Abstract
Objectives:
KRAS mutations, which occur in approximately 25% of lung adenocarcinoma cases, represent a major unmet clinical need in thoracic oncology. Preclinical studies have demonstrated that KRAS mutant NSCLC cell lines and xenografts with additional alterations in either TP53 or CDKN2A (INK4A/ARF) loci are sensitive to focal adhesion kinase (FAK) inhibition. Defactinib (VS-6063) is a selective oral inhibitor of FAK.
Materials and Methods:
Patients with previously treated advanced KRAS mutant NSCLC were prospectively assigned to one of four molecularly defined cohorts based on the presence or absence of TP53 or CDKN2A alterations and received treatment with defactinib 400 mg orally BID until disease progression or intolerable toxicity. The primary endpoint was progression-free survival (PFS) at 12 weeks.
Results:
Fifty-five patients were enrolled. Mean age was 62 years; 51% were female. The median number of prior lines of therapy was 4 (range 1–8). Fifteen (28%) patients met the 12-week PFS endpoint, with one patient achieving a partial response. Median PFS was 45 days. Clinical efficacy did not correlate with TP53 or CDKN2A status. The most common adverse events were fatigue, gastrointestinal, and increased bilirubin, and were generally grade 1 or 2 in severity.
Conclusion:
In heavily pretreated patients with KRAS mutant NSCLC, defactinib monotherapy demonstrated modest clinical activity. Efficacy was not associated with TP53 and CDKN2A status. Defactinib was generally well tolerated.
Keywords: adenocarcinoma, targeted therapy, tyrosine kinase inhibitor, KRAS, TP53, CDKN2A
1.0. Introduction
Despite years of research, KRAS mutant non-small cell lung cancer (NSCLC) remains a major unmet clinical need. KRAS mutations, the second most common genomic alteration in human cancer after TP53 mutations, occur in 25–30 percent of lung adenocarcinomas.[1] [2] High affinity binding of KRAS to its GTP substrate has hindered the development of therapeutic agents that directly inhibit KRAS. Only recently have KRAS-specific inhibitors have been developed.[3, 4] Among them, direct, covalent KRASG12C inhibitors have demonstrated encouraging efficacy and appear well tolerated.[5, 6] However, this subtype represents only one-third of KRAS mutant NSCLC. Other KRAS treatment strategies under investigation include inhibition of post-translational modification, inhibition of effector pathways, and targeting synthetic lethality interactions.[7–9]
Focal adhesion kinase (FAK) may represent a new therapeutic target in KRAS mutant lung cancer. FAK is a highly conserved non-receptor tyrosine kinase that has roles in cellular adhesion, migration, and stem cell proliferation.[10–13] Additionally, studies in xenograft and transgenic models of KRAS mutant lung adenocarcinoma have demonstrated that the RHOA-FAK pathway provides a critical signaling axis for these cancers.[14] Specifically, FAK inhibition with RNA interference and with small molecules attenuated growth of KRAS mutant but not KRAS wild adenocarcinomas and provided a radiosensitizing effect.[15] Importantly, concomitant alteration of tumor suppressors TP53 and/or CDKN2A lead to increase activation of the RHOA-FAK pathway and were requisite for efficacy.
Defactinib (VS-6063; Verastem, Needham, MA) is a potent and selective ATP-competitive small molecular inhibitor of FAK and proline-rich tyrosine kinase 2 (Pyk2), each with IC50 <0.6 nM. Clinically, the drug is well tolerated, with minimal drug-drug interactions, consistent bioavailability independent of food intake, and principal toxicities of low-grade nausea, fatigue, headache, and a reversible Gilbert’s-type isolated unconjugated hyperbilirubinemia.[16] In a phase 1 study in subjects with advanced solid tumors, doses up to 750 mg twice daily were administered. Exposure was found not to increase substantially above doses of 425 mg twice daily, with no maximum tolerated dose established. Based on these data and preclinical pharmacodynamic studies, the recommended phase 2 dose was established as 400 mg orally twice daily.
Given the ongoing unmet clinical need, strong preclinical rationale, and the availability of a well-tolerated orally bioavailable FAK inhibitor, we conducted a single-arm, open-label, multi-cohort trial of defactinib in previously treated advanced KRAS mutant NSCLC. Following the preclinical observations that co-alterations in TP53 and CDKN2A were associated with benefit from FAK inhibition in KRAS-driven tumors, these genomic alterations were determined up-front in enrolled patients and incorporated into cohort assignment.
2.0. Materials and Methods
2.1. Patients
Written, informed consent was obtained from all enrolled patients. Eligible patients were recruited at nine U.S. medical centers and had inoperable advanced (stage IVA/IVB, AJCC 7th edition) NSCLC harboring a documented KRAS mutation, had received at least one prior line of chemotherapy that included at least one platinum-based chemotherapy doublet for metastatic or locally recurrent disease, had measurable disease by Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 guidelines, and no prior treatment with a FAK inhibitor. There was no limit to the number of prior lines of therapy. Adequate bone marrow, cardiovascular (including ECG QTc interval <480 msec), liver, and renal function, and performance status (ECOG 0–1) were required. Previously treated brain metastases with no evidence of progression were permitted. In addition, radiographically and clinically stable brain metastases not requiring steroids were allowed without requirement for prior radiation or surgical resection. Leptomeningeal metastasis was excluded. Submission of archival and/or fresh tumor tissue biopsy samples for molecular analysis was required. For patients with inadequate archival tissue, a study-specific biopsy could be performed.
2.2. Study Design and Treatment
Enrolled patients were initially assigned to one of four cohorts: (A) KRAS mutation with wild type CDKN2A and wild type TP53; (B) KRAS mutation with CDKN2A alteration and wild type TP53; (C) KRAS mutation with wild type CDKN2A and TP53 mutation; (D) KRAS mutation with both CDKN2A alteration and TP53 mutation. Pre-screening of patients for molecular profile while they were receiving earlier lines of therapy was permitted. During the course of the study, molecular characterization was modified to include CDKN2A deletions by FISH as well as CDKN2A mutations among CDKN2A alterations. A complete list of TP53 mutations and CDKN2A alterations examined is provided in Supplemental Table 1 and Supplemental Table 2. Cases with wild type CDKN2A for which sufficient tissue was not available for CDKN2A FISH were assigned to a fifth cohort (undetermined; E).
All patients were treated with defactinib (provided as 200 mg tablets) 400 mg orally twice daily continuously in 21-day cycles until disease progression or unacceptable toxicity. Prophylactic antiemetics and antidiarrheals were not routinely administered. Radiographic assessment of response was performed every two cycles. For patients with previously untreated brain metastases, CNS imaging was performed at the time of other disease imaging throughout treatment. Safety assessments included monitoring of adverse events, clinical laboratory parameters, vital sign measurements, physical examinations, and ECGs. In selected cases with evidence of clinical benefit by investigator judgment, defactinib could be continued beyond progression.
Dose reduction from 400 mg twice daily to 200 mg twice daily was performed for Grade 3 or 4 neutropenia or thrombocytopenia, recurrent Grade 3 and initial Grade 4 non-hematological adverse events (except alopecia and nausea, vomiting, or diarrhea not managed by optimal medical interventions), and for the first episode of Grade 3 or 4 hyperbilirubinemia with an increase in AST and/or ALT above the baseline grade. Defactinib was permanently discontinued for ≥ Grade 2 hyperbilirubinemia with AST and/or ALT ≥ Grade 2.
2.3. Molecular Analysis
For enrollment, KRAS mutation positive status was defined as codon 12, 13, or 61 mutations detected by sequencing by a CLIA-certified assay. Central KRAS testing was not required. Submission of archival and/or fresh tumor or DNA samples was mandated for additional molecular analysis. Specific requirements included 10–15 unstained slides (10-micron preferred) or 110–120 ng of archival DNA (by Nanodrop quantitation) or a minimum of 20 ng (by Qubit analysis). If DNA was submitted for analysis, an additional five unstained 5-micron slides were required for biomarker testing. This tissue underwent NextGeneration sequencing using the Ion PGM™ system and the Ion PGM™ Sequencing 200 v2 Kit (Life Technologies) according to the manufacturer’s instructions, which was performed centrally at the Baylor College of Medicine Molecular Genetics Laboratory. A detailed description of library construction, template preparation, and sequencing and data analysis is provided in Supplemental Materials.
After study activation, central FISH analysis was added to evaluate for CDKN2A deletions. Fluorescence in situ hybridization with the dual color CDKN2A probe set (CEP9 SpectrumGreen/Vysis LSI CDKN2A SpectrumOrange) was performed on the FFPE slides (Vysis, Downers Grove, IL). Slides were hybridized to the CDKN2A probe according to manufacturer’s instructions with minor modification. Loss of one or two red signals was interpreted as heterozygous or homozygous CDKN2A deletion, respectively. A complete listing of TP53 mutations and CDKN2A alterations examined is provided in Supplemental Table 1 and Supplemental Table 2. Cases with wild type CDKN2A for which sufficient tissue was not available for CDKN2A FISH were assigned to a fifth cohort (undetermined; E).
Planned exploratory biomarkers included FAK and phospho-FAK expression, CDKN2A methylation status, SKT11/LKB1 sequencing by ion torrent, and TP53-equivalent alterations (ATM mutation, nuclear ATM expression, BIM expression, Bcl-2 expression), although tissue availability limited performance to only some of these assays.
2.4. Endpoints and Statistical Considerations
The primary endpoint of this study was 12-week progression-free survival (PFS) rate. A landmark PFS rate was selected because (1) at the time of trial design and execution, objective responses had not been noted in clinical trials of single-agent targeted therapy for KRAS mutant NSCLC, and (2) landmark PFS rate is less subject to bias than PFS. PFS was measured from the first day of study treatment until radiographic disease progression per RECIST 1.1 criteria, or death of any cause. Secondary endpoints included PFS, overall survival (OS), overall response rate (ORR), and safety. Radiographic assessment of response was based on investigator assessment. Pharmacodynamic endpoints included the association between tumor biomarkers and clinical outcome measures (ORR, PFS, OS). Adverse events were classified based on the National Cancer Institute Common Terminology Criteria for Adverse Events (CTCAE) version 3.0 and were assessed on Days 1 and 8 during Cycle 1 and on Day 1 during subsequent cycles.
PFS and OS in each cohort were estimated using Kaplan-Meier product limit estimates. For binary outcomes (including the primary endpoint of 12-week PFS) the Fisher’s exact test or logistic regression was conducted. For time-to-event outcomes, the Cox proportional hazard model was used. Cox proportional hazards models were used to explore the association of time-to-event and covariates (patient characteristics, clinical characteristics, and biomarkers). Fisher’s exact test or logistic regression was used to evaluate the association between tumor markers and efficacy outcomes.
Two efficacy populations (intent-to-treat and efficacy evaluable) and one safety population were defined. The intent-to-treat population and safety populations were defined as all patients administered at least one dose of study drug. The efficacy evaluable population was defined as either (1) patients administered at least 12 weeks of study drug and at least one follow-up objective disease assessment, or (2) patients receiving at least 14 days of study drug who discontinued therapy due to toxicity or disease progression and underwent at least one follow-up objective disease assessment, or (3) patients receiving at least 14 days of study drug who died from any cause. If additional efficacy data was required in a cohort, non-evaluable patients were replaced (Supplemental Figure 1).
The trial was developed using a Simon’s two-stage optimal design.[17] In the first stage of each cohort, enrollment of 11 patients was planned. If three or fewer patients were progression-free at 12 weeks, enrollment in the cohort would be stopped. If four or more patients were progression-free at 12 weeks, 23 additional patients would be accrued, for a total of 34 evaluable patients in the cohort. This interim analysis was not completed, as study enrollment was closed early and the study did not proceed to stage 2 due to sponsor prioritization of alternative treatment strategies (such as defactinib combination regimens).
The sample size was calculated based on improvement of 12-week PFS rate from a historical control of 25 percent (derived from the KRAS mutation-positive population in the international phase 3 INTEREST trial[18, 19]) to 50 percent. Thirty-four patients provided 85% power at one-sided significance of 0.05 to demonstrate this difference. All efficacy analyses were conducted on the efficacy evaluable population. Each cohort was analyzed independently. Additionally, Cohorts B, C, and D (KRAS mutation with CDKN2A alteration and/or TP53 alteration) were each compared with Cohort A (KRAS mutation with wild type CDKN2A and wild type TP53), and Cohorts B, C, and D were pooled together and compared with Cohort A for PFS and OS endpoints.
2.5. Study Oversight
This study was designed by the investigators with input from Verastem. Data collection was monitored and maintained in a central electronic database. Analyses were performed by Verastem. Local institutional review boards approved the study at all sites. The study was registered at clinicaltrials.gov () prior to enrollment of any patients. Written informed consent from each patient was obtained prior to the performance of any study-related procedures. Verastem provided funding for the study.
3.0. Results
3.1. Patients
A total of 75 subjects were screened, of whom 55 (73%) were enrolled on the study between September 2013 and June 2016 and received at least one dose of study therapy (Table 1 and Supplemental Figure 1). Mean age was 62 years, 51% were female, and 91% were white. Patients were assigned to individual cohorts as follows: A (12 patients) - KRAS mutation with wild type CDKN2A and wild type TP53, B (13 patients) - KRAS mutation with CDKN2A alteration and wild type TP53, C (12 patients) - KRAS mutation with wild type CDKN2A and TP53 alteration, D (12 patients) - KRAS mutation with CDKN2A alteration and TP53 alteration, E (6 patients) - KRAS mutation with undetermined CDKN2A status.
Table 1.
Demographic and Baseline Characteristics
| Category | Cohort A N=12 |
Cohort B N=13 |
Cohort C N=12 |
Cohort D N=12 |
Cohort E N=6 |
Overall N=55 |
|---|---|---|---|---|---|---|
| Wild type INK4a/ARF and wild type TP53 | INK4a/ARF alteration and wild type TP53 | Wild type INK4a/ARF and TP53 mutation | INK4a/ARF alteration and TP53 mutation | Unknown | ||
| Sex, n (%) | ||||||
| Male | 7 (58) | 7 (54) | 5 (42) | 5 (42) | 3 (50) | 27 (49) |
| Female | 5 (42) | 6 (46) | 7 (58) | 7 (58) | 3 (50) | 28 (51) |
| Race, n (%) | ||||||
| White | 12 (100) | 12 (92) | 11 (92) | 11 (92) | 5 (83) | 50 (92) |
| Black | 0 | 0 | 1 (8) | 1 (8) | 0 | 2 (4) |
| Asian | 0 | 1 (8) | 0 | 0 | 0 | 1 (2) |
| Native Hawaiian or Pacific Islander | 0 | 0 | 0 | 0 | 1 (17) | 1 (2) |
| Age, mean (SD) (y) | 63 (9.2) | 63 (12.3) | 67 (7.7) | 59 (12.6) | 58 (12.0) | 62 (10.9) |
| ECOG performance status, n (%) | ||||||
| 0 | 1 (8.3) | 2 (15.4) | 3 (25.0) | 1 (8.3) | 1 (16.7) | 8 (14.5) |
| 1 | 11 (91.7) | 11 (84.6) | 8 (66.7) | 11 (91.7) | 5 (83.3) | 46 (83.6) |
| 2 | 0 | 0 | 1 (8.3) | 0 | 0 | 1 (1.8) |
| Smoking history, n (%) | ||||||
| Yes | 10 (83) | 9 (69) | 12 (100) | 10 (83) | 4 (67) | 45 (82) |
| No | 2 (17) | 4 (31) | 0 | 2 (17) | 2 (33) | 10 (18) |
| Number of prior systemic therapies | ||||||
| Median (range) | 5.5 (3–11) | 6.0 (2–10) | 3.0 (1–8) | 3.0 (2–9) | 4.0 (2–6) | 4.0 (1–11) |
ECOG = Eastern Cooperative Oncology Group; SD = standard deviation
KRAS mutation type is shown in Table 2. Over 90% were codon 12 mutations, with codon 13 and codon 61 mutations together accounting for only 7% of cases. We did not observe an association between type of KRAS mutation and TP53 or CDKN2A status.
Table 2.
KRAS mutations
| KRAS mutation | Cohort A N=12 |
Cohort B N=13 |
Cohort C N=12 |
Cohort D N=12 |
Cohort E N=6 |
Overall N=55 |
|---|---|---|---|---|---|---|
| Wild type INK4a/ARF and wild type TP53 | INK4a/ARF alteration and wild type TP53 | Wild type INK4a/ARF and TP53 mutation | INK4a/ARF alteration and TP53 mutation | Unknown | ||
| G12A | 0 | 1 | 2 | 2 | 0 | 5 |
| G12C | 3 | 2 | 4 | 4 | 1 | 14 |
| G12D | 4 | 3 | 1 | 2 | 2 | 12 |
| G12F | 1 | 0 | 0 | 0 | 0 | 1 |
| G12R | 1 | 1 | 0 | 0 | 0 | 2 |
| G12S | 0 | 0 | 2 | 0 | 0 | 2 |
| G12V | 2 | 2 | 0 | 1 | 2 | 7 |
| G13D | 0 | 1 | 0 | 0 | 0 | 1 |
| G13R | 0 | 1 | 0 | 0 | 0 | 1 |
| Q61H | 0 | 0 | 1 | 0 | 0 | 1 |
| Q61L | 0 | 0 | 0 | 1 | 0 | 1 |
| Not listed | 1 | 2 | 2 | 2 | 1 | 8 |
3.2. Treatment Exposure
All enrolled patients terminated treatment before the study ended. The most common reasons for termination of treatment included disease progression (n=35, 64%), symptomatic deterioration (n=7, 13%), and unacceptable AEs (n=5, 9%). The most common reason for discontinuation from the study was death (n=42, 76%). Overall, patients were exposed to defactinib for a mean of 89 days. Patients in Cohorts A and B had the highest durations of exposure (mean 120 and 115 days, respectively), while patients in Cohorts C, D, and E had mean exposure ranging 64 to 70 days. A total of 18 patients had dose interruptions (mean 1.4), with a mean duration of 12 days. A single patient had a dose reduction during the study.
3.3. Efficacy
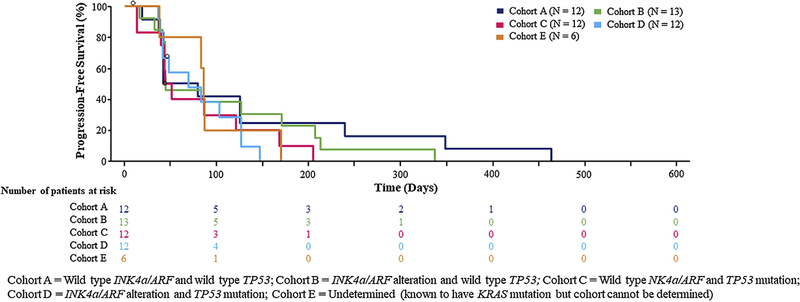
The efficacy evaluable population included 43 patients (Cohort A, n=11; Cohort B, n=11; Cohort C, n=10; Cohort D, n=11). Overall, 17 patients (31%) were progression free at 12 weeks as follows (Cohort A, n=5 [42%]; Cohort B, n=5 [39%]; Cohort C, n=3 [25%]; Cohort D, n=3 [25%], Cohort E, n=1 [17%]). There was no statistically significant difference in the risk of disease progression or death between arms (overall P>0.05) (Figure 1).
Figure 1.
Progression-free survival
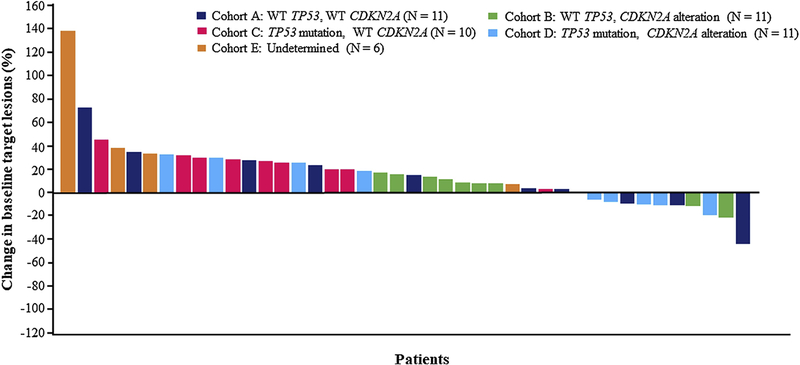
One patient achieved a partial response (PR) during the study. Eight patients (33%) achieved a best overall response of stable disease (SD) and 11 subjects (46%) achieved a best overall response of progressive disease (PD) (Figure 2). Median progression-free survival (PFS) for Cohorts B, C, and D combined was 47 (90% CI, 43–102) days, compared to 41 days (90% CI, 36–126) days for Cohort A. According to pre-specified criteria, cohorts A and B met threshold for cohort expansion (≥ 4 subjects out of 11 with ≥ 12 weeks of PFS). However, because the study failed to identify a candidate molecular subset or biomarker to predict benefit from defactinib, the trial sponsor and investigators decided not to pursue additional enrollment but instead devote effort and resources to possible future combination trials. PFS analysis in the ITT population approximated that in the efficacy evaluable population (Supplemental Table 2).
Figure 2.
Best radiographic response
We also examined efficacy according to type of KRAS mutation. No patient with a KRAS codon 13 or 61 mutation reached the 12-week PFS endpoint. Among codon 12 mutations, there was not a significant difference in 12-week PFS between patients with G12C or G12V mutations, which activate the RAL pathway, versus G12D which activates the RAF/MAPK and PI3K pathways.[20]
Median overall survival across cohorts is shown in Figure 3. There was no statistically significant increase in overall survival in Cohorts B, C, and D each versus Cohort A or Cohorts B, C, and D combined versus Cohort A.
Figure 3.
Overall survival
Next generation sequencing covering 50 unique cancer-associated genes was performed on 47 patients in the evaluable cohort (see Supplemental Materials). We evaluated whether co-current alterations which occurred in three or more patients were more or less commonly observed in two populations: 1) patients with a PFS ≥200 days (n=7) or patients with an OS ≥1 year (n=14). Four cases had occurring STK11 mutations although this alteration was only observed in patients with a PFS < 200 days. Similarly, four GNAS mutations were identified and was only seen in patients with a PFS <200 days. In addition, three cases had PIK3CA mutations; however, this alteration was not observed in any patient with a PFS ≥200 days or OS ≥1 year. Conversely, STK11 alterations were seen in two patients with an OS ≥1year. GNAS mutations were more common in patients with a shorter OS (n=3); however, one patient with a GNAS mutation was alive at one year despite rapid progression on study. Interestingly, all three patients with a concurrent PIK3CA mutation had an OS≤ 1 year.
3.4. Safety
Overall, 54 patients (98%) experienced at least one treatment-emergent adverse event (AE) during the study. In total, 44 patients (80%) experienced at least one AE considered related by the investigator to study therapy (Table 3A). Fifteen patients (27%) experienced a ≥ Grade 3 treatment-related AE (Table 3B). Overall, five patients discontinued treatment due to an AE, of which two (cerebrovascular accident and respiratory failure) were considered possibly related to study treatment. Twenty-seven patients (49%) experienced a total of 56 serious adverse events, of whom seven patients had serious adverse events considered by the investigator to be related to study therapy. Eleven patients (20%) experienced fatal adverse events (including eight cases of cancer progression, two cases of respiratory failure, and one case of myocardial infarction). Only one fatal event (respiratory failure occurring on day 5 of study therapy) was considered by the investigator to be related to treatment.
Table 3A.
Treatment-related Adverse Events Occurring in >10% of the safety population (N=55)
| Term | Number (%) |
|---|---|
| Fatigue | 19 (35) |
| Nausea | 12 (22) |
| Diarrhea | 11 (20) |
| Vomiting | 10 (18) |
| Hyperbilirubinemia | 9 (16) |
| Decreased appetite | 6 (11) |
| Peripheral edema | 6 (11) |
Table 3B.
Grade ≥3 treatment-related adverse events in the safety population (N=55)
| Term | Number (%) |
|---|---|
| Nausea | 4 (7) |
| Vomiting | 3 (6) |
| Hyperbilirubinemia | 3 (6) |
| Anemia | 1 (2) |
| Fatigue | 1 (2) |
| QTc prolongation | 1 (2) |
| Increased lipase | 1 (2) |
| Cerebrovascular accident | 1 (2) |
| Dizziness | 1 (2) |
| Intracranial hemorrhage | 1 (2) |
| Acute renal failure | 1 (2) |
| Respiratory failure | 1 (2) |
Notes: CTCAE, Version 4.03 was used for severity grading. If a subject experienced more than 1 event with a given preferred term, that subject was counted only once for that preferred term. Subjects were tabulated based on their most severe occurrence of a preferred term. Adverse events with a missing severity were analyzed as Grade 3 (severe).
The most common treatment-related (investigator assessment) AEs (incidence ≥ 5%) were fatigue (35%), nausea (22%), diarrhea (20%), vomiting (18%), hyperbilirubinemia (16%), decreased appetite (11%), peripheral edema (11%), dizziness (7%), and headache (6%). In total, 32 subjects (58%) experienced ≥ Grade 3 AEs. The most common were malignant neoplasm progression (16%), dyspnea (7%), nausea (7%), hyperbilirubinemia (6%), respiratory failure (6%), and vomiting (6%).
Nineteen patients developed total bilirubin values greater than CTCAE Grade 1 (17 with Grade 2; two with Grade 3), all of which subsequently decreased to CTCAE Grade 1 or lower. The elevations were transient, appearing between 14 and 21 days after the initiation of study drug and generally resolving within approximately 25 days. The elevations did not lead to discontinuation from the study or study drug, or to any dose modifications.
4.0. Discussion
Based on evidence that FAK inhibition inhibits growth of transgenic and xenograft KRAS mutant lung adenocarcinoma models and the availability of a well-tolerated FAK inhibitor, we conducted this multi-cohort single-arm phase 2 trial of defactinib in previously treated advanced KRAS mutant NSCLC. To our knowledge, this was the first trial in KRAS mutant NSCLC that prospectively identified patient cohorts according to concurrent genomic alterations. Specifically, we determined status of the tumor suppressors CDKN2A and TP53 in addition to documenting KRAS mutations.
In preclinical models, efficacy of FAK inhibition was limited to those KRAS mutant tumors with concurrent CDKN2A and/or TP53 alterations.[14] Accordingly, in this clinical trial we hypothesized that patients with tumors harboring either CDKN2A and/or TP53 alterations would demonstrate improved clinical outcomes compared to the dual wild-type population. However, this trend was not observed, with 12-week PFS rates comparable across all cohorts and only one patient achieving a radiographic response.
That this trial did not confirm preclinical observations may reflect small patient numbers, biologic differences between laboratory models and human patients (in whom co-mutation of TP53 and KRAS may predict deleterious effects of adjuvant chemotherapy[21]), unmeasured biomarkers, or the complexity of characterizing the two genes of interest. For instance, we initially performed gene sequencing of both TP53 and CDKN2A. However, further discussion after trial initiation led to the decision to include CDKN2A FISH to enhance detection of CDKN2A deletions. However, by that point archival tissue for some patients had been exhausted. Indeed, despite requesting 10–15 unstained slides (or 110–120 mg of archival DNA plus 5 slides) per patient, we were not able to perform a number of planned secondary biomarker analyses on any of the cases due to insufficient residual tissue. Whether those proposed biomarkers would have yielded additional predictive information is not clear. Of note, numerous recent studies suggest that the dual KRAS mutant and LKB1/STK11 altered subset appears to represent a biologically distinct entity.[22] Molecular characterization in this study was limited to examining LKB1/STK11 mutations, which were identified in only six cases. As LKB1/STK11 is often deleted or epigenetically silenced, it remains to be explored whether this molecular subtype may predict benefit from FAK inhibition.
The prevalence of CDKN2A alterations and TP53 mutations in this trial largely resembled that reported in other KRAS mutant populations. In a preliminary retrospective analysis of cases from Memorial Sloan Kettering Cancer Center, approximately 50% of KRAS mutant NSCLC harbored either INK4a or TP53 alterations. Approximately half of cases had an INK4a alteration, half had a TP53 mutation, and co-existence of both gene alterations was a rare event.[14, 23, 24]
In the present trial, we did not observe a significant difference in efficacy according to type of codon 12 mutation. Clinical outcomes were similar between patients with KRAS G12C or G12V mutations (which activate RAL pathway) and patients with G12D mutations (which activate RAF/MAPK leading to RHO/FAK activation).[20] [25] Similarly, in early-stage NSCLC, specific KRAS codon 12 mutations do not have prognostic effects.[26] In contrast, among patients with KRAS mutant NSCLC treated with docetaxel plus the MEK inhibitor selumetinib, there were trends toward greater improvement in OS, PFS, and ORR among those with KRAS G12C or G12V mutations.[27]
How does the efficacy of defactinib compare to other treatment strategies investigated for KRAS mutant NSCLC? Across cohorts in this relatively heavily pretreated population (median 4 lines of prior therapy), the disease control rate was approximately 50%, similar to that of sorafenib reported in KRAS mutant cases in the Biomarker-Integrated Approaches of Targeted Therapy for Lung cancer Elimination (BATTLE) trial.[28] Historically, response rates to single-agent molecularly targeted therapies in KRAS mutant NSCLC have been low. For the MEK inhibitors trametinib and selumetinib, response rates are 12% and 0%.[29, 30] KRASG12C inhibitors represent an important exception to these relatively disappointing results, with partial responses in more than half of lung cancer cases.[6] By comparison, in the current trial, approximately 20% of cases had evidence of radiographic shrinkage, although only one met criteria for PR. Approximately 10% of cases had disease control for 9 months or longer.
Defactinib was generally safe and well tolerated in this study. Principal toxicities were fatigue and gastrointestinal events, most of which were low grade. Higher-grade events responded to supportive care, resolved upon withholding of treatment, and did not recur after modification of prophylactic regimens and/or dose modification. Hyperbilirubinemia was predominantly unconjugated, not accompanied by transaminitis, and was not associated with clinical symptoms. The single case of grade 5 respiratory failure had no clinical or radiographic features of pneumonitis, and high-grade treatment-related respiratory events were not reported in the phase 1 defactinib trial or a phase 2 trial of maintenance defactinib in mesothelioma.[31, 32]
The main limitations of this trial are small sample size and incomplete molecular characterization of cases. In comparing efficacy across relatively small patient cohorts, differences arising from tumor molecular characteristics may be masked by clinical heterogeneity of cases (such as the range in number of lines of prior treatment and unmeasured variables such as response to prior therapies and burden of disease[33]). Despite relatively stringent requirements for submission of archival tissue, we were not able to complete assessment of several biomarkers. Finally, the single-arm design of the trial limits conclusions regarding the efficacy of single-agent FAK inhibition in this population.
5.0. Conclusions
In conclusion, this trial demonstrates that expanded and timely molecular characterization of KRAS mutant lung cancer cases for patient selection and treatment assignment is feasible. Single-agent defactinib, an orally bioavailable and well-tolerated FAK inhibitor, has only modest activity in KRAS mutant NSCLC. In contrast to preclinical studies, this activity does not appear dependent on CDKN2A or TP53 status. Recently, defactinib failed to improve outcomes as maintenance therapy after standard first-line chemotherapy for mesothelioma, where FAK inhibition had been shown to selectively kill mesothelioma cells expressing low levels of moesin-ezrin-radixin-like protein (merlin).[32] Based on the drug’s favorable toxicity profile and preclinical data supporting combination therapy (including taxane chemotherapy,[34] PI3K inhibitors,[35] mTOR inhibitors,[36] and the multi-targeted kinase inhibitor cabozantinib[37]), any future defactinib studies in KRAS mutant NSCLC will likely be focused on multi-drug regimens.
Supplementary Material
Highlights:
KRAS mutant non-small cell lung cancer (NSCLC) is a major unmet clinical need
RHOA-focal adhesion kinase (FAK) pathway is a signaling axis for KRAS mutant NSCLC
KRAS mutant NSCLC models with TP53 or CDKN2A alterations respond to FAK inhibition
Defactinib (VS-6063) is a selective oral inhibitor of FAK
Defactinib had modest effects in KRAS mutant NSCLC, independent of TP53 or CDKN2A
Acknowledgements:
The authors thank Ms. Dru Gray for assistance with manuscript preparation. The authors thank Ms. Stephanie Lustgarten for assistance with data collection and figure generation.
Funding:
This work was supported by Verastem, Inc., by a National Cancer Institute (NCI) Midcareer Investigator Award in Patient-Oriented Research (K24 CA201543-01, to D.E.G.), and by the Biostatistics Shared Resource of the Harold C. Simmons Comprehensive Cancer Center, which is supported in part by National Cancer Institute Cancer Center Support Grant 1P30CA142543-01 and by the Cancer Prevention and Research Institute of Texas (RP150596).
Abbreviations:
- FAK
Focal adhesion kinase
- merlin
moesin-ezrin-radixin-like protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Summary Conflict of Interest Statement:
David T. Weaver, Louis Vaikus, Mitchell Keegan, and Joanna C. Horobin were employees of Verastem, Inc., at the time of study design and conduct. David E. Gerber received research funding from Verastem, Inc.
Prior Presentations:
Presented in abstract form at the 16th World Conference on Lung Cancer, Denver, CO, September 6–9, 2015.
References
- 1.Kris MG, et al. , Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA, 2014. 311(19): p. 1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlesi F, et al. , Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet, 2016. [DOI] [PubMed] [Google Scholar]
- 3.Ostrem JM, et al. , K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature, 2013. 503(7477): p. 548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim SM, et al. , Therapeutic Targeting of Oncogenic K-Ras by a Covalent Catalytic Site Inhibitor. Angew Chem Int Ed Engl, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janes MR, et al. , Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell, 2018. 172(3): p. 578–589 e17. [DOI] [PubMed] [Google Scholar]
- 6.AMG 510 First to Inhibit “Undruggable” KRAS. Cancer Discov, 2019. [DOI] [PubMed]
- 7.Fiordalisi JJ, et al. , High affinity for farnesyltransferase and alternative prenylation contribute individually to K-Ras4B resistance to farnesyltransferase inhibitors. J Biol Chem, 2003. 278(43): p. 41718–27. [DOI] [PubMed] [Google Scholar]
- 8.Janne PA, et al. , Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol, 2013. 14(1): p. 38–47. [DOI] [PubMed] [Google Scholar]
- 9.Westcott PM and To MD, The genetics and biology of KRAS in lung cancer. Chin J Cancer, 2013. 32(2): p. 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunton VG and Frame MC, Src and focal adhesion kinase as therapeutic targets in cancer. Curr Opin Pharmacol, 2008. 8(4): p. 427–32. [DOI] [PubMed] [Google Scholar]
- 11.Schlaepfer DD and Mitra SK, Multiple connections link FAK to cell motility and invasion. Curr Opin Genet Dev, 2004. 14(1): p. 92–101. [DOI] [PubMed] [Google Scholar]
- 12.Luo M, et al. , Mammary epithelial-specific ablation of the focal adhesion kinase suppresses mammary tumorigenesis by affecting mammary cancer stem/progenitor cells. Cancer Res, 2009. 69(2): p. 466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibue T, et al. , The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov, 2012. 2(8): p. 706–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konstantinidou G, et al. , RHOA-FAK Is a Required Signaling Axis for the Maintenance of KRAS-Driven Lung Adenocarcinomas. Cancer Discov, 2013. 3(4): p. 444–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang KJ, et al. , Focal Adhesion Kinase Regulates the DNA Damage Response and Its Inhibition Radiosensitizes Mutant KRAS Lung Cancer. Clin Cancer Res, 2016. 22(23): p. 5851–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Infante JR, et al. , Safety, pharmacokinetic, and pharmacodynamic phase I dose-escalation trial of PF-00562271, an inhibitor of focal adhesion kinase, in advanced solid tumors. J Clin Oncol, 2012. 30(13): p. 1527–33. [DOI] [PubMed] [Google Scholar]
- 17.Simon R, Optimal two-stage designs for phase II clinical trials. Control Clin Trials, 1989. 10(1): p. 1–10. [DOI] [PubMed] [Google Scholar]
- 18.Kim ES, et al. , Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet, 2008. 372(9652): p. 1809–18. [DOI] [PubMed] [Google Scholar]
- 19.Douillard JY, et al. , Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol, 2010. 28(5): p. 744–52. [DOI] [PubMed] [Google Scholar]
- 20.Ihle NT, et al. , Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst, 2012. 104(3): p. 228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shepherd FA, et al. , Pooled Analysis of the Prognostic and Predictive Effects of TP53 Comutation Status Combined With KRAS or EGFR Mutation in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol, 2017. 35(18): p. 2018–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skoulidis F, et al. , Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov, 2015. 5(8): p. 860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding L, et al. , Somatic mutations affect key pathways in lung adenocarcinoma. Nature, 2008. 455(7216): p. 1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imielinski M, et al. , Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell, 2012. 150(6): p. 1107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinidou G, et al. , RHOA-FAK is a required signaling axis for the maintenance of KRAS-driven adenocarcinomas. Cancer Discov, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepherd FA, et al. , Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol, 2013. 31(17): p. 2173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janne PA, et al. , Impact of KRAS codon subtypes from a randomised phase II trial of selumetinib plus docetaxel in KRAS mutant advanced non-small-cell lung cancer. Br J Cancer, 2015. 113(2): p. 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim ES, et al. , The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov, 2011. 1(1): p. 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blumenschein GR Jr., et al. , A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)dagger. Ann Oncol, 2015. 26(5): p. 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter CA, Rajan A, and Szabo E, Two parallel randomized phase II studies of selumetinib (S) and erlotinib (E) in advanced non-small cell lung cancer selected by KRAS mutations. J Clin Oncol, 2013. 31: p. 8026. [Google Scholar]
- 31.Jones SF, et al. , A phase I study of VS-6063, a second-generation focal adhesion kinase inhibitor, in patients with advanced solid tumors. Invest New Drugs, 2015. 33(5): p. 1100–7. [DOI] [PubMed] [Google Scholar]
- 32.Fennell DA, et al. , Maintenance Defactinib Versus Placebo After First-Line Chemotherapy in Patients With Merlin-Stratified Pleural Mesothelioma: COMMAND-A Double-Blind, Randomized, Phase II Study. J Clin Oncol, 2019. 37(10): p. 790–798. [DOI] [PubMed] [Google Scholar]
- 33.Gerber DE, et al. , Baseline tumour measurements predict survival in advanced non-small cell lung cancer. Br J Cancer, 2013. 109(6): p. 1476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang Y, et al. , Role of focal adhesion kinase in regulating YB-1-mediated paclitaxel resistance in ovarian cancer. J Natl Cancer Inst, 2013. 105(19): p. 1485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavazzoni A, et al. , Enhanced efficacy of AKT and FAK kinase combined inhibition in squamous cell lung carcinomas with stable reduction in PTEN. Oncotarget, 2017. 8(32): p. 53068–53083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francois RA, et al. , Targeting Focal Adhesion Kinase and Resistance to mTOR Inhibition in Pancreatic Neuroendocrine Tumors. J Natl Cancer Inst, 2015. 107(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee YC, et al. , Identification of Bone-Derived Factors Conferring De Novo Therapeutic Resistance in Metastatic Prostate Cancer. Cancer Res, 2015. 75(22): p. 4949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.