular-type structure, cubes could require greater deformation in order to successfully bind to PgP. While PgP-bound NANPs may undergo some transient deformation, we determined that the NANPs remain intact and reassume their original shapes when released.
We administered our polyplexes systemically via tail-vein injection into mice to evaluate the role of the NANP structure in their biodistribution. We determined that the polyplexes accumulated mainly in the liver and spleen and that such accumulation was independent of NANP shape. However, perhaps the most promising result from our biodistribution studies was that all structures were found, at least to some extent, to traverse the BBB into the brain (Figs. 7A and E). The delivery of bioactive compounds to the brain is a major challenge in the treatment of various maladies as the BBB is highly selective, which limits access by most nanoparticles and conventional therapeutics. As such, our results suggest that that PgP, an amphiphilic micelle-forming block copolymer, might be suitable as a nucleic acid delivery carrier to administer NANPs of various shapes across the BBB to the CNS.
A major obstacle to the therapeutic use of nanoparticles with various chemical compositions is the sensitivity of the CNS to off-target/immunotoxic effects as resident glial cells initiate and propagate immune responses in the CNS. As such, optimizing a therapeutic to enhance efficacy while limiting immunotoxicity is paramount for the generation of a successful platform. Here, we transfected hμglia cells with NANPs using L2K or PgP and measured the secretion of key immune mediators. We observed that NANPs delivered using L2K stimulated the release of both IL-6 and IFN-β. Similar to our previous studies with NANPs composed of RNA, we primarily observed a robust type I IFN response with only modest inflammatory cytokine responses (45). In contrast, we observed no statistically significant release of IL-6 or IFN-β in response to NANPs delivered using PgP. Only cubes delivered with PgP stimulated a detectable release of immune mediators. Consistent with this finding, cubes were demonstrated to be the only NANP capable of stimulating TLR7 when delivered via PgP. The ability of cubes, but not other structures, to elicit immune mediator release has been observed previously (6,46) and could be attributable to the globular nature of these nucleic acid nanostructures as opposed to the planar or fibrous structures of rings and fibers. Additionally, our data suggests that the delivery of TNAs via PgP reduces the immunogenicity of such complexes, raising the exciting possibility that this carrier could limit the detrimental immune responses previously associated with nanoparticle administration. Alternatively, several groups have used immunostimulatory nucleic acids to purposefully provoke an immune response for therapeutic action. Unmethylated cytosine-phosphate-guanosine (CpG) motifs have been delivered using nanoparticles to elicit a response via TLR 9 for targeted immune responses (47,48). Other research has used NANPs with chemical modificaitons of various shapes to modulate the immune response (49).This additional control over not only the therapeutic payload but also the immunostimulant potential lends another unforeseen layer of control over their application, allowing for desired immunostimulation with specific gene silencing on a structure and carrier dependent basis.
CONCLUSION
Here we have demonstrated and evaluated the use of functional NANPs complexed to polymeric carriers for the efficient delivery of TNAs. We show uptake and silencing efficacy in a manner that is not significantly impacted by the shape or size of the conjugated nucleic acid nanoparticle. Additionally, we show that the immunostimulatory activity of these polyplexes is markedly lower than that seen with other complexes employing the same NANPs. Expanding the arsenal of TNAs complexed with PgP to include miRNAs, aptamers, and siRNAs directed against other genes could prove highly useful in the treatment of currently intractable diseases and injuries. The present demonstration that nanostructured nucleic acids complexed to PgP are effective, non-toxic, and non-immunogenic support their further development as pharmaceuticals.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM120487 (to K.A.A). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. L.R. was supported in part by funding provided by NSF-REU and DOD-ASSURE under NSF Grant No. CHE 1460867. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E (M.V.). J.S.L. was partly supported by National Institute of Neurological Disorders and Strokes (NINDS) under Grant number 1R01 NS111037-01 and by the NIGMS under Grant No. 5P20GM103444-07 of the NIH. J.S.L was also supported by South Carolina - Spinal Cord Injury Research Fund (SC-SCIRF) under Grant number SCIRF 2017 B-01. The authors would like to thank Christian Macks for his assistance in animal studies. The authors would also like to thank Alexander Lushnikov and Alexey Krasnoslobodtsev for performing AFM imaging of the nanoparticles at the Nanoimaging Core Facility at the University of Nebraska Medical Center.
Footnotes
DISCLAIMER
All authors declare no financial conflict of interest associated with this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERNCES
- 1.Adams D, Gonzalez-Duarte A, O'Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL et al. (2018) Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N Engl J Med, 379, 11–21. [DOI] [PubMed] [Google Scholar]
- 2.Bramsen JB and Kjems J (2011) Chemical modification of small interfering RNA. Methods In molecular biology (Clifton, N.J, 721, 77–103. [DOI] [PubMed] [Google Scholar]
- 3.Bramsen JB and Kjems J (2012) Development of Therapeutic-Grade Small Interfering RNAs by Chemical Engineering. Frontiers in genetics, 3, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G and Sood AK (2011) RNA interference in the clinic: challenges and future directions. Nature Reviews Cancer, 11, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE and Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 6.Hong EP, Halman JR, Shah AB, Khisamutdinov EF, Dobrovolskaia MA and Afonin KA (2018) Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Letters, 18, 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo P (2010) The emerging field of RNA nanotechnology. Nature nanotechnology, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasinski D, Haque F, Binzel DW and Guo P (2017) Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano, 11, 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binzel DW, Shu Y, Li H, Sun M, Zhang Q, Shu D, Guo B and Guo P (2016) Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Molecular therapy: the journal of the American Society of Gene Therapy, 24, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla GC, Haque F, Tor Y, Wilhelmsson LM, Toulme JJ, Isambert H, Guo P, Rossi JJ, Tenenbaum SA and Shapiro BA (2011) A Boost for the Emerging Field of RNA Nanotechnology. ACS Nano, 5, 3405–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bui MN, Brittany Johnson M, Viard M, Satterwhite E, Martins AN, Li Z, Marriott I, Afonin KA and Khisamutdinov EF (2017) Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomedicine, 13, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart JM, Viard M, Subramanian HK, Roark BK, Afonin KA and Franco E (2016) Programmable RNA microstructures for coordinated delivery of siRNAs. Nanoscale, 8, 17542–17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parlea L, Puri A, Kasprzak W, Bindewald E, Zakrevsky P, Satterwhite E, Joseph K, Afonin KA and Shapiro BA (2016) Cellular Delivery of RNA Nanoparticles. ACS combinatorial science, 18, 527–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA and Jaeger L (2010) In vitro assembly of cubic RNA-based scaffolds designed in silico. Nature nanotechnology, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA and Jaeger L (2011) Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc, 6, 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haque F, Shu D, Shu Y, Shlyakhtenko LS, Rychahou PG, Evers BM and Guo P (2012) Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano today, 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khisamutdinov EF, Jasinski DL and Guo P (2014) RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano, 8, 4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khisamutdinov EF, Li H, Jasinski DL, Chen J, Fu J and Guo P (2014) Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Res, 42, 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Lytton-Jean AKR, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M et al. (2012) Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nature nanotechnology, 7, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osada E, Suzuki Y, Hidaka K, Ohno H, Sugiyama H, Endo M and Saito H (2014) Engineering RNA-protein complexes with different shapes for imaging and therapeutic applications. ACS Nano, 8, 8130–8140. [DOI] [PubMed] [Google Scholar]
- 21.Halman JR, Satterwhite E, Roark B, Chandler M, Viard M, Ivanina A, Bindewald E, Kasprzak WK, Panigaj M, Bui MN et al. (2017) Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic acids research, 45, 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, Kashlev M and Shapiro BA (2014) In silico design and enzymatic synthesis of functional RNA nanoparticles. Acc. Chem. Res, 47, 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilczewska AZ, Niemirowicz K, Markiewicz KH and Car H (2012) Nanoparticles as drug delivery systems. Pharmacological Reports, 64, 1020–1037. [DOI] [PubMed] [Google Scholar]
- 24.De Jong WH and Borm PJA (2008) Drug delivery and nanoparticles: Applications and hazards. International Journal of Nanomedicine, 3, 133–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egusquiaguirre SP, Igartua M, Hernandez RM and Pedraz JL (2012) Nanoparticle delivery systems for cancer therapy: advances in clinical and preclinical research. Clinical & Translational Oncology, 14, 83–93. [DOI] [PubMed] [Google Scholar]
- 26.Cruz-Acuna M, Halman JR, Afonin KA, Dobson J and Rinaldi C (2018) Magnetic nanoparticles loaded with functional RNA nanoparticles. Nanoscale, 10, 17761–17770. [DOI] [PubMed] [Google Scholar]
- 27.Cutlar L, Zhou DZ, Hu XJ, Duarte B, Greiser U, Larcher F and Wang WX (2016) A nonviral gene therapy for treatment of recessive dystrophic epidermolysis bullosa. Experimental Dermatology, 25, 818–820. [DOI] [PubMed] [Google Scholar]
- 28.Mintzer MA and Simanek EE (2009) Non-viral Vectors for Gene Delivery. Chemical Reviews, 109, 259–302. [DOI] [PubMed] [Google Scholar]
- 29.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR and Anderson DG (2014) Non-viral vectors for gene-based therapy. Nature Reviews Genetics, 15, 541–555. [DOI] [PubMed] [Google Scholar]
- 30.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B and Behr JP (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gwak SJ, Macks C, Bae S, Cecil N and Lee JS (2017) Physicochemical stability and transfection efficiency of cationic amphiphilic copolymer/pDNA polyplexes for spinal cord injury repair. Scientific Reports, 7, 11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gwak SJ, Nice J, Zhang J, Green B, Macks C, Bae S, Webb K and Lee JS (2016) Cationic, amphiphilic copolymer micelles as nucleic acid carriers for enhanced transfection in rat spinal cord. Acta Biomaterialia, 35, 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonin KA, Viard M, Kagiampakis I, Case CL, Dobrovolskaia MA, Hofmann J, Vrzak A, Kireeva M, Kasprzak WK, KewalRamani VN et al. (2015) Triggering of RNA Interference with RNA-RNA, RNA-DNA, and DNA-RNA Nanoparticles. ACS Nano, 9, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonin KA, Kasprzak W, Bindewald E, Puppala PS, Diehl AR, Hall KT, Kim TJ, Zimmermann MT, Jernigan RL, Jaeger L et al. (2014) Computational and experimental characterization of RNA cubic nanoscaffolds. Methods, 67, 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Afonin KA, Kireeva M, Grabow WW, Kashlev M, Jaeger L and Shapiro BA (2012) Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett, 12, 5192–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA and Jaeger L (2011) Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett, 11, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Afonin KA, Viard M, Koyfman AY, Martins AN, Kasprzak WK, Panigaj M, Desai R, Santhanam A, Grabow WW, Jaeger L et al. (2014) Multifunctional RNA nanoparticles. Nano Lett, 14, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonin KA, Viard M, Tedbury P, Bindewald E, Parlea L, Howington M, Valdman M, Johns-Boehme A, Brainerd C, Freed EO et al. (2016) The Use of Minimal RNA Toeholds to Trigger the Activation of Multiple Functionalities. Nano Lett, 16, 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sajja S, Chandler M, Fedorov D, Kasprzak WK, Lushnikov A, Viard M, Shah A, Dang D, Dahl J, Worku B et al. (2018) Dynamic Behavior of RNA Nanoparticles Analyzed by AFM on a Mica/Air Interface. Langmuir, 34, 15099–15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rackley L, Stewart JM, Salotti J, Krokhotin A, Shah A, Halman JR, Juneja R, Smollett J, Lee L, Roark K et al. (2018) RNA Fibers as Optimized Nanoscaffolds for siRNA Coordination and Reduced Immunological Recognition. Advanced Functional Materials, 28, 1805959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gwak SJ, Macks C, Jeong DU, Kindy M, Lynn M, Webb K and Lee JS (2017) RhoA knockdown by cationic amphiphilic copolymer/siRhoA polyplexes enhances axonal regeneration in rat spinal cord injury model. Biomaterials, 121, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jovic M, Sharma M, Rahajeng J and Caplan S (2010) The early endosome: a busy sorting station for proteins at the crossroads. Histology and histopathology, 25, 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weigel PH and Oka JA (1981) Temperature-dependence of endocytosis mediated by the asialoglycoprotein receptor in isolated rat hepatocytes - evidence for 2 potentially rate-limiting steps. Journal of Biological Chemistry, 256, 2615–2617. [PubMed] [Google Scholar]
- 44.Afonin KA, Viard M, Martins AN, Lockett SJ, Maciag AE, Freed EO, Heldman E, Jaeger L, Blumenthal R and Shapiro BA (2013) Activation of different split functionalities on reassociation of RNA-DNA hybrids. Nature nanotechnology, 8, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson MB, Halman JR, Satterwhite E, Zakharov AV, Bui MN, Benkato K, Goldsworthy V, Kim T, Hong E, Dobrovolskaia MA et al. (2017) Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling. Small, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong E, Halman JR, Shah A, Cedrone E, Truong N, Afonin KA and Dobrovolskaia MA (2019) Toll-Like Receptor-Mediated Recognition of Nucleic Acid Nanoparticles (NANPs) in Human Primary Blood Cells. Molecules (Basel, Switzerland), 24, 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Givens BE, Geary SM and Salem AK (2018) Nanoparticle-based CpG-oligonucleotide therapy for treating allergic asthma. Immunotherapy, 10, 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanagata N (2012) Structure-dependent immunostimulatory effect of CpG oligodeoxynucleotides and their delivery system. International Journal of Nanomedicine, 7, 2181–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khisamutdinov EF, Li H, Jasinski DL, Chen J, Fu J and Guo PX (2014) Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic Acids Research, 42, 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.