Abstract
Introduction
The role of Escherichia coli in the pathogenesis of inflammatory bowel disease (IBD) is still controversial. The study aimed to investigate the pathotypes and the phylogenetic groups of E. coli in Egyptian patients with IBD in an attempt to find an association between any type or group with the severity of the disease.
Methods
Thirty ulcerative colitis (UC), 30 Crohn’s disease (CD), and 20 control subjects with normal colonoscopy were included in a cross-sectional study. E. coli were isolated from stool samples by culture. Eight intestinal virulence genes coding for diarrheagenic E. coli were investigated using multiplex PCR. Phylogenetic grouping was performed by a triplex PCR. Antimicrobial susceptibility of all isolates was done using disc diffusion method.
Results
Enteroaggregative E. coli (EAEC) were identified in 25% (15/60) of IBD cases and in none of the controls (p=0.013). Out the 60 IBD cases, 30 (50%) were from phylogenetic group B2. No statistically significant differences in the distribution of E. coli phylogenetic groups were found between study groups. However, 80% of EAEC were assigned to group B2 and D. No statistically significant differences in calprotectin level or in disease severity scores were reported between the four phylogenetic groups. E. coli from both UC and CD patients showed a high rate of resistance to most antimicrobials when compared to the control group.
Conclusions
The identification of EAEC belonging mainly to group B2 and D in IBD cases may indicate the importance of this pathotype in the pathogenesis of IBD in Egyptian patients.
Keywords: E. coli, enteroaggregative, IBD, phylogenetic group
Introduction
Inflammatory bowel disease (IBD) constitutes a group of inflammatory gastrointestinal relapsing and remitting clinical conditions, of which Crohn’s disease (CD) and ulcerative colitis (UC) are the most common forms. The pathogenesis of IBD remains controversial. Both genetic and environmental factors are involved.1
Although Escherichia coli are common colonizers of the human gastrointestinal tract (GIT), some intestinal E. coli pathotypes have acquired virulence factors, increasing their ability to cause GIT disease.2 Other IBD-associated E. coli isolates display strong adherence and invasion properties, and usually do not bear the virulence characteristics of typical E. coli strains and they are referred to as ExPEC (extra-intestinal pathogenic E. coli).3 E. coli of IBD demonstrated multidrug-resistance4 and mostly belonged to B2 and D phylogenetic groups.5 However, it is not known whether E. coli are involved in the early inflammatory process or are secondary in the disease progression of IBD.6
This study aimed to determine the pathotypes and the phylogenetic groups of E. coli stool isolates from IBD in a cohort of Egyptian patients, as well as to assess a possible association between the phylogenetic group and the severity of the disease. Additionally, the study aimed to evaluate the antimicrobial susceptibility of such isolates in order to avoid treatment failure.
Methods
This cross-sectional study included 80 subjects (30 consecutive subjects fulfilling the diagnosis of UC and 30 subjects fulfilling the diagnosis of CD, as well as 20 consecutive subjects with normal colonoscopic findings) recruited from the outpatient clinic, or admitted to the Internal Medicine Department at Alexandria Main University Hospital (AMUH), Alexandria, Egypt, and scheduled for colonoscopy. The study was conducted during a six months period, from July to December 2018. The study subjects were divided into the following groups: Group I included 30 patients with UC divided into 2 subgroups (A: 15 active UC patients, and B: 15 UC inactive patients in remission). Group II included 30 patients with CD divided into 2 subgroups (A: 15 active CD patients and B: 15 CD inactive patients in remission). The diagnosis of UC or CD was based on medical history, clinical presentation, laboratory, endoscopic and histopathological investigations. Group III was the control group. Patients with other GIT diseases, recent intestinal interventions, infectious diarrhea, sepsis, chronic medical conditions, and those with use of non-steroidal anti-inflammatory drugs or antibiotics in the previous three months were excluded from the study.
A written informed consent was taken from all subjects included in the study. The study protocol was approved by the local ethics committee of Alexandria Faculty of Medicine.
History taking and clinical data collection
All patients were subjected to detailed history taking with emphasis on GIT symptoms as well as symptoms of extra-intestinal manifestations of IBD. Thorough systemic physical examination was also done. UC disease activity was assessed clinically by the Simple clinical colitis activity index (SCCAI),7 while CD activity was assessed by the CD activity index (CDAI).8
Sample collection and transport
Stool samples were collected from all study subjects (patients and controls) in sterile containers and were immediately transferred to AMUH Microbiology Laboratory for processing.
Determination of fecal calprotectin level
Quantitative assessment of fecal calprotectin was performed using ELISA (Calprest NG, Eurospital SpA, Trieste, Italy) according to the manufacturer's instructions. Briefly, fecal samples were homogenized in extraction buffer. The fecal extracts were further diluted before testing. Values >50 mg/kg were considered positive.
Culture of stool samples for isolation of E. coli
An amount equivalent to 10 μL of stool samples was suspended in two mL phosphate buffered saline. The suspension was properly mixed and 10 μL of the resulting suspension was inoculated onto MacConkey's agar (Thermo Fisher, Oxoid, Hampshire, UK) and incubated at 37°C overnight. The plates were examined for E. coli colonies. At least 20 different E. coli colonies were further identified according to standard microbiological biochemical identification methods including sugar fermentation in triple sugar iron, negative citrate, negative urease, motility, positive indole, positive ornithine decarboxylation and positive methyl red tests.9
Detection of intestinal virulence genes of E. coli using multiplex PCR
For extraction of bacterial DNA, bacterial sweep of all confirmed E. coli colonies was inoculated in Luria broth (Thermo Fisher) and incubated overnight at 37°C. This was further diluted 1:40 in sterile water, followed by boiling at 100°C for 20 minutes, and centrifugation at 9500 rpm for 10 minutes. One half μL of the resulting supernatant was used in PCR reactions.
For analysis of E. coli pathotypes [EPEC (enteropathogenic E. coli), EAEC (enteroaggregative E. coli), ETEC (enterotoxigenic E. coli), EHEC (enterohemorrhagic E. coli), STEC (Shiga toxin E. coli), EIEC (enteroinvasive E. coli)], isolates were screened by a sequential multiplex PCR for the presence the following genes: bundle-forming pili (bfp), intimin (eae) for attachment/effacement on intestinal epithelial cells, plasmid of aggregative adhesin (CVD432), heat labile and stable enterotoxins (elt, estA1, estA2-4), Shiga toxins (VTcom; Stx1 and Stx2), and invasion-associated locus of the invasion plasmid (ial). The PCR protocol was similar to that described previously by Tobias et al.10 with some modifications: we added ial gene primers to the original multiplex PCR reaction 1 described by Tobias et al. The primers used in the study are listed in Table 1.
Table 1. Sequences of primers used in the multiplex PCRs for the detection of E. coli intestinal genes and phylogenetic groups.
| Pathotypes | Gene | Sequence | Amplified product length in base pairs |
|---|---|---|---|
| Typical EPEC | bfp |
bfp F: GGAAGTCAAATTCATGGGGGTAT bfp R: GGAATCAGACGCAGACTGGTAGT |
300 bp |
| EPEC or EHEC | eae |
eae F: TCAATGCAGTTCCGTTATCAGTT eae R: GTAAAGTCCGTTACCCCAACCTG |
482 bp |
| ETEC | elt |
elt F: ACGGCGTTACTATCCTCTC elt R: TGGTCTCGGTCAGATATGTG |
273 bp |
| EAEC | CVD 432 | CVD F: CTGGCGAAAGACTGTATCATCVD R: AAATGTATAGAAATCCGCTGTT | 630 bp |
| ETEC | estA1 |
estA1 F: TCTTTCCCCTCTTTTAGTCAG estA1 R: ACAGGCAGGATTACAACAAAG |
166 bp |
| ETEC | estA2-4 |
estA2 F: TTCACCTTTCCCTCAGGATG estA2 R: CTATTCATGCTTTCAGGACCA |
120 bp |
| STEC | VTcom (Stx1 + Stx2) |
VT F: GAGCGAAATAATTTATATGTG VT R: TGATGATGGCAATTCAGTAT |
518 bp |
| EIEC | ial |
Ial F: CTGGTAGGTATGGTGAGG Ial R: CCAGGCCAACAATTATTTCC |
320 bp |
| Phylogenetic group | chuA |
chuA F: GACGAACCA ACGGTCAGGAT chuA R: TGCCGCCAGTACC AAAGACA |
279 bp |
| YjaA |
YjaA F: TGAAGTGTCAGGAGACGCTG YjaA R: ATGGAGAATGCGTTCCTCAAC |
211 bp | |
| TspE4C2 | Tsp F: GAGTAATGTCGGGGCATTCA Tsp R: CGCGCCAACAAAGTATTACG |
152 bp |
EAEC – enteroaggregative E. coli; EHEC – enterohemorrhagic E. coli; EIEC – enteroinvasive E. coli; EPEC – enteropathogenic E. coli; ETEC – enterotoxigenic E. coli; STEC – Shiga toxin E. coli.
The criteria for identification were as follows: the presence of bfp and eae defined typical EPEC, but the presence of only eae confirmed atypical EPEC, the presence of pCVD confirmed the identification of EAEC, the presence of elt and/or estA1, and/or estA2-4 defined ETEC, the presence of Stx1 and/or Stx2 defined STEC (the additional presence of eae defined a typical EHEC isolate) and finally the presence of ial defined EIEC.
Phylogenetic group analysis of E. coli isolates
E. coli isolates were assigned to one of the phylogenetic groups (A, B1, B2 and D) by performing a triplex PCR, using primers to amplify chuA (required for heme transport in EHEC O157:H7), yjaA (found in E. coli K-12 with unknown function), and TSPE4.C2 (anonymous DNA fragment) genes. The interpretation of the combination of the amplified PCR products allowed the phylogenetic grouping as described by Clermont et al.11
Antimicrobial susceptibility testing
All isolated E. coli were subjected to antimicrobial susceptibility testing by Kirby-Bauer disc diffusion method, according to Clinical Laboratory Standards Institute (CLSI) guidelines, using the following antibiotic discs (Thermo Fisher): ampicillin (AMP; 10 μg), piperacillin (PRL; 100 μg), cefazolin (KZ; 30 μg), cefuroxime (CXM; 30 μg), ceftriaxone (CRO; 30 μg), cefotaxime (CTX; 30 μg), ceftazidime (CAZ; 30 μg), cefepime (FEP; 30 μg), aztreonam (ATM; 30 μg), amoxicillin-clavulanic acid (AMC; 20/10 μg), ampicillin-sulbactam (SAM; 10/10 μg), cefoperazone-sulbactam (SCF; 75/10 μg), piperacillin-tazobactam (TZP; 100/10 μg), imipenem (IPM; 10 μg), meropenem (MEM; 10 μg), ertapenem (ETP; 10 μg), levofloxacin (LEV; 5 μg), ciprofloxacin (CIP; 5 μg), tetracycline (TE; 30 μg), doxycycline (DO; 30 μg), gentamicin (CN; 10 μg), amikacin (AK; 30 μg), sulfamethoxazole-trimethoprim (SXT; 1.25/23.75 μg).12
Isolates that were resistant to one or more third-generation cephalosporins were further tested for production of extended spectrum beta-lactamase (ESBL) by the CLSI combined disc method ceftazidime and ceftazidime/clavulanic acid. A ≥5 mm increase in inhibition zone diameter of combined disc, when compared to ceftazidime disc alone, was interpreted as ESBL producer.12
In addition, all carbapenem resistant or intermediate isolates were subjected to modified carbapenem inactivation method (CIM) to test for carbapenemase production according to CLSI recommendations. Carbapenemase production was defined as a meropenem zone diameter of 6-15 mm or presence of pinpoint colonies within a 16-18 mm zone.12
Statistical analysis
Qualitative data was described as number and percent. Quantitative data was described as range (minimum and maximum), mean, standard deviation and median. Chi-square test, Mann Whitney test, and Kruskal Wallis tests were used as indicated. Correlation was performed using Spearman coefficient. Significance was judged at the 5% level. Analysis was performed using IBM statistical package SPSS software version 20.0 (IBM Corp, Armonk, NY, USA).
Results
Clinical and laboratory data results
Group IA (Active UC) included eight males (53.3%) and seven females (46.7%), with a mean age of 34.27±12.43 years. Group IB (UC in remission) included nine males (60%) and six females (40%), their mean age was 36.60±11.43 years. Group IIA (Active CD) included three males (20%) and 12 females (80%), their mean age was 38.40±11.38 years. Group IIB (CD in remission) included 12 males (80%) and three females (20%), with a mean age of 37.60±9.18 years. The control group included 12 males (60%) and eight females (40%), their age ranged between 18 and 50 years with a mean age of 33.55±10.28 years. There were statistically significant differences between the four studied groups and the control group regarding sex (p=0.021).
In respect to fecal calprotectin, higher levels were observed in IBD patients than in controls (p≤0.001). Its mean level was 837.87±743.1 mg/kg in group IA, 198.47±164.0 mg/kg in group IB, 413.87±716.1 mg/kg in group IIA, 41.80±40.79 mg/kg in group IIB, and finally 33.20±12.39 mg/kg in the control group.
E. coli pathotypes and phylogenetic groups results
The results of multiplex PCR confirmed the presence of 15 EAEC (15/60; 25%), two ETEC (2/60; 3.3%) in IBD cases (17/60; 28.3%) and only one atypical EPEC (1/20; 5%) in the control group. Out of the 15 EAEC, six were from UC patients (three active, three inactive UC), while nine were positive from CD patients (six active, three inactive CD). Typical EPEC, EIEC, EHEC, STEC were negative in all cases and controls. All IBD cases were negative for atypical EPEC and none of the controls was positive for EAEC. Chi-square test demonstrated a significant difference between all IBD cases and controls regarding the presence of EAEC (p=0.013), and a non-statistically significant difference between active or inactive cases in the UC group as well as in the CD group (Table 2).
Table 2. Comparison between the study groups according to frequency of E. coli pathotypes and phylogenetic groups.
| Active UC (n = 15) | Inactive UC (n = 15) | Active CD (n = 15) | Inactive CD (n = 15) | Controls (n = 20) | χ2 | P value | |
|---|---|---|---|---|---|---|---|
| E. coli pathotype | |||||||
| EAEC | 3a (20%) | 3a (20%) | 6a (40%) | 3a (20%) | 0b (0%) | 9.108 | 0.058 |
| ETEC | 0 (0%) | 1 (6.7) | 0 (0%) | 1 (6.7%) | 0 (0%) | 3.217 | 0.522 |
| EPEC (atypical) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (5%) | 2.887 | 0.577 |
| E. coli phylogenetic group | |||||||
| A | 6 (40%) | 4 (26.7%) | 5 (33.3%) | 6 (40%) | 10 (50%) | 2.372 | 0.668 |
| B1 | 0 (0%) | 2 (13.3%) | 0 (0%) | 0 (0%) | 0 (0%) | 8.487 | 0.075 |
| B2 | 6 (40%) | 7 (46.7%) | 8 (53.3%) | 9 (60%) | 10 (50%) | 2.628 | 0.622 |
| D | 3 (20%) | 2 (13.3%) | 2 (13.3%) | 0 (0%) | 0 (0%) | 6.523 | 0.163 |
CD – Crohn's disease; EAEC – enteroaggregative E. coli; EPEC – enteropathogenic E. coli; ETEC – enterotoxigenic E. coli; UC – ulcerative colitis. The Chi square test was used for comparing the different groups. In the first row, different letters indicate statistical significance.
Regarding the phylogenetic groups of E. coli, out of the 60 IBD cases, 30 (50%) were from group B2, 21 (35%) from group A, seven (11.7%) from group D, and two (3.3%) from group B1. Out of the control group, 50% were from group A, and 50% were from group B2. None of the controls had group B1 or D and none of the CD patients had group B1. The distribution of the phylogenetic groups of E. coli in UC, CD and control groups is shown in Table 2. No statistically significant differences in the distribution of E. coli phylogenetic groups were found between UC, CD (whether active or inactive), and controls (Figure 1 and Table 2).
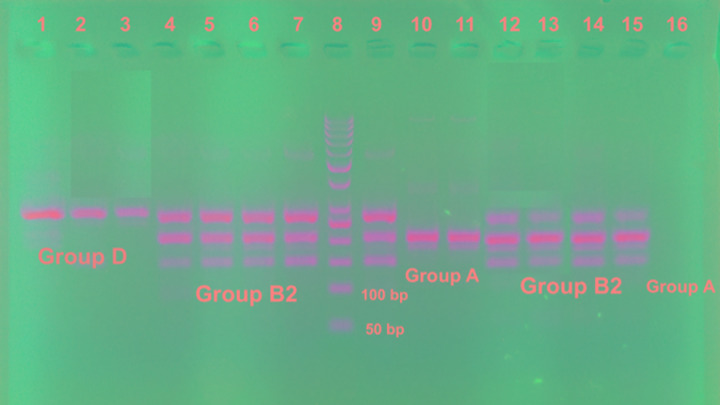
Figure 1. Gel stained with ethidium bromide for triplex PCR of phylogenetic groups of E. coli.
Lanes 1, 2, 3 show Group D; lanes 4-7, 9, 12-15 show Group B2; lanes 10, 11, 16 show Group A; lane 8 shows 50 bp DNA ladder.
A very interesting finding is that 80% (12/15) of EAEC were assigned to group B2 (7 isolates) and D (5 isolates), while only three (20%) EAEC isolates were assigned to group A. One ETEC was from group B2 and the other was from group A.
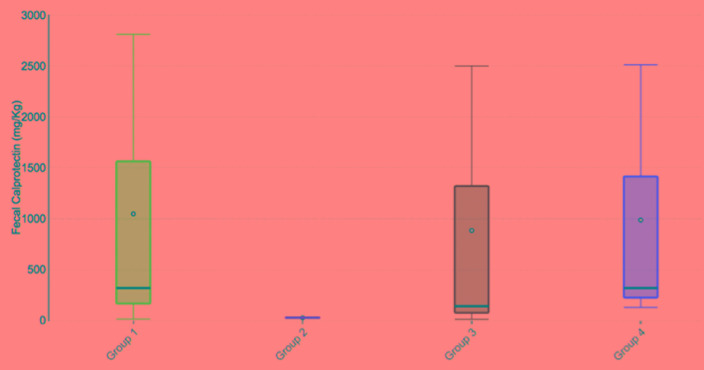
The mean level of fecal calprotectin was assessed in the four phylogenetic groups of IBD cases. Calprotectin mean level was the highest (644.3±867.6) for group D, followed by 410.6±600.5 for group A, 341.5±529 for group B2, and 28±4.24 for group B1. This difference was not statistically significant (p=0.133, Figure 2).
Figure 2. Relation between calprotectin level and different E. coli phylogenetic groups in IBD cases.
Group 1 = phylogenetic group A; Group 2 = phylogenetic group B1; Group 3 = phylogenetic group B2; Group 4 = phylogenetic group D. The median (min - max) was 320 (15 - 2810), 28 (25 - 31), 142 (13 - 2500), 320 (130 - 2512) for Groups A, B1, B2, and D respectively.
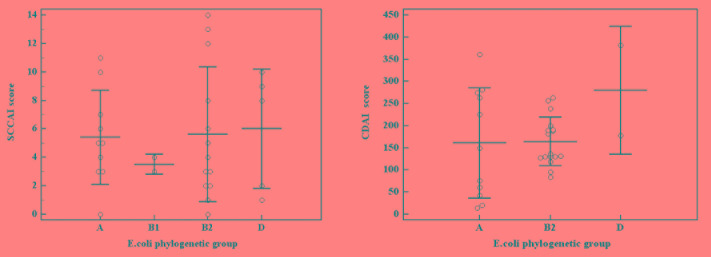
The relation between different E. coli phylogenetic groups and the severity of IBD was investigated. When comparing the severity of UC in the four phylogenetic groups, the highest SCCAI mean score was found in group D (6±4.2), followed by group B2 (5.6±4.7), group A (5.4±3.3), then group B1 (3.5±0.7). Similarly, when assessing the CDAI scores, group D showed the highest score (280±144.3) followed by group B2 164.3±54.38), and group A (160.6±124.5). Although our results confirmed a more severe disease with both group D and B2 in UC and CD, this was not statistically significant when performing Kruskal Wallis test (p=0.929 and p=0.455, respectively, Figure 3).
Figure 3. The relation between E. coli phylogenetic groups and (A) Simple clinical colitis activity index (SCCAI) of ulcerative colitis (B) Crohn’s disease activity index (CDAI).
Antimicrobial susceptibility results
E. coli isolates from UC and CD patients showed a high rate of antibiotic resistance, where 93.3% and 86.7% were ESBL producers, respectively. Additionally, 13.3% of UC and CD E. coli isolates were carbapenemase producer organisms (CPO). The detailed antimicrobial profile of UC, CD and controls is shown in Table 3.
Table 3. Antimicrobial resistance profile of E. coli isolates from UC, CD, and control groups.
| Antibiotics | Resistant E. coli isolates | ||
|---|---|---|---|
| UC (n=30) | CD (n=30) | Controls (n=20) | |
| N (%) | N (%) | N (%) | |
| Ampicillin | 30 (100%) | 30 (100%) | 20 (100%) |
| Piperacillin | 28 (93.3%) | 21 (70%) | 16 (80%) |
| Cefazolin | 30 (100%) | 30 (100%) | 20 (100%) |
| Cefuroxime | 30 (100%) | 28 (93.3%) | 9 (45%) |
| Ceftriaxone | 28 (93.3%) | 26 (86.7%) | 9 (45%) |
| Cefotaxime | 28 (93.3%) | 26 (86.7%) | 9 (45%) |
| Ceftazidime | 28 (93.3%) | 26 (86.7%) | 7 (35%) |
| Cefepime | 18 (60%) | 26 (86.7%) | 4 (20%) |
| Aztreonam | 26 (86.7%) | 26 (86.7%) | 7 (35%) |
| Amoxicillin/clavulanic acid | 9 (30%) | 6 (20%) | 2 (10%) |
| Ampicillin/sulbactam | 18 (60%) | 13 (43.3%) | 7 (35%) |
| Cefoperazone/sulbactam | 4 (13.3%) | 9 (30%) | 2 (10%) |
| Piperacillin/tazobactam | 9 (30%) | 6 (20%) | 2 (10%) |
| Imipenem | 4 (13.3%) | 4 (13.3%) | 0 |
| Meropenem | 4 (13.3%) | 4 (13.3%) | 0 |
| Ertapenem | 4 (13.3%) | 4 (13.3%) | 0 |
| Levofloxacin | 21 (70%) | 11 (36.7%) | 4 (20%) |
| Ciprofloxacin | 23 (76.7%) | 13 (43.3%) | 4 (20%) |
| Tetracycline | 26 (86.7%) | 21 (70%) | 9 (45%) |
| Doxycycline | 19 (63.3%) | 21 (70%) | 4 (20%) |
| Gentamicin | 12 (40%) | 11 (36.7%) | 4 (20%) |
| Amikacin | 5 (16.7%) | 4 (13.3%) | 2 (10%) |
| Sulfamethoxazole/trimethoprim | 26 (86.7%) | 19 (63.3%) | 9 (45%) |
| ESBL positive | 28 (93.3%) | 26 (86.7%) | 9 (45%) |
| CPO positive | 4 (13.3%) | 4 (13.3%) | 0 |
CD – Crohn's disease; ESBL – extended spectrum beta-lactamase, CPO – carbapenemase producing organism; UC – ulcerative colitis.
All EAEC (100%) from CD (6 isolates) and UC patients (9 isolates) were ESBL and CPO positive. Similarly, the two ETEC isolates as well as the only atypical EPEC were ESBL positive. In respect to antibiotic resistance of different phylogenetic groups, all group A, B2, B1 and D isolates of UC patients were ESBL positive except two isolates from group A. Three UC CPO (3/13; 23%) isolates were from group B2 and one isolate (1/5; 20%) was from group D. Additionally, out of 30 CD isolates, all group B2, D and A were ESBL positive except four isolates from group A. The four CPO CD isolates were from group B2. Concerning the control group, 7 out of 10 (70%) B2 isolates, and two out of 10 (20%) A isolates were ESBL positive.
Discussion
Several studies have linked bacteria to the intestinal inflammation that characterizes CD and UC. There is growing evidence suggesting the contribution of E. coli in GIT dysbiosis, which is considered an essential factor for initiation, or aggravation of the lesions and/or symptoms of IBD.
The detection of an increased number of E. coli in the stool and in the gut mucosa of IBD patients as well as the elevation of titers of serum antibodies against E. coli antigens suggests its involvement in the pathogenesis of IBD.5,13 The abundance of adherent-invasive E. coli has been found in ileal biopsies from patients with CD.14 Active E. coli has also been shown very predominant in mucosa of UC patients.15
In the present study, out of all screened E. coli pathotypes, EAEC and ETEC were detected in 17 (28.3%) IBD patients, where CVD432 was identified in 15 (25%) patients while elt and estA1 were identified in two (3.3%) patients. None of the controls harbored EAEC or ETEC. The prevalence of intestinal E. coli pathotypes reported in our study was higher than that reported in previous studies; one study reported the detection of eae and aggR in 5 out of 67 (7.5%) samples tested.16 Another study found that CVD432 of EAEC represented only 2.5%, EPEC 1.2% and ETEC 1.8% of all isolated E. coli strains. EIEC and STEC were not found in collected samples.17 Moreover, EAEC was detected in 2.5%, EPEC in 1.3% and ETEC in 1.5% of all IBD E. coli isolates in the study of Curová et al. The authors concluded that their low incidence was in agreement with the fact that they are associated with diarrhea, not to other GIT diseases,18 while other studies found that all E. coli IBD isolates were negative for intestinal genes.5,6 They justified the very low detection rate of classical E. coli intestinal genes by the previous observations confirming that E. coli from IBD patients are more closely related to ExPEC. Furthermore, de Souza et al. suggested that the population of E. coli in their study presented a low prevalence of virulence genes and was found in the ileum of CD and the rectum and the sigmoid colon of both UC and CD patients, which are already the common sites of IBD lesions. Therefore, they concluded that they are not related to the disease process.16
In agreement with the results of the present study, a Brazilian study, conducted on 131 E. coli isolates cultured from rectal biopsies of UC and CD patients, suggested that the increased population of E. coli, especially those detected with UC, was mainly of aggregative adherent (EAEC) type.14 Similarly, a high prevalence of EAEC was observed in the distal colon of both UC and CD as well as in the stool samples of CD patients in the study of Watanabe et al.19 EAEC is known by its ability to attach to the intestinal epithelium with subsequent biofilm formation and induction of mucus secretion by goblet cells, which provide a suitable environment for the proliferation in the inflamed colonic mucosa of IBD patients. In addition, EAEC can induce the secretion of interleukin-8 by the mucosal cells, which is a potent attractant of neutrophils to the intestinal mucosa.19
An interesting study, conducted by da Silva Santos et al.,20 reported the isolation of an E. coli strain in the ileum and stool of a CD patient, which combine the invasiveness property as well as the features of EHEC (eae and stx1) and EAEC (aggR) pathotypes. They concluded that the detection of an E. coli combining multiple virulence factors of intestinal pathotypes points to its relevance to the pathogenesis of the disease.
Many studies demonstrated that E. coli in IBD patients belong to the phylogenetic groups B2 and D.5,21 It was found that E. coli harboring the chuA gene and belonging to the phylogenetic groups B2 and D are mostly able to colonize the GIT mucosa and to survive inside the cells especially in CD patients.21 Moreover, a strong correlation was found between the isolation of group B2 E. coli and IBD.13 Our results are not consistent with the previous reports, since we found no statistically significant differences between IBD isolates and control isolates in respect to phylogenetic groups. However, 80% (12/15) of EAEC isolated from UC and CD patients in our study were assigned to group B2 (7 isolates) and D (5 isolates). In agreement with our results, Jensen et al.6 reported that all UC patients were colonized with E. coli phylogenetic group B2, however, CD patients were colonized with mixed phylogenetic groups: A, B2 and D. Similarly, a recent study found that the majority of E. coli isolates from patients with IBD were in group B2 (36.5%), followed by group D (30.3%), A (24.2%), and B1 (3.9%) with no statistically significant difference between IBD E. coli isolates and stool commensal isolates.22
Further, a previous study confirmed that the mean levels of calprotectin were significantly increased in IBD patients colonized with E. coli of B2 phylogenetic group. It also demonstrated that UC patients colonized with E. coli B2 had significantly increased Colitis Activity Index scores as compared to patients colonized with E. coli of group D and A.21 Similarly, Petersen et al.13 found an association between the presence of B2 E. coli and active colitis. On the other hand, De la Fuente et al.23 found no correlation between the presence of E. coli and CD activity or intestinal area affected, however, they reported an association between the presence of E. coli and disease activity in UC patients. These results are not in agreement with the results of the present study where the highest calprotectin level was found with group D, followed by group A, group B2, and then group B1. Although the results of our study demonstrated higher disease severity scores in both UC and CD with group D and B2, this finding was not statistically significant.
The role of antibiotics in IBD is controversial. They have been used to treat IBD primary disease process or its complications. They might play a role in treating bacterial overgrowth, or targeting specific bacteria thought to be implicated in the disease process. Ciprofloxacin and aminoglycosides have been used against pathogenic E. coli.24 Additionally, a meta-analysis proved a better clinical response (2.3 times) for IBD patients receiving broad-spectrum antibiotics.25 In the present study, E. coli from both UC and CD patients showed a high rate of resistance to most antimicrobials when compared to the control group; 76.7% of UC and 43.3% of CD isolates are resistant to ciprofloxacin, which is a commonly used empiric treatment in IBD. In accordance with our results, Dogan et al.4 found that 2/3 of CD isolates were resistant to one or more antimicrobials and that ciprofloxacin-resistant isolates were more common in CD than in healthy subjects.
The current study has several limitations. The study investigated fecal E. coli isolates of IBD patients and not mucosa-associated isolates. The study also focused on classical intestinal pathovars of E. coli. Further studies should target mucosa-associated E. coli especially adherent-invasive E. coli pathotype and extra-intestinal genes of E. coli and their contribution to the inflammatory process in IBD.
Conclusions
The presence of EAEC belonging mainly to group B2 and D in IBD cases and its absence from all controls may indicate the importance of this pathotype in the pathogenesis of IBD in Egyptian patients. It also raises the question whether these bacteria play a primary or a secondary role in IBD etiopathogenesis.
Footnotes
Authors’ contributions statement: MM designed the study, performed the bacterial culture, and genetic analysis of virulence genes and of phylogenetic groups in the laboratory, interpreted the results, analyzed the data and drafted the manuscript. KA and DH performed sample and data collection and statistical analysis; All authors critically revised the manuscript, approved the final version to be published, and agree to be accountable for all aspects of the work.
Conflicts of interest: The authors declare that there are no conflicts of interest.
Funding: None to declare.
REFERENCES
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–78. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.Barnich N, Carvalho FA, Glasser AL, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–74. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dogan B, Scherl E, Bosworth B, et al. Multidrug resistance is common in Escherichia coli associated with ileal Crohn’s disease. Inflamm Bowel Dis. 2013;19:141–50. doi: 10.1002/ibd.22971. [DOI] [PubMed] [Google Scholar]
- 5.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–75. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen SR, Mirsepasi-Lauridsen HC, Thysen AH, et al. Distinct inflammatory and cytopathic characteristics of Escherichia coli isolates from inflammatory bowel disease patients. Int J Med Microbiol. 2015;305:925–36. doi: 10.1016/j.ijmm.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Walmsley RS, Ayres RC, Pounder RE, Allan RN. A simple clinical colitis activity index. Gut. 1998;43:29–32. doi: 10.1136/gut.43.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Palma JA, Farraye FA. Crohn's disease: the first visit. Gastroenterol Hepatol. 2011;7:163–9. [PMC free article] [PubMed] [Google Scholar]
- 9.Tille PM. Bailey & Scott's Diagnostic Microbiology. 13th edition. Philadelphia, PA, USA: Mosby Elsevier; 2013. [Google Scholar]
- 10.Tobias J, Vutukuru SR. Simple and rapid multiplex PCR for identification of the main human diarrheagenic Escherichia coli. Microbiol Res. 2012;167:564–70. doi: 10.1016/j.micres.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing; 28th ed. CLSI supplement M100. Wayne, Pennsylvania, USA: CLSI; 2018. [Google Scholar]
- 13.Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, Krogfelt KA. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC Microbiol. 2009;9:171. doi: 10.1186/1471-2180-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomazini CM, Samegima DA, Rodrigues MA, Victoria CR, Rodrigues J. High prevalence of aggregative adherent Escherichia coli strains in the mucosa-associated microbiota of patients with inflammatory bowel diseases. Int J Med Microbiol. 2011;301:475–9. doi: 10.1016/j.ijmm.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 15.Sokol H, Lepage P, Seksik P, Dore J, Marteau P. Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol. 2006;44:3172–7. doi: 10.1128/JCM.02600-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Souza HL, de Carvalho VR, Romeiro FG, Sassaki LY, Keller R, Rodrigues J. Mucosa-associated but not luminal Escherichia coli is augmented in Crohn's disease and ulcerative colitis. Gut Pathog. 2012;4:21. doi: 10.1186/1757-4749-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kmetova M, Curova K, Gombosova L, et al. Escherichia coli pathotypes isolated in inflammatory bowel diseases and oncological diseases of gastrointestinal tract. Int J Infect Dis. 2008;12:e454. [Google Scholar]
- 18.Curová K, Kmetová M, Sabol M, Gombosová L, Lazúrová I, Siegfried L. Enterovirulent E. coli in inflammatory and noninflammatory bowel disease. Folia Microbiol (Praha) 2009;54:81–6. doi: 10.1007/s12223-009-0012-y. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe E, Romeiro F, Sassaki L, Keller R, Rodrigues J. Enteroaggregative Escherichia coli seems to proliferate in the terminal portion of the colon of IBD patients. Inflamm Bowel Dis. 2012;18:S112. [Google Scholar]
- 20.da Silva Santos AC, Gomes Romeiro F, Yukie Sassaki L, Rodrigues J. Escherichia coli from Crohn's disease patient displays virulence features of enteroinvasive (EIEC), enterohemorragic (EHEC), and enteroaggregative (EAEC) pathotypes. Gut Pathog. 2015;7:2. doi: 10.1186/s13099-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirsepasi-Lauridsen HC, Halkjaer SI, Mortensen EM, et al. Extraintestinal pathogenic Escherichia coli are associated with intestinal inflammation in patients with ulcerative colitis. Sci Rep. 2016;6:31152. doi: 10.1038/srep31152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Micenková L, Frankovičová L, Jaborníková I, et al. Escherichia coli isolates from patients with inflammatory bowel disease: ExPEC virulence- and colicin-determinants are more frequent compared to healthy controls. Int J Med Microbiol. 2018;308:498–504. doi: 10.1016/j.ijmm.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 23.De la Fuente M, Franchi L, Araya D, et al. Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int J Med Microbiol. 2014;304:384–92. doi: 10.1016/j.ijmm.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nitzan O, Elias M, Peretz A, Saliba W. Role of antibiotics for treatment of inflammatory bowel disease. World J Gastroenterol. 2016;22:1078–87. doi: 10.3748/wjg.v22.i3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan KJ, Ullman TA, Ford AC, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106:661–73. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]