Abstract
Purpose:
We hypothesized that integrated analysis of cancer types from different lineages would reveal novel molecularly defined subgroups with unique therapeutic vulnerabilities. Based on the molecular similarities between subgroups of breast and ovarian cancers, we analyzed these cancers as a single cohort to test our hypothesis.
Experimental design:
Identification of transcriptional subgroups of cancers and drug sensitivity analyses were performed using mined data. Cell line sensitivity to heat shock protein 90 inhibitors (Hsp90i) was tested in vitro. The ability of a transcriptional signature to predict Hsp90i sensitivity was validated using cell lines, and cell line- and patient-derived xenograft models. Mechanisms of Hsp90i sensitivity were uncovered using immunoblot and RNAi.
Results:
Transcriptomic analyses of breast and ovarian cancer cell lines uncovered two mixed subgroups comprised primarily of triple-negative breast and multiple ovarian cancer subtypes. Drug sensitivity analyses revealed that cells of one mixed subgroup are significantly more sensitive to Hsp90i compared to cells from all other cancer lineages evaluated. A gene expression classifier was generated that predicted Hsp90i sensitivity in vitro, and in cell line- and patient-derived xenografts. Cells from the Hsp90i-sensitive subgroup underwent apoptosis mediated by Hsp90i-induced upregulation of the pro-apoptotic proteins Bim and PUMA.
Conclusions:
Our findings identify Hsp90i as potential therapeutic strategy for a transcriptionally defined subgroup of ovarian and breast cancers. This study demonstrates that gene expression profiles may be useful to identify therapeutic vulnerabilities in tumor types with limited targetable genetic alterations, and to identify molecularly definable cancer subgroups that transcend lineage.
Keywords: breast, ovarian, gene expression, therapeutic
Introduction
Technical advances in DNA sequencing and molecular profiling have ushered in an age of “precision medicine,” where the genetics of a patient’s tumor can be accurately interrogated to identify therapeutically targetable alterations. Tumor-targeted therapeutics are often selected based on somatic DNA alterations in a drug target, such as anti-HER2 therapies for HER2/ERBB2-amplified breast cancer (BRCA), and BRAF inhibitors for BRAFV600-mutant lung cancers and melanomas. In such cases, drug development strategies have been straightforward. However, there are many cancer subtypes for which targeting individual somatic genetic alterations has not translated into therapeutic success, including triple-negative breast cancer [TNBC, which lacks expression of estrogen receptor α (ER), progesterone receptor (PR), and HER2] and ovarian cancer (OVCA). TNBC comprises 10-15% of BRCA cases, and ~1/3 of the patients (~50,000-75,000/year worldwide) treated for early-stage TNBC later experience recurrence (1,2). Most of the ~239,000 new OVCA cases/year worldwide ultimately recur, and OVCA has the highest mortality rate among female cancers (3). The current standard treatments for both diseases in the primary and metastatic settings include anthracyclines, platinum agents, and microtubule-targeted chemotherapies (4,5), highlighting a need to discover new tumor-targeted therapeutic opportunities in these cancer subtypes. Deficiencies in homologous recombination-mediated DNA repair, such as those caused by germline mutations in BRCA1 or BRCA2 in ~10% of TNBC and OVCA, sensitize cancer cells to poly[adenosine diphosphate (ADP)-ribose] polymerase (PARP) inhibitors (6,7), which are now approved for these cancers in patients with germline BRCA1/2 alterations. However, advanced TNBC and OVCA typically develop resistance to all approved therapies.
Two obstacles to the development of effective tumor-targeted therapies for TNBC and OVCA have been A) heterogeneity within cancer subtypes, and B) intrinsic drug resistance. TNBC and OVCA may contain as many as 6 and 4 molecular subtypes, respectively (8–10). Thus, development of a broadly effective “pan-TNBC therapy” or “pan-OVCA therapy” is unlikely, and dissection of oncogenic pathways within subgroups of TNBC and OVCA to identify therapeutic targets is warranted. BRCA and OVCA have been shown to have similar (epi)genetic and transcriptional profiles (11,12), which led us to hypothesize that analyzing these two cancer types as a single cohort may reveal novel molecularly identifiable mixed subgroups that are uniquely sensitive to certain drugs.
Materials and Methods
Clustering of gene and (phospho)protein expression data
Robust Multi-array Average (RMA)-normalized gene expression data for 1,074 cancer cell lines were downloaded from the Cancer Cell Line Encyclopedia (CCLE), and for 623 cancer cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) database (13,14). Morpheus software (Broad Institute) was used to collapse gene expression data to one probe set per gene using a maximum-mean collapsing strategy (15).
Level 4 normalized expression data from reverse-phase protein arrays (RPPA) for 452 (phospho)proteins across 651 cell lines were downloaded from the MD Anderson Cell Lines Project (MCLP), and were filtered manually using a complete-case-analysis approach (16).
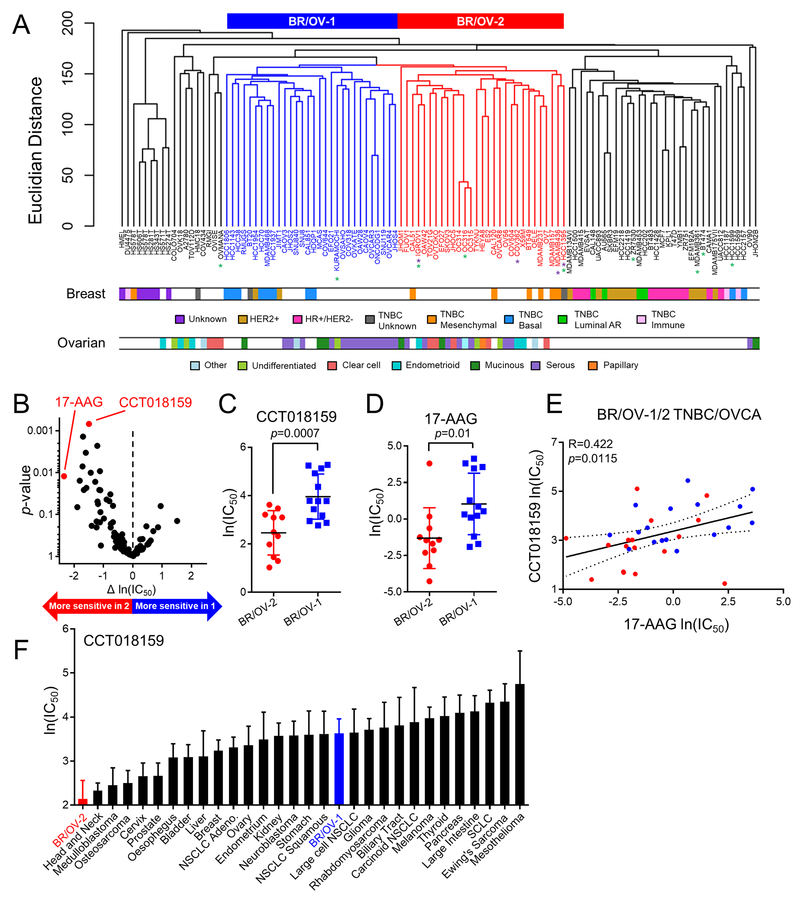
Hierarchical clustering (Euclidean distance) of gene and (phospho)protein expression profiles from BRCA and OVCA cell lines was performed using package ‘gplots’, and heatmaps and dendrograms were generated with R software (17). We identified two mixed subgroups containing primarily triple-negative BRCA and OVCA cell lines, termed BR/OV-1 and -2 (Fig. 1A).
Fig. 1.
Clustering of breast and ovarian cancer cell lines reveals a mixed subgroup with sensitivity to Hsp90 inhibition. (A) Hierarchical clustering of BRCA and OVCA cell lines as a single cohort. BRCA and OVCA subtype designations were obtained from refs. (10,47,48). Mixed subgroups of BRCA and OVCA cell lines are indicate by blue (BR/OV-1) and red (BR/OV-2) dendrograms, respectively. Purple and green asterisks denote cell lines with BRCA1 or BRCA2 mutations, respectively. (B) Volcano plot comparing the sensitivity of BR/OV-1 vs. -2 cell lines to 99 drugs in GDSC database v5. Each point represents one drug. p-values were calculated by two-tailed t-test comparing ln(IC50) of BR/OV-1 vs. -2 cell lines. (C/D) t-test comparison of ln(IC50) for CCT018159 and 17-AAG between BR/OV-1 vs. -2 cells. Horizontal lines represent mean ± SD. (E) Sensitivity data for the Hsp90 inhibitors CCT018159 and 17-AAG were correlated for BR/OV-1/2 cell lines by Pearson’s method. (F) Comparison of CCT018159 sensitivity of BR/OV-1/2 subgroups and cell lines from all other lineages in GDSC. Cell lines in BR/OV-1/2 subgroups were also included in the Breast or Ovarian subtypes as appropriate. Data are shown as mean + SD.
Generation and validation of a BR/OV-1/2 gene expression classifier
BRCA and OVCA cell lines (Table S1) were assigned to the BR/OV-1 or -2 subgroup based on CCLE gene expression data (Fig. 1A). Differentially expressed genes between BR/OV-1 vs. -2 cell lines in the CCLE dataset were used to generate a BR/OV-1/2 gene expression classifier using two-sided t-test and Bonferroni multiple-comparison adjusted p≤0.05. The t-statistic was calculated using the formula ,where and s are mean and standard deviation, respectively. The classifier was applied to GDSC gene expression data, and clustering of cell lines as BR/OV-1 and -2 was validated.
Support vector machine (SVM) regression (SVR) was used to classify cell lines as BR/OV-1 or -2 in the GDSC gene expression datasets using genes from the BR/OV-1/2 classifier as features. One hundred iterations of Monte Carlo cross-validation were implemented to evaluate model performance: half of cell lines were randomly selected to train the classifier, which was then used to predict BR/OV-1 or -2 status in the remaining cell lines. After cross-validation, model accuracy was evaluated by calculating the Area under the Receiver Operating Characteristic Curve. One hundred iterations of Monte Carlo cross-validation were then performed 10,000 times using cell lines randomly assigned to BR/OV-1 or -2 subgroups to generate a p-value.
Drug sensitivity database analyses
BRCA and OVCA cell lines in GDSC were classified as BR/OV-1 or -2 based on CCLE gene expression data. Sensitivity of BR/OV-1 vs. -2 cell lines [n=24; ln(IC50)] was compared in response to 99 drugs (GDSC v5) to generate p-values by two-sided t-test adjusted for multiple comparisons using the Benjamini-Hochberg FDR procedure. As a post hoc validation analysis, sensitivity of BR/OV-1 vs. -2 cell lines [n=36; area under the curve (AUC)] was compared in response to 211 drugs (GDSC v7).
Ovarian cancer xenograft studies
These studies were approved by the Dartmouth College IACUC and performed on-site. Luciferase-expressing OVCA cells (106) were injected i.p. into 4-wk-old female NOD-scid/IL2Rγ−/− (NSG) mice (obtained from Norris Cotton Cancer Center Mouse Modeling Shared Resource). Tumor burden was serially measured by bioluminescence imaging. When bioluminescent flux reached ≥103 photons/second, mice were randomized to twice weekly treatment by intraperitoneal injection of AT13387 [70 mg/kg; provided by Astex Therapeutics, Ltd.] or vehicle (n=6-8 mice/group). For molecular analyses, COV644 tumors were harvested 24 h after drug treatment on Day 36, and were snap-frozen for immunoblot analysis, or formalin-fixed and paraffin-embedded (FFPE) for histologic analyses. Due to near-complete tumor regression in most AT13387-treated mice bearing JHOC5 xenografts, half of the vehicle-treated mice at Day 36 were treated with AT13387 for 24 h prior to harvesting of tumors to provide specimens from vehicle- and AT13387-treated mice.
Breast cancer PDX studies
Expression levels of the 275 genes in the BR/OV-1/2 gene expression classifier were extracted from whole-transcriptome RNA sequencing data (complete dataset being reported elsewhere) obtained from 88 BRCA PDX models from 20 patients (18). Z-scores were generated for each gene across the 88 models and weighted based on the gene expression fold-change from the classifier. The BR/OV-1/2 Subgroup Score for each PDX model represents the difference between the average weighted z-score for BR/OV-2 genes vs. the average weighted z-score for BR/OV-1 genes.
These studies were approved by the Mayo Clinic IACUC and performed on-site. PDX tumor fragments were s.c. implanted into the flanks of 6-8-week-old female NOD-scid (NOD.CB17-Prkdcscid/NCrCrl) mice (Charles River Labs). Tumor dimensions were measured twice weekly using calipers (volume = length x width2/2). When average tumor volume reached ~200 mm3, mice were randomized to treatment with AT13387 or vehicle (n=9-13 mice/group). Tumors were harvested after 4-8 wk of treatment.
Statistical analyses
For comparisons of cell growth, ln(IC50), AUC, mitochondrial depolarization, and gene and protein expression levels, data were analyzed by two-tailed t-test. For IHC and TUNEL assays, data were analyzed by ANOVA with Bonferroni multiple comparison-adjusted post hoc testing between groups. To estimate progression/regression of tumors, the following linear mixed model was employed: log10(tumor volumeit) = ai + b*t + eit, where i represents the ith mouse, t represents the time of tumor volume measurement, ai represents the mouse-specific log tumor volume at baseline (t=0), slope b represents rate of tumor volume growth (or reduction), and eit represents deviation of measurements from the model over time (19). The variance of ai is interpreted as mouse heterogeneity, and b*loge(10)*100 estimates percent tumor volume increase per wk. The computation was carried out in R using function ‘lme’ from library ‘nlme.’ Treatment groups were compared using Z-test for slopes with standard error derived from lme.
Additional details are provided in Supplemental Methods.
Results
Transcriptomic analysis of breast and ovarian cancer cell lines reveals mixed BRCA/OVCA subgroups.
An overview of our experimental approach is presented in Fig. S1. We first performed hierarchical clustering of gene expression profiles from 63 BRCA cell lines from the Cancer Cell Line Encyclopedia (CCLE) (13). BRCA cell lines mainly clustered by receptor status: receptor-positive lines (ER+, HER2+, and Luminal Androgen Receptor (AR)-positive TNBC) clustered separately from receptor-negative TNBC lines (Fig. S2A). Similar analysis of 47 OVCA cell lines revealed only loose clustering by histologic subtype (e.g., clear cell, serous) (Fig. S2B). Integrated hierarchical clustering of both BRCA and OVCA cell lines as a single cohort revealed two novel mixed clusters containing predominantly OVCA and receptor-negative TNBC; these subgroups were termed BR/OV-1 and -2 (Fig. 1A). The strength of the clusters was confirmed using the Partitioning Around Medoids (PAM) algorithm (k=4; Rand Index = 0.79; Fig. S3). Clusters were further verified by multiscale bootstrap resampling (p=0.10).
Drug sensitivity analysis of BR/OV-1 and -2 TNBC/OVCA cells.
Drug sensitivity (IC50) for 99 compounds was compared between subgroups of BRCA and OVCA cell lines in the Genomics of Drug Sensitivity in Cancer (GDSC) database v5 (14) (Fig. 1B and Table S2). Since mutations in BRCA1 or BRCA2 are associated with sensitization to agents targeting DNA repair [e.g., PARP inhibitors (7)], and BRCA1/2 mutations are more frequent among BR/OV-2 cell lines (Fig. 1A), BRCA1/2-mutant cell lines were excluded from these analyses to focus on cancer subgroups lacking known targetable alterations. We assessed sensitivity of 13 BR/OV-1 cell lines and 11 BR/OV-2 cell lines to the 99-compound panel. Among the top 8 drugs with significantly different ln(IC50) values between BR/OV-1 vs. -2 cells, two Hsp90i (CCT018159 and 17-AAG) were more effective against BR/OV-2 cells (t-test p=0.0007 and p=0.01, respectively; FDR-adjusted p=0.067 and p=0.149, respectively; Fig. 1C/D and Table S2). However, a third Hsp90i (AUY922) was not significantly more effective in either subgroup, possibly due to differences in cellular sensitivity to structurally distinct HSP90i across cell lines (20). These findings were confirmed using AUC sensitivity data for CCT018159 and 17-AAG in the updated v7 of GDSC with 211 drugs tested in 19 BR/OV-1 and 17 BR/OV-2 cell lines (p=0.002 and p=0.007, respectively; Fig. S4). Given the correlation between CCT018159 and 17-AAG drug sensitivity profiles across BR/OV-1/2 cell lines (Fig. 1E), the >20-year history of clinical data demonstrating limited efficacy of Hsp90i in unselected populations of cancer patients (21,22), and the lack of a biomarker to identify tumors likely to respond to Hsp90i, we focused on the therapeutic potential of Hsp90i for the BR/OV-2 subgroup of BRCA/OVCA in functional validation studies.
To test the specificity of the Hsp90i sensitivity phenotype, the ln(IC50) values of BR/OV-1 and -2 cell lines were compared to those of all other histologic cancer subtypes represented in GDSC. BR/OV-2 cells were more sensitive to CCT018159 than any other established cancer subtype (Fig. 1F), making the BR/OV-2 subgroup of BRCA/OVCA an attractive candidate for the development of Hsp90-directed therapeutics.
Generation of a BR/OV-1/2 gene expression classifier.
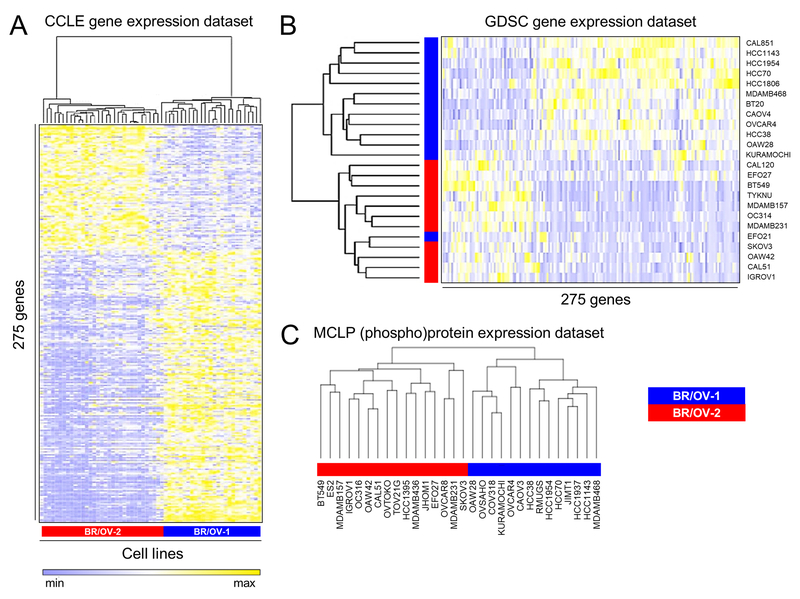
Genes differentially expressed (adjusted p≤0.05) between BR/OV-1 cell lines (10 BRCA and 19 OVCA) and BR/OV-2 cell lines (7 BRCA and 22 OVCA) were determined using transcriptomic data from CCLE, which yielded a 275-gene classifier (Fig. 2A and Table S3). Gene Ontology (GO) classification of these genes indicated enrichment for growth/proliferation-related phenotypes in the BR/OV-2 subgroup, and enrichment for epithelial phenotypes (e.g., cell-cell adhesion) in the BR/OV-1 subgroup (Table S4). Prior studies identified mesenchymal-like and epithelial/basal-like subtypes of TNBC and OVCA based on gene expression profiles (8,9,23–25). Lehmann et al. assigned 6/7 BR/OV-2 TNBC cell lines to mesenchymal or mesenchymal stem-like subtypes (8). However, a reanalysis using the TNBCtype tool (26) on the same BRCA gene expression datasets, and using CCLE and GDSC gene expression datasets, did not indicate significant enrichment for mesenchymal-like TNBC cell lines in the BR/OV-2 subgroup (Table S5). Similarly, we found only minimal overlap between BR/OV-2-upregulated genes and published mesenchymal gene signatures (Table S6). Furthermore, a prior report classified 10/20 OVCA cell lines of the BR/OV-2 subgroup as mesenchymal-like, while the remaining 10 cell lines were classified as epithelial or stem-like (9). Thus, the BR/OV-1 and -2 subgroups represent unique mixed groups within BRCA and OVCA.
Fig. 2.
Generation of gene and (phospho)protein expression classifiers that predict BR/OV-1 vs. -2 status. (A) Genes differentially expressed between BR/OV-1 vs. -2 cells were determined using CCLE data to derive a classifier. (B) The BR/OV-1/2 classifier was validated by application to the GDSC gene expression dataset. (C) Hierarchical clustering of RPPA data. Blue and red stripes denote respective BR/OV-1 and -2 assignments from Fig. 1A.
The ability of the BR/OV-1/2 classifier (Table S3) to create two mixed subgroups of BRCA/OVCA was tested using clustering of independent gene expression data from GDSC (14). Based on the BR/OV-1/2 assignments from Fig. 1A (from CCLE data), one GDSC cluster contained 12 BR/OV-1 cell lines, and the other cluster contained 11 BR/OV-2 cell lines and one BR/OV-1 line, demonstrating that the classifier distinguished BR/OV-1 from -2 cell lines with 96% (23/24) accuracy in an independent gene expression dataset (Fig. 2B). We further evaluated the robustness of this classifier in a supervised setting using support vector machine (SVM) regression (SVR) to predict BR/OV-1/2 status using expression of genes in the classifier as features. After Monte Carlo cross-validation, we found that the classifier could distinguish BR/OV-1 from -2 cell lines with high accuracy in the GDSC dataset (AUC=0.87; p=5.51×10−89; Fig. S5).
To confirm that these BRCA/OVCA subgroups also differed at the level of (phospho)protein expression, hierarchical clustering was performed for BR/OV-1 and -2 cell lines (n=30) using RPPA data from the MCLP dataset (16). Clustering confirmed classification of cell lines into BR/OV-1 and -2 subgroups with 100% accuracy (Figs. 2C and S6, and Table S7). TMT-based mass spectrometry was also performed to quantitatively profile the proteomes of 3 BR/OV-1 and 3 BR/OV-2 TNBC cell lines. Analysis revealed 588 proteins significantly differentially expressed between BR/OV-1 vs. -2 cells (p≤0.05; Table S8 and Fig. S7A/B). Differentially expressed proteins were classified by GO, revealing functions classically associated with epithelial (e.g., epidermis and ectoderm development) or mesenchymal (e.g., transforming growth factor β and bone morphogenetic protein signaling) phenotypes in BR/OV-1 or BR/OV-2 cells, respectively (Fig. S7C/D and Table S9).
BR/OV-2 TNBC and OVCA cells are sensitized to Hsp90 inhibition.
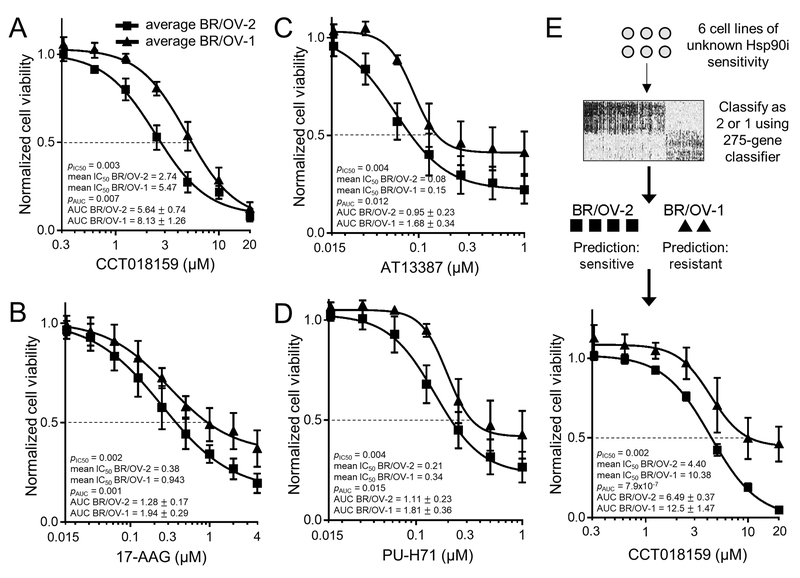
Hsp90i sensitivity was tested in growth experiments using 3 TNBC and 3 OVCA cell lines from each of the BR/OV-1 and -2 subgroups (total of 12 cell lines). BR/OV-2 cells showed overall increased sensitivity to CCT018159 compared to BR/OV-1 cells [pln(IC50)=0.003; Figs. 3A and S8]. The same differential sensitivity was apparent when analyzing cell lines from a single tissue of origin (breast or ovary), indicating that sensitivity to CCT018159 is associated with BR/OV-1/2 status and not tissue of origin (Fig. S9).
Fig. 3.
BR/OV-2 TNBC and OVCA cells exhibit sensitization to Hsp90 inhibition. (A-D) Cells were treated with CCT018159, 17-AAG, AT13387, or PU-H71. Relative viable cell numbers were measured after 5 d. Mean of triplicates was used to provide a single value at each dose for each cell line, and these values were averaged across BR/OV-1 or -2 lines [6 lines per subgroup in (A); 4 lines per subgroup in (B-D)]. Data are shown as mean ± SD for each subgroup. ln(IC50) and AUC values between subgroups were compared by t-test. (E) Gene expression profiles of 2 TNBC and 4 OVCA cell lines were used to assign BR/OV-1/2 status. Cells were treated and analyzed as in (A).
To confirm that differential sensitivity to Hsp90i was not compound-specific, we similarly tested sensitivity of 8 cell lines (2 per BR/OV-1/2 subgroup and tissue of origin) to 3 other Hsp90i: 17-AAG, AT13387 (onalespib), and PU-H71. BR/OV-2 cells consistently showed increased sensitivity to these Hsp90i compared to BR/OV-1 cells (all pln(IC50)<0.01; Fig. 3B–D).
We then selected 2 TNBC and 4 OVCA cell lines for which gene expression profiles were available (from CCLE), but drug sensitivity data were not (i.e., cell lines were not in GDSC v5). A BR/OV-1/2 Subgroup Score was generated for each cell line, classifying 4 of these cell lines (OVTOKO, JHOC5, MDA-MB-436, ES2) as BR/OV-2, and 2 lines (COV644, HDQ-P1) as BR/OV-1. Growth assays revealed that the cell lines classified as BR/OV-2 are more sensitive to Hsp90i than those classified as BR/OV-1, confirming the ability of the BR/OV-1/2 classifier to predict Hsp90i sensitivity in TNBC and OVCA cell lines (pln(IC50)=0.002; Figs. 3E and S10).
Hsp90i induces Bim- and PUMA-dependent intrinsic apoptosis in BR/OV-2 but not BR/OV-1 TNBC and OVCA cells.
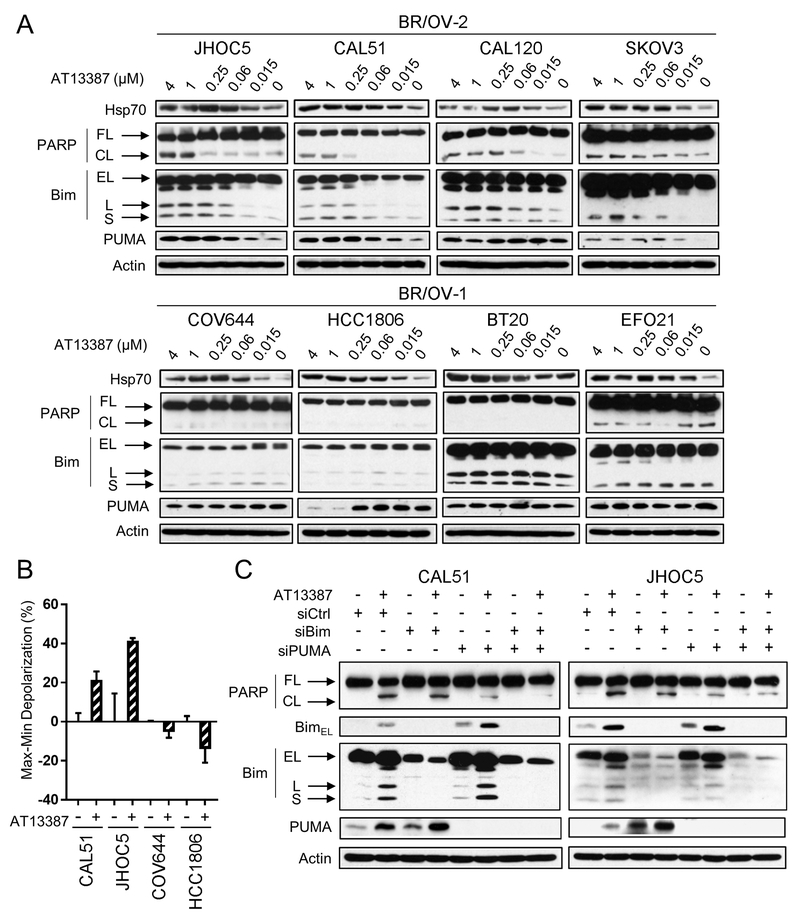
To discover mechanisms underlying differential Hsp90i sensitivity between BR/OV-1 and -2 cells, 4 BR/OV-1 and 4 BR/OV-2 cell lines (balanced for breast and ovarian origin) were treated with a dose range of AT13387 [chosen for further studies for its clinical potential (27)]. Compensatory upregulation of Hsp70, inhibition of major signal transduction pathways including PI3K/AKT/mTOR, MEK/ERK, STAT1/3, and TGFβ, and downregulation of cell cycle proteins, which are known effects of Hsp90i (28–32), occurred consistently in both BR/OV-1 and -2 cells in response to AT13387 (Figs. S11–S12). The most notable differences between BR/OV-1 and -2 cells in signaling response to AT13387 occurred at the level of apoptosis, where only BR/OV-2 cells demonstrated drug-induced increases in PARP cleavage, which coincided with upregulation of the pro-apoptotic BH3-only proteins Bim [particularly the potent BimL and BimS isoforms (33)] and PUMA (Figs. 4A, S12, and S13). Differential apoptotic sensitivity to Hsp90i was confirmed by Bim BH3 profiling, where AT13387 increased mitochondrial outer membrane depolarization in BR/OV-2 cells, but not BR/OV-1 cells (Fig. 4B).
Fig. 4.
BR/OV-2 TNBC and OVCA cells require Bim and PUMA for apoptotic response to Hsp90 inhibition. (A) Cells were treated ± AT13387 for 24 h. Lysates were analyzed by immunoblot. (B) Cells were treated ± 1 μM AT13387 for 24 h, and Hsp90i-induced mitochondrial membrane depolarization relative to control was calculated. Data are shown as mean of triplicates ± SD. *p<0.05 by t-test. (C) Cells were transfected with siRNA targeting Bim, PUMA, both, or non-silencing control. After 48 h, cells were treated ± 1 μM AT13387 for 24 h. Lysates were analyzed by immunoblot. FL-Full-Length. CL-Cleaved. EL-Extra-Long. L-Long. S-Short.
RNAi-mediated knockdown of Bim, PUMA, and the combination was performed in 2 BR/OV-2 cell lines (CAL51 TNBC cells and JHOC5 OVCA cells) to functionally test whether Bim and/or PUMA modulate Hsp90i-induced apoptosis. Interestingly, knockdown of either Bim or PUMA induced compensatory upregulation of the other (Fig. 4C), which may be due to increased availability of binding sites on anti-apoptotic Bcl-2 family proteins to stabilize BH3-only proteins (34,35). Dual Bim/PUMA knockdown effectively prevented Hsp90i-induced apoptosis in BR/OV-2 cells. These data collectively suggest that BH3-only proteins are differentially regulated between BR/OV-1 and -2 cells in response to Hsp90i, which may underlie differences in drug sensitivity between the BR/OV-1 and -2 subgroups of BRCA and OVCA.
BR/OV-2 tumors are more sensitive to Hsp90 inhibition than BR/OV-1 tumors.
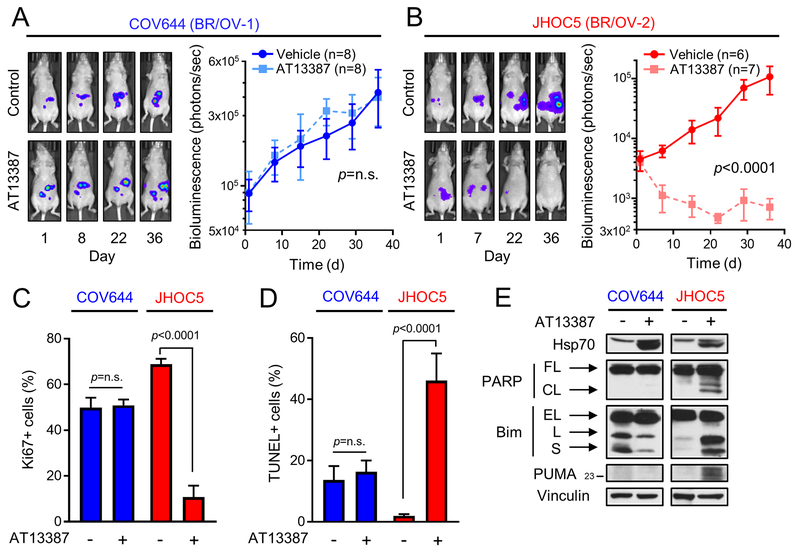
COV644 and JHOC5 OVCA cells were assigned to the BR/OV-1 and -2 subgroups, respectively, based on the BR/OV-1/2 classifier (Fig. 3E). Luciferase-labeled cells were injected intraperitoneally into mice to provide xenograft models of advanced OVCA. Relative tumor burden was serially measured by bioluminescence imaging. When bioluminescent flux reached ≥103 photons/sec, mice were randomized to treatment with AT13387 or vehicle. COV644 (BR/OV-1) tumors were completely resistant to Hsp90i, while JHOC5 (BR/OV-2) tumors were exquisitely sensitive (Figs. 5A/B and S14), confirming the predicted responses of BR/OV-1 and -2 tumors to Hsp90i. AT13387 significantly decreased tumor cell proliferation [assessed by Ki67 immunohistochemistry (IHC)] and increased apoptosis (assessed by TUNEL) in JHOC5 tumors but not COV644 tumors (Figs. 5C/D and S15). Due to small tumor volumes, tumor lysates from each treatment group were pooled for immunoblot analysis. AT13387 blocked Hsp90 activity (assessed by increased Hsp70) in both tumor models, while PARP cleavage and upregulation of Bim and PUMA occurred only in JHOC5 (BR/OV-2) tumors (Fig. 5E).
Fig. 5.
BR/OV-2 OVCA tumors are exquisitely sensitive to Hsp90 inhibition. (A/B) Peritoneal models of advanced OVCA were established using luciferase-labeled OVCA cell lines predicted to be BR/OV-1 (COV644) or BR/OV-2 (JHOC5). Tumor burden was serially measured by bioluminescence imaging. Tumor-bearing mice were randomized to treatment with AT13387 or vehicle. Bioluminescence values were analyzed by linear mixed modeling to compare treatment groups. Exposure-matched images from one representative mouse per group are shown. (C/D) Tumors (n=3/group) were analyzed by Ki67 IHC or TUNEL. Data are shown as mean + SE, and were analyzed by multiple comparison-adjusted Bonferroni post hoc test. (E) Pooled tumor lysates were analyzed by immunoblot. FL-Full-Length. CL-Cleaved. EL-Extra-Long. L-Long. S-Short.
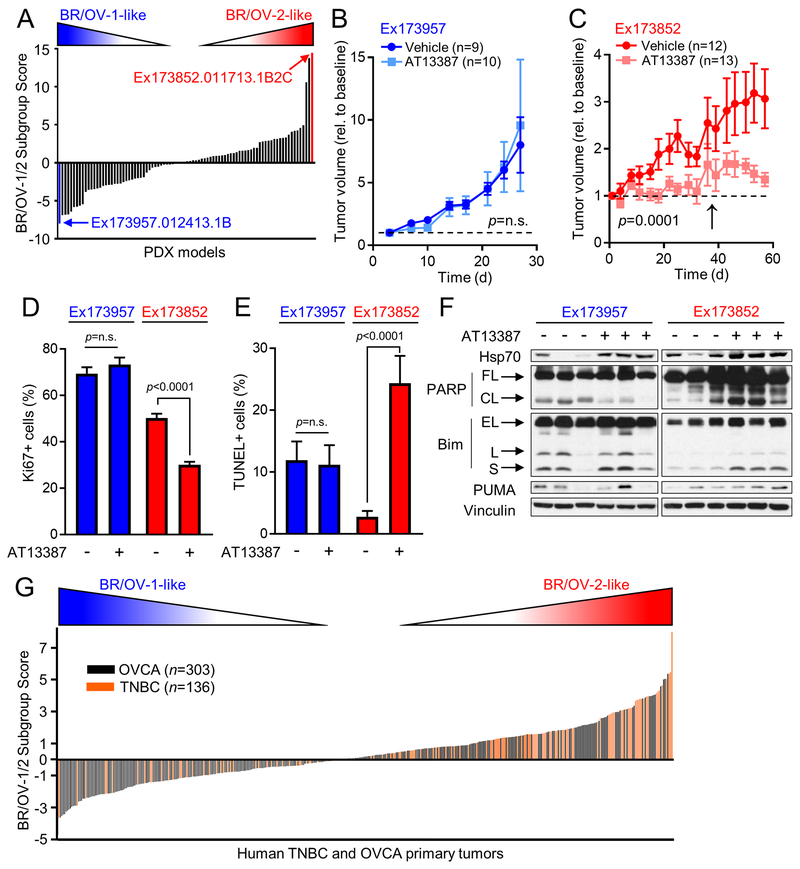
Expression profiles for the 275 genes in the BR/OV-1/2 classifier were then extracted from RNA sequencing data from 88 BRCA PDX models, which were generated from 20 patients enrolled in the BEAUTY study (18). A BR/OV-1/2 Subgroup Score was calculated for each PDX model (Table S10), and the models with the highest score [most BR/OV-2] and the lowest score [most BR/OV-1] were selected for functional analysis (Fig. 6A). Mice bearing subcutaneous tumors were randomized to treatment with AT13387 or vehicle. Ex173957 (BR/OV-1) tumors were insensitive to AT13387, while the growth of Ex173852 (BR/OV-2) tumors was significantly suppressed by AT13387 (Figs. 6B/C and S16). Toxicity (weight loss) was detected in 7/10 mice bearing Ex173852 (BR/OV-2) tumors after one month of drug treatment (Fig. S17), leading to a change in treatment schedule on Day 36 from twice weekly to once weekly in this model that may have blunted treatment efficacy. Such toxicity was not observed in mice bearing Ex173957 (BR/OV-1) PDX tumors, likely due to shorter duration of study (Figs. 6B and S17). In PDX models, AT13387 treatment significantly decreased tumor cell proliferation and increased apoptosis in Ex173852 (BR/OV-2) tumors but not Ex173957 (BR/OV-1) tumors (Figs. 6D/E and S18). Immunoblot analysis of PDX lysates showed that AT13387 blocked Hsp90 activity (assessed by increased Hsp70) in both models, while PARP cleavage and upregulation of Bim and PUMA consistently occurred only in Ex173852 (BR/OV-2) tumors (Fig. 6F).
Fig. 6.
BR/OV-2 TNBC PDX tumors are sensitive to Hsp90 inhibition. (A) The BR/OV-1/2 classifier was used to calculate a BR/OV-1/2 Subgroup Score for each of 88 BRCA PDX models. The models with the highest (BR/OV-2-like, red) and lowest (BR/OV-1-like, blue) Scores were selected for functional testing. (B/C) Tumors were implanted s.c. into mice. Tumor dimensions were serially measured using calipers. Tumor-bearing mice were randomized to treatment as indicated. Tumor volumes were analyzed by linear mixed modeling to compare treatment groups. Arrow in (C) denotes time point when mice were switched from twice-weekly to once-weekly treatment. (D/E) Tumors (n=4/group) were analyzed by Ki67 IHC or TUNEL. Data are shown as mean + SE, and were analyzed by multiple comparison-adjusted Bonferroni post-hoc test. (F) Tumor lysates were analyzed by immunoblot. FL-Full-Length. CL-Cleaved. EL-Extra-Long. L-Long. S-Short. (G) RNA-seq profiles from TNBC and OVCA tumors from TCGA were used to calculate a BR/OV-1/2 Subgroup Score for each tumor.
Homologous recombination deficiency (HRD), often involving mutations in BRCA1 or BRCA2, is known to increase sensitivity to DNA-damaging chemotherapy in BRCA and OVCA (36,37). To rule out HRD-related alterations as a contributor to differences in drug sensitivity between BR/OV-1 and -2 tumors, DNA mutation and copy number data were evaluated for xenograft models. Analysis of common HRD-related genes revealed BRCA1 amplification in the JHOC5 (BR/OV-2) cell line, and PARP1 amplification in the BR/OV-1 PDX model, which are not known to be associated with phenotypes of drug sensitivity or resistance, respectively (Fig. S19).
Finally, RNA sequencing data from TNBC (n=136) and OVCA (n=303 serous cystadenocarcinoma) cases from The Cancer Genome Atlas (TCGA) were used to calculate a BR/OV-1/2 Subgroup Score for each primary tumor. A substantial proportion of TNBC and OVCA tumors had relatively high Scores, suggesting alignment with the BR/OV-2 subgroup (Fig. 6G); such tumors would be predicted to exhibit increased sensitivity to Hsp90i. Based on our finding that 29/41 TNBC/OVCA cell lines with a Score >0 were in the BR/OV-2 like cluster in Fig. 1A, we predict that 71% (95% CI: 55-84%; exact binomial test p=0.012) of primary human TNBC/OVCA cases with a Score >0 are BR/OV-2-like; thus, approximately 49% and 35% of all TNBC and OVCA cases, respectively, are predicted to be BR/OV-2-like.
Discussion
We present an integrated strategy to uncover new therapeutic opportunities in cancer types with limited targetable genetic alterations, such as TNBC and OVCA. To our knowledge, this is the first study to generate gene and protein expression classifiers, and integrate drug sensitivity analyses for molecularly identifiable subgroups that transcend cancer lineage. In so doing, we identified two mixed subgroups of OVCA and BRCA (consisting of mostly TNBC). We discovered that BR/OV-2 subgroup cell lines were sensitized to Hsp90i, which we validated with multiple cell lines and several Hsp90 inhibitors. We developed a gene expression classifier that predicted sensitivity to Hsp90i in cultured cells, cell line-derived xenografts, and PDX models. Hsp90 thus represents a viable therapeutic opportunity in subgroups of TNBC and OVCA. Furthermore, this cancer lineage-transcendent strategy may be applied more broadly to discover other novel cancer subgroups with unique therapeutic vulnerabilities.
Hsp90 is a family of ATP-dependent molecular chaperones consisting of multiple isoforms with high sequence identity that are expressed in different subcellular compartments. The two major isoforms, Hsp90α (stress-inducible, major isoform) and Hsp90β (constitutive, minor isoform), share a conserved amino-terminal ATP-binding domain targeted by most Hsp90 inhibitors (38). BR/OV-1 and -2 cells expressed similar levels of Hsp90α, while BR/OV-1 cells expressed more Hsp90β (Fig. S13); however, an association between Hsp90 isoform levels and Hsp90i sensitivity in cancer cells has not been reported. Hsp90 chaperones are involved in the stabilization and activation of many client proteins essential for signal transduction and stress responses, including oncogenic proteins such as Src, Bcr/Abl, ErbB receptors, p53, and nuclear hormone receptors (39). As such, Hsp90 has been recognized as an important target for anti-cancer drug discovery. Most Hsp90 inhibitors have been developed to inhibit ATP binding, which decreases the affinity of Hsp90 for client proteins and results in their subsequent ubiquitination and degradation (40). Although Hsp90 is expressed in most mammalian cells, cancer cells are frequently more sensitive to Hsp90i than non-cancer cells, providing a therapeutic window and further highlighting the clinical appeal of Hsp90i (41). Several Hsp90 inhibitors have been tested clinically, but results have been disappointing with modest response rates in unselected patient populations (21,22). Hsp90 inhibitors are being tested in ongoing clinical trials that include patients with BRCA or OVCA (e.g., NCT02474173, NCT01962948); our findings suggest that such trials evaluating broad patient populations will have mixed success due to differential Hsp90i sensitivity between cancers that are BR/OV-2-like vs. other, which could be tested through retrospective molecular analysis of tumors. Future Hsp90i clinical trials may benefit from prospectively screening for BR/OV-2-like TNBC or OVCA to identify patients more likely to benefit.
In agreement with prior observations (29,32), Hsp90i induced compensatory upregulation of Hsp70, inhibition of major signal transduction pathways (e.g., PI3K/AKT/mTOR, MEK/ERK, STAT, TGFβ), and downregulation of cell cycle proteins in both BR/OV-1 and -2 cells (Figs. 4A, 5E, 6F, and S11–S13); thus, such effects do not define the mechanism of enhanced response to Hsp90i seen in BR/OV-2 cells. On the other hand, we found major differences in apoptosis, where BR/OV-2 cells and tumors showed Hsp90i-induced apoptosis while BR/OV-1 cells and tumors did not (Figs. 4, 5D/E, 6E/F, and S11–S13). Hsp90i-induced apoptosis coincided with upregulation of Bim and PUMA in BR/OV-2 cells but not BR/OV-1 cells, implicating differential response to Hsp90i at the level of pro-apoptotic BH3-only protein effectors. To confirm that these BH3-only proteins mediate response to Hsp90i, we showed that knockdown of Bim and PUMA abrogated apoptosis in BR/OV-2 cells (Fig. 4C). Bim and PUMA mediate apoptosis by A) sequestering anti-apoptotic Bcl-2 family proteins (e.g., Bcl-2, Bcl-xL) away from Bax and Bak, which prevents Bax/Bak pore formation in the mitochondrial outer membrane (MOM) and cytochrome C release, and/or B) binding Bax/Bak directly in the MOM (42,43). Mechanisms regulating Bim and PUMA expression have been partly elucidated. Bim is destabilized by ERK phosphorylation, and inhibition of MEK/ERK signaling leads to an abundance of Bim (44). PUMA is downregulated by PI3K/AKT/FOXO signaling, and PI3K/AKT inhibition leads to nuclear translocation of FOXO transcription factors to drive PUMA transcription (45,46). Our data show differential modulation of Bim and PUMA in BR/OV-2 vs. -1 cells despite consistent downregulation of MEK/ERK and PI3K/AKT upon Hsp90i, suggesting additional uncharacterized mechanisms regulating Bim and PUMA that warrant further investigation.
Our approach to identify novel multi-lineage cancer subgroups and subgroup-specific therapeutic vulnerabilities has broad implications for precision oncology in subtypes that do not have obvious targetable genetic alterations, such as TNBC and OVCA without germline BRCA1/2 mutations. Our study is proof of the concept that transcriptional/protein classifier generation and drug sensitivity analyses in cell lines could provide the basis for future umbrella clinical trials where patients will get different drugs depending on tumor expression-based predictors of drug sensitivity. This approach may prove especially useful in trials involving drug targets without an obvious target patient population, as is the case for current Hsp90i trials. Furthermore, our initial results warrant additional post hoc analyses of human tumors from completed Hsp90i clinical trials, which may provide further insight into the clinical utility of a transcriptional/protein classifier to predict drug sensitivity.
Supplementary Material
Statement of Translational Relevance.
Precision oncology seeks to integrate molecular characteristics of a patient’s tumor to tailor anti-cancer therapy. While there exists an array of instances in which genomic DNA alterations in a drug target have provided therapeutic opportunities, there are many cancer types that do not have obvious targetable genomic DNA alterations. We demonstrate that integrated analyses of transcriptional and drug sensitivity data from cancer cell lines can be used to reveal therapeutic vulnerabilities that transcend cell lineage. We discovered that a novel, transcriptionally-definable mixed subgroup containing primarily triple-negative breast and ovarian cancer cells is vulnerable to treatment with heat shock protein 90 inhibitors (Hsp90i). Furthermore, a gene expression signature was used to predict sensitivity to Hsp90i in cell line- and patient-derived xenografts. These findings suggest that transcriptional signatures may be useful to identify cancer subgroups that transcend lineage and are sensitive to a given class of therapeutics.
Acknowledgements:
We thank Alan Eastman for sharing antibodies and suggestions, Jonathan Marotti for pathology interpretation, and Norris Cotton Cancer Center Shared Resources for assistance (Mouse Modeling, Genomics and Molecular Biology, Pathology, Microscopy).
Financial Support: NIH (F30CA216966 to KS; R01CA200994, R01CA211869, and a pilot grant via P20GM113132 to TWM; R35GM119455 to ANK; Dartmouth College Norris Cotton Cancer Center Support Grant P30CA023108; Mayo Clinic Breast SPORE P50CA116201-9 to MPG, LW, KRK; Mayo Clinic Cancer Center Support Grant P50CA015083); American Cancer Society (RSG-13-292-01-TBE to TWM); Susan G. Komen (CCR1533084 to TWM); Mayo Clinic Center for Individualized Medicine; Nadia’s Gift Foundation; John P. Guider; The Eveleigh Family; George M. Eisenberg Foundation for Charities; Prospect Creek Foundation; Randy Shaver Cancer Research and Community Fund.
Footnotes
Conflicts of interest: none.
References Cited
- 1.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med 2010;7(5):e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzger-Filho O, Sun Z, Viale G, Price KN, Crivellari D, Snyder RD, et al. Patterns of Recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: results from international breast cancer study group trials VIII and IX. J Clin Oncol 2013;31(25):3083–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136(5):E359–86. [DOI] [PubMed] [Google Scholar]
- 4.Cardoso F, Harbeck N, Fallowfield L, Kyriakides S, Senkus E, Group EGW. Locally recurrent or metastatic breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2012;23 Suppl 7:vii11–9. [DOI] [PubMed] [Google Scholar]
- 5.Ledermann JA, Raja FA, Fotopoulou C, Gonzalez-Martin A, Colombo N, Sessa C, et al. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24 Suppl 6:vi24–32. [DOI] [PubMed] [Google Scholar]
- 6.Buys SS, Sandbach JF, Gammon A, Patel G, Kidd J, Brown KL, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 2017;123(10):1721–30. [DOI] [PubMed] [Google Scholar]
- 7.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434(7035):917–21. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121(7):2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan TZ, Miow QH, Huang RY, Wong MK, Ye J, Lau JA, et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med 2013;5(7):1051–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longacre M, Snyder NA, Housman G, Leary M, Lapinska K, Heerboth S, et al. A Comparative Analysis of Genetic and Epigenetic Events of Breast and Ovarian Cancer Related to Tumorigenesis. Int J Mol Sci 2016;17(5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483(7391):603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41(Database issue):D955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, et al. Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinformatics 2011;12:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Zhao W, Akbani R, Liu W, Ju Z, Ling S, et al. Characterization of Human Cancer Cell Lines by Reverse-phase Protein Arrays. Cancer Cell 2017;31(2):225–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R_Core_Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; (Vienna, Austria; wwwR-projectorg) 2019 [Google Scholar]
- 18.Goetz MP, Kalari KR, Suman VJ, Moyer AM, Yu J, Visscher DW, et al. Tumor Sequencing and Patient-Derived Xenografts in the Neoadjuvant Treatment of Breast Cancer. J Natl Cancer Inst 2017;109(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demidenko E Mixed Models: Theory and Applications with R. Wiley; (Hoboken, NJ: ) 2013 [Google Scholar]
- 20.Mayor-Lopez L, Tristante E, Carballo-Santana M, Carrasco-Garcia E, Grasso S, Garcia-Morales P, et al. Comparative Study of 17-AAG and NVP-AUY922 in Pancreatic and Colorectal Cancer Cells: Are There Common Determinants of Sensitivity? Transl Oncol 2014;7(5):590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jhaveri K, Miller K, Rosen L, Schneider B, Chap L, Hannah A, et al. A phase I dose-escalation trial of trastuzumab and alvespimycin hydrochloride (KOS-1022; 17 DMAG) in the treatment of advanced solid tumors. Clinical Cancer Research 2012;18(18):5090–8. [DOI] [PubMed] [Google Scholar]
- 22.Modi S, Stopeck A, Linden H, Solit D, Chandarlapaty S, Rosen N, et al. HSP90 Inhibition Is Effective in Breast Cancer: A Phase II Trial of Tanespimycin (17-AAG) Plus Trastuzumab in Patients with HER2-Positive Metastatic Breast Cancer Progressing on Trastuzumab. Clinical Cancer Research 2011;17(15):5132–9. [DOI] [PubMed] [Google Scholar]
- 23.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 2015;21(7):1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 2008;14(16):5198–208. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474(7353):609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, et al. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform 2012;11:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro GI, Kwak E, Dezube BJ, Yule M, Ayrton J, Lyons J, et al. First-in-human phase I dose escalation study of a second-generation non-ansamycin HSP90 inhibitor, AT13387, in patients with advanced solid tumors. Clin Cancer Res 2015;21(1):87–97. [DOI] [PubMed] [Google Scholar]
- 28.Graham B, Curry J, Smyth T, Fazal L, Feltell R, Harada I, et al. The heat shock protein 90 inhibitor, AT13387, displays a long duration of action in vitro and in vivo in non-small cell lung cancer. Cancer Sci 2012;103(3):522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth T, Van Looy T, Curry JE, Rodriguez-Lopez AM, Wozniak A, Zhu M, et al. The HSP90 inhibitor, AT13387, is effective against imatinib-sensitive and -resistant gastrointestinal stromal tumor models. Mol Cancer Ther 2012;11(8):1799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White PT, Subramanian C, Zhu Q, Zhang H, Zhao H, Gallagher R, et al. Novel HSP90 inhibitors effectively target functions of thyroid cancer stem cell preventing migration and invasion. Surgery 2016;159(1):142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazenby M, Hills R, Burnett AK, Zabkiewicz J. The HSP90 inhibitor ganetespib: A potential effective agent for Acute Myeloid Leukemia in combination with cytarabine. Leuk Res 2015;39(6):617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zitzmann K, Ailer G, Vlotides G, Spoettl G, Maurer J, Goke B, et al. Potent antitumor activity of the novel HSP90 inhibitors AUY922 and HSP990 in neuroendocrine carcinoid cells. Int J Oncol 2013;43(6):1824–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. Embo J 1998;17(2):384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurtulus S, Tripathi P, Moreno-Fernandez ME, Sholl A, Katz JD, Grimes HL, et al. Bcl-2 allows effector and memory CD8+ T cells to tolerate higher expression of Bim. Journal of immunology 2011;186(10):5729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callus BA, Moujallad DM, Silke J, Gerl R, Jabbour AM, Ekert PG, et al. Triggering of Apoptosis by Puma Is Determined by the Threshold Set by Prosurvival Bcl-2 Family Proteins. J Mol Biol 2008;384(2):313–23. [DOI] [PubMed] [Google Scholar]
- 36.Yang D, Khan S, Sun Y, Hess K, Shmulevich I, Sood AK, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA 2011;306(14):1557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res 2016;22(15):3764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer MP, Le Breton L. Hsp90: breaking the symmetry. Mol Cell 2015;58(1):8–20. [DOI] [PubMed] [Google Scholar]
- 39.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer 2005;5(10):761–72. [DOI] [PubMed] [Google Scholar]
- 40.Blagg BS, Kerr TD. Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med Res Rev 2006;26(3):310–38. [DOI] [PubMed] [Google Scholar]
- 41.Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 2003;425(6956):407–10. [DOI] [PubMed] [Google Scholar]
- 42.Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 2013;152(3):519–31. [DOI] [PubMed] [Google Scholar]
- 43.Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell 2000;103(6):839–42. [DOI] [PubMed] [Google Scholar]
- 44.Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK½ signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 2003;278(21):18811–6. [DOI] [PubMed] [Google Scholar]
- 45.You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M, et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. The Journal of experimental medicine 2006;203(7):1657–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amente S, Zhang J, Lavadera ML, Lania L, Avvedimento EV, Majello B. Myc and PI3K/AKT signaling cooperatively repress FOXO3a-dependent PUMA and GADD45a gene expression. Nucleic Acids Res 2011;39(22):9498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollestelle A, Nagel JH, Smid M, Lam S, Elstrodt F, Wasielewski M, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res Treat 2010;121(1):53–64. [DOI] [PubMed] [Google Scholar]
- 48.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006;10(6):515–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.