Abstract
The receptor kinase c-MET has emerged as a target for glioblastoma therapy. However, treatment resistance emerges inevitably. Here, we performed global metabolite screening with metabolite set enrichment coupled with transcriptome and gene set enrichment analysis and proteomic screening, and identified substantial reprogramming of tumor metabolism involving oxidative phosphorylation and fatty acid oxidation (FAO) with substantial accumulation of acyl-carnitines accompanied by an increase of PGC1α in response to genetic (shRNA and CRISPR/Cas9) and pharmacological (crizotinib) inhibition of c-MET. Extracellular flux and carbon tracing analyses (U-13C-Glucose, U-13C-Glutamine and U-13C-Palmitic acid) demonstrated enhanced oxidative metabolism, which was driven by FAO and supported by increased anaplerosis of glucose carbons. These findings were observed in concert with increased number and fusion of mitochondria and production of reactive oxygen species (ROS). Genetic interference with PGC1α rescued this oxidative phenotype driven by c-MET inhibition. Silencing and chromatin immunoprecipitation experiments demonstrated that CREB regulates the expression of PGC1α in the context of c-MET inhibition. Interference with both oxidative phosphorylation (metformin, oligomycin) and beta-oxidation of fatty acids (etomoxir) enhanced the anti-tumor efficacy of c-MET inhibition. Synergistic cell death was observed with with c-MET inhibition and gamitrinib treatment. In patient-derived xenograft models, combination treatments of crizotinib and etomoxir, and crizotinib and gamitrinib were significantly more efficacious than single treatments and did not induce toxicity. Collectively, we have unraveled the mechanistic underpinnings of c-MET inhibition and identified novel combination therapies that may enhance its therapeutic efficacy.
Keywords: c-MET, glioblastoma, tumor cell metabolism, PGC1α, patient-derived xenograft
Introduction
The kinase c-MET has evolved as a therapeutic target for cancer therapy of some glioblastomas (GBM), the most common malignant primary brain tumors, displaying high levels of c-MET expression through several mechanisms, including MET amplification (1–4). When c-MET is activated through its cognate ligand, HGF, a signal cascade is activated, leading to activation of the AKT and ERK signaling pathway, which are commonly dysregulated in cancer, including glioblastoma. Several c-MET inhibitors have been designed, such as crizotinib, which has been tested in glioblastoma.
Akin to other monotherapies, primary or secondary resistance prevails and therefore drug combination therapies are warranted to address this problem. In this context, we targeted tumor cell metabolism following acute and chronic c-MET inhibition. By utilizing proteomic and transcriptomic analysis coupled with untargeted polar and non-polar metabolite analysis by liquid chromatography/mass spectrometry, we identified a specific metabolic program elicited by c-MET inhibition. Interference with c-MET drives oxidative metabolism by increasing fatty acid oxidation (FAO) and glucose anaplerosis, which was orchestrated by the master-regulator, PGC1α. Based on a drug screen (5), we further found that the mitochondrial matrix chaperone inhibitor, gamitrinib, along with c-MET inhibition causes synergistic cell death, which was mechanistically related to the ability of gamitrinib to suppress oxidative metabolism. In alignment with these findings, FAO inhibitor, etomoxir, enhanced the anti-proliferative effects of c-MET inhibition as well. Both combination therapies were active in vivo, suggesting two novel potential combination therapies, involving c-MET inhibitors.
Materials and Methods
Reagents
Crizotinib, Etomoxir, Foretinib, Metformin and Oligomycin were purchased from Sellekchem. Gamitrinib-TPP (GTPP) was kindly provided by Dr. Dario Altieri (Wistar Institute, Philadelphia, PA). The compounds were dissolved in DMSO.
Cell cultures and growth conditions
U87MG, LN229 and A172 human glioblastoma cell lines were obtained from the American Type Culture Collection (Manassas, VA). The respective cell line depository authenticated the cells. Cells were maintained in DMEM with 10% FBS, 4.5 g/L glucose, 4 mM L-glutamine, 1 mM pyruvate and primocin. Patient-derived xenograft, GBM12 and GBM14 cells were obtained from Dr. Jann Sarkaria. All cell lines were obtained between 2015–2017. The U87CrizoR, A172CrizoR, GBM14CrizoR and U87ForeR cells have been exposed to 1 μM crizotinib/foretinib for 14 days.
Cell viability assays
Viability assays were performed as previously described (6). For synergism analysis, the CompuSyn software (ComboSyn, Inc., Paramus, NJ) was used for the determination of the combination index (CI).
Flow cytometry analysis of apoptosis, mitochondrial membrane potential, mitotracker and cellRox
Annexin V Apoptosis Detection Kit (BD Pharmingen), Mitochondrial Membrane Potential (TMRE) (Cell Signaling Technology), Mitotracker (Thermo Fisher) and CellRox (Thermo Fisher) stainings were performed according to the manufacturer’s instructions and analyzed on a LSRII flow cytometer (BD Pharmingen). The data were analyzed with the FlowJo software (version 8.7.1; Ashland, OR).
Transfections of siRNAs
Transfections were performed with Lipofectamine RNAiMaX (Invitrogen, Carlsbad, CA) according to manufacturers’ instructions.
Lentiviral particle transduction (shRNA and CRISPR/Cas9)
c-MET shRNAs were purchased from Santa Cruz Biotech (Santa Cruz, CA). The CRISPR/Cas9 MET knockout lentivirus was purchased from ABMGOOD. Cells were infected in the presence of 8 ug/mL polybrene and were selected with puromycin.
Western blot analysis and capillary electrophoresis
Specific protein expression in cell lines was determined by western blot analysis or capillary electrophoresis as described before (6). The following antibodies were applied: p-MET (Tyr1234/1235) (Cell Signaling Technology (CST) 3077S; 1:250), MET (CST 8198S;1:1000), Mcl-1 (CST 94296S; 1:500), Bcl-2 (CST 15071S; 1:500), BIM (CST 2933S; 1:500), Bcl-xL (CST 2764; 1:500), Noxa (Millipore OP180; 1:500), β-actin (Sigma Aldrich A1978, clone AC15; 1:2,000). The HRP linked secondary antibodies were from Santa Cruz Biotechnology Inc. Western blot signals were detected by using a CCD – camera system (Azure C300 imager). For capillary electrophoresis (Wes instrument, Proteinsimple) the following antibodies were used: p-MET (Tyr1234/1235) (CST 3077S, 1:25), MET (CST 8198, 1:25), p-mTOR (Ser2448) (CST 5536S, 1:25), p-ERK (Thr202/Tyr204) (CST 4370S, 1:25), ERK (CST 4695, 1:25), mTOR (CST 2983, 1:25), p-CREB (Ser133) (CST 9198S, 1:25), CREB (CST 9197S, 1:25), PGC1α (CST2178, 1:25), Vinculin (Abcam ab129002, 1:200).
Microarray and gene set enrichment analysis
Transcriptome and gene set enrichment analysis (GSEA) were performed as previously described (6). The experiment was deposited online ID: U87:GSE113961, GBM14:GSE134676.
Reverse phase protein array
Glioblastoma cell lysates were diluted and arrayed on nitrocellulose-coated slides for the generation of sample spots. Sample spots were exposed to antibodies, involving a tyramide-based signal amplification approach. For detection, a colorimetric system with DAB was used. Stained slides were digitalized by the Huron TissueScope scanner, resulting in a 16-bit tiff image. The Array-Pro Analyzer was used to localize and quantify the digitalized antibody spots. The relative protein levels are determined though interpolation of the different dilution curves. The standard curve is obtained by a script. The relative protein levels are then provided as log2 values and normalized to protein loading, after which they were transformed to linear values.
Real-time PCR analysis
RNA was extracted with the miRNAeasy Mini Kit (QIAGEN). mRNA was reverse transcribed (cDNA synthesis kit, Origene). For qPCR sample preparation, the SYBR green RT-PCR kit (Applied Biosystems) was used. The reactions were run on a qPCR cycler (Quantabio). Primer sequences are in the Table 1 (Supplementary Table S1).
Liquid Chromatography and Mass Spectrometry (LC/MS) Analysis
Analysis and extraction was performed as described before (7).
Seahorse extracellular flux analysis
The Seahorse XFe24 instrument (Agilent Inc.) was used for the assessment of the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) (8). 3×104 glioblastoma cells were seeded. The assay media was Seahorse XF base medium supplemented with 10 mM glucose, 2 mM glutamine, 1 mM pyruvate. The following compounds were injected in a sequential order: 2 μM oligomycin, 2 μM FCCP, and 0.5 μM rotenone/antimycin. For the evaluation of glycolysis, GBM cells were exposed to Seahorse XF base medium supplemented with 1 mM l-glutamine in a CO2-free incubator for 1 hour preceding the assay. During the course of the assay, 10 mM glucose, 1 μM oligomycin, and 50 mM 2-DG were added sequentially.
Metabolomics and isotope tracing
Cells were incubated with DMEM without glucose, glutamine, and phenol red (Thermo Fisher) for 1 hour. Thereafter, the cells were exposed to media, containing either 25mM D-Glucose (U-13C6) (Cambridge Isotope Laboratories, Inc), 4mM L-Glutamine (U-13C5) or 100 μM (U-13C16) palmitic acid (Cambridge Isotope Laboratories, Inc) for 24 h in the presence of 10% dialyzed FBS (Thermo Fisher). Analysis and extraction was performed as described before (7).
Subcutaneous xenograft model
A solvent mixture consisting of Cremophor EL (Sigma, St. Louis, MO), Ethanol (Pharmco-Aaper, Brookfield, CT) and PBS was used to solubilize the drugs. Cells were implanted subcutaneously into the flanks of 6–8 week-old Nu/Nu mice. A caliper is used to measure the tumor sizes based on the formula: (length x width2) x 0.5. Treatments were given three times a week (Gamitrinib: 5 mg/kg, Crizotinib: 50 mg/kg, Etomoxir: 20 mg/kg).
Orthotopic glioblastoma xenograft model
300.000 U87 GBM cells were implanted into the right striatum of Nu/Nu mice (2mm lateral of the bregma). Starting on day 5, animals were randomly assigned to four treatment groups and a total of nine treatments was given (crizotinib: 30–50 mg/kg, etomoxir: 20 mg/kg; between day 5 and 20). Survival was the primary endpoint and was defined as death of an animal, a moribund state or the appearance of neurological symptoms in accordance with the IACUC guidelines.
TUNEL and Ki67 staining
The paraffin-embedded sections were dewaxed, rehydrated, and incubated in proteinase K (Agilent DAKO) 5 min at 37°C. Following rinsing the slides were exposed to TUNEL reaction mixture (1:10 in label solution) for 1 h at 37oC. Following termination of the reaction converter-POD solution was added for 30 min at 37°C. TUNEL staining was highlighted with diaminobenzidine and hematoxylin was used for non-specific nuclear staining. The sections were rehydrated by using different concentration of alcohol. For Ki67 staining, the antigen retrieval was performed using citrate buffer and heating. Incubate the Ki67 (Dako, Cat# GA626) for 90 min at RT. The slides were incubated with horse anti mouse IgG (1:200) for 30 min at RT. Then, the slides were incubated in ABC-Peroxidase Solution (1:50) for 30 min at RT.
Statistical analysis
Statistical analysis involved the two-tailed Student’s t-test or ANOVA (for multiple comparisons) using Prism version 8.00 (GraphPad, La Jolla, CA). Statistical outliers were removed by applying the Grubbs’ test. A p ≤ 0.05 was set as the level of statistical significance. *p < 0.05; **p < 0.01; ***/****p < 0.001. For drug synergism analysis, the CompuSyn software (ComboSyn, Inc., Paramus, NJ) was used to compute the combination index (CI) (CI < 1 synergistic, CI = 1 additive and CI > 1 as antagonistic).
Study approval
All procedures were in accordance with Animal Welfare Regulations and approved by the Institutional Animal Care and Use Committee at the Columbia University Medical Center.
Results
Interference with c-MET signaling elicits evidence of activation of mitochondrial oxidative energy metabolism
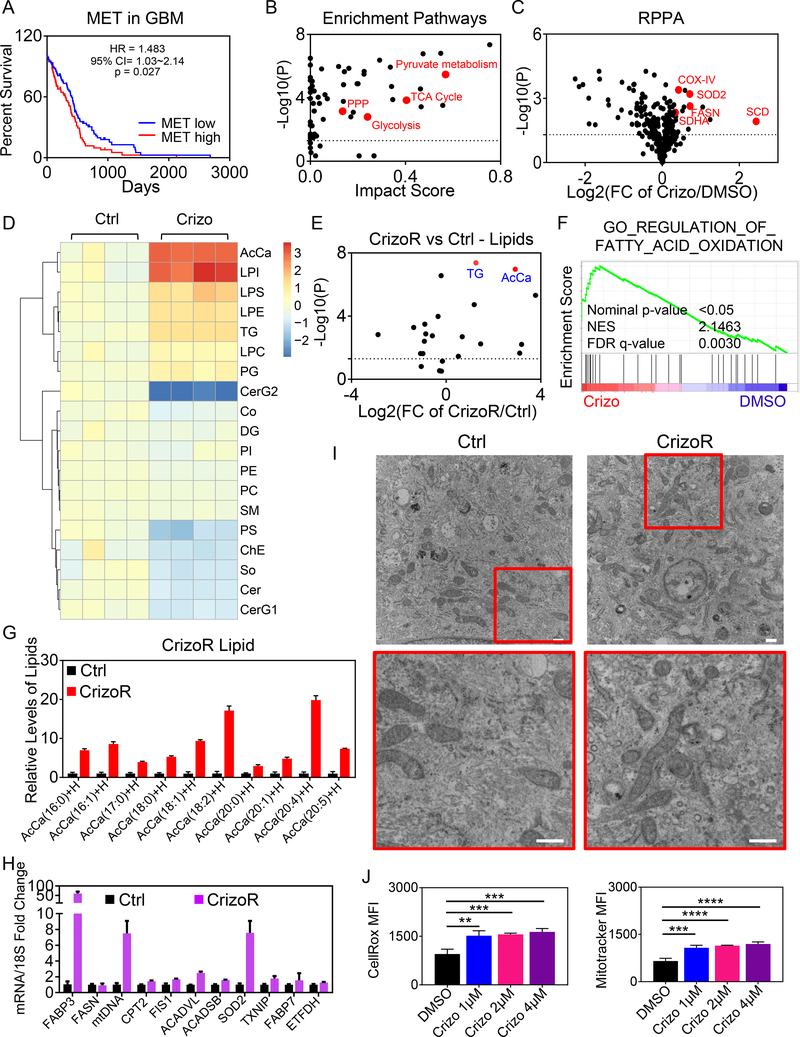
MET kinase signaling is important in glioblastoma and other malignancies. To demonstrate a survival impact, we interrogated the TCGA database and found that high-levels of MET mRNA correlate with an unfavorable survival in patients (Fig. 1A). Similar results were seen in the low grade glioma database (Supplementary Fig. S1A). We confirmed that crizotinib and another c-MET inhibitor, foretinib, interferes with c-MET signaling in our applied model systems, including established glioblastoma and patient-derived xenograft cells (Supplementary Fig. S1B–C). In this context, western blot analysis showed a strong expression of phosphorylated c-MET in glioblastoma cells, which was abrogated by the FDA-approved c-MET inhibitor, crizotinib, and foretinib (Supplementary Fig. S1B–C). We also generated CrizoR cells that were chronically exposed to crizotinib to mimic repeated dosing of the relevant compound which also resulted in a slight resistance towards crizotinib (Supplementary Fig. S1D–G). These cells were bigger and displayed a more complex cytosolic architecture (Supplementary Fig. S1H–J). We initiated our studies by performing a combined screen, involving transcriptome, proteome and metabolomic analyses (polar and non-polar) in the context of chronic c-MET inhibition with crizotinib (Fig. 1B–H and Supplementary Fig. S1D–G). Our metabolite analysis demonstrated an accumulation of carnitine related metabolites. Enrichment pathway analysis revealed an increase in oxidative metabolism pathways, such as the TCA cycle and pyruvate metabolism (Fig. 1B), which was associated with up-regulation of enzymes related to oxidative metabolism based on a proteomic reverse phase protein array (RPPA) screen, including COX-IV, SDHA and SOD2 (Fig. 1C). Aside from oxidative metabolism, there was an enrichment of carbohydrate metabolism, involving glycolysis and the pentose phosphate pathway (PPP) (Fig. 1B). Given the pronounced changes in carnitine-derivatives and up-regulation of genes and proteins related to fatty acid metabolism/oxidation, including FASN and SCD (Fig. 1C and 1H), we extended our analyses to non-polar metabolites, involving a broad-spectrum lipidomic screen. The main findings arising from this screen was a substantial increase in acyl-carnitines and triglycerides, suggesting an increase in beta-oxidation (Fig. 1D, 1E and 1G and Supplementary Fig. S2A–2C). Within the group of acyl-carnitines, we detected an increase in palmitoyl-carnitine (C16-carbon) as well as stearoyl-carnitine (C18-carbon), which are considered as major substrates for beta-oxidation (Fig. 1G). Interestingly, several polyunsaturated fatty acids (PUFAs) were substantially increased (Fig. 1E), which may be related to the observation that SCD is up-regulated by crizotinib. We also noted an increase in lysophospholipids (Fig. 1D), a phenomenon often observed in settings of enhanced FAO. Real-time PCR analysis of chronically crizotinib exposed cells demonstrated an increase in transcripts related to fatty acid transporters and slightly elevated levels of enzyme related to FAO (Fig. 1H). With regards to the general oxidative phenotype are the up regulation of mtDNA, SOD2 and TXNIP, which includes also the A172 GBM cells (Fig. 1H, 4E and Supplementary Fig. S2D–S2G). Coupled with this oxidative signature, electron microscopy demonstrated an increase in mitochondria that revealed a tubulated phenotype in U87 cells chronically exposed to crizotinib (Fig. 1I). Consistently, crizotinib treated cells showed an enhancement in ROS production and an increase mitochondrial mass as assessed by flow cytometric analysis (Fig. 1J and Supplementary Fig. S2H and S2I).
Fig. 1. Inhibition of c-MET leads to reprogramming of fatty acid metabolism.
A, The TCGA database was interrogated to correlate mRNA levels of MET with glioblastoma patients’ survival. Shown is a Kaplan-Meier-survival curve. B, U87 GBM cells chronically exposed to crizotinib 2 μM and parental cells were harvested for LC/MS analysis for polar metabolites. Metabolite set enrichment analysis was performed to identify dysregulated metabolic pathways through crizotinib treatment. Shown are the corresponding enrichment pathways. C, RPPA analysis of U87 cells treated with DMSO or 2 μM crizotinib for 24h. Shown are p-values vs. fold changes (FC) (n=2). D, E, The LC/MS analysis of lipids fraction from U87 GBM cells chronically exposed to crizotinib and parental cells. AcCa: Acyl Carnitine; LPI: lysophosphatidylinositol; LPS: lysophosphatidylserine; LPE lysophosphatidylethanolamine; TG: triglyceride; LPC: lysophosphatidylcholine; PG: phosphatidylglycerol; CerG2: Neutral Glycosphingolipid; Co: Coenzyme; DG: diglyceride; PI: phosphatidylinositol; PE: phosphatidylethanolamine; PC: phosphatidylcholine; SM: sphingomyelin; PS: phosphatidylserine; ChE: Cholesterol Ester; So: Sphingosine; Cer: Ceramides; CerG1: Neutral Glycosphingolipids. A heat map of the two conditions is shown. In panel E, the non-polar metabolite groups are plotted as indicated. Highlighted are triglycerides (TG) and acyl carnitines (AcCa) (n=4, biological replicates) F, Transcriptome and gene set enrichment analysis of U87 GBM cells with DMSO or 2 μM crizotinib for 24h. NES: normalized enrichment score. FDR: false discovery rate. G, LC/MS analysis of the lipid fraction from U87 GBM cells chronically exposed to crizotinib and parental cells. Shown are the individual fractions of acyl carnitines (AcCa). H, Real-time PCR analysis of U87 GBM cells chronically exposed to crizotinib and parental cells (n=2–3). I, Electron microscopy of U87 GBM cells chronically exposed to crizotinib and parental cells. High magnification images are provided from areas with mitochondria. Scale bar: 500 nm. J, U87 GBM cells were treated with indicated concentration of Crizotinib (Crizo) for 24h, labeled with either the CellROX or mitotracker dye and analyzed by flow-cytometry (n=3). Shown are means and SD. **p < 0.01; ***/****p < 0.001.
Chronic MET inhibition favors enhanced carbon incorporation in TCA cycle metabolites
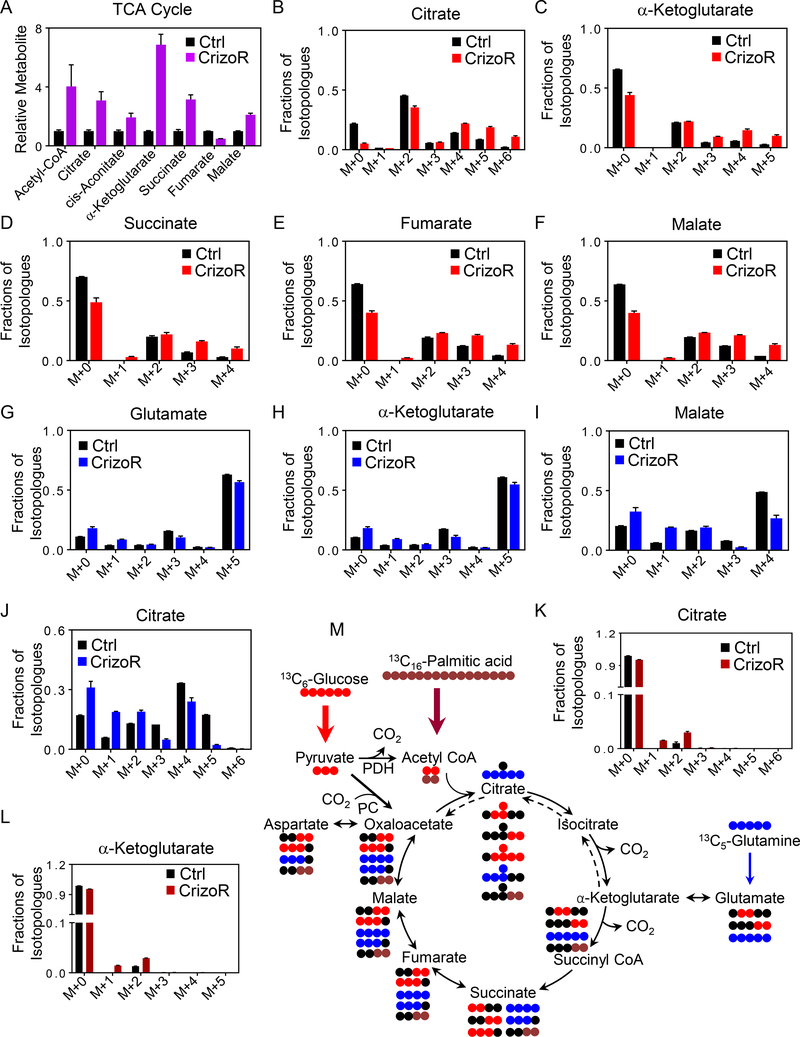
From our polar LC/MS analysis, we noted an increase in metabolites of the TCA cycle (Fig. 2A and Supplementary Fig. S3A–C). To address the questions about how chronically MET-inhibitor treated glioblastoma cells alter their fuel utilization and how the increases in TCA cycle metabolites are mediated, we conducted glucose, glutamine, and palmitic acid tracing experiments (Figure 2B–M). Glucose carbons (U-13C-Glucose) are metabolized in glycolysis to pyruvate. Pyruvate can either be converted to acetyl-CoA via pyruvate dehydrogenase (PDH) reaction or to oxaloacetate via pyruvate carboxylase (PC). This results in either m+2 citrate (PDH reaction) or m+3 citrate (PC reaction) (Figure 2M). In crizotinib treated cells, we noted a substantial enhancement of carbon labeling throughout all the major metabolites of the TCA cycle (Fig. 2B–2F). However, it is noteworthy that we detected a reduction of m+2 citrate, while the m+3, m+4, m+5 and m+6 isotopologes of citrate were all increased, suggesting a reduction of PDH and an increase in PC activity, which is consistent with enhanced anaplerosis to fuel the TCA-cycle (Fig. 2B). Similar observations were made with regards to the other TCA cycle metabolites (Fig. 2C–2F). In addition to the TCA cycle metabolites we detected increased labeling of glutamate, aspartate and glutathione from glucose carbons (Supplementary Fig. S3D–S3F).
Fig. 2. MET inhibition increases palmitic acid and glucose labeling, but decreases glutamine labeling of TCA-cycle metabolites and associated non-essential amino acids.
A, U87 parental or chronically crizotinib treated cells were isolated for LC/MS analysis for TCA-cycle metabolites (n=2–4, biological replicates). B-F, U87 GBM cells were cultured in DMEM media devoid of glucose, glutamine and pyruvic acid (phenol-free) in the presence of 25 mM U-13C-glucose and 4 mM glutamine supplemented with 10% dialyzed FBS. Shown are relative percentages of the isotopologues for each metabolite (n=3). G-J, U87 GBM cells were cultured in DMEM media devoid of glucose, glutamine and pyruvic acid (phenol-free) in the presence of 25 mM glucose and 4 mM U-13C-glutamine supplemented with 10% dialyzed FBS. Shown are relative percentages of the isotopologues for each metabolite (n=3). K, L, U87 GBM cells were cultured in DMEM media devoid of glucose, glutamine and pyruvic acid (phenol-free) in the presence of 5 mM glucose, 1 mM glutamine and 100 μM U-13C-palmitic acid supplemented with 10% dialyzed FBS. Shown are relative percentages of the isotopologues for each metabolite (n=3). M, Summary of key reactions related to the tracer experiment. Blue circles indicate 13C carbons from glutamine, brown circles indicate 13C carbons from palmitic acid, whereas red circles highlight 13C carbons from glucose. Black circles are used to display 12C carbons. Glucose is metabolized to pyruvic acid (m+3) (three carbons labeled) before entering to the TCA cycle. Citric acid (m+2) (two carbons labeled) is produced when glucose is oxidized and citric acid (m+3) (three carbons labeled) is produced when glucose is used for anaplerosis. Glucose carbons are harnessed for the biosynthesis of glutathione either through the serine/glycine pathway or through the TCA cycle via oxoglutaric acid and glutamate. Glutamine reacts to α-ketoglutarate and either is oxidized (clockwise turn) or metabolized under reductive carboxylation (counter clockwise). Palmitic acid is oxidized to acetyl-CoA that condenses with oxaloacetate to yield citric acid. The graphical presentation is representative for only one turn of the TCA cycle. Bold arrows indicate enhanced utilization of glucose (red arrow) and palmitic acid (brown arrow) by the TCA-cycle.
Aside from glucose, glutamine is known to have similar functions. Through glutaminase and glutamate dehydrogenase glutamine is converted to α-ketoglutarate (m+5). In turn, α-ketoglutarate is oxidized to citric acid (m+4) via succinate, fumarate, malate and oxaloacetate (Figure 2M). However, via reductive carboxylation α-ketoglutarate may be converted to citrate (m+5). In the setting of chronic MET inhibitor treated cells, we noted an overall decrease in carbon labeling of TCA metabolites originating from glutamine (Fig. 2G–2J, Supplementary Fig. S3G–3K). Consistently, both glutamine oxidation as well as reductive carboxylation were significantly reduced. Consequently, glutamine driven carbon labeling of aspartate and glutathione was reduced as well (Supplementary Fig. S3G–S3I). In contrast, oxidation of palmitic acid was increased with enhanced labeling of citric acid, α-ketoglutarate, L-acetylcarnitine, succinate and malate by palmitic acid derived carbons (Fig. 2K and 2L and Supplementary Figure 3L–N). These findings highlight an overall dependence of the TCA cycle on glucose carbons in the context of chronic MET inhibition. A summary of the metabolite pathways described above is shown in Fig. 2M. Moreover, given the enhancement of glucose anaplerosis, we noted an increase in phosphorylation of pyruvate dehydrogenase (PDH) at 24 hours after crizotinib treatment which indicates inhibition of PDH (Supplementary Fig. S3O).
Inhibition of MET signaling leads to an activation of oxidative metabolism and an increase in glycolytic intermediates
Having noticed an increase in oxidative metabolism, we then assessed the role of glucose metabolism in the context of c-MET inhibition since our pathway analysis suggested a dysregulation of glycolysis and the pentose phosphate pathway (Supplementary Fig. S3A–S3C). To this end, we utilized extracellular flux analysis on the seahorse analyzer. We found that chronic crizotinib treated cells revealed enhanced extracellular acidification rate (ECAR) glycolysis with lactate production accompanied by a reduced coupling efficiency (Supplementary Fig. S4A–C). In support of the functional extracellular flux analysis, we noted a significant increase in glycolytic metabolites (Supplementary Fig. S3A), in keeping with an activation of this pathway. Aside from glycolysis, the PPP is pivotal for cancer cells to maintain their metabolic requirements. In the context of c-MET inhibition, our screening analysis suggested a potential involvement of this pathway. In line with this notion, we found that crizotinib treated cells exhibit an up-regulation of key metabolites of the PPP, including 6-phospho-gluconolactone and others (Supplementary Fig. S3A–S3C). Accordingly, this leads to an increase in reduction equivalents in the form of NADPH2 that in turn may be used for ROS scavenging due to a global increase in oxidative metabolism. These findings position glucose metabolism as a potential liability of crizotinib treated cells that may be exploited for combination therapies.
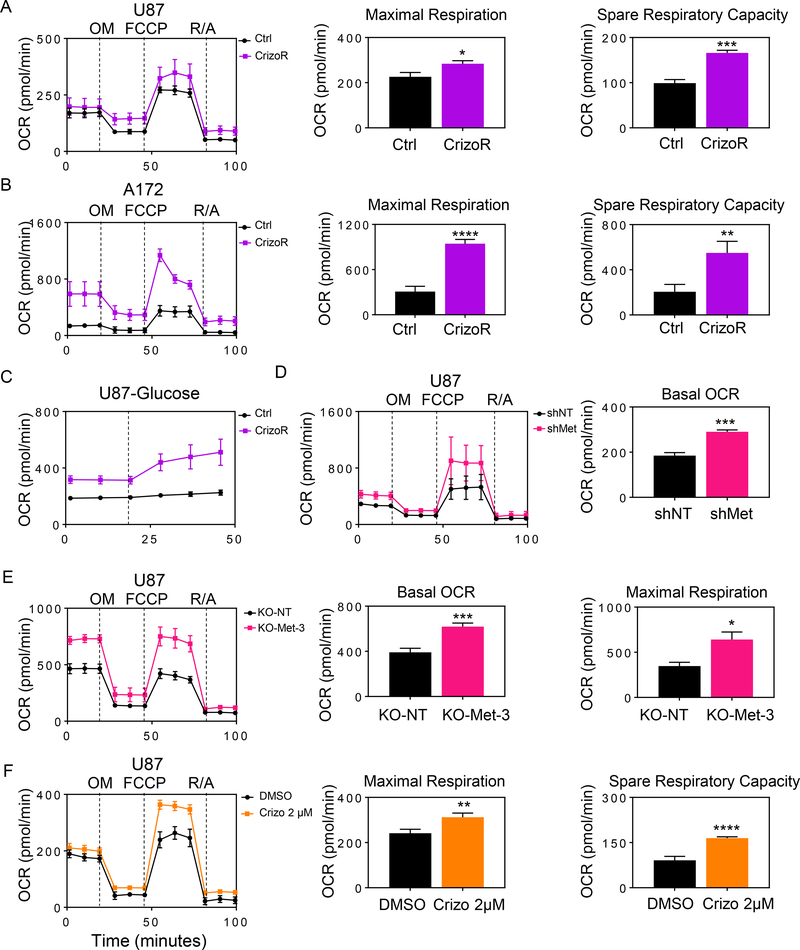
To assess oxidative metabolism, we performed extracellular flux analysis in chronically exposed crizotinib cells and found that they had an elevated oxygen consumption rate (OCR) with an increased spare respiratory capacity and maximal respiration (Fig. 3A). Similarly, we observed the same findings in A172 CrizoR cells (Fig. 3B and Supplementary Fig. S4D–S4F). Given that the TCA-cycle intermediates displayed an increase in labeling from glucose carbons, we asked whether or not glucose contributes to enhanced oxygen consumption rate, acknowledging that glucose in itself is not oxidized, but through anaplerosis contributes to the oxidation of other substrates, e.g. fatty acid (Fig. 3C).
Fig. 3. Chronic MET inhibition favors oxidative energy metabolism.
A, Seahorse mitochondrial stress assay of parental or chronically crizotinib treated U87 GBM cells. OM: Oligomycin, F: FCCP, R/A: Rotenone/Antimycin. Maximal respiration and spare respiratory capacity were interpolated from the time course plot (n=3–4, biological replicates) B, Seahorse mitochondrial stress assay of parental or chronically crizotinib treated A172 GBM cells. Maximal respiration and spare respiratory capacity were derived from the time course plot (n=4, biological replicates). C, Parental or chronically crizotinib treated U87 GBM cells were glucose starved and oxygen consumption rate were analyzed on the seahorse analyzer with acute glucose (10 mM) injection (n=3–4, biological replicates). D, Seahorse mitochondrial stress assay of U87 GBM cells with non-targeting or c-MET specific shRNA. The basal respiration was derived from the time course plot (n=3, biological replicates). E, Seahorse mitochondrial stress assay of U87 GBM cells transduced with a non-targeting or c-MET specific CRISPR-Cas9 constructs. Basal OCR and maximal respiration were plotted (n=3–4, biological replicates). F, Seahorse mitochondrial stress assay of U87 GBM cells treated with DMSO or crizotinib for 24h. Maximal respiration and spare respiratory capacity are plotted (n=3–5, biological replicates). Shown are means and SD. *p < 0.05; **p < 0.01; ***/****p < 0.001.
Because crizotinib is a pharmacological inhibitor, which may elicit off-target effects, we silenced the expression of c-MET in both U87 and GBM14 cells, using shRNA or alternatively with CRISPR/Cas9. Next, we conducted extracellular flux analysis to determine the oxygen consumption in the setting of silenced MET. Genetic interference enhanced oxidative metabolism in both established as well as patient-derived xenograft cell cultures (Fig. 3D, 3E and Supplementary Fig. S4G–S4H and S5A–S5L). We also measured the extracellular acidification rate and found that in like manner it was increased in cells with silenced MET (Supplementary Fig. S5D–S5G).
Given the findings of chronic c-MET inhibition on energy metabolism, we also interrogated early events upon acute MET inhibition. U87 GBM cells were treated with crizotinib for 24h and thereafter we determined the oxygen consumption rate and related parameters. We noted an increase in oxygen consumption rate, especially the maximal respiration and spare respiratory capacity (Fig. 3F). We validated these results in patient-derived xenograft cells, GBM14. Akin to the established cell culture, crizotinib mediated an increase in oxygen consumption rate (Supplementary Fig. S4G) and an increase in ECAR (Supplementary Fig. S4H). An increased oxygen consumption rate was observed following chronic foretinib inhibition as well (Supplementary Fig. S4I).
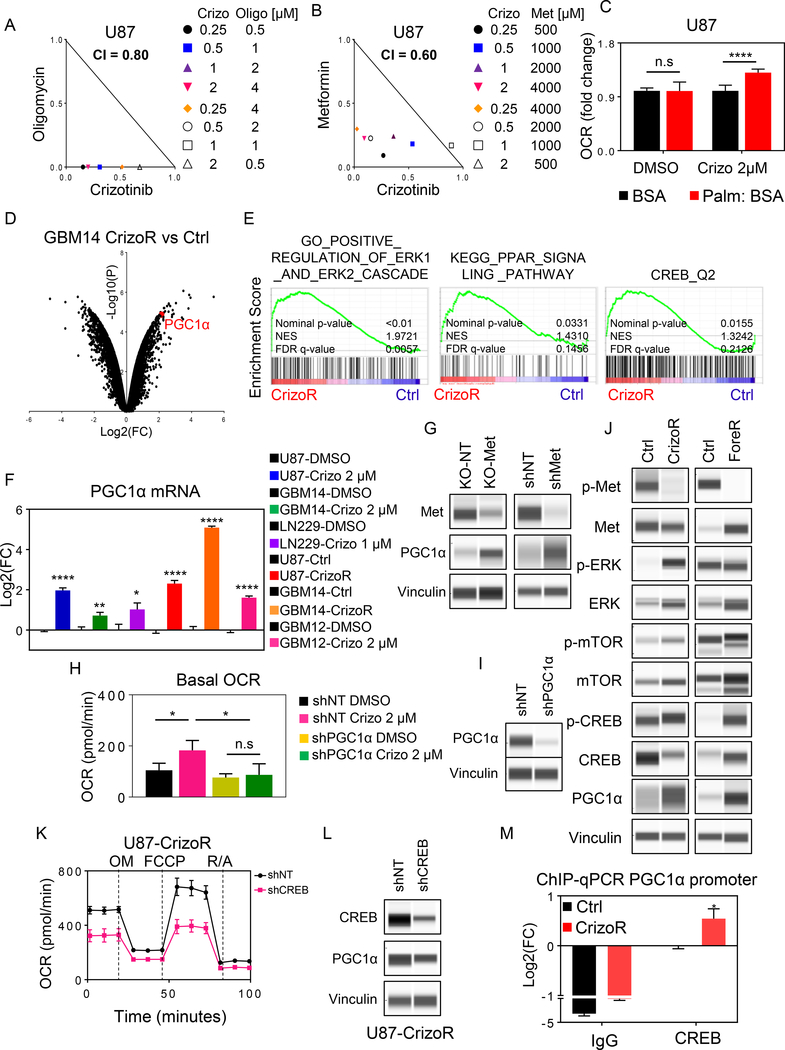
To further corroborate the importance of oxidative phosphorylation in the context of MET inhibition, we utilized two inhibitors, Metformin, an FDA-approved drug that blocks complex I of the respiratory chain, and Oligomycin, an ATP-synthase inhibitor (complex V). Combining crizotinib with either metformin or oligomycin leads to synergistic reduction in proliferation of tumor cells (Fig. 4A, 4B, Supplementary Fig. S6A–G), suggesting that oxidative phosphorylation operates as a pro-survival pathway in the context of MET-inhibition. Given our earlier observation of an increase in polar and non-polar metabolites related to FAO and an up-regulation of transporters related to fatty acid transport we hypothesized that MET regulates FAO and that MET inhibition will facilitate beta-oxidation, which in turn will increase the oxygen consumption rate. To this end, we took advantage of measuring oxidation of palmitic acid through extracellular flux analysis. Our findings revealed that MET inhibition enhanced the oxidation of palmitic acid (Fig. 4C), confirming our observation from the metabolite and transcriptome analysis and further suggesting that fatty acids are a major fuel for the observed enhanced oxygen consumption rate.
Fig. 4. MET-inhibition increases oxidative metabolism through PGC1α depending on the transcription factor CREB.
A, B, U87 GBM cells were treated with crizotinib in the presence or absence of oligomycin or metformin for 72h. Cellular viability was assessed and synergism analysis was performed. Isobolograms are shown. C, U87 GBM cells were treated with DMSO or 2 μM crizotinib for 24h. Thereafter, cells were starved, incubated in medium with albumin stabilized palmitic acid and oxygen consumption rate was measured (n=15–20). D, Shown is a volcano plot of chronically crizotinib treated GBM14 with its parental cells. Highlighted is the mRNA level of PGC1α. fc: fold changes, p: p-value (n=2). E, Parental or chronically crizotinib treated U87 GBM cells were subjected to transcriptomic analysis, followed by gene set enrichment analysis. Shown are enrichment plots. FDR: false discovery rate, NES: normalized enrichment score. F, U87, GBM14, GBM12 and LN229 cells were treated with DMSO or crizotinib for 24h. Alternatively, chronically crizotinib treated U87 or chronically crizotinib treated GBM14 and their respective parental cells were used for the analysis. The mRNA levels of PPARGC1a (PGC1α) were analyzed. Values are given in log2 fold-changes (n=3–4). G, U87 GBM cells were transduced with non-targeting or MET specific sgRNA (KO). Alternatively, cells were transduced with shRNAs. Cells were extracted and analyzed by capillary electrophoresis for the indicated proteins. H, I, U87 GBM cells were transduced with non-targeting or PGC1α specific shRNAs. Stable cell lines were treated with DMSO or 2 μM crizotinib. Thereafter, cells were analyzed for oxygen consumption rate on the seahorse analyzer (n=3–5). Silencing is validated by capillary electrophoresis in I. J, Capillary electrophoresis of the indicated markers from U87 parental or chronically crizotinib/foretinib exposed cells. K, L, U87 GBM cells were transduced with non-targeting or CREB specific shRNA. The transduced cells were subjected to a mitochondrial stress assay. OM: Oligomycin, F: FCCP, R/A: Rotenone/Antimycin (n=4). Silencing is validated by capillary electrophoresis in L. M, Parental or chronically crizotinib exposed U87 GBM cells were subjected to CHIP with an isotype IgG control or a CREB specific antibody. The promoter region of PPARGC1a was amplified. Shown are log2 fold changes (n=6–8). Shown are means and SD. *p < 0.05; **p < 0.01; ***/****p < 0.001.
Our transcriptome analysis demonstrated that MET-inhibition led to an increase of a master-regulator of oxidative metabolism, PGC1α, which was confirmed by real-time PCR analysis as well as on the protein level in several GBM model systems (Fig. 4D–4I and Supplementary Figure 6C and 6G). In agreement, gene set enrichment analysis of chronically crizotinib treated cells revealed an increase for the PPAR signaling pathway, which was accompanied by an activation of ERK and CREB signaling, which are known pathways to be up-stream of PGC1α suggesting their potential involvement in the transcriptional regulation of PGC1α through MET-inhibition (Fig. 4E).
We hypothesized that the increase of PGC1α is a key event in the metabolic reprogramming elicited by MET inhibition. To this purpose, we silenced the expression of PGC1α to assess its impact on oxidative metabolism by extracellular flux analysis (Fig. 4H and 4I). Whereas non-targeting glioblastoma cells responded with an increase in oxygen consumption rate upon crizotinib treatment, we found an almost complete abrogation of this effect when PGC1α is silenced, positioning PGC1α as a major regulator of metabolism in the context of MET-inhibition. Since CREB is the transcription factor that binds to the proximal promoter region of PGC1α to drive its transcription, we hypothesized that CREB may be involved in the regulation of PGC1α expression in the context of MET inhibition. We even noted an increase in phosphorylation of CREB after chronic exposure to crizotinib and foretinib in U87 cells (Fig. 4J). To assess the role of CREB on the oxygen consumption rate in chronically crizotinib exposed cells, we silenced the expression of CREB by shRNA and analyzed these cells on the seahorse analyzer (Fig. 4K and 4L). We detected a substantial suppression of OCR in CREB silenced cells compared to non-targeting transduced cells. Moreover, silencing of CREB suppressed the expression levels of PGC1α in chronically crizotinib treated cells (Fig. 4K). Overall, these findings support the hypothesis that CREB is implicated in the regulation of oxidative phosphorylation in chronically crizotinib treated cells, and this occurs dependent on the transcription. To demonstrate that there is enhanced binding of CREB to the promoter region of PGC1α we conducted CHIP-qPCR. We observed an increased binding of CREB to the promoter region of PGC1α, in keeping with an up-regulation of PGC1α (Fig. 4M).
MET inhibition elicits survival dependency on fatty acid oxidation
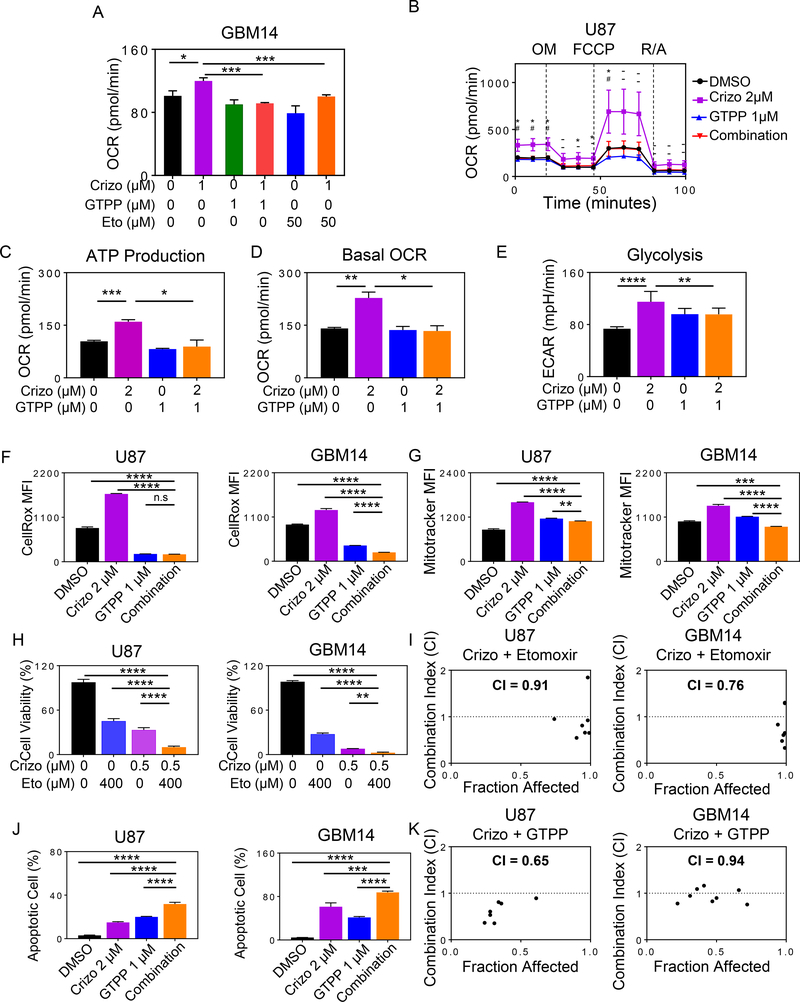
In connection with earlier findings that crizotinib treated cells displayed an enhanced oxidation of fatty acids, we hypothesized that inhibition of FAO by etomoxir and gamitrinib will have an impact on oxygen consumption rate, mitochondrial size, ROS production and survival of tumor cells in the context of MET inhibition (Fig. 5A–5G). To this purpose, glioblastoma cells were treated with crizotinib in the presence or absence of etomoxir, a clinical validated drug compound inhibiting CPT1A. CPT1A is one of the key transport proteins that facilitates the translocation of fatty acid into the mitochondria. We found that the combination treatment of crizotinib and etomoxir synergistically reduced the proliferation rate of established and patient-derived xenograft cell cultures, suggesting that FAO exerts a pro-survival function in the context of MET inhibition and that crizotinib and etomoxir might be a potential drug combination therapy (Fig. 5H, 5I and Supplementary Fig. S7A). Consistent with this effect on proliferation, we also confirmed that etomoxir reverses the increase in oxygen consumption rate in the context of MET inhibition (Fig. 5A, 5H, and 5I).
Fig. 5. Dual inhibition of CPT1A or TRAP1 and c-MET elicits synergistic reduction in cellular proliferation of glioblastoma cells.
A, GBM14 cells were treated with crizotinib (Crizo), gamitrinib (GTPP), etomoxir (Eto) singly or in combination as indicated for 24h. Thereafter, extracellular flux analysis in the context of a mito stress assay was performed on the seahorse analyzer (n=6–12). B, U87 GBM cells were treated with 2 μM crizotinib, 1 μM gamitrinib or the combination of both for 24h. Thereafter, extracellular flux analysis was performed as in A (n=3). OM: Oligomycin, F: FCCP, R/A: Rotenone, Antimycin. C, D, ATP production and basal oxygen consumption rate (OCR) were calculated from the plot in B (n=3). E, Shown are ECAR measurements from the experiment in B (n=9). F, G, Cells were treated with 2 μM crizotinib, 1 μM gamitrinib or the combination of both for 24h. Cells were labeled with CellROX or Mitotracker dye. Thereafter, they were analyzed by flow cytometry and quantified (n=3). H, I, GBM14 or U87 cells were treated with etomoxir (Eto), crizotinib (Crizo) or the combination of both for 72h. Thereafter, cell viability was evaluated and synergism analysis was performed (n=3). CI values below 1 indicate a synergistic interaction. J, GBM14 and U87 cells were treated with crizotinib, gamitrinib or the combination of both. Shown are apoptotic cells obtained by flow cytometric analysis (n=3). K, U87 or GBM14 cells were treated with crizotinib, gamitrinib or the combination of both for 72h. Cellular viability analysis was performed and synergism effects were computed. CI values below 1 indicate a synergistic interaction. Shown are means and SD. *p < 0.05; **p < 0.01; ***/****p < 0.001.
Based on an earlier drug screen, involving the TRAP1 inhibitor, gamitrinib (GTPP) (5), we found that MET inhibition is synthetically lethal with gamitrinib. Therefore, we confirmed that crizotinib/foretinib and gamitrinib reduce cellular proliferation in a synergistic manner, and this is mediated by enhanced apoptosis in U87, LN229, GBM14 and GBM12 cells (Fig. 5J, 5K, Supplementary Fig. S7B–I, S8A–C, S9A–F and S10A). Similar observations were made when foretinib was employed in lieu of crizotinib, demonstrating synergistic reduction cell growth following the combination treatment with gamitrinib/and or etomoxir in several model systems tested (Supplementary Fig. S7D–S7I). Following treatment with the drug combination of gamitrinib and crizotinib, we noted a loss of mitochondrial membrane potential and cleavage of initiator and effector caspases as well as cleavage of PARP (Supplementary Fig. S8A), consistent with a cell death with apoptotic features (Supplementary S9E and S9F). Because of the engagement of apoptosis, we assessed the protein levels of down-stream regulators of apoptosis, Bcl-2, Bcl-xL, Mcl-1, BIM and Noxa. The results of this analysis showed a mixed regulation with Mcl-1 being the most consistent member to be down-regulated (Supplementary Fig. S8B). To specifically confirm the role of c-MET, we evaluated the efficacy of gamitrinib in the context of silenced c-MET and found an enhancement of gamitrinib mediated growth reduction (Supplementary Fig. S8C).
Related to this finding, we hypothesized that this synergistic interaction is based on the fact that TRAP1 regulates oxidative phosphorylation at the level of succinate dehydrogenase (complex II of the respiratory chain). To this purpose, we conducted extracellular flux analysis in U87 GBM cells and found that crizotinib increases oxygen consumption rate and ATP production, which is almost completely abrogated in the presence of gamitrinib (Fig. 5B–5D). Similar data was observed in GBM14 PDX cell line (Fig. 5A). Earlier research has demonstrated that gamitrinib interferes with aerobic glycolysis by interference with hexokinase-II (9) Therefore, we tested whether or not crizotinib driven glycolysis is attenuated in the presence of gamitrinib. In keeping with our hypothesis, we found that suboptimal dosages of gamitrinib suppress MET inhibitor driven glycolysis (Fig. 5E). Consistently, we noted a suppression of crizotinib mediated ROS production, a decrease in mitochondrial mass and ATP production by gamitrinib (Fig. 5F, 5G and Supplementary Fig. S10B–D).
In addition, it is noteworthy that the combination treatment of gamitrinib and crizotinib was more efficient to elicit synergistic apoptosis induction than various combination treatments, involving kinase inhibitors of the MEK, PI3K and mTOR pathway (Supplementary Fig. S10E–J).
Combined inhibition of FAO and MET results in an enhanced reduction of tumor sizes and extension of overall survival in GBM model systems
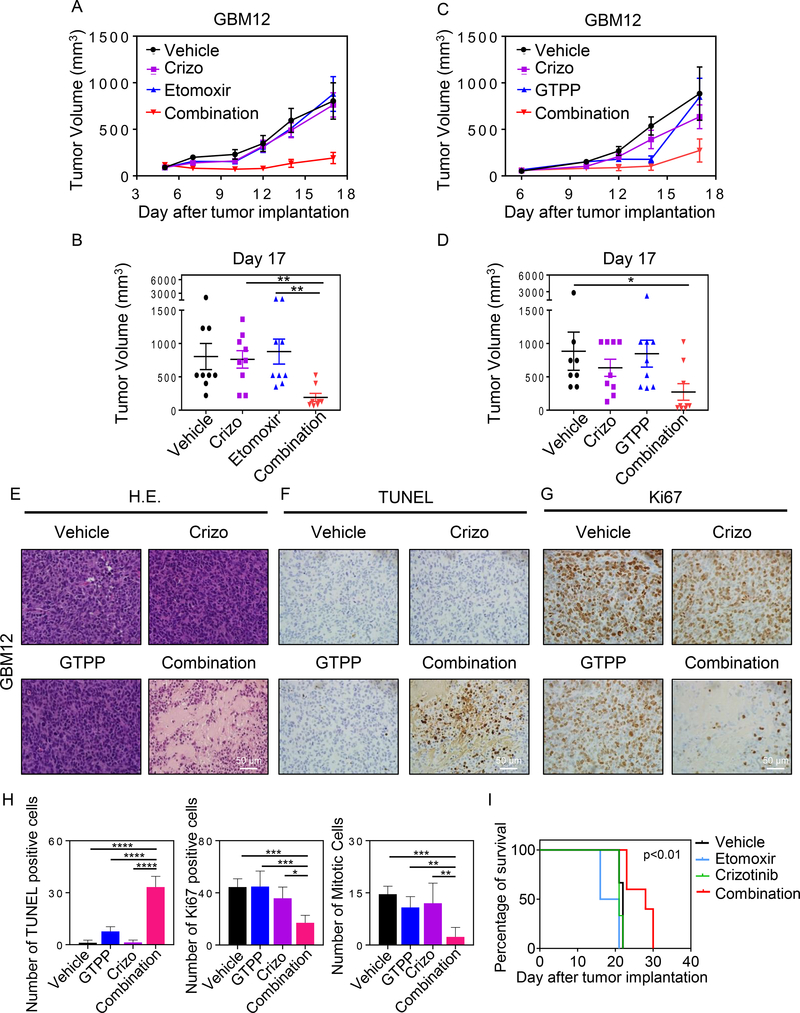
Finally, we tested whether or not our findings bear translational relevance. To assess this, we used two glioblastoma patient derived xenograft models, GBM12 and GBM14. We initiated our studies with GBM12. After cell injection and tumor formation, mice were randomly divided in the indicated groups (control, crizotinib, etomoxir and the combination of etomoxir and crizotinib). While vehicle, crizotinib and etomoxir treatment groups did not differ in tumor growth, we noted a substantial and statistical significant suppression in tumor growth in animals that received the combination treatment of crizotinib and etomoxir (Fig. 6A and 6B). Next, we tested the drug combination of gamitrinib and crizotinib in vivo. To this end, we established GBM12 tumors in mice, formed treatment groups and treated them with the indicated drug compounds. We noted a statistical significant reduction of tumor growth in the combination treatment with single treatments and controls (Fig. 6C and 6D). The same experiments as performed in the GBM12 model were subsequently conducted in GBM14. Akin to the GBM12 model, we found that both combination treatments (crizotinib and gamitrinib; crizotinib and etomoxir) led to a more pronounced reduction in tumor growth than each single treatment or control (Supplementary Fig. S11A and S11B).
Fig. 6. Dual inhibition of CPT1A or TRAP1 and c-MET elicits growth reduction of patient-derived xenografts.
A, B, GBM12 PDX grafts were serially transplanted into mice. Following establishment of tumors, four treatment groups were randomly assigned. Thereafter, treatment was initiated with vehicle, crizotinib, etomoxir or the combination of both by i.p. injections three times a week. On day 17, the experiment was terminated and a final measurement was taken, which was statistically evaluated (B) (n=9; 2–3 tumors per animal). C, D, GBM12 PDX grafts were serially transplanted into mice. Following establishment of tumors, four treatment groups were randomly assigned. Thereafter, treatment was initiated with vehicle, gamitrinib (GTPP), crizotinib or the combination of both by i.p. injections three times a week. On day 17, the experiment was terminated and a final measurement was taken, which was statistically evaluated (D) (n=8–9; 2–3 tumors per animal). E-G, Tumors from experiment C were harvested, fixed and embedded in paraffin followed by staining with standard H&E or be subjected to immunohistochemistry for TUNEL or Ki67. H, Sections from the experiment in 6C were quantified for apoptotic bodies, number of TUNEL positive cells and number of mitosis per multiple high-power fields (n=4–7). Shown are means and SD. *p < 0.05; **p < 0.01; ***/****p < 0.001. Scale bar: 50 μm. I, U87 GBM cells were implanted in the right striatum of nude mice. Starting on day 5, four groups were formed: Vehicle, etomoxir, crizotinib, etomoxir and crizotinib. In total, nine treatments were administered and animal survival is provided (Kaplan-Meier-curve). The log rank test was used to assess statistical significance (n=2–5 animals per group). Crizotinib and etomoxir vs etomoxir (p<0.01), crizotinib and etomoxir vs crizotinib (p<0.01) and crizotinib and etomixr vs control (p<0.01).
To confirm how the combination treatment (crizotinib and gamitrinib) reduced tumor growth we performed histological analysis of tumors with H&E, TUNEL and Ki67 staining (Fig. 6E–6H). Through TUNEL staining, we found that overall there is more cell death in tumor slides from the combination treatment. The proliferation index (Ki67) was reduced in the combination treatment as well. These findings suggest both a reduced proliferation of tumors cells as well as an enhanced induction of cell death which provides an explanation for the reduced size of the tumors in the combination treatment. Similarly, a reduction of cellularity and a reduced proliferation index were found in the combination treatment of crizotinib and etomoxir (Supplementary Fig. S11C–11G). Regarding the toxicity of the combination treatment, we assessed the integrity of organs (brain, heart, intestine, kidney, liver, lung, pancreas and spleen) and did not find histological evidence of organ toxicity following the combination treatment (Supplementary Fig. S11H).
Finally, we validated whether the combination treatment of c-MET and FAO inhibition is effective in glioblastoma cells engrafted into the brain of nude mice. To this purpose, U87 cells were utilized since these cells bear highest levels of c-Met and have been previously utilized by others to assess impact of c-MET inhibition (10). Following engraftment and after completion of a treatment regimen of either vehicle, single (crizotinib, etomoxir) or the combination treatment, we noted a small, but significant survival extension of animals receiving the combination treatment, suggesting that this treatment concept may have potential following optimization of delivery or inhibitor modification (Figure 6I).
Discussion
The integration of tumor cell metabolism as an opportunity to design novel therapies is receiving considerable attention in the cancer research community (11–15). First, with the wealth of novel technical opportunities to measure metabolomic changes, it has become increasingly easier to characterize tumor cell metabolism (16). Second, sophisticated high-throughput genetic screening methods and large data sets obtained from deep sequencing have permitted the community to identify key genetic alterations in metabolism. Examples include the mutation of IDH1, amplifications in the PHGDH gene or loss of succinate dehydrogenase (17)(18). Third, through advancement in drug discovery and synthesis tumor cell metabolism becomes a more suitable target. In this vein, PHGDH inhibitors have been recently developed (11,13,15,19–21).
Targeting aberrant active kinases is a central dogma in cancer therapy with certainly some, but limited success (22). In this work, we have focused on the MET kinase pathway, which is active in human glioblastoma and has been identified as a therapeutic target before by others (10,23). Consequently, several drugs have been designed, including crizotinib. Since c-MET inhibition displays limited anti-tumor effects, we intended to identify means that improve the efficacy of c-MET inhibition in the current work. To accomplish this goal, we utilized several screening methods, involving gene set enrichment and proteome analysis coupled with metabolite screening by LC/MS. This approach has resulted in the identification of substantial metabolic reprogramming elicited by c-MET inhibition that to the best of our knowledge has not been demonstrated before. Short and long exposure leads to activation of oxidative metabolism in established and patient-derived xenograft cells, which was morphologically exemplified by an increase in mitotracker, reactive oxygen species production, enhanced oxygen consumption rate with related indices (as determined by extracellular flux analysis) and an increase in mitochondria as demonstrated by electron microcopy. These findings were accompanied by up-regulation of the master-regulator transcription factor, PGC1α (24–26), which was a major mediator of these effects. To the best of our knowledge, our group is the first describing these metabolic aberrations upon c-MET inhibition. The present findings are in keeping with an earlier report demonstrating that BRAF-kinase inhibition in BRAF V600E mutated melanomas activates oxidative energy metabolism (27). Similar findings had be seen in the context of CDK4 inhibition in pancreatic tumor cells, which resulted in an increase of both glycolysis and oxidative metabolism (28).
Coupled with the enhanced oxidative metabolism is our observation that c-MET inhibition drives FAO, which we validated in extracellular flux analysis since the CPT1a inhibitor, etomoxir, attenuated c-MET inhibition mediated oxygen consumption rate. Whereas, fatty acids were oxidized, glucose carbons appeared to be mainly necessary to provide its carbons for anaplerosis of the TCA cycle. In addition, etomoxir and crizotinib reduced proliferation of glioblastoma cells in a synergistic manner in vitro and in vivo, further corroborating the translational implication of our findings. Enhancement of both oxidative phosphorylation and FAO has been seen in the context of treatment of myeloid leukemia by cytarabine. Similarly, to our findings, etomoxir enhanced the effects of cytarabine (29).
Gamitrinib is a novel drug compound that will enter clinical testing. It elicits anti-tumor activity primarily by dampening oxidative metabolism through inhibition of mitochondrial matrix chaperones. In connection with an earlier high-throughput drug screen (5), we found that the c-MET inhibitor synergizes with gamitrinib in vitro and in vivo. As predicted, gamitrinib potently attenuated the pro-oxidative effects by c-MET inhibition.
Another recent paper that used similar model systems like in our study demonstrated that crizotinib resistance activated several central signaling pathways, such as mTOR, EGFR, FGFR1, STAT3 and COX2 (10). They showed that interference with these pathways enhanced the efficacy of crizotinib in vivo (10). It is tempting to speculate whether due to the poor brain penetration of crizotinib the in vivo effects in orthotopic models fall below expectations. While our study made similar observations with regards to mTOR signaling our focus rested on tumor cell metabolism and how c-MET inhibition mediated reprogrammed tumor cell metabolism can be targeted for therapy. From a broader perspective it is possible that c-MET inhibition driven mTOR and EGFR signaling might unleash a glycolytic and oxidative phenotype in glioblastoma cells since these pathways are inherently linked to drive tumor cell metabolism.
Taken together, our work has unraveled two novel drug combination therapies by integrating transcriptome, proteome and metabolite analyses.
Supplementary Material
Statement of Significance.
c-MET inhibition causes profound metabolic reprogramming that can be targeted by drug combination therapies.
Acknowlegement
M.D. Siegelin: NIH NINDS R01NS095848, R01NS102366, K08NS083732, Louis V. Gerstner, Jr. Scholars Program (2017–2020) and American Brain Tumor Association Discovery Grant 2017 (DG1700013). Trang Nguyen: American Brain Tumor Association Basic Research Fellowship (BRF1900018). Transcriptome analysis was supported by the CTSA grant UL1-TR001430 to the Boston University Microarray and Sequencing Resource Core Facility. These studies used the resources of the Cancer Center Flow Core Facility funded in part through center grant P30CA013696 and S10RR027050. Metabolomics shown in Fig. 1, 2A and Supplementary S2A, S2B, S3A–C were performed by the Whitehead Institute Metabolite Profiling Facility (Cambridge, MA).
Footnotes
Conflict of interest statement: The authors have declared that no conflict of interest exists.
References
- 1.Farrell PJ, Matuszkiewicz J, Balakrishna D, Pandya S, Hixon MS, Kamran R, et al. MET Tyrosine Kinase Inhibition Enhances the Antitumor Efficacy of an HGF Antibody. Molecular cancer therapeutics 2017;16:1269–78 [DOI] [PubMed] [Google Scholar]
- 2.Boccaccio C, Comoglio PM. The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer research 2013;73:3193–9 [DOI] [PubMed] [Google Scholar]
- 3.Jahangiri A, De Lay M, Miller LM, Carbonell WS, Hu YL, Lu K, et al. Gene expression profile identifies tyrosine kinase c-Met as a targetable mediator of antiangiogenic therapy resistance. Clinical cancer research : an official journal of the American Association for Cancer Research 2013;19:1773–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Q, Bradley R, Kang L, Koeman J, Ascierto ML, Worschech A, et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proceedings of the National Academy of Sciences of the United States of America 2012;109:570–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh JC, Siegelin MD, Vaira V, Faversani A, Tavecchio M, Chae YC, et al. Adaptive mitochondrial reprogramming and resistance to PI3K therapy. J Natl Cancer Inst 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpel-Massler G, Ishida CT, Bianchetti E, Zhang Y, Shu C, Tsujiuchi T, et al. Induction of synthetic lethality in IDH1-mutated gliomas through inhibition of Bcl-xL. Nat Commun 2017;8:1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen TTT, Ishida CT, Shang E, Shu C, Torrini C, Zhang Y, et al. Activation of LXRbeta inhibits tumor respiration and is synthetically lethal with Bcl-xL inhibition. EMBO Mol Med 2019:e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishida CT, Zhang Y, Bianchetti E, Shu C, Nguyen TTT, Kleiner G, et al. Metabolic Reprogramming by Dual AKT/ERK Inhibition through Imipridones Elicits Unique Vulnerabilities in Glioblastoma. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24:5392–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chae YC, Caino MC, Lisanti S, Ghosh JC, Dohi T, Danial NN, et al. Control of tumor bioenergetics and survival stress signaling by mitochondrial HSP90s. Cancer Cell 2012;22:331–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruickshanks N, Zhang Y, Hine S, Gibert M, Yuan F, Oxford M, et al. Discovery and Therapeutic Exploitation of Mechanisms of Resistance to MET Inhibitors in Glioblastoma. Clin Cancer Res 2019;25:663–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJ, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nature chemical biology 2016;12:452–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gui DY, Sullivan LB, Luengo A, Hosios AM, Bush LN, Gitego N, et al. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab 2016;24:716–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 2011;43:869–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016;532:255–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 2015;47:1475–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang C, Chen L, Rabinowitz JD. Metabolomics and Isotope Tracing. Cell 2018;173:822–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell 2011;19:17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B, Zheng A, Hydbring P, Ambroise G, Ouchida AT, Goiny M, et al. PHGDH Defines a Metabolic Subtype in Lung Adenocarcinomas with Poor Prognosis. Cell Rep 2017;19:2289–303 [DOI] [PubMed] [Google Scholar]
- 20.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011;476:346–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med 2018;24:1036–46 [DOI] [PubMed] [Google Scholar]
- 22.Zhang G, Frederick DT, Wu L, Wei Z, Krepler C, Srinivasan S, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest 2016;126:1834–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proceedings of the National Academy of Sciences of the United States of America 2011;108:9951–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo C, Lim JH, Lee Y, Granter SR, Thomas A, Vazquez F, et al. A PGC1alpha-mediated transcriptional axis suppresses melanoma metastasis. Nature 2016;537:422–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 2013;23:287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruns I, Sauer B, Burger MC, Eriksson J, Hofmann U, Braun Y, et al. Disruption of peroxisome proliferator-activated receptor gamma coactivator (PGC)-1alpha reverts key features of the neoplastic phenotype of glioma cells. J Biol Chem 2019;294:3037–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer cell 2013;23:302–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franco J, Balaji U, Freinkman E, Witkiewicz AK, Knudsen ES. Metabolic Reprogramming of Pancreatic Cancer Mediated by CDK4/6 Inhibition Elicits Unique Vulnerabilities. Cell Rep 2016;14:979–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov 2017;7:716–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.