Abstract
Triple negative breast cancer (TNBC) is a heterogeneous disease enriched for mutations in PTEN and dysregulation of innate immune signaling. Here we demonstrate that Rab7, a recently identified substrate of PTEN phosphatase activity, is also a substrate of the innate immune signaling kinases TBK1/IKKε on the same serine-72 site. An unbiased search for novel TBK1/IKKε substrates using stable isotope labeling with amino acids in cell culture (SILAC) phosphoproteomic analysis identified Rab7 serine-72 (S72) as a top hit. PTEN-null TNBC cells expressing a phosphomimetic version of Rab7-S72 exhibited diffuse cytosolic Rab7 localization and enhanced innate immune signaling, in contrast to a kinase-resistant version, which localized to active puncta that promote lysosomal-mediated STING degradation. Thus, convergence of PTEN loss and TBK1/IKKε activation on Rab7-S72 phosphorylation limited STING turnover and increased downstream production of IRF3 targets including CXCL10, CCL5, and IFN-β. Consistent with this data, PTEN-null TNBC tumors expressed higher levels of STING, and PTEN-null TNBC cell lines were hyper-responsive to STING agonists. Together these findings begin to uncover how innate immune signaling is dysregulated downstream of TBK1/IKKε in a subset of TNBCs and reveals previously unrecognized cross-talk with STING recycling that may have implications for STING agonism in the clinic.
Introduction
Triple negative breast cancers (TNBCs) are typically aggressive and account for a disproportionate number of metastatic cases and breast cancer deaths (1-3). TNBCs are also heterogeneous, with diverse somatic mutations, gene amplifications, and deletions as reflected by multiple subtypes defined by different gene signatures (4); however, PTEN loss is a common event (5-9). In addition, a significant proportion of TNBCs also exhibit a high amount of immune cell infiltration and elevated cytokine production, which we previously linked to aberrant expression of IκB kinase ε (IKKε), which promotes feedforward production of NF-κB associated cytokines with its homologue TANK-binding kinase 1(TBK1) (10).
Ras-related protein Rab-7a (Rab7) is a member of a larger family of Ras GTPases and has been shown to be an important modulator of phagocytosis (11), endosomal sorting (12), and the biogenesis of lysosome-related organelles (13). While Rab7 has been extensively studied for its role in endosomal trafficking and maturation, recent studies have highlighted the role of Rab7 in attenuating receptor signaling in tumors (14,15). The attenuation of receptor signaling by Rab7 occurs for outer membrane receptors such as epidermal growth factor receptor (EGFR) (14) as well as intracellular signaling adaptors such as stimulator of interferon genes (STING) (15). In each case, Rab7 is directly responsible for protein degradation by trafficking receptor/adaptor containing vesicles to the lysosome. Notably, the tumor suppressor PTEN was also recently identified to regulate Rab7 function by dephosphorylation of serine-72 (S72), promoting its mislocalization; PTEN loss or constitutively phosphorylated Rab7-S72 thus increased intracellular EGFR activation as the receptor was internalized but its degradation was impaired (14).
Here we performed integrated phosphoproteomic studies to search for novel TBK1/IKKε substrates, which yielded Rab7-S72 as a top hit. Through subsequent studies in PTEN null TNBC cells, we identify TBK1/IKKε mediated phosphorylation of this site as a key regulator of Rab7 mislocalization, which sustains levels of the upstream TBK1 adaptor STING, and thus promotes hyperactive innate immune signaling. These findings begin to uncover a key molecular event that de-regulates innate immune signaling in PTEN null TNBC cells, with potentially important therapeutic implications.
Material and Methods
Cell culture
HEK 293T and breast cancer cell lines (HCC70, HCC1143, HCC1187, HCC1937, MDA-MB-231, MDA-MB-468, MCF7, MCF10A, T-47D, SKBr3, ZR-751) used in this study were obtained from American Type Culture Collection (ATCC). HEK 293T and MDA-MB-231 cells were cultured in DMEM (ThermoFisher Scientific) whereas all other cell lines were grown in RPMI-1640 (ThermoFisher Scientific) with 10% FBS (Gemini Bio-products) and 1X penicillin-streptomycin (Gemini Bio-products). Jurkat T-cells expressing CXCR3 were generated as previously described (16) and grown in RPMI-1640 with 10% FBS. All cell lines were confirmed by short tandem repeat profiling, tested mycoplasma negative by PCR as recent as 2 months prior to last experiment, and used between passage 3-15.
Plasmids, plasmid construction, and generation of lentivirus
All plasmids were generated using Gateway Cloning (Invitrogen). Mutant plasmids, kinase-dead TBK1, catalytically inactive PTEN mutant (PTEN-C124S), Rab7-S72E, and Rab7-S72A were generated with PCR-based site-directed mutagenesis. Rab7 mutants were cloned into the V5-tagged pLX304 plasmid. TBK1 clones, kinase-dead TBK1, wild-type TBK1, and control EGFP were cloned into the pLX980 plasmid. PTEN clones, wild-type PTEN (PTEN-WT), and PTEN-C124S were cloned into the pLX307 plasmid. HEK 293T cells were used for the generation of lentivirus for the establishment of Rab7 and PTEN mutant cell lines. Transfections occurred over 48 h with 1μg plasmid using X-tremeGENE ™ 9 DNA transfection reagent (Sigma-Aldrich). Post transfection, viral media was collected and combine with 8μg ml−1 polybrene (Santa Cruz Biotech) prior to addition to MDA-MB-468 cells. PTEN mutants were selected with puromycin whereas all other mutants were selected with blasticidin.
SILAC assay and analysis
To determine TBK1 specific phosphorylation sites, we utilized a previously published protocol for stable isotope labeling in cell culture (17,18). In brief, HEK 293T cells were cultured for 1 week in low-glucose DMEM supplemented with either light [L-arginine (R0) and L-lysine (K0)], medium [L-arginine 13C6-HCL (R6) and L-lysine-4,4,5,5d4 (K4)], or heavy [L-arginine 13C6-15N4-HCl (R10) and L-lysine 13C6, 15N2-HCl (K8)] amino acids (Sigma-Aldrich). Cells were transfected using FuGENE (Promega) with the plasmids control EGFP, wild-type TBK1, or kinase-dead TBK1 and then selected with blasticidin. Lysates were digested with trypsin (Promega) overnight to generate peptides. For in vitro kinase assays, peptides were generated after amino acid culturing, incubated first with calf intestinal alkaline phosphatase (New England BioLabs) and then 0.5μg of the following recombinant kinases for 1 h at 37˚C: TBK1 (Millipore), IKKε (Millipore), and control polo-kinase 2 (ThermoFisher Scientific).
All lysates were prepared for mass spectrometry as previously described (17) and then purified using immobilized metal affinity chromatography (IMAC) (19). Enriched phosphopeptides were analyzed using the Agilent 1200 LC coupled online to an LTQ Orbitrap Velos mass spectrometer (ThermoFisher Scientific) (18). Xcalibur 2.1 software was used for data-dependent acquisition. Mass spectra was processed using the MaxQuant software package and the Mascot search engine (Matrix Science) (17). Peptides were identified with a concatenated forward and reversed IPI protein sequence database (version 3.60). Median SILAC ratios for identified peptides were generated for all iterations of the same peptide with the highest scoring versions reported (Supplemental Table S1 and S2). Ratios were used to generate a z-score for each assay with a cut off value of 2.5. TBK1/IKKε in vitro core hits were analyzed by Gene Set Enrichment Analysis with the Molecular Signature Databases collection C5 Gene Ontology Biological Processes. Motif-x software was used to identify common peptide motifs (20,21) and then queried in Ras GTPase family members using Basic Local Alignment Search Tool.
In vitro enzymatic kinetic assay
Enzymatic quantification was performed using the ADP Glo Assay (Promega) with recombinant TBK1, synthetic peptide substrates and ATP, as previously described (22). Synthetic peptides were as follows: IKKε-tide (ADDDYDSLDWDAKKK), IKKε-tide L8A (ADDDYDSADWDAKKK), Rab7-tide (IWDTAGQERFQSLGVAFYRGADCC), and Rab7-tide L8A(IWDTAGQERFQSAGVAFYRGADCC). Data is representative of 8 experiments with 2 biological replicates.
Immunofluorescence staining and analysis
After being grown on chamber slides (CellTreat) and subjected to various treatment conditions, cells were fixed and permeabilized according to standard protocols. Individual proteins were visualized using specific antibodies listed in Supplemental Table S3, and slides were imaged with the Nikon Eclipse 80i™ microscope. Three blinded investigators each counted 100 consecutive cells for the presence of perinuclear STING foci. Graphs depict the mean and SEM of counts from 3 different investigators and 2 experiments for a total of 6 data points.
Cytokine analysis
Cytokine and chemokine protein expression were analyzed by multiplex assay using the Human Cytokine/Chemokine Magnetic Bead Panel (Luminex, #HCYTMAG-60K-PX30) on a MAGPIX system (Merck). Fold changes relative to the corresponding control were calculated and plotted as log2FC. Lower and upper limits of quantitation (LLOQ/ULOQ) were inputted from standard curves for cytokines above or below detection. The media from control cells and those treated with ADU-S100 (Chemietek) were subjected to CXCL10 ELISA assay (R&D systems, #DIP100) at 24 h, whereas in TBK1/IKKε studies supernatants were analyzed at 48 h. Additionally, supernatants from untreated and ADU-S100 treated samples were subjected to CCL5 and IFNβ ELISA assays (R&D systems, #DRN00B, #DIFNB0) at 72 h. Assays are representative of 3 experiments in duplicate.
Quantitative RT-PCR
Total RNA was isolated using RNeasy Mini Kit (Qiagen). Following extraction, 1 μg of RNA was used for cDNA generation with SuperScript® III First-Strand Synthesis SuperMix (ThermoFisher Scientific, #1683483). qRT-PCR was performed using Power SYBR Green PCR Master Mix (ThermoFisher Scientific, #4367659) and the Applied Biosystems 7300 Fast real-time PCR system and software. The relative expression of each gene was normalized to the expression of the housekeeping gene 36B4. Primer sequences are listed in Supplemental Table S3.
Rab7 and TBK1/IKKε inhibition studies
For all inhibitor studies, cells were pretreated for 30 min with each inhibitor +/− 1μg ml−1 poly(dA:dT) (Invivogen) prepared with X-tremeGENE™ 9. Rab7 was inhibited with 100nM of Ras Superfamily GTPases Binding Antagonist (CID 1067700, Millipore Sigma) and stimulated with poly(dA:dT) for 6 h prior to analysis. TBK1/IKKε inhibition immunofluorescence studies were performed with Compound 1 (Gilead Sciences), MRT67307 (Sigma-Aldrich), and CYT387 (MedChemExpress). Pretreatment or stimulation times were determined through a time course looking at STING perinuclear foci formation with 6 h, 8 h, and 3 h selected for each of the inhibitors respectively. Similarly, treatment times with the inhibitors were the same with the addition of poly(dA:dT).
STING agonist studies
Using the STING agonist ADU-S100 at 10 μM, Rab7 mutants were stimulated and resultant cytokine (CXCL10, CCL5, and IFNβ) expression was measured with ELISA. Similarly, breast cancer cell lines were treated with ADU-S100 at 10 μM and CXCL10 ELISA was performed at 24 h. In addition, a growth inhibition assay was performed with twice weekly 10 μM ADU-S100 treatment over 6 weeks. Cells were then stained for viability with 1% crystal violet.
Immunoblotting and antibodies
Protein isolation was performed with RIPA buffer containing phosphatase and protease inhibitors (Roche) and quantified by bicinchoninic acid assay (Pierce Biotechnology). Twenty micrograms of total protein lysate was loaded in 4%–12% NuPAGE gels and electrophoresed (Invitrogen), transferred to nitrocellulose membranes (Millipore), and immunoblotted. Membranes were blocked for 1 h with the Odyssey blocking buffer (LI-COR) and probed for various proteins overnight in tris buffered saline solution with 0.05% Tween-20. Membranes were then washed for 10 minutes in TBST (3X) and incubated with the corresponding secondary antibody for 1 h. See Supplemental Table S4 for all primary and secondary antibodies. Immunoblots were visualized using the LI-COR Odyssey. Supplementary figures (S1, S2, and S3) contains uncropped images of the blots.
Immunohistochemistry
Paraffin-embedded TNBC blocks were obtained under Dana-Farber Cancer Institute/Harvard Cancer Center (DF/HCC) IRB protocol #93-085. After deparaffinizing tissue blocks, slides were processed per standard protocol. These slides along with the Biomax Breast Cancer Tumor Microarray slides were then stained for STING and PTEN (Cell Signaling Technology) as previously described (23) with antibodies listed in Supplemental Table S4. Visualization was achieved using EnVision+/HRP Rabbit (Dako), diaminobenzidine (Dako), and hematoxylin counterstain. A semi-quantitative score of STING and PTEN expression for each microarray core was determined by a pathologist and negative staining was determined if the percentage was less than or equal to 15%.
T-cell migration assay
MDA-MB-468 cells were plated in ultra-low attachment plates to allow for the formation of spheroids, which were then embedded into an AIM chip microfluidic device as previously described (23,24). Media was then added to the exterior channels with or without 10μM ADU-S100 overnight. In the morning, Jurkat T-cells were stained with CellTrace™ Violet (Invitrogen) and added to one exterior chamber of the device. Co-cultures were incubated at 37˚C for 48 h, then fixed and counter-stained with DAPI (Invitrogen). Wells were visualized with a Nikon Eclipse 80i™ microscope, and all resultant images were quantified for Jurkat T-cell migration by ImageJ.
Statistical analysis
GraphPad Prism 7.0 was used for statistical analysis, data processing, and graph generation. Values reported are the mean and SEM. When comparing only two groups, a Student’s t-test was applied, otherwise an ANOVA multivariate analysis was performed with a post hoc modification as described in the figure legends. Statistical significance was determined as p<0.05. Peptide ratios between groups were analyzed with a Pearson’s correlation.
Data availability
The raw SILAC data are available from the corresponding author upon reasonable request.
Results
Integrated phosphoproteomic studies to identify novel TBK1/IKKε substrates
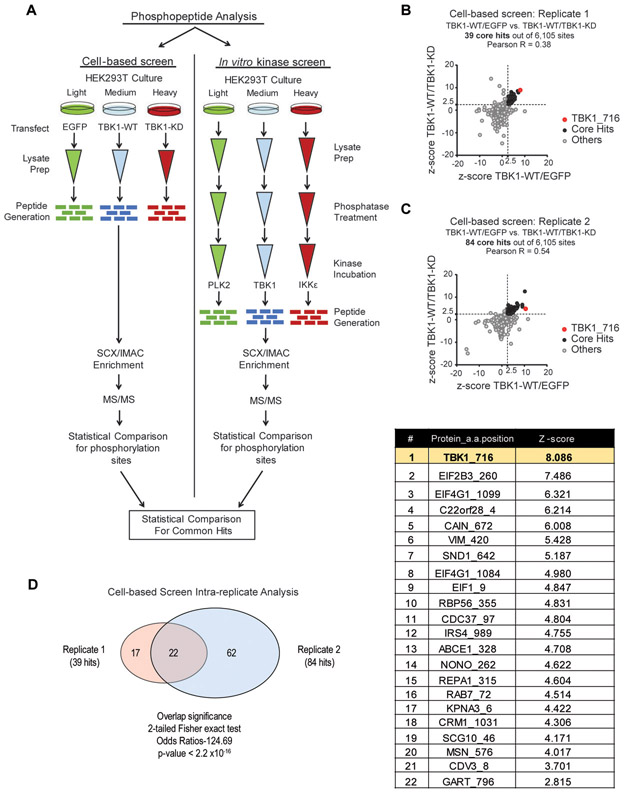
To uncover novel downstream targets of TBK1/IKKε kinase activity, we developed an integrated phosphoproteomics method using stable isotope labeling with amino acids in cell culture (SILAC) to identify particularly robust and specific substrates. We compared results obtained from a cellular phosphoproteomics screen following transfection of TBK1 constructs in HEK 293T cells with those of a biochemical assay using recombinant TBK1/IKKε kinases and dephosphorylated HEK 293T protein lysates (Fig. 1A). Thus, integration of the phosphosite hits from these complementary approaches identifies putative substrates that both occur in cells and are direct in nature. For the cell-based screen, HEK 293T cells were cultured with light, medium, or heavy amino-acid containing media, and then transfected with control EGFP, wild-type TBK1 (TBK1-WT), or kinase dead (K38M) TBK1 (TBK1-KD) constructs respectively (Fig. 1A). Protein lysates at the 24-hour timepoint post-transfection were prepared, mixed in equal proportions, and subjected to tandem mass spectrometry. Resulting peptides were evaluated for TBK1 target specificity by comparison of z-scores of differential phosphopeptide abundance for both TBK1-WT/EGFP and TBK1-WT/TBK1-KD conditions, to normalize against both control vector expression and absence of kinase activity. Employing this technique in two independent experiments yielded 39 core hits from experimental replicate 1 and 84 core hits from experimental replicate 2 (Fig. 1B and 1C, Supplemental Fig. S4A, Table S1). As expected, TBK1-WT/EGFP and TBK1-WT/TBK1-KD hits were largely concordant with each other, with the serine-716 TBK1 autophosphorylation site representing a top hit in both experiments. Of note, peptides containing the serine-172 TBK1 activation site were not generated by the tryptic digestion and were thus not identified (Supplemental Table S1). After determination of the individual experiment core hits from both replicates, a secondary analysis was performed to identify overlapping peptides. From both experiments, there were 22 common core hits of which the TBK1 autophosphorylation site was the top ranked phosphopeptide (Fig. 1D).
Fig. 1.
Stable isotope labeling with amino acids in cell culture (SILAC) determination of novel TBK1 substrates. A, Schematic of SILAC screen analysis for the determination of TBK1 substrates. Left panel: cell-based screen where HEK 293T cells were cultured in the presence of light, medium, or heavy isotopes then transfected with three plasmids containing the following proteins for expression: EGFP, TBK1-WT, or TBK1-KD. Right panel: in vitro kinase screen where following isotope culturing, HEK 293T lysates were generated and dephosphorylated. Light labeled lysates were left untreated while heavy and medium-labeled lysates were incubated in one of the following recombinant kinases: PLK2, TBK1 or IKKε. Each screen was then evaluated for kinase specific phosphorylation sites by tandem mass spectrometry. B and C, Graphical representation of the phosphopeptides common in TBK1-WT/EGFR and TBK1-WT/TBK1-KD analyses from cell-based screen experimental replicate 1 (B) and replicate 2 (C). D, Venn diagram of the comparison of phosphopeptides from replicate 1 and replicate 2 for common core hits. List of phosphopeptides that were significant if their respective z-scores were >2.5 in both conditions.
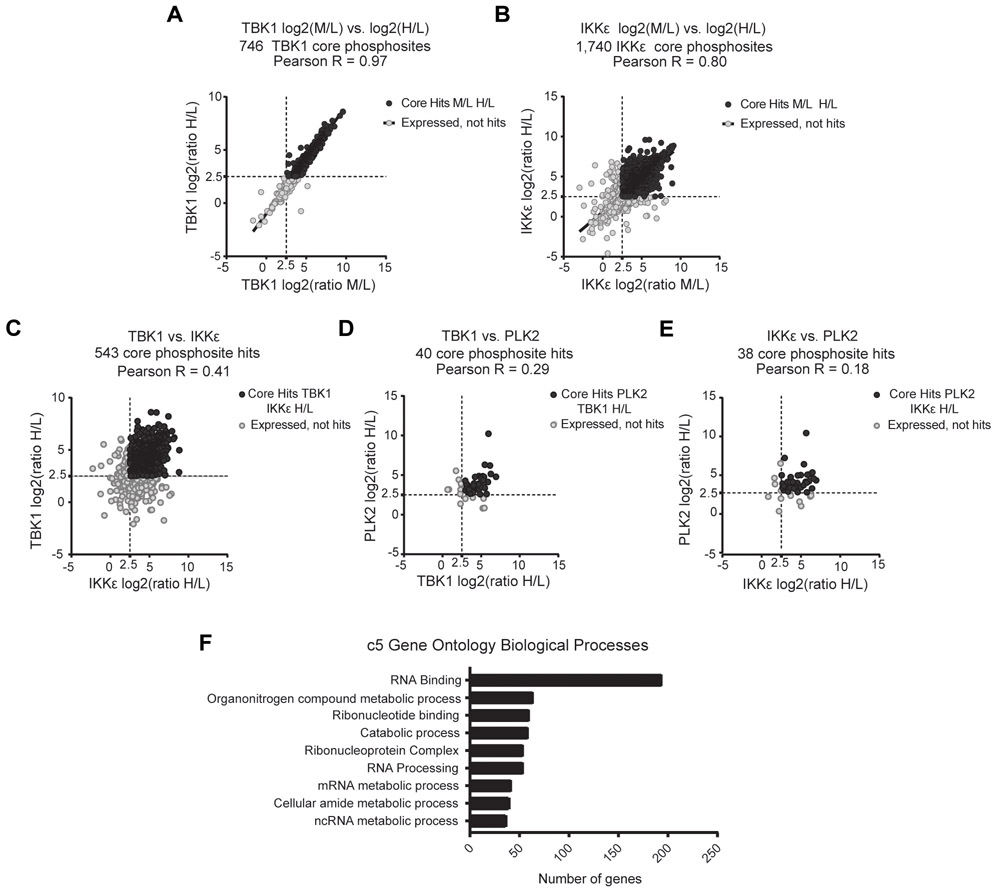
In parallel, to identify which of these hits might represent bona fide direct TBK1/IKKε substrates, we conducted the in vitro phosphoproteomics screen (Fig. 1A). In this case, dephosphorylated SILAC labeled HEK 293T lysates were incubated with purified recombinant TBK1 or IKKε and compared with polo-like kinase 2 (PLK2) as an unrelated control enzyme. As for the cell-based screen, in vitro generated phosphopeptides were identified after purification and tandem mass spectrometry. For these experiments, medium and heavy labeled lysates were incubated with either recombinant TBK1, IKKε, or PLK2 and light labeled lysates were left untreated, then ratios between the heavy and light (H/L) or medium and light (M/L) labeled lysate conditions were determined. Intra-replicate concordance for both TBK1 and IKKε was high and reinforced phosphopeptide specificity (Fig. 2A and Fig. 2B). Cross enzyme concordance, for the subset of phosphosites that overlapped between TBK1 and IKKε, consisted of 543 statistically significant phosphopeptides, whereas there were less than 50 phosphopeptides identified for the subset that included PLK2 and TBK1 or PLK2 and IKKε (Fig. 2C-2E, Supplemental Fig. S4B-S4I, Table S2). Utilizing the Molecular Signatures Database (MSigDB) (25), we interrogated cellular pathways associated with these in vitro identified TBK1/IKKε phosphopeptides. Amongst the most enriched pathways were those involved in the catabolic processes, consistent with the known role of TBK1 in autophagy (Fig. 2F) (26,27). Additionally, motif analysis of TBK1/IKKε recovered peptides revealed x-pS-L/I/F/M-x, consistent with a previously identified IKKε substrate peptide motif (Table 1) (28). Together, despite the artificial nature of this in vitro SILAC screen, these findings highlight specificity related to TBK1/IKKε substrate targets.
Fig. 2.
In vitro SILAC screen analysis of phosphopeptides for the determination of novel TBK1 substrates. Enzyme phosphopeptide ratios were determined from peptides identified from medium or heavy labeled lysates incubated with either IKKε, TBK1, or PLK2 over no enzyme light-labeled lysates. A and B, Intra replicate concordance for TBK1(A) and IKKε (B). C-E, Cross enzyme concordance between TBK1 and IKKε (C), TBK1 and PLK2 (D), and IKKε and PLK2 (E). In all phosphopeptide analyses, hits were considered positive if z-scores were >2.5 in both conditions. F, Pathway analysis for phosphopeptides from TBK1/IKKε core hits from (C) utilizing Molecular Signature Database C5 Gene Ontology Biological Processes Collection.
Table 1.
Common phosphomotifs present in in vitro SILAC phosphopeptides determined with motif-x.
| # | Motif | Motif Score |
Foreground Matches |
Foreground Size |
Background Matches |
Background Size |
Fold Increase |
|---|---|---|---|---|---|---|---|
| 1 | ......SL...... | 16 | 221 | 470 | 111211 | 1100344 | 4.65 |
| 2 | .......SI...... | 16 | 76 | 249 | 40290 | 989133 | 7.49 |
| 3 | .......SF...... | 16 | 48 | 173 | 40975 | 948843 | 6.42 |
| 4 | ......SM..... | 16 | 27 | 125 | 18075 | 907868 | 10.85 |
Identification of Rab7-S72 as a bona fide TBK1/IKKε substrate
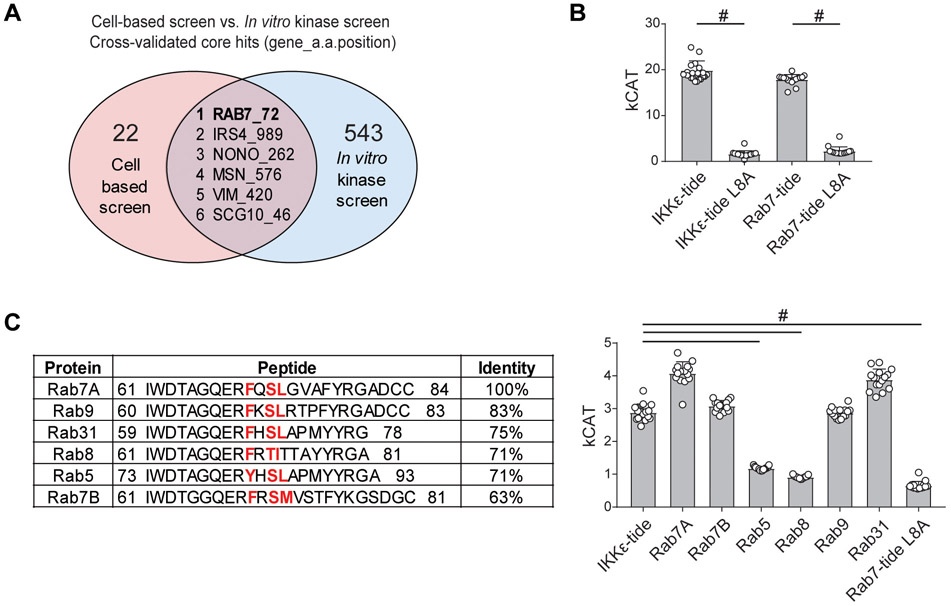
In order to identify the most robust candidate substrates from these screens, we integrated both the cellular and the in vitro SILAC phosphoproteomics data. Cross validation of the core hits resulting from the cell-based screen and the in vitro kinase screen revealed 6 common phosphopeptides as highly likely bona fide TBK1 substrates, including Rab7-S72, Irs4-S989, Nono-S262, Msn-S576, Vim-S420, and Scg10-S46 (Fig. 3A). Of note, many of these novel targets (Msn, Vim, and Scg10) are involved in cytoskeletal reorganization, while the top hit, Rab7-S72, was also recently identified in a separate phosphoproteomics screen as a TBK1 target in the context of mitophagy (29). Additionally, Rab7 was included within the enriched catabolic processes category of our pathway analysis (Fig. 2F), and we also previously linked Rab7 with negative regulation of TBK1 signaling (27), prompting us to evaluate this hit further.
Fig. 3.
Determination and confirmation of Rab7-S72 as a TBK1/IKKε substrate target. A, Venn diagram showing the six common phosphorylation sites resulting from analysis of cell-based screen and in vitro kinase screen core hits (sites were selected based on z-score >2.5). B, Short peptide sequences were generated for the positive control TBK1 substrate, IKKε-tide and surrounding the predicted substrate, Rab7-S72. Peptide sequences were unaltered (IKKε-tide or Rab7-tide) or contained a negative control +1 mutation, where leucine was mutated to alanine (IKKε-Tide L8A or Rab7-Tide L8A). Synthetic peptides were incubated with recombinant TBK1 kinase and ATP, and kCat was calculated based upon the ADP generated (bars, n=16, #p<0.0001 One-way ANOVA with Tukey post-test). C, Selected Rab GTPase family members containing the conserved TBK1/IKKε motif, identified by BLAST analysis, and used for TBK1 enzymatic analysis. Small peptides were generated for Rab proteins identified and analyzed by biochemical assay as in B (bars, n=16, #p<0.0001 One-way ANOVA with Tukey post-test).
As a positive and negative control, we utilized the previously identified IKKε-tide which contains the F-x-pS-L motif (Table 1) (28) or a mutated version in which the +1 leucine is mutated to an alanine (IKKε-Tide L8A). We also generated a peptide flanking the S72 site with the corresponding +1 leucine to alanine mutation (Rab7-Tide L8A). As expected, we observed significant in vitro phosphorylation of the positive control IKKε-Tide and the wild type Rab7-Tide, but not of the IKKε-Tide L8A or Rab7-Tide L8A negative controls (Fig. 3B). Since this Rab7 peptide region contained the more specific IKKε F-x-pS-L motif and is a member of a larger family of Rab GTPases, we next used protein BLAST to identify additional candidate Rab family members that could be phosphorylated by TBK1 or IKKε. This analysis identified Rab9, Rab31, Rab8, Rab5, and Rab7B as additional potential substrates. We utilized the same TBK1 in vitro kinase assay to assess activity against these peptides, which identified Rab7A, Rab7B, Rab9, and Rab31 as capable of being phosphorylated in vitro (Fig. 3C). However, while these other Rab family members can be phosphorylated in vitro, only Rab7A was identified as being a target of TBK1 activity in HEK 293T cells.
Interrogation of the functional consequences of TBK1 phosphorylation of Rab7 mutants
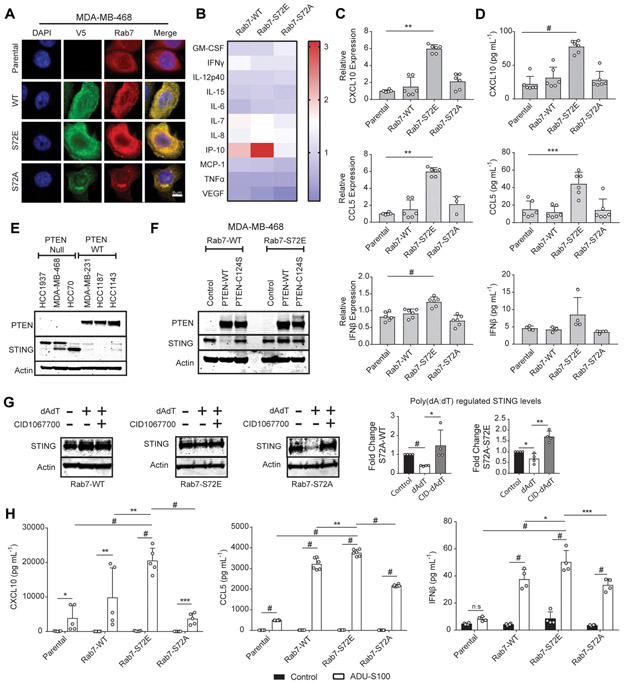
Since PTEN-mediated dephosphorylation of Rab7-S72 was shown to promote fusion of endosomes to lysosomes, we considered the possibility that TBK1/IKKε mediated phosphorylation of this site might similarly impair Rab7 localization to these endolysosome-associated puncta (14). We utilized the PTEN null TNBC cell line MDA-MB-468, which contains elevated IKKε levels (10), and examined baseline Rab7 localization, compared with stable over-expression of WT (V5-Rab7-WT), a phospho-mimetic mutant (V5-Rab7-S72E), or phospho-resistant mutant (V5-Rab7-S72A). Consistent with prior work, over-expression of the Rab7-S72E mutant resulted in a diffuse staining pattern, in contrast to the Rab7-S72A mutant which localized to puncta, which we confirmed both by V5 and Rab7 indirect immunofluorescence (Fig. 4A, Supplemental Fig. S5) (14).
Fig. 4.
Regulation of STING turnover and innate immune signaling by Rab7-S72 phosphorylation. A, Representative immunofluorescent image showing localization of Rab7 or V5-Rab7 in parental and mutant MDA-MB-468 cells under basal conditions (n=3, scale=6μm). B, Multiplexed cytokine analysis of conditioned medium from Rab7 mutant cell lines cultured under basal conditions for 24 h. (n=6). C, qRT-PCR measurement of CXCL10, CCL5 and IFNβ levels in MDA-MB-468 Rab7 mutant cell lines under basal conditions at 24 h (CXCL10) or 72 h (CCL5 and IFNβ) (bars, n=6; **p<0.01, #p<0.0001 by One-way ANOVA with Tukey post-test). D, Cytokine expression of CXCL10, CCL5, and IFNβ in supernatants from cells cultured for 24 h (CXCL10) or 72 h (CCL5 and IFNβ) and analyzed by ELISA (bars, n=6; ***p<0.001, #p<0.0001 by One-way ANOVA with Tukey post-test). E, Immunoblot of STING and PTEN expression in a panel of TNBC cell lines under basal conditions. F, Immunoblot of STING and PTEN expression in MDA-MB-468 cells expressing the indicated PTEN and Rab7 constructs. G, Immunoblots of STING levels in MDA-MB-468 Rab7 cell lines stimulated with poly(dA:dT) (1μg mL−1) +/− 100nM CID1067700 for 6 h. Densitometry quantification of enhanced poly(dA:dT) STING-degradation by Rab7-S72A and sensitivity to CID106770 (bars, n=4; *p<0.5, **p<0.01, #p<0.0001 by unpaired t-test). H, Expression of CXCL10, CCL5, and IFNβ by MDA-MB-468 Rab7 cell lines cultured for either 24 h (CXCL10) or 72 h (CCL5 and IFNβ) in the presence or absence of 10μM ADU-S100 (bars, n=6; *p<0.05, **p<0.01, ***p<0.001, #p<0.0001 by Two-way ANOVA with Tukey post-test).
We previously demonstrated that MDA-MB-468 cells required IL-1β priming to secrete NF-κB associated cytokines including IL-6 and CCL5 downstream of TBK1/IKKε (10). Interestingly, when we examined 24 h conditioned media from each of these stably derived MDA-MB-468 Rab7 over-expressing cell lines using multiplexed cytokine/chemokine profiling, we identified baseline upregulation of CXCL10 (IP-10) by Rab7-S72E in contrast to Rab7-S72A, as compared to Rab7-WT (Fig. 4B). Indeed, this finding was confirmed both by quantitative RT-PCR and by ELISA and was highly significant for the TBK1/IKKε interferon regulatory factor 3 (IRF3) regulated chemokines CXCL10 and CCL5, with a modest effect on baseline IFNβ expression (Fig. 4C and 4D). Taken together, these data revealed that mislocalization of Rab7 by over-expression of the S72 phosphomimetic mutant promotes dysregulated innate immune signaling and specifically increases IRF3 target chemokines downstream of TBK1/IKKε.
Since STING bridges TBK1 to IRF3, and was recently shown to undergo Rab7 mediated trafficking to the lysosome (15), we considered whether dysregulation of CXCL10, CCL5, and IFNβ expression might be related to altered regulation of Rab7 mediated STING degradation. We first compared baseline STING protein levels between triple negative breast cancer cell lines with wildtype PTEN (MDA-MB-231, HCC1187, HCC1143) versus those with impaired Rab7-S72 phosphatase activity due to PTEN null status (HCC1937, MDA-MB-468, HCC70). Indeed, PTEN null cell lines exhibited higher basal expression of STING, in contrast to PTEN wild-type cell lines with intact Rab7-S72 phosphatase activity, as it is capable of maintaining Rab7 in an active dephosphorylated state (Fig. 4E, Supplemental Fig. S1). To assess the causality of this relationship, we next over-expressed PTEN-WT or the catalytically inactive mutant PTEN-C124S in Rab7-WT or Rab7-S72E expressing MDA-MB-468 cells. PTEN downregulated STING levels in a phosphatase dependent manner in the setting of Rab-7 WT expression, but not in the context of Rab7-S72E (Fig. 4F, Supplemental Fig. S2). Together, these findings reveal that hyperphosphorylated Rab7-S72 in the setting of PTEN loss and TBK1/IKKε activation enhances STING stability.
We next examined the consequences of stimulating STING activity, which promotes rapid feedback STING degradation (30). Notably, transfection with the dsDNA mimic, poly(dA:dT) failed to promote feedback STING downregulation following Rab7-WT or Rab7-S72E over-expression, in contrast to the TBK1/IKKε phospho-resistant Rab7-S72A mutant (Fig. 4G, Supplemental Fig. S3). Furthermore, degradation of STING following Rab7-S72A over-expression required intact Rab7 function, since this effect was abrogated by co-treatment with the Rab7 inhibitor CID 1067700. Promotion of STING degradation by Rab7-S72A led us to postulate that expression of this mutant would also confer reduced sensitivity to STING agonism. Indeed, quantification of cytokine/chemokine expression after treatment with the direct STING agonist ADU-S100 revealed that Rab7-WT and especially Rab7-S72E over-expressing cells exhibited significantly higher levels of CCL5 and CXCL10, as well as IFNβ as compared to Rab7-S72A (Fig. 4H). These data thus provide evidence that phosphorylation of Rab7-S72 by TBK1/IKKε in PTEN null TNBC cells limits Rab7 mediated degradation of STING (15) and enhances innate immune cytokine production, especially following STING agonism.
Rab7-S72E is resistant to pharmacologic inhibition by TBK1/IKKε inhibitors
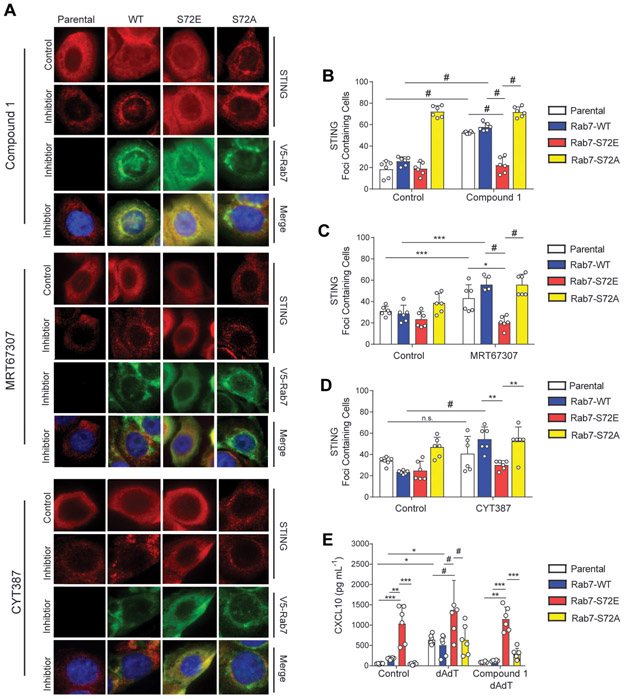
We next directly interrogated the effect of TBK1/IKKε inhibition on Rab7 mediated trafficking by using three well characterized TBK1/IKKε inhibitors--Compound 1, MRT67307, and CYT387 (10,24,31). Given that phosphorylation of Rab7 promoted STING dispersal into the cytosol, we hypothesized that TBK1/IKKε inhibition would promote focal perinuclear STING localization in MDA-MB-468 cells. Indeed, treatment with Compound 1 led to formation of STING foci in parental MDA-MB-468 cells, which colocalized with active Rab7 puncta (Fig. 5A and 5B). Perinuclear STING localization was also observed in Rab7-WT and Rab7-S72A expressing cell lines, but not in Rab7-S72E cells where it remained more dispersed, confirming the dominant negative effect of this mutant (Fig. 5A and 5B). Similarly, TBK1/IKKε inhibition with MRT67307 or CYT387 also resulted in the formation of STING perinuclear foci in parental, Rab7-WT, and Rab7-S72A expressing MDA-MB-468 cells, but not in Rab7-S72E cells (Fig. 5A, 5C, and 5D). Since PTEN also modulates EGFR late endocytic trafficking by dephosphorylation of Rab7 (14), we interrogated the effects of TBK1/IKKε inhibitor treatment on EGFR localization with the Rab7 mutants. We found that the dominant negative effect of Rab7-S72E on STING localization was also seen for EGFR localization with TBK1/IKKε inhibition (Supplemental Fig. S6A).
Fig. 5.
Rab7 S72E is resistant to TBK1/IKKε inhibition on STING localization and immune signaling in Rab7 mutants. A, Immunofluorescence imaging of parental and Rab7 cell lines: WT, S72E, and S72A, treated with either 1μM Compound 1 (6 h), 5μM MRT67307 (8 h) or 5μM CYT387 (3 h) alone (representative, n=2; scale=10μm). B, Quantification of STING foci containing cells treated with Compound 1 as in A (bars, n=6; #p<0.0001 by Two-way ANOVA with Tukey post-test). C, Quantification of STING foci in containing cells treated with MRT67307 as in A (bars, n=6; *p<0.05, ***p<0.001, #p<0.0001 by One-way ANOVA with a Tukey post-test). D, Quantification of images from CYT387 treated cells A (bars, n=6; **p<0.01, ***p<0.001, #p<0.0001 by One-way ANOVA with a Tukey post-test). E, Analysis of conditioned media from Rab7 expressing cells following 30 min pretreatment with 1μM Compound 1, then stimulated for 48 h with poly(dA:dT) (Compound 1 dA:dT) or poly(dA:dT) alone (dA:dT) for CXCL10 expression by ELISA (bars, n=6; *p<0.05, **p<0.01, ***p<0.001, #p<0.0001 by Two-way ANOVA with Tukey post-test).
We next explored the impact of TBK1/IKKε inhibition on cytokine/chemokine production by measuring CXCL10 production in conditioned medium at baseline or following poly(dA:dT) stimulation. Notably, even following concurrent Compound 1 and poly(dA:dT) treatment, STING was dispersed in the context of Rab7-S72E over-expression (Supplemental Fig. 6B and 6C). Consistent with this finding, Compound 1 treatment inhibited poly(dA:dT) stimulated CXCL10 in parental, Rab7-WT and Rab7-S72A cell lines, but was less effective in Rab7-S72E cells, indicative of sustained STING/TBK1 downstream signaling in this setting (Fig. 5E). Taken together, these data support the model that TBK1/IKKε regulated phosphorylation of Rab7-S72 impairs STING trafficking in TNBC cells, sustaining its levels and promoting hyperactive innate immune signaling in the context of PTEN loss.
PTEN loss results in sensitivity to STING agonism
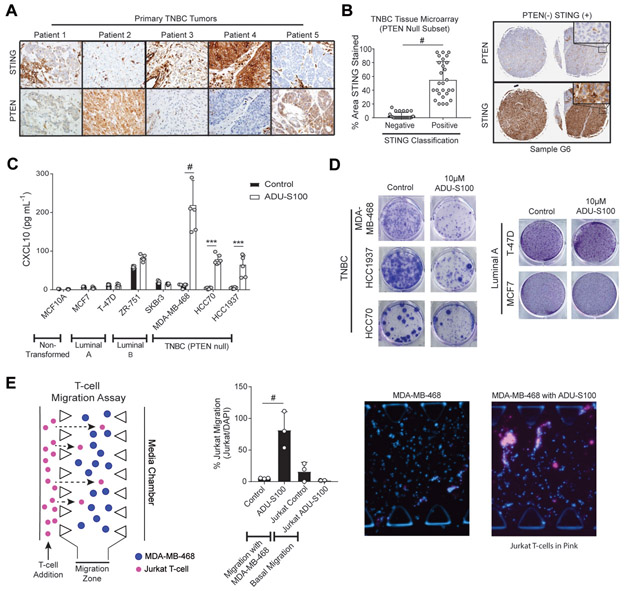
Our findings suggest that the PTEN null state limits STING mobilization to perinuclear foci where it is degraded as a negative feedback mechanism following STING agonism. As such this may serve as a biomarker for enhanced STING agonist activity in TNBC, given that this subtype of breast cancer is frequently associated with PTEN loss (5,9,32) which results in a worse prognosis and more rapid tumor proliferation (33,34). We first investigated this relationship by immunohistochemical (IHC) staining for PTEN and STING using five primary TNBC tumors derived from patients who were treatment naïve. Consistent with our in vitro results, samples with negative PTEN staining exhibited strong STING positivity (Fig. 6A). To further validate whether this PTEN null/STING high state is prevalent in the clinical setting among patients with TNBCs, we performed IHC for STING and PTEN expression using a commercially available breast cancer tumor microarray. STING levels were also significantly increased in PTEN null cases (Fig. 6B, Supplemental Table S5). This data suggests that PTEN null TNBC cancers are particularly poised for STING agonism as the result of aberrant STING trafficking.
Fig. 6.
PTEN loss increases TNBC STING expression in vivo and enhances STING agonist response. A, Patient-derived TNBC histology sections stained for PTEN or STING by immunohistochemistry. B, Commercially available tissue microarray (TMA) of breast cancer carcinomas were stained for STING and PTEN. Left panel: Quantitative analysis of percent STING staining of PTEN null TNBC cores (bar, n=26; #p<0.001 by unpaired t-test). Right panel: representative images of PTEN negative and STING positive staining from duplicate TNBC cores. C, Expression of CXCL10 in supernatants from either non-transformed (MCF-10A), Luminal A (MCF7 and T-47D), Luminal B (ZR-751 and SKBr3) or TNBC cell lines (MDA-MB-468, HCC1937, and HCC70) following 10μM ADU-S100 treatment for 24 h (bars, n=6; ***p<0.001, #p<0.0001 by Two-way ANOVA with Tukey post-test). D, Breast cancer cell line growth inhibition following twice weekly treatment with 10μM ADU-S100 for 6 weeks. Colony formation was visualized with crystal violet staining. E, First panel: Schematic of T-cell migration assay, utilizing a 3D microfluidic device with MDA-MB-468 spheroids embedded in a central collagen-filled channel, co-cultured with CXCR3 expressing Jurkat T-cells. Second panel: Quantification of Jurkat T-cell migration towards MDA-MB-468 cells −/+ ADU-S100 treatment (bars, n=3; #p<0.0001 by One-way ANOVA with Tukey post-test). Third and Fourth Panels: Representative images of T-cell migration towards MDA-MB-468 cells −/+ ADU-S100 treatment.
We therefore tested whether this relationship promotes STING agonist responsiveness. Indeed, when we stimulated PTEN null TNBC cell lines (MDA-MB-468, HCC70, and HCC1937) with the direct STING agonist ADU-S100 at 10μM for 24 h and measured the resultant supernatant for CXCL10 expression by ELISA, TNBC cell lines exhibited a 10-fold increase in CXCL10 expression as compared to control. In contrast, luminal A cell lines (MCF7 and T-47D), luminal B cell lines (ZR-751 and SKBr3), and the non-transformed breast cancer cell line (MCF-10A) showed no significant induction of CXCL10 (Fig. 6C). Given the responsiveness of PTEN null TNBC cell lines to ADU-S100 stimulation, we further characterized the cellular response by treating cells plated at a low density and then proceeded with twice weekly ADU-S100 treatment for 6 weeks. Whereas chronic treatment inhibited TNBC cell line proliferation, there were no differences seen with the luminal A breast cancer cell lines (Fig. 6D). These findings further reveal that STING agonism of PTEN null TNBCs also induces cell intrinsic cytotoxicity.
We next interrogated the potential in vivo implications of the enhanced CXCL10 secretion using a previously described 3D microfluidic T-cell migration assay (16). MDA-MB-468 spheroids were embedded in collagen within the central region/migration zone of the 3D microfluidic device, then treated −/+ ADU-S100 at 10μM, and co-cultured with CXCR3 expressing Jurkat T-cells through the side media chamber. As expected, Jurkat T-cell migration was only seen in the presence of MDA-MB-468 cells that were pretreated with ADU-S100. As an additional control, no cells were plated in the migration zone and with the introduction of Jurkat T-cells in the media chamber, no significant migration of T-cells was seen. Quantification of T-cell migration identified a 10-fold increase in T-cell infiltration in the ADU-S100 pretreated MDA-MB-468 spheroids as compared to control pretreated or Jurkat alone cultures (Fig. 6E). Together, this data confirms that CXCL10 induction by STING agonism promotes T-cell recruitment, supporting the idea that targeting STING agonists to PTEN null TNBCs also promotes immunogenicity.
Discussion
Through integrated SILAC phosphoproteomic studies using cell-based and in vitro kinase screens, our study has identified 6 common phosphopeptides as high confidence TBK1/IKKε substrates, including Rab7-S72 as the top ranked peptide. Interestingly, the exact same site on Rab7 was also recently identified through phosphoproteomic studies evaluating TBK1 targets involved in mitophagy (29). We confirmed in vitro that specific phosphorylation of this site requires the known IKKε substrate motif, and that the phosphomimetic Rab7-S72E mutant rescues the cellular impact of TBK1/IKKε inhibition, validating this site as a bona fide substrate of these kinases. Furthermore, under basal conditions Rab7-S72E expression resulted in increased CXCL10 production and was resistant to dsDNA stimulated STING degradation, in contrast to the active Rab7 dephosphorylated mutant, Rab7-S72A. Together, these data suggest a working model, in which TBK1/IKKε through phosphorylation of Rab7-S72 prevents the endolysosomal degradation of STING; thereby creating a sustained dsDNA sensing mechanism in the cytosol (Supplemental Fig. S7).
As a response to pathogens and aberrant cellular DNA detected in the cytosol, the cGAS-STING pathway produces an interferon mediated response that is tightly regulated. More recently, it has been shown that Rab7 regulates this innate immune response through trafficking-mediated degradation of the STING protein to endolysosomes (15), suggesting that targeting this pathway for blockade may help improve the therapeutic potential of STING agonists. Consistent with our results, inhibition of lysosome acidification was found to increase STING protein levels and enhance STING signaling, resulting in CXCL10, CCL5, and IFNβ mRNA expression (15). Here we show that increased STING signaling is directly correlated with the phosphorylation status of Rab7-S72. Specifically, phosphorylation of S72 deactivates Rab7 preventing STING trafficking to the endolysosome, resulting in downstream activation of TBK1-IRF3 regulated chemokines such as CXCL10, CCL5, and IFNβ. Importantly, prior studies of STING signaling have determined that it remains active throughout its trafficking from the endoplasmic reticulum to the endolysosome, at which point it is inactivated as the vesicle acidifies (15,35,36). Indeed, in our studies, constitutively active Rab7 (S72A) enhanced STING protein turnover following dsDNA stimulation and continually co-localized with STING in perinuclear puncta, impairing CXCL10, CCL5, and IFNβ production.
Notably, PTEN mediated dephosphorylation of Rab7-S72 was also shown to be necessary for its recruitment to the endolysosome and to promote EGFR recycling (14). As such, it was postulated that in tumors that are PTEN deficient, degradation of EGFR by endosomes is blunted, resulting in enhanced EGFR signaling and enhanced tumor progression. As predicted, higher levels of EGFR were also maintained following EGF stimulation in MDA-MB-468 cells, and reversed with reconstitution of wild-type PTEN (14). Consistent with impaired degradation as a result of PTEN loss, our studies also showed elevations of basal STING expression in PTEN null TNBC cell lines which was recapitulated in primary TNBC tumor histology. Recent work has also showed that EGFR signaling can be immunosuppressive, decreasing both MHC-1 and IFN-γ expression (37). Furthermore, in TNBCs, IL-1 within the tumor microenvironment also promotes IKKε associated NF-κB signaling, inducing CCL5 and IL-6 which are immune suppressive and can recruit myeloid suppressor cells (10). IL-1 induced CCL5 and IL-6 downstream of TBK1/IKKε are further enhanced by impaired autophagy and endolysosome maturation (27). Thus, in addition to elevated STING levels, these additional factors could also promote an immunosuppressed tumor microenvironment in the context of defective Rab7 trafficking.
Currently STING agonism is a developing area of therapeutic focus for various types of cancers. Our studies show that TNBCs are primed for STING agonist treatment, given this breast cancer subtype is associated with high rates of PTEN loss and elevated basal STING levels. Similar to the findings of our T-cell migration assay and growth inhibition assay, injection of STING agonist into the tumor microenvironment of the TNBC murine model 4T1 resulted in regression of both primary and distant tumors while inducing T-cell activation and memory (38-40). Furthermore, emerging findings from a Phase 1 study of ADU-S100 in combination with anti-PD-1 therapy for patients with advanced metastatic solid tumors or lymphomas show that treatment response was largely limited to patients with TNBCs (41). As these trials are moving forward with dose escalation it is likely that only a subset of patients will respond and identifying biomarkers will be critical. Thus, our findings that PTEN loss and TBK1/IKKε signaling converge to promote dysregulated STING trafficking identifies a specific genomic context in TNBC that enhances STING agonist activity. Additional optimization is underway to improve STING retention in the tumor bed (42) and enhance membrane permeability (43-45) to further increase the clinical effectiveness of STING agonist therapy. Regardless, our findings suggest that PTEN null, IKKε high TNBCs may represent an optimal target for STING agonism, converting an immunosuppressive tumor microenvironment into an immunogenic one.
Supplementary Material
Significance: Findings identify RAB7 as a substrate for TBK1 for regulation of innate immune signaling, thereby providing important insight for strategies aimed at manipulating the immune response to enhance therapeutic efficacy in TNBC.
Acknowledgements
We thank Dr. Gabriela Alexe (Broad Institute and Dana Farber Cancer Institute, Boston, MA) for assistance with computational analysis of the SILAC data. This work was funded in part through the generosity of an anonymous donor (to T. Barbie), Department of Defense Award #W81XWH-13-1-0029 (to T. Barbie), and the BWH Department of Surgery Robert T. Osteen Fellowship (to T. Barbie); NCI R01 CA190394 (to D. Barbie); National Cancer Institute Clinical Proteomic Tumor Analysis Consortium grants NIH/NCI U24-CA210986 and NIH/NCI U01 CA214125 (to S. Carr.).
Footnotes
Conflict of Interest Statement. T.U.B. is a consultant for N-of-One/Qiagen. D.A.B. is consultant for N-of-One/Qiagen, Tango Therapeutics, and MADALON consulting. D.A.B. is cofounder of Xsphera Biosciences. D.A.B has received research funding from Novartis and Bristol Myers Squibb.
Reference
- 1.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 2007;13:2329–34 [DOI] [PubMed] [Google Scholar]
- 2.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 2008;26:1275–81 [DOI] [PubMed] [Google Scholar]
- 3.Lin NU, Vanderplas A, Hughes ME, Theriault RL, Edge SB, Wong YN, et al. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the National Comprehensive Cancer Network. Cancer 2012;118:5463–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phuah SY, Looi LM, Hassan N, Rhodes A, Dean S, Taib NA, et al. Triple-negative breast cancer and PTEN (phosphatase and tensin homologue) loss are predictors of BRCA1 germline mutations in women with early-onset and familial breast cancer, but not in women with isolated late-onset breast cancer. Breast Cancer Res 2012;14:R142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu M, Guo S, Stiles JK. The emerging role of CXCL10 in cancer. Oncology Letters 2011;2:583–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012;486:395–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbie TU, Alexe G, Aref AR, Li S, Zhu Z, Zhang X, et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J Clin Invest 2014;124:5411–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra F, Bucci C. Multiple Roles of the Small GTPase Rab7. Cells 2016;5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem 2009;284:12110–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Numrich J, Ungermann C. Endocytic Rabs in membrane trafficking and signaling. Biol Chem 2014;395:327–33 [DOI] [PubMed] [Google Scholar]
- 14.Shinde SR, Maddika S. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat Commun 2016;7:10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonugunta VK, Sakai T, Pokatayev V, Yang K, Wu J, Dobbs N, et al. Trafficking-Mediated STING Degradation Requires Sorting to Acidified Endolysosomes and Can Be Targeted to Enhance Anti-tumor Response. Cell Rep 2017;21:3234–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, Sundararaman SK, et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell 2011;147:853–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choudhary A, Hu He K, Mertins P, Udeshi ND, Dancik V, Fomina-Yadlin D, et al. Quantitative-proteomic comparison of alpha and Beta cells to uncover novel targets for lineage reprogramming. PLoS One 2014;9:e95194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature Protocols 2008;3:1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou MF, Schwartz D. Biological Sequence Motif Discovery Using motif-x. Current Protocols in Bioinformatics 2011;35:13.5.1–.5.24 [DOI] [PubMed] [Google Scholar]
- 21.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nature Biotechnology 2005;23:1391. [DOI] [PubMed] [Google Scholar]
- 22.Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, et al. Phosphorylation of the Tumor Suppressor CYLD by the Breast Cancer Oncogene IKKɛ Promotes Cell Transformation. Molecular Cell 2009;34:461–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med 2018;24:1143–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov 2018;8:196–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad L, Mashbat B, Leung C, Brookes C, Hamad S, Krokowski S, et al. Human TANK-binding kinase 1 is required for early autophagy induction upon herpes simplex virus 1 infection. J Allergy Clin Immunol 2019;143:765–9 e7 [DOI] [PubMed] [Google Scholar]
- 27.Yang S, Imamura Y, Jenkins RW, Canadas I, Kitajima S, Aref A, et al. Autophagy Inhibition Dysregulates TBK1 Signaling and Promotes Pancreatic Inflammation. Cancer Immunol Res 2016;4:520–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutti JE, Porter MA, Cheely AW, Cantley LC, Wang X, Kireev D, et al. Development of a High-Throughput Assay for Identifying Inhibitors of TBK1 and IKKε. PLOS ONE 2012;7:e41494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heo JM, Ordureau A, Swarup S, Paulo JA, Shen K, Sabatini DM, et al. RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK-PARKIN pathway. Science Advances 2018;4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013;155:688–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov 2014;4:452–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones N, Bonnet F, Sfar S, Lafitte M, Lafon D, Sierankowski G, et al. Comprehensive analysis of PTEN status in breast carcinomas. Int J Cancer 2013;133:323–34 [DOI] [PubMed] [Google Scholar]
- 33.Beg S, Siraj AK, Prabhakaran S, Jehan Z, Ajarim D, Al-Dayel F, et al. Loss of PTEN expression is associated with aggressive behavior and poor prognosis in Middle Eastern triple-negative breast cancer. Breast Cancer Res Treat 2015;151:541–53 [DOI] [PubMed] [Google Scholar]
- 34.Li S, Shen Y, Wang M, Yang J, Lv M, Li P, et al. Loss of PTEN expression in breast cancer: association with clinicopathological characteristics and prognosis. Oncotarget 2017;8:32043–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe 2015;18:157–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009;461:788–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lizotte PH, Hong RL, Luster TA, Cavanaugh ME, Taus LJ, Wang S, et al. A High-Throughput Immune-Oncology Screen Identifies EGFR Inhibitors as Potent Enhancers of Antigen-Specific Cytotoxic T-lymphocyte Tumor Cell Killing. Cancer Immunol Res 2018;6:1511–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, et al. Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep 2015;11:1018–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demaria O, De Gassart A, Coso S, Gestermann N, Di Domizio J, Flatz L, et al. STING activation of tumor endothelial cells initiates spontaneous and therapeutic antitumor immunity. Proc Natl Acad Sci U S A 2015;112:15408–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baird JR, Friedman D, Cottam B, Dubensky TW Jr., Kanne DB, Bambina S, et al. Radiotherapy Combined with Novel STING-Targeting Oligonucleotides Results in Regression of Established Tumors. Cancer Res 2016;76:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meric-Bernstam F, Shandu S, Hamid O, Spreafico A, Kasper S, Dummer R, et al. Phase 1b study of MIW815 (ADU-S100) in combination with spartalizumab (PDR001) in patients (pts) with advanced/metastatic solid tumors or lymphomas. ASCO 2019. Abstract 2019 [Google Scholar]
- 42.Park CG, Hartl CA, Schmid D, Carmona EM, Kim H-J, Goldberg MS. Extended release of perioperative immunotherapy prevents tumor recurrence and eliminates metastases. Science Translational Medicine 2018;10:eaar1916. [DOI] [PubMed] [Google Scholar]
- 43.Koshy ST, Cheung AS, Gu L, Graveline AR, Mooney DJ. Liposomal Delivery Enhances Immune Activation by STING Agonists for Cancer Immunotherapy. Adv Biosyst 2017;1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 2012;36:1073–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.STINGing Antitumor Immunity into Action. Cancer Discov 2018;8:259–60 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw SILAC data are available from the corresponding author upon reasonable request.