Abstract
Natural Killer (NK) cells are cytotoxic lymphocytes targeting virus-infected cells and cancer cells. Specific pro- and anti-killing signals modulate the overall ability of NK cells to kill cancer cells, however, several immune-escape mechanisms can be enacted by cancer cells to avoid NK-mediated killing. Recently, increasing evidence has shown that extracellular vesicles (EVs) released by NK cells carry proteins and microRNAs (miRs) able to exert an anti-tumoral effect, even within a highly immune-suppressive tumor microenvironment. These recent findings suggest a possible use of NK-derived EVs as anticancer agents, and propel the development of new strategies to enrich EVs with the most effective anti-cancer cargo as a promising new anti-cancer approach.
Keywords: exosomes, microRNAs, Natural Killer cells, cancer, neuroblastoma
Introduction
Natural Killer (NK) cell-mediated cytotoxicity: receptors, effectors and formation of the immunological synapsis
Natural Killer (NK) cells are innate immunity lymphocytes, specialized to perform cytotoxic killing of cancer cells and virally infected cells, with no requirement for any previous activation (1). Human NK cells are negative for CD3 and express the CD56 neural cell adhesion molecule. According to the levels of CD56 they can be classified in CD56bright and CD56dim. The former group represents about 5–20% of circulating NK cells in humans and a subset with a reduced cytotoxic activity against cancer cells, since they have low expression of CD16 (a receptor for the fragment crystallizable region (Fc) of immunoglobulins); conversely, the latter CD56dim NK cell group (which are the majority of NK cells in the blood) expresses high surface levels of CD16 and elicits a strong anti-cancer response (2,3).
NK cells express different surface receptors that either promote or inhibit NK killing. The overall prominence of activating vs inhibiting receptor signaling is responsible for the induction of NK-mediated killing. The opposite outcome (inhibition of NK killing) will occur in case of a prominent activation of inhibitory receptors. Briefly, activating receptors include the CD16 receptor, which is able to bind to antibody opsonizing the surface of infected/transformed cells through a mechanism called antibody-dependent cell cytotoxicity (ADCC) (4), and natural cytotoxicity receptors (NCR), which belong to the immunoglobulin superfamily and can recognize hemagglutinins and neuraminidases expressed by virally-infected cells, as well as PCNA expressed by cancer cells (5). Among the inhibitory receptors the Killer-cell immunoglobulin-like receptors (KIRs) are able to recognize MHC class I and HLA-G in primates and represent the dominant way for NK cells to recognize and preserve self, healthy cells (6–8). One of the most common mechanisms of immune-escape by cancer cells is to down-regulate MHC class I surface expression in order to avoid T-cell killing. However, by doing so, they become vulnerable to NK-cell killing, through the lack of binding of MHC-I to inhibitory KIRs which represent a very powerful inducer of the NK-killing response (9). Ly49 receptors have been identified in mice, whereas in humans only the pseudogene variant KLRA1 has been identified (10–12). The Ly49 receptors are C-type lectins with inhibitory functions when bound to MHC class I. The NKG2 receptor family also is a C-type lectin family which dimerizes with CD94 (harboring a cytoplasmic domain responsible for signal transduction) that includes both activating and inhibiting receptors. These receptors bind to HLA-E in humans and while NKG2A and NKG2B have inhibitory functions, NKG2C, NKG2E and NKG2H have activating functions. NKG2D is quite different from the other members of this receptor family, since it does not dimerize with CD94 but with DAP10 and binds to MHC class I homologues MICA, MICB and ULBP expressed on the surface of stressed, inflamed and cancerous epithelial and endothelial cells and leads to activation of NK-mediated killing (13). Finally, the ILT or LIR receptors also belong to the family of immunoglobulin-like receptors and harbor activating functions (14). ADCC and receptor-mediated NK killing mechanisms are summarized in Figure 1 (mechanisms 1 and 2).
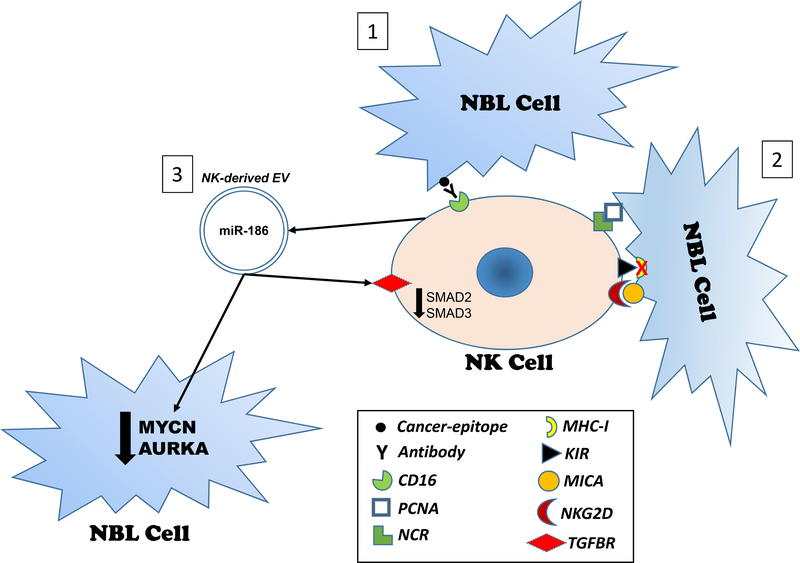
Figure 1. Overview of NK-mediated killing mechanisms.
1) Antibody-dependent cell cytotoxicity (ADCC) is based on the opsonization of cancer cells by an antibody, whose Fc segment can bind to CD16 on the surface of NK cells. This creates an immunological synapse that allows NK degranulation and cytotoxicity; 2) Receptor-mediated cytotoxicity. The figure indicates 3 possible activating signals for NK-mediated degranulation and cytotoxicity: the binding of NCRs to the cancer cell surface antigen PCNA, the lack of expression of MHC-I on the surface of cancer cells, preventing the KIR-mediated inhibition of NK-killing activity, and the binding of MICA (on the surface of cancer cells) to the NK activating receptor NKG2D; 3) EV-mediated cytotoxicity. The figure indicates the secretion of EVs containing miR-186 by NK cells. These EVs can shuttle miR-186 back to NK cells where it silences receptors for TGF-β (TGFBR1 and TGFBR2) and induces downregulation of downstream signaling proteins SMAD2 and SMAD3 overall leading to resistance of NK cells to the immune-suppression mediated by TGF-β, present at high concentrations in the Tumor Microenvironment. Moreover, EVs can shuttle miR-186 to neuroblastoma (NBL) cells located also at a distance from the actual NK cell and miR-186 can directly silence MYCN and AURKA, two key oncogenes for neuroblastoma, eliciting a paracrine anti-cancer effect.
Regardless of how NK cells recognize their target cell and which types of ligand-receptor interaction is involved, once the NK-cell immunological synapsis is formed, NK cells ultimately elicit their cytotoxic activity through 3 main mechanisms of action: receptor-mediated, perforin-granzyme-mediated and granulysin-mediated interaction. The first type of cytotoxicity involves the FasL/Fas interaction. NK cells express FasL, whereas the target cells express its receptor Fas (or CD95). As a consequence of the binding of FasL to its receptor, the death-inducing signaling complex (DISC) is triggered (15), leading to activation of caspase-8 and −10 and apoptosis both by activation of downstream caspases (such as caspase-9, −3 and −7), and by formation of the apoptosome, a quaternary protein platform whose formation is triggered by the mitochondrial release of cytochrome C and which leads to activation of caspase-3 and −7, ultimately leading to cell death (16). The second mechanism of NK killing is mediated by a pore-forming protein called perforin, a glycoprotein able to polymerize in presence of Ca++ and forming holes in the plasma-membrane of the target cell through which cytotoxic proteins can be introduced (17). Two of these cytotoxic proteins are granzyme A and granzyme B. Granzyme A triggers caspase-independent cell death by targeting the SET complex in the endoplasmic reticulum and releasing the NM23-H1 DNAse, which causes DNA damage (18–20). Granzyme A also impairs the transmembrane potential of the mitochondria increasing the production of Reactive Oxygen Species (ROS) (20), however the mitochondrial damage is not leading to the release of mitochondrial cytochrome C. Therefore, the mechanism of cell death mediated by granzyme A is believed to be entirely caspase-independent. Granzyme B is a powerful activator of the caspase cascade both by promoting a direct cleavage of initiator caspases-8, −9, and −10 and by triggering mitochondrial cytochrome C release as a consequence of its proteolytic activity of the BID protein (16,18,19,21). The third known mechanism of NK-mediated killing involves granulysin, a member of the saposin-like protein (SAPLIP) family able to electrostatically bind to the surface of target cells because of its positive charge at the N-terminus (21). Granulysin can also enter the target cell cytoplasm through pores formed by perforin. Granulysin causes ion fluxes (Ca++ enters the cytoplasm and K+ exits) (22) that severely damage the mitochondria, leading to release of cytochrome C and activation of the caspase apoptotic cascade (23).
Classification of Extracellular Vesicles (EVs)
It is very well documented that all cells, including NK cells, release in their surroundings multi-sized vesicles, generally referred to as extracellular vesicles (EVs). After an initial confusion in the field due to lack of standardization in the nomenclature of the different types of EVs, the International Society of Extracellular Vesicles (ISEV) has recently published and regularly updated, a minimal requirement consensus document for the classification of EVs (24,25). In general, we can identify 4 main types of EVs, roughly based on size and the expression of surface markers. Exosomes are EVs whose size range spans from 30 to 150 nm and whose surface is enriched for tetraspanins such as CD63, CD81, CD9. Exosomes are mostly derived from a precursor intracellular organelle called “multivesicular body” and their genesis occurs through the endosomal compartment of the cell (26). While there is no ultimate and specific surface marker for exosomes, it is a combination of expressed markers and lack of expression of specific intracellular proteins that meets the current minimal requirement for the definition of an exosome. Microvesicles, are EVs between 50 nm and about 1micron in size, mostly expressing CD40 ligand, integrins and surface selectins. They mostly derive from direct budding of the cellular plasma membrane (27). Apoptotic bodies are larger in size (they span from 500 nm to 2 microns) and express large amount of surface phosphatidylserine, due to their genesis from apoptotic cells (28). A 4th group of EVs includes large oncosomes (ranging from 1 micron to 10 microns in size) generated especially by highly metastatic cancer cells, whose biological properties are multi-faceted and harbor fascinating implications for the Tumor Microenvironment (TME) (29).
EVs carry a functional cargo which includes microRNAs
All types of EVs carry a cargo of multiple macromolecules which includes proteins and nucleic acids (DNA and RNA both coding and non-coding). A recent report has questioned the nucleic acid content of small EVs (30). However, a large proportion of studies supports the presence of functional DNA and RNAs in EVs. Therefore, further studies are necessary to clarify this important aspect of EV biology. Among the most studied cargo components of EVs there are microRNAs.
MicroRNAs (miRs) are single stranded short non-coding RNAs (ncRNAs) whose most well understood function is to regulate translation of mRNA transcripts by binding to a complementary sequence mostly located in the 3’-untranslated region (3’-UTR) of the transcript (31). While in the majority of cases the complementarity is imperfect and leads to translational repression, in some cases (especially in plants) the miR exactly matches a sequence in the target mRNA leading to degradation of the transcript (32). In both cases, the binding of a miR to a target mRNA results in a reduced expression of the protein encoded by that mRNA. It has also been documented that the binding of miRs to the target mRNA 5’-UTR region can lead to up-regulation of the protein (33), however such mechanism does not seem to be predominant. Alternative mechanisms of action of miRs include their ability to directly bind to proteins, preventing their transcriptional regulation of genes and their binding as ligands to receptors, triggering downstream signaling. A list of the miRs discussed in this paper, with their essential genomic, functional, biomarker and therapeutic data, is provided in Table 1. The first example was discovered by Eiring et al, showing that miR-328 can directly bind to hRNPE2, preventing its binding to the transcription factor CEBPA, involved in promoting myeloid differentiation during hematopoiesis (34). Downregulation of miR-328, which occurs in leukemias, does not prevent hRNPE2 inhibiting interaction with CEBPA, leading to a block in granulocytic differentiation, which is at the very core of the leukemic blastic crisis (34). The second mechanism of action was discovered by our group in 2012 (35). We showed that lung cancer cells secrete miR-21 and miR-29a within EVs and these miRs are uptaken by surrounding Tumor-Associated Macrophages (TAMs) expressing Toll-like Receptor 8 (TLR8) in their endosomal compartment. We proved that miR-21 and −29a can reach the endosomal compartment in TAMs and bind to TLR8, leading to the MyD88-mediated downstream activation of NF-κB which promotes secretion of Interleukin-6 (IL-6) and Tumor Necrosis Factor alpha (TNF-α) by TAMs, with the overall outcome of creating a TME conducive to cancer growth and increased metastatic potential (35). More recently, we provided experimental evidence that the binding of miR-21 to TLR8 in TAMs is relevant also for the TME biology of neuroblastoma, where it induces the NF-κB-mediated secretion of miR-155 within TAM-derived EVs, that are then up-taken by the cancer cells, where miR-155 silences the expression of TERF1, an inhibitor of telomerase, with the overall effect of increasing telomerase activity and resistance to chemotherapy (36). In this work we also provided evidence that the miR-21/TLR8/miR-155/TERF1 axis is relevant in several different types of cancers including breast, lung, melanoma, thyroid cancer and medulloblastoma (36).
TABLE 1.
Genomic Location, Function, role as Biomarker (B) or as Therapeutics (T) for the miRs discussed in this manuscript.
| miRNA | Genomic location | Function | Biomarker(B)/Therapeutics(T) | Reference |
|---|---|---|---|---|
| miR-328–3p | 16q22.1 | Binds to hRNPE2 preventing inactivation of CEBPA and allowing normal granulocytic differentiation | T: Restoration to overcome block of granulocytic differentiation in CML-BC. | (34) |
| miR-21–5p miR-29a-3p |
17q23.1 7q32.3 |
Bind to human Toll-like Receptor 8 (TLR8) triggering a pro-tumoral inflammatory response | T: Inhibition of miR-TLR8 interaction. | (35) (36) |
| miR-155–5p | 21q21.3 | Targets TERF1 and increases telomerase activity and resistance to chemotherapy | T: Inhibition to restore drug sensitivity. B: Possible biomarker of TAM infiltration in primary tumor. |
(36) |
| miR-186–5p | 1p31.1 | Directly inhibits MYCN, AURKA, TGFBR1, TGFBR2; indirectly inhibits SMAD2 and SMAD3 | T: Nanoparticles to deliver mimic to cancer cells (neuroblastoma) anti-cancer effect. T: Nanoparticles to deliver mimic to NK cells to increase their cytotoxic activity. |
(49) |
Overall, miRs orchestrate the biology of the TME by affecting growth, dissemination and drug resistance of cancer cells.
NK cells secrete cytotoxic Extracellular Vesicles
The first evidence that NK-derived EVs elicit cytotoxic effect against cancer cells was provided by Fais et al. (37), who showed that both resting and activated NK cells isolated from healthy donors are able to kill several types of cancer cell lines and extend their cytotoxicity also to activated (but not to resting) immune cells (37). They also showed for the first time that NK-derived EVs express FasL and perforin molecules and hypothesized that EVs might be involved in a paracrine and systemic modulation of the immune response within the TME. Following this initial report, our group confirmed the cytotoxic activity of NK-derived EVs against neuroblastoma (38), and showed that in addition to perforin also granzyme A, granzyme B and granulysin could be detected as cargo of these EVs and that the NK surface marker CD56 could also be used to isolate NK-derived EVs. In 2017, Ahn et al also published a study showing cytotoxic effects of CD63+ and ALIX+ EVs derived from the NK92-MI cell line against melanoma cells both in vitro and in vivo (39). Intriguingly, no cytotoxicity was observed when these NK-derived EVs were used against normal cells. They provided evidence that these EVs contain perforin and express surface FasL. More recently, the same group used a similar approach to target glioblastoma in a xenograft murine model, and showed that the systemic administration of NK92-MI-derived EVs increased cancer cell killing especially if animals were pre-treated with dextran sulfate 2h before the EV treatment (40). They also studied the biodistribution of the EVs and reported that these systemically administered NK-derived EVs localize especially to the liver and the spleen. Interestingly, they also did not observe toxicity in animals even after repeated intravenous injections of the vesicles (40). The importance of the Tumor Microenvironment (TME) in NK-mediated cytotoxicity has been addressed by Lozupone et al., showing that NK cell treatment of human melanoma xenografts is more efficient than treatment with both γδ1 and γδ2 γ/δ T-lymphocytes in controlling tumor growth (41). This is in part due to the fact that NK cells survive longer than T cells in the TME. Specifically, lymphocytes are very sensitive to the acidic conditions of the TME (42,43), whereas NK cells might be more resistant and survive longer in an acidic microenvironment. Moreover, EV concentration and cargo is affected by the pH of the surrounding microenvironment (44–46). Neutralizing TME acidity with proton pump inhibitors and/or buffer treatment appears to be a promising approach to reduce cancer immune-escape, as reported by recent pre-clinical and clinical evidence (47,48). Overall, these studies confirmed a cytotoxic role of NK-derived EVs against different types of cancer cells, and provided the preclinical rationale for their use as anti-cancer agents.
Mechanisms of NK-derived EV-mediated killing
NK-derived EVs have proven to be cytotoxic against a variety of cancer cells. Progressively increasing concentrations of EVs elicit increasing cytotoxicity, however while a plateau seems to be reached for certain cell lines of neuroblastoma, such a limit does not seem to occur with other types of neuroblastoma cell lines (49). The first studies showing this cytotoxic effect have also assessed whether the typical mediators of NK cell killing were part of their EV cargo. As expected, surface FasL and cargo of perforin, granzyme A, granzyme B and granulysin were confirmed by several independent studies supporting a role for the “usual suspects” as effectors of the observed anti-cancer cytotoxicity (38,50). Recently, we correlated cytotoxicity and content of cytotoxic proteins from NK-derived EVs by ELISA. This study reported that the killing of target cancer cells is not mediated by only one effector molecule but by a multitude of mechanisms occurring simultaneously and involving both the caspase cascade activation and caspase-independent cell death pathways (51).
We also investigated a possible role for miRs in the killing mediated by NK-derived EVs. We performed a systematic analysis of the miR-content in the cargo of EVs isolated from human NK cells, taking advantage of a protocol that we perfected and that allows us to generate pure and functional human NK cells starting from peripheral blood mononuclear cells exposed to engineered K562 cells expressing recombinant IL-21 in their surface (38). Among the top represented miRs, we decided to focus on miR-186–5p (from now on referred to as miR-186) since it was among the top represented miRs as cargo of cytotoxic EVs, it was predicted to target key oncogenes in neuroblastoma (such as MYCN, and AURKA), key genes in the TGF-β pathway (such as TGFBR1, TGFBR2, SMAD2, SMAD3) and it had been previously observed as down-regulated in high-risk neuroblastomas (52). Despite the obvious interest in silencing a gene such as MYCN, which is still considered an “undruggable gene” together with its close “cousin” MYC, and the fact that miR-186 is predicted to silence MYCN both directly and indirectly (by targeting AURKA, a stabilizer of MYCN and also an important oncogene in neuroblastoma itself), there is an additional intriguing aspect in the predicted group of target genes for miR-186: effectors of the TGF-β network. The TME of several (if not all) types of cancers in imbued with TGF-β, mostly derived from TAMs and cancer-associated fibroblasts (53). TGF-β elicits an important immune-suppressive function for NK cells by binding to the TGF-β receptors (TGFBR1 and TGFBR2) on the surface of NK cells and triggering a SMAD2 and SMAD3 downstream signaling (54) ultimately leading to a block in the killing activity of NK cells. The fact that miR-186 is possibly silencing the expression of TGFBR1, TGFBR2, SMAD2 and SMAD3 suggests a central role for this miR (and for the NK-derived EVs which contain a huge amount of miR-186) in preventing the TGF-β-mediated block of NK cell killing activity. Therefore, we performed a series of experiments showing that indeed miR-186 directly targets MYCN, AURKA, TGFBR1 and TGFBR2. While we observed direct targeting of MYCN and AURKA, and down-regulation of SMAD2 and SMAD3 in cells treated with miR-186, we did not observe a direct silencing of SMAD2 and SMAD3, suggesting a possible indirect mechanism through which miR-186 reduces the expression of these 2 genes (49). We confirmed the tumor suppressor role of miR-186 in neuroblastoma cells in vitro. Next, we were able to generate anionic lipopolyplex nanoparticles containing miR-186 and coated with anti-GD2 antibody (GD2 is a well-known surface marker for neuroblastoma) and we showed that these nanoparticles, systemically administered, were able to significantly reduce the growth and prolong the overall survival of mice in an orthotopic murine model of neuroblastoma (49), confirming feasibility and safety of miR-enriched nanoparticles as anti-cancer agents. One of the most interesting aspects of this study, which is the first to show that miRs are involved in the killing of cancer cells mediated by NK-derived EVs, was that NK-derived EVs continued to be cytotoxic even in presence of TGF-β, while NK cells were unable to kill (49,55). The study also confirmed the lack of toxicity of NK-derived EVs on normal cells (49). Limitations of this study include the fact that it was focused on one miR, not taking into consideration the effects of other miRs (and of their combination) contained in NK-EVs, and the fact that no assessment of the role of miR-186 and other (non-miR) molecules in the cargo of NK-EVs was performed. Most importantly, the study did not provide a systematic analysis of the dose of NK-EVs to be administered in order to achieve the most efficient (and still non-toxic) anti-tumoral effect in vivo.
Despite these limitations, this study provides the rationale for the use of miRs in NK-derived EVs as new anti-cancer agents. The EV-mediated killing by NK cells is summarized in Figure 1, mechanism 3.
Future Perspectives
NK cells are currently tested as anti-cancer agents in clinical trials (e.g. , from ClinicalTrial.gov). However, the fact that NK-derived EVs also exert anti-cancer properties is of great interest since we can envision at least three main advantages of using EVs over NK cells as therapeutics. First, it is well known that every cell-based therapy (including NK cell based-infusion) in patients harbors the risk of triggering the so-called “cytokine storm”, a massive systemic release of cytokines that can force the patient to suspend the treatment or could even be life-threatening in some cases. It is conceivable that the use of NK-derived EVs may not be accompanied by this severe side effect and certainly the preliminary results of the in vivo experiments with NK-derived EVs are encouraging in terms of safety against this and other possible non-specific side effects. Secondly, NK cells have a hard time to reach the so-called “pharmacologic sanctuaries”, where cancer cells can nest and escape the immune response. The most typical examples are the blood-brain barrier and the blood-testis barrier. Increasing evidence suggests that EVs are able to cross these anatomical barriers, providing an advantage over the use of cell-based therapies (56,57). Finally, as we have previously described, several immune-escape mechanisms (such as inhibitory NK receptors and TGF-β in the TME) can prevent NK cell-mediated killing. Such mechanisms are likely not able to prevent the killing mediated by NK-derived EVs, such as we observed in the case of NK-EVs in presence of TGF-β (49). However, tumor cells release EVs expressing ligands for death receptors (e.g. FasL and TRAIL) that are involved in tumor immune-escape in a non-tumor specific manner (58,59). The possibility that tumor-derived EVs may impair the killing mediated by NK-EVs through their death receptor ligands should be further explored. Clearly, the future holds promise for the use of NK-derived EVs as therapeutics, most likely in combination with existing anti-cancer treatments. However, much efforts need to be geared towards improving EVs as therapeutics. For instance, while the use of nanoparticles is helpful to study the role of a specific cargo molecule, it is clear that they will not recapitulate the complexity and possibly the efficacy of the plethora of cargo molecules in an actual EV. Therefore, we can envision a future in which the use of EVs as a whole (possibly enriched for specific anti-cancer molecules) will be used as a drug. In other words, the creation of a “super-exosome” or a “super-EV”, containing the basal cargo plus enrichment for specific killing proteins and/or miRs should be explored in the near future. The other key aspect to be further improved is how to reach specificity of delivery of therapeutic EVs to cancer cells (or to immune cells in order to overcome immune-escape mechanisms). This is also of paramount importance to be able to administer higher doses of EVs and keeping negligible risks of side effects. Clearly these aspects also require the development of improved strategies to load a specific cargo molecule in EVs and to equip these EVs with signals able to direct them to specific target cells within the TME. I would like to conclude with a provocative observation. The increasing evidence that some of the cytotoxic effects of NK cells are actually mediated by NK-derived EVs suggests a possible paracrine and more systemic role of NK cells (to make use of the words by Fais et al (37)). In addition to limiting this effect of NK-EVs on immune cells, it can be speculated that this paracrine/systemic function applies also to cancer cells, suggesting that the cytotoxic activity of one NK cell may be broader than we thought before: not only limited to the cancer cell involved in the immune synapsis with the NK cell but also extended to cancer cells at a more distant site. In this sense it will be interesting to study the impact of one NK cell to a number of cancer cells within the TME. Our increasing knowledge of the biology of NK cells, miRs and EVs enriches our understanding of the neoplastic process. While these three fields have developed and grown independently, it is now clear the need for their full integration, in order to empower our strategies to develop new and most effective drugs to defeat cancer.
Acknowledgments
Dr. Fabbri is supported by the NIH/NCI grants R01CA215753 and R01CA219024.
Financial Support: Dr. Fabbri is supported by the NIH/NCI grants R01CA215753 and R01CA219024.
Footnotes
Conflict of Interest: M Fabbri has ownership interest in a patent application to be filed.
REFERENCES
- 1.Sirisinha S Evolutionary insights into the origin of innate and adaptive immune systems: different shades of grey. Asian Pac J Allergy Immunol 2014;32:3–15 [PubMed] [Google Scholar]
- 2.Michel T, Poli A, Cuapio A, Briquemont B, Iserentant G, Ollert M, et al. Human CD56bright NK Cells: An Update. J Immunol 2016;196:2923–31 [DOI] [PubMed] [Google Scholar]
- 3.Dubois S, Conlon KC, Müller JR, Hsu-Albert J, Beltran N, Bryant BR, et al. IL15 Infusion of Cancer Patients Expands the Subpopulation of Cytotoxic CD56. Cancer Immunol Res 2017;5:929–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandelboim O, Malik P, Davis DM, Jo CH, Boyson JE, Strominger JL. Human CD16 as a lysis receptor mediating direct natural killer cell cytotoxicity. Proc Natl Acad Sci U S A 1999;96:5640–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudspeth K, Silva-Santos B, Mavilio D. Natural cytotoxicity receptors: broader expression patterns and functions in innate and adaptive immune cells. Front Immunol 2013;4:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yawata M, Yawata N, Abi-Rached L, Parham P. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol 2002;22:463–82 [PubMed] [Google Scholar]
- 7.Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet 2006;7:277–300 [DOI] [PubMed] [Google Scholar]
- 8.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 2002;20:217–51 [DOI] [PubMed] [Google Scholar]
- 9.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol 2002;38:1007–21 [DOI] [PubMed] [Google Scholar]
- 10.Nylenna O, Naper C, Vaage JT, Woon PY, Gauguier D, Dissen E, et al. The genes and gene organization of the Ly49 region of the rat natural killer cell gene complex. Eur J Immunol 2005;35:261–72 [DOI] [PubMed] [Google Scholar]
- 11.Gays F, Aust JG, Reid DM, Falconer J, Toyama-Sorimachi N, Taylor PR, et al. Ly49B is expressed on multiple subpopulations of myeloid cells. J Immunol 2006;177:5840–51 [DOI] [PubMed] [Google Scholar]
- 12.Barten R, Trowsdale J. The human Ly-49L gene. Immunogenetics 1999;49:731–4 [DOI] [PubMed] [Google Scholar]
- 13.Gunturi A, Berg RE, Forman J. The role of CD94/NKG2 in innate and adaptive immunity. Immunol Res 2004;30:29–34 [DOI] [PubMed] [Google Scholar]
- 14.Colonna M, Nakajima H, Cella M. Inhibitory and activating receptors involved in immune surveillance by human NK and myeloid cells. J Leukoc Biol 1999;66:718–22 [DOI] [PubMed] [Google Scholar]
- 15.Lavrik IN, Krammer PH. Regulation of CD95/Fas signaling at the DISC. Cell Death Differ 2012;19:36–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi YH, Jin N, Kelly F, Sakthivel SK, Yu T. Elevation of Alanine Aminotransferase Activity Occurs after Activation of the Cell-Death Signaling Initiated by Pattern-Recognition Receptors but before Activation of Cytolytic Effectors in NK or CD8+ T Cells in the Liver During Acute HCV Infection. PLoS One 2016;11:e0165533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osińska I, Popko K, Demkow U. Perforin: an important player in immune response. Cent Eur J Immunol 2014;39:109–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 2003;112:659–72 [DOI] [PubMed] [Google Scholar]
- 19.Cullen SP, Brunet M, Martin SJ. Granzymes in cancer and immunity. Cell Death Differ 2010;17:616–23 [DOI] [PubMed] [Google Scholar]
- 20.Martinvalet D, Dykxhoorn DM, Ferrini R, Lieberman J. Granzyme A cleaves a mitochondrial complex I protein to initiate caspase-independent cell death. Cell 2008;133:681–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cullen SP, Martin SJ. Mechanisms of granule-dependent killing. Cell Death Differ 2008;15:251–62 [DOI] [PubMed] [Google Scholar]
- 22.Anderson DH, Sawaya MR, Cascio D, Ernst W, Modlin R, Krensky A, et al. Granulysin crystal structure and a structure-derived lytic mechanism. J Mol Biol 2003;325:355–65 [DOI] [PubMed] [Google Scholar]
- 23.Okada S, Li Q, Whitin JC, Clayberger C, Krensky AM. Intracellular mediators of granulysin-induced cell death. J Immunol 2003;171:2556–62 [DOI] [PubMed] [Google Scholar]
- 24.Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 2018;75:193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tricarico C, Clancy J, D’Souza-Schorey C. Biology and biogenesis of shed microvesicles. Small GTPases 2017;8:220–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SJ, Kim JM, Kim J, Hur J, Park S, Kim K, et al. Molecular mechanisms of biogenesis of apoptotic exosome-like vesicles and their roles as damage-associated molecular patterns. Proc Natl Acad Sci U S A 2018;115:E11721–E30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol 2015;40:41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of Exosome Composition. Cell 2019;177:428–45.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J 2008;14:1–6 [DOI] [PubMed] [Google Scholar]
- 32.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 2013;12:847–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007;318:1931–4 [DOI] [PubMed] [Google Scholar]
- 34.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 2010;140:652–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A 2012;109:E2110–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst 2015;107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lugini L, Cecchetti S, Huber V, Luciani F, Macchia G, Spadaro F, et al. Immune surveillance properties of human NK cell-derived exosomes. J Immunol 2012;189:2833–42 [DOI] [PubMed] [Google Scholar]
- 38.Jong AY, Wu CH, Li J, Sun J, Fabbri M, Wayne AS, et al. Large-scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells. J Extracell Vesicles 2017;6:1294368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L, Kalimuthu S, Gangadaran P, Oh JM, Lee HW, Baek SH, et al. Exosomes Derived From Natural Killer Cells Exert Therapeutic Effect in Melanoma. Theranostics 2017;7:2732–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu L, Oh JM, Gangadaran P, Kalimuthu S, Baek SH, Jeong SY, et al. Targeting and Therapy of Glioblastoma in a Mouse Model Using Exosomes Derived From Natural Killer Cells. Front Immunol 2018;9:824. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Lozupone F, Pende D, Burgio VL, Castelli C, Spada M, Venditti M, et al. Effect of human natural killer and gammadelta T cells on the growth of human autologous melanoma xenografts in SCID mice. Cancer Res 2004;64:378–85 [DOI] [PubMed] [Google Scholar]
- 42.Calcinotto A, Filipazzi P, Grioni M, Iero M, De Milito A, Ricupito A, et al. Modulation of microenvironment acidity reverses anergy in human and murine tumor-infiltrating T lymphocytes. Cancer Res 2012;72:2746–56 [DOI] [PubMed] [Google Scholar]
- 43.Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, Russell S, Weber AM, Luddy K, et al. Neutralization of Tumor Acidity Improves Antitumor Responses to Immunotherapy. Cancer Res 2016;76:1381–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Logozzi M, Spugnini E, Mizzoni D, Di Raimo R, Fais S. Extracellular acidity and increased exosome release as key phenotypes of malignant tumors. Cancer Metastasis Rev 2019;38:93–101 [DOI] [PubMed] [Google Scholar]
- 45.Logozzi M, Mizzoni D, Angelini DF, Di Raimo R, Falchi M, Battistini L, et al. Microenvironmental pH and Exosome Levels Interplay in Human Cancer Cell Lines of Different Histotypes. Cancers (Basel) 2018;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem 2009;284:34211–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gillies RJ, Pilot C, Marunaka Y, Fais S. Targeting acidity in cancer and diabetes. Biochim Biophys Acta Rev Cancer 2019;1871:273–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pillai SR, Damaghi M, Marunaka Y, Spugnini EP, Fais S, Gillies RJ. Causes, consequences, and therapy of tumors acidosis. Cancer Metastasis Rev 2019;38:205–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neviani P, Wise PM, Murtadha M, Liu CW, Wu CH, Jong AY, et al. Natural Killer-Derived Exosomal miR-186 Inhibits Neuroblastoma Growth and Immune Escape Mechanisms. Cancer Res 2019;79:1151–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen C, Seeger RC, Fabbri M, Wang L, Wayne AS, Jong AY. Biological roles and potential applications of immune cell-derived extracellular vesicles. J Extracell Vesicles 2017;6:1400370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu CH, Li J, Li L, Sun J, Fabbri M, Wayne AS, et al. Extracellular vesicles derived from natural killer cells use multiple cytotoxic proteins and killing mechanisms to target cancer cells. J Extracell Vesicles 2019;8:1588538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res 2007;67:976–83 [DOI] [PubMed] [Google Scholar]
- 53.Bottino C, Dondero A, Bellora F, Moretta L, Locatelli F, Pistoia V, et al. Natural killer cells and neuroblastoma: tumor recognition, escape mechanisms, and possible novel immunotherapeutic approaches. Front Immunol 2014;5:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han G, Wang XJ. Roles of TGFβ signaling Smads in squamous cell carcinoma. Cell Biosci 2011;1:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmittgen TD. Exosomal miRNA Cargo as Mediator of Immune Escape Mechanisms in Neuroblastoma. Cancer Res 2019;79:1293–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen CC, Liu L, Ma F, Wong CW, Guo XE, Chacko JV, et al. Elucidation of Exosome Migration across the Blood-Brain Barrier Model In Vitro. Cell Mol Bioeng 2016;9:509–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zagrean AM, Hermann DM, Opris I, Zagrean L, Popa-Wagner A. Multicellular Crosstalk Between Exosomes and the Neurovascular Unit After Cerebral Ischemia. Therapeutic Implications. Front Neurosci 2018;12:811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andreola G, Rivoltini L, Castelli C, Huber V, Perego P, Deho P, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med 2002;195:1303–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huber V, Fais S, Iero M, Lugini L, Canese P, Squarcina P, et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology 2005;128:1796–804 [DOI] [PubMed] [Google Scholar]