Abstract
Mitochondria are essential organelles within the cell where most ATP is produced through oxidative phosphorylation (OXPHOS). A subset of the genes needed for this process are encoded by the mitochondrial DNA (mtDNA). One consequence of OXPHOS is the production of mitochondrial reactive oxygen species (ROS), whose role in mediating cellular damage, particularly in damaging mtDNA during ageing, has been controversial. There are subsets of neurons that appear to be more sensitive to ROS-induced damage, and mitochondrial dysfunction has been associated with several neurodegenerative disorders. In this review, we will discuss the current knowledge in the field of mtDNA and neurodegeneration, the debate about ROS as a pathological or beneficial contributor to neuronal function, bona fide mtDNA diseases, and insights from mouse models of mtDNA defects affecting the central nervous system.
Keywords: mitochondrial DNA, neurodegeneration, reactive oxygen species
Most cellular ATP is produced in the mitochondria through oxidative phosphorylation (OXPHOS). The majority of the genes needed to make up the five OXPHOS complexes are encoded in the nuclear DNA (nDNA). On the other hand, mitochondrial DNA (mtDNA) encodes for a small number of genes, all OXPHOS related. Damaged mtDNA has been implicated in normal ageing and many neurodegenerative disorders. Furthermore, patients with bona fide mitochondrial diseases frequently have neurological impairments. There are many pathological insults which can damage mtDNA, but one which has been more commonly discussed is oxidative damage through mitochondrial reactive oxygen species (ROS). Although ROS can cause cellular damage, and there is a link between increased ROS and oxidative damage in neurodegenerative diseases, the physiological roles for ROS in cellular health and viability are unclear.
Studies with cultured cells showed that some mtDNA mutations can be associated with higher ROS levels, whereas others are not [1,2]. In postmortem brains from aged patients or patients with neurodegenerative disease low levels of mtDNA damage have been reported; however, there is no direct correlation between these low levels of mtDNA mutations and elevated ROS [3]. There are now several mouse models of mtDNA damage which affect the central nervous system (CNS) available, which are helping better understand the link between mtDNA damage, ROS and neurodegeneration.
Mitochondrial DNA
Structure and function
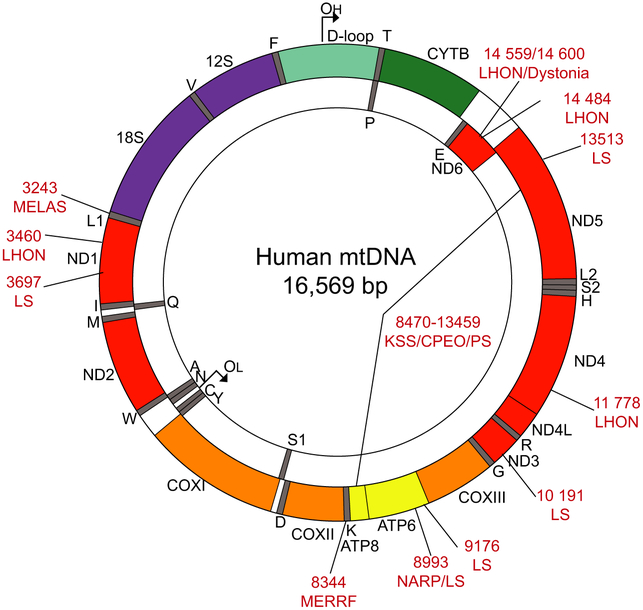
Human mtDNA, which was first discovered in 1963, is a 16 569 bp circular, double-stranded, supercoiled molecule which encodes for 37 genes, essential for OXPHOS and mitochondrial protein synthesis [4,5]. The OXPHOS system is comprised of five multisubunit enzymatic complexes located on the inner mitochondrial membrane. MtDNA encodes for 13 subunits, one or more of the essential subunits for the NADH-ubiquinone oxidoreductase (Complex I), ubiquinone-cytochrome c oxidoreductase (Complex III), cytochrome c oxidase (Complex IV) and the ATP synthase (Complex V), while the entirety of the succinate-ubiquinone oxidoreductase (Complex II) is encoded by the nDNA [6]. In addition to these 13 subunits, the mtDNA also encodes for 22 tRNAs and 2 rRNAs (Fig. 1). The mtDNA strands are termed the heavy strand (H-strand) and light strand (L-strand), where the former is characterized as being guanine rich and the latter is cytosine rich [7]. Twenty-eight genes are encoded on the H-strand, while the remaining nine are encoded on the L-strand.
Fig. 1.
Schematic representation of human mitochondrial DNA. The figure represents the 16 569 bp human mtDNA. The regulatory region, also known as the D-loop containing OH is depicted in teal; Complex I genes in red; Complex III gene in green; Complex IV genes in orange, Complex V genes in yellow; tRNAs in grey and rRNAs in purple. Mutations and deletions and their associated disorders which are discussed in the text are annotated with red text.
Mitochondrial DNA exists in cells in multiple copies
In most cells, there are approximately 1000 mitochondrial genomes [8]. Comparatively, there are only two copies of the nuclear genome in a cell. The levels of mtDNA molecules are generally dependent on the cellular energy demands of a cell. MtDNA replication is independent of the cell cycle, and there are few enzymes that are known participants in this process [9]. The mitochondrial polymerase, POLG, and the mtDNA helicase, Twinkle, are two of these players; mutations in both of these genes have been implicated in mtDNA abnormalities and mitochondrial diseases [10–13].
Due to this high copy number, often mutated and wild-type mtDNA molecules exist together in a single cell (mtDNA heteroplasmy) [14]. Because mtDNA replication is cell-cycle independent, and mtDNA can be segregated during replication, heteroplasmy levels are dynamic, and can change during a lifetime in both mitotic and postmitotic cells/tissues [9].
Along with the nature of the mutation, the percentage of heteroplasmy is the major factor which determines the clinical severity of mitochondrial diseases. There is a biochemical threshold associated with mutant mtDNA percentage, that must be surpassed for decreased mitochondrial function and phenotype development [15]. While this threshold is dependent on the mutation, the cell- and tissue-type, heteroplasmy threshold levels can be between 70% and 90% for an detectable phenotype to present [16].
Mitochondrial DNA damage
Damage or replication errors to mtDNA result in point mutations or rearrangements, which can either be inherited or sporadic. MtDNA mutations had a reported prevalence ranging between 1 : 5000 and 1 : 500 000, and affect mitochondrially encoded proteins, tRNAs, rRNAs, and therefore eventually ATP production [14,17].
Point mutations are generally maternally inherited, but can occur sporadically [18]. Most pathogenic mtDNA point mutations are heteroplasmic [18]. The mechanism of point mutation formation is likely through inefficiency of the mtDNA repair system [19]. In comparison, mtDNA rearrangements, like large-scale deletions (1.8–8 kb) remove large portions of mtDNA. This ablates several mitochondrial genes encoding for proteins, tRNAs or rRNAs, depending on the size and location of the deletion. MtDNA rearrangements are almost exclusively sporadic and invariably exist in heteroplasmy [20]. The mechanism for the formation of mtDNA rearrangements is somewhat controversial, but it is thought they could form from errors in replication, double-strand breaks or inefficiency of the mtDNA repair systems [19]. MtDNA deletions have also been found to accumulate in postmitotic tissues during normal ageing [21–25].
Pathological and physiological consequences of reactive oxygen species
Generation of reactive oxygen species
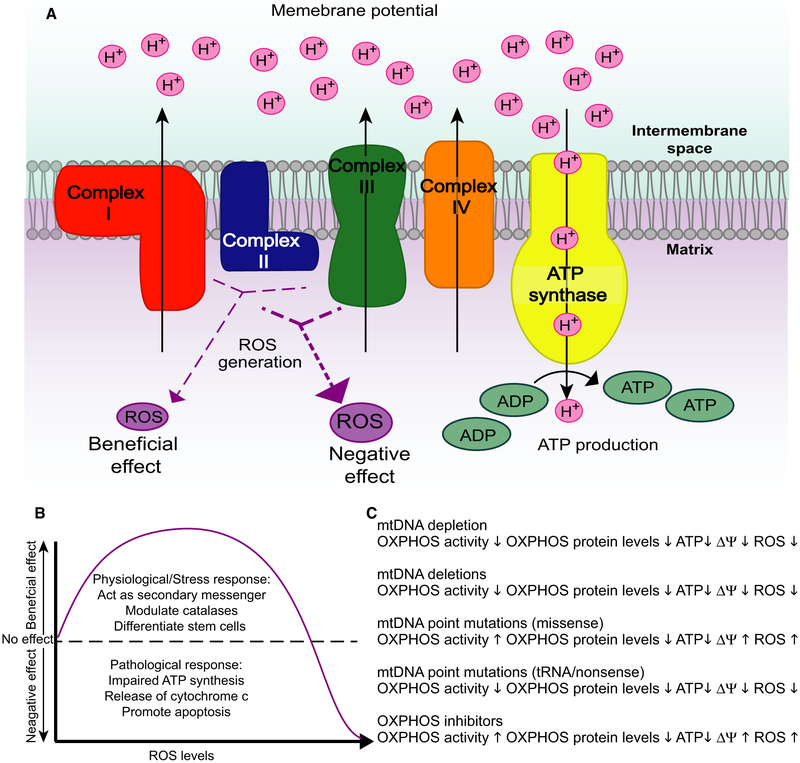
While ROS can be generated in multiple cellular compartments, the vast majority of cellular ROS, approximately 90%, are generated in the mitochondria during the production of ATP through OXPHOS (Fig. 2A) [26]. Mitochondrial ROS are primarily produced at Complexes I and III of the electron transport chain, with electrons that either derive from NADH or FADH2 to react with O2 [27]. There is evidence of ROS production at Complex II, but relative to the rates of production at Complexes I and III, this contribution is negligible [28,29]. Superoxide () is the source of most ROS and is generated by the single electron reduction in O2 [26]. Superoxide molecules are further reduced to hydrogen peroxide (H2O2) by mitochondrial superoxide dismutase 2 (SOD2) [26]. Following the catalase reaction to convert H2O2 into H2O and O2, the O2 can then be reduced again by electrons escaping the respiratory chain [29]. Generally, the single-electron reactions predominate in ROS production, but there is evidence for two-electron reactions which allow for the direct reduction from molecular O2 to H2O2 [30]. Complex I also produces large amounts of superoxide through reverse electron transport (RET). RET occurs when a reduced CoQ pool forces electrons back from CoQH2 into Complex I, and reduce NAD+ to NADH at the FMN site [31].
Fig. 2.
Schematic representation of the oxidative phosphorylation system and downstream consequences. (A) The respiratory chain is composed of four complexes (I–IV) and the ATPase, which together transfer electrons in a step-wise manner to reduce O2 to H2O. This electron transfer is coupled with creating a proton gradient across the mitochondrial inner membrane, and this electrochemical gradient drives ATP synthesis as protons re-enter the mitochondrial matrix through ATP synthase. One consequence of oxidative phosphorylation is the production of ROS, which occurs at Complexes I and III. (B) When ROS generation is low, there it can serve as a physiological signalling molecule or in response to low levels of cellular stress, however, when ROS generation becomes too great, there are pathological consequences of ROS, including signalling apoptosis. (C) Different mtDNA defects have different downstream consequences which affect OXPHOS activity, OXPHOS protein levels, ATP production, membrane potential, and ROS generation.
There are at least eight known sites of mitochondrial superoxide production; of these, only one is known to deposit in the mitochondrial intermembrane space, while the other sites are only known to deposit into the mitochondrial matrix [32,33]. One could speculate that the superoxide from this single site would contribute more to cytosolic signalling than the other seven sites, as those molecules would have the additional step of exiting the matrix into the intermembrane space, however, the physiological relevance of ROS production at each of these sites has not yet been determined [32].
Pathological consequences of ROS
Historically ROS production was thought to solely be the upstream step in oxidative damage to mitochondrial proteins, membranes and mtDNA [29]. This is due in part to ROS generation occurring in the mitochondrial matrix, where mtDNA resides. Additionally, ROS can impair the ability of mitochondria to synthe-size ATP and carry out their wide range of metabolic functions including fatty acid oxidation, the TCA cycle, the urea cycle, amino acid metabolism and haeme synthesis [34]. Oxidative damage can increase the tendency of mitochondria to release cytochrome c through the mitochondrial permeability transition pore leading to activation of the apoptosis cascade [35]. Potential pathological outcomes of ROS production also include the formation of mutations or deletions to mtDNA, oxidative damage to the respiratory chain, lipid peroxidation and overall mitochondrial dysfunction [36].
In the brain, as with most organs, the response to oxidative damage is not uniform [37]. While there are many neuronal subtypes that are able to deal with this rise in oxidative stress, there are select neuronal populations which have a higher susceptibility to elevated ROS [38]. Neurons which undergo dopamine metabolism are more susceptible to ROS-induced damage as well as neurons which have higher metal content [39]. The Surmeier group has shown that a specific oxidant stress in dopaminergic neurons, due to the engagement of L-type calcium channels, causes a mild mitochondrial uncoupling, leading to mitochondrial dysfunction [40].
ROS as a physiological signalling molecule
More recently there has been a growing field of research into the physiological roles for ROS. There is evidence against the singular role of ROS as a mediator of cellular damage. While high levels of ROS are associated with cellular dysfunction it is now known that ROS is necessary for physiological cellular function. Additionally, ROS is an important regulator of intracellular signalling pathways, including but not limited to controlling proliferation, cell death, and senescence [41,42]. The cyclic oxidation/reduction in cysteine residues in kinases, phosphatases, and other regulatory factors by ROS modulates the strength and duration of signalling through redox-sensitive signalling pathways. ROS acts as a second messenger, modulating cytokines, growth factors whose activity regulates classical signalling cascades under physiological and stress-conditions, including the ERK, JNK, and MAPK cascades, and the JAK/STAT pathway [43]. Additionally, ROS can modulate the activities of enzymes such as catalase, GPxs and Prdx which are regulated by kinases and phosphatases, leading to a regulatory network [44]. Finally, ROS has been found to play a significant role as one of the molecular mechanisms which guides stem cells to either differentiate or renew. [45–47].
Free radical theory of ageing versus the gradual ROS response hypothesis
The free radical theory of ageing which was first proposed in the 1950s by Harman, and later refined to the mitochondrial free radical theory of ageing in the 1970s was based on the idea that somatic mtDNA mutations would impair OXPHOS complexes, particularly Complexes I and III [48,49]. With the impairment of Complexes I and III a corresponding increase in ROS production was hypothesized, leading to a vicious cycle of further damaging mitochondrial proteins, lipids and mtDNA. Studies done in aged cortical samples showed a concomitant decrease in mitochondrial function and an increase in mtDNA point mutations and deletions, which is compatible with the free radical theory of ageing. Other evidence that supports this theory includes the fact that: (a) the respiratory chain is an inherent source of superoxides, (b) there is an increased production of ROS in aged tissues, and (c) the assumption that mtDNA is more susceptible to oxidative damage than nDNA.
While there is circumstantial evidence that supports the mitochondrial free radical theory of ageing, there is an increasing number of studies which do not support it. In a mouse model of an exonuclease-deficient Polg, while there is an increase in mtDNA point mutations and deletions and the mice age prematurely, there is no evidence of increased oxidative damage to mitochondrial proteins, lipids or mtDNA [50–53]. In a different study done in mice lacking one copy of the enzyme Mclk1, which is necessary for the synthesis of the antioxidant ubiquinone, there is an increase in mitochondrial oxidative stress, but these mutant mice have an extended lifespan over their wild-type siblings [54]. Additionally, these Mclk1+/− mice have a slower decline in mitochondrial function and a slower development of an aged phenotype. In two models of mutations in mitochondrial respiratory subunits isp-1 (Complex III) and nuo-6 (Complex I) in Caenorhabditis elegans, there is an increase in lifespan of the mutant worms compared to the wild-type, even with an increase in superoxides [55]. Contrary to the expected results, the addition of the antioxidants NAC and vitamin C reduced the lifespan in the mutants, indicating a lifespan role for the superoxide molecules. Mouse models with a Complex IV conditional KO showed no increase in oxidative stress [56,57].
Based on these observations, an alternative theory (Mitohormesis), which was proposed in 2011 by Hekimi and colleagues, states that there is a gradual ROS response between ROS and ageing [58]. ROS can at as a signalling molecules to modulate the stress response pathway, and small increases in ROS levels can extend lifespan. In yeast, the reduction in TORC1 signalling has been demonstrated to extend chronic lifespan [59]. This extension is directly associated with a cell-intrinsic regulation of mitochondrial respiratory coupling which elevates mitochondrial membrane potential and ROS. Based on this, the gradual ROS response hypothesis proposes that cellular insults trigger protective stress responses where ROS would act as a secondary messenger. However, at a certain point this age-dependent damage would increase past a certain threshold where ROS signalling would be sustained and maladaptive (Fig. 2B). The mitohormesis hypothesis addresses some of the inconsistencies that are seen with the mitochondrial free radical theory of ageing. There is also a growing body of research that suggests that oxidative damage increases in the mammalian brain during ageing [37].
Mitochondrial DNA diseases affect the central nervous system
Studies done over the last decades have demonstrated that the link between elevated ROS and mtDNA damage is not clear. There are several mitochondrial disorders which have a CNS-dysfunction component, but the levels of heteroplasmy are variable. Additionally, there are several neurodegenerative disorders that are characterized by mitochondrial dysfunction, many with mutated or damaged mtDNA, and some with generalized oxidative damage. In the next sections, we will be discussing neurodegenerative diseases and disorders that have a mtDNA component in the pathogenesis, and how the mtDNA mutations may affect ROS levels.
Biochemical consequences of mitochondrial DNA mutations
There are over 330 pathogenic point mutations that have been identified in the human mtDNA in the last 30 years [6]. These mutations are located across the mtDNA molecule. A number of these point mutations lead to mitochondrial encephalopathies (Fig. 1). The generation and consequences of such mutations are still being investigated.
Leber’s hereditary optic neuropathy (LHON) was the first reported inheritable mtDNA disease, and is thought to be one of the most prevalent of the mitochondrial diseases [60]. There are a number of mutations leading to LHON, but the three most common mutations in European populations are: G11778A in ND4, G3460A in ND1, and T14484C in ND6; these mutations tend to present as homoplasmic mutants (Fig. 1) [60–62]. There are, however, other point mutations which lead to a more severe Complex I deficiency, which present as heteroplasmic mutants, including G14459A or G14600A, both in ND6 (Fig. 1) [63,64]. Cybrid cells, cell lines generated from LHON patientderived platelet mitochondria fused with human ρ0 (mtDNA-less) cells, have revealed Complex I deficiencies, Site I respiration defects, reduced ATP production, increased sensitivity of the mitochondrial permeability transition pore, and increased ROS production [65].
Neuropathy, ataxia, and retinitis pigmentosa (NARP) is a mitochondrial disorder which most commonly originates from the point mutation T8993G/C in ATPase 6 (Fig. 1) [16,66]. When the levels of the mutant mtDNA population is high, infants present with Leigh syndrome (LS), a subacute neuronal degeneration of structures in the basal ganglia [67]. Patient-derived cells with 50% heteroplasmy or homoplasmic for the T8993G mutation showed increases in ROS (via H2O2) through CMH2-DCFDA measurements, and mitochondrial SOD (MnSOD/SOD2), but cytoplasmic SOD (CuSOD/SOD1) [1]. A negative correlation between ATP synthesis and increasing T8993G mutation load has also been reported [2,68].
Mitochondrial myopathy, encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS) is a mitochondrial disorder where about 80% of patient cases are caused by the point mutation A3243G in the mitochondrial tRNALeu(UUR) gene (Fig. 1) [6,69,70]. Patient cybrids with 90% of the A3243G mutation showed increased ROS through MitoSOX Red, which was reduced upon incubation with CoQ or Riboflavin [71].
Compared to other mitochondrial diseases, LS is unique in that is associated with both mutations in the mtDNA and the nDNA [72–74]. LS mtDNA mutations have been found in genes encoding for Complexes I, IV, and V, as well as tRNAs, and are commonly found in heteroplasmy [75,76]. Many of the point mutations associated with LS are found in Complex I genes: G3697A in ND1, T10191C in ND3, and G13513A in ND5, while some are also found in other genes including T9176C in ATPase 6, T8993G/C in ATPase 6 (also associated with NARP) (Fig. 1) [77–79]. Patient-derived cells with the three Complex I point mutations described above showed increased ROS by CMH2-DCFDA as well as decreased SOD2 protein expression [80].
There are three common phenotypes associated with large mtDNA deletions: Pearson syndrome (PS), chronic progressive external ophthalmoplegia (CPEO), and Kearns-Sayre syndrome (KSS), all of which are multisystemic disorders [81,82]. Of the large deletions that are seen in patients, the most common is a 4977 bp deletion (Δ4977) known as the common deletion (Fig. 1) [83]. Cybrid cells harboring 99% of Δ4977 mtDNA showed an increase in ROS using the dye HPF [84]. Although cybrid studies with certain pathogenic mtDNA mutations suggest an increase in ROS, there are no compelling reports showing oxidative damage as a prominent pathological finding in mitochondrial encephalopathies [85].
Mitochondrial DNA defects and ROS in ageing and age-related neurodegeneration
Ageing
MtDNA damage, mutations and deletions have been reported to increase in an age-dependent manner in both the human and rodent CNS. Whether the accumulation of mutated mtDNA has a causal role in ageing, it has been suggested that mutated mtDNA can serve as a biomarker of ageing, independent of the lifespan of the organism [86]. The levels of mutant mtDNA often vary substantially between different cells in the same tissue of an affected patient [87]. There are reports which show the levels of deleted or mutated mtDNA is variable in different regions of the adult human brain, with the highest levels in the substantia nigra, putamen and cerebral cortex [87]. While the levels of these damaged mtDNA molecules is very low in normal ageing, around 1%, in patients with neurodegenerative diseases such as Parkinson’s disease (PD; reviewed in more detail below) can have higher levels of the mutant mtDNA species [88].
Oxidative damage has been associated with CNS ageing as well, however, the link between oxidative damage and age-related mtDNA damage is still poorly understood [89,90].
Parkinson’s disease
Parkinson’s disease is the second most common neurodegenerative disorder, affecting about 1% of the population over the age of 60 [91]. PD is characterized by rigidity, tremor and postural instability, as well as bradykinesia. Histology studies done in postmortem brain slices show that, while there is neuronal loss in many brain areas, the dopaminergic neurons of the substantia nigra (SN) pars compacta are preferentially depleted [22,92]. Another histological hallmark of PD is seen in the surviving neurons where there is an accumulation of α-synuclein aggregates [93]. The A53T a-synuclein mutation in humans is associated with familial PD, and impairs vesicular dopamine storage, leading to increased cytosolic dopamine, which interacts with iron, generating ROS [94,95].
While the etiology of PD is still unknown, mitochondrial dysfunction, particularly dysfunction of Complex I has been proposed as a contributing factor [96]. Rotenone and 1-metyl-4phenyl-1,2,3,6-tetrahydropyridine, both Complex I inhibitors, promote PD like symptoms and the formation of α-synuclein containing protein aggregates [97]. Two provocative studies in 2006 showed that dopaminergic neurons of aged humans have high levels of mtDNA deletions [22,24]. This has been further observed in early-stage PD patients [98]. These observations suggested that dopaminergic neurons are close to a threshold for a biochemical defect and cell death.
Complex I defects have been reported in different tissues of PD patients [99]. Additionally, relatively high levels of mtDNA deletions have been found in the substantia nigra of ageing patients and PD individuals. These relatively high levels (40–60%) of mtDNA deletions were associated with a cytochrome c oxidase (COX) deficiency in individual neurons [22,24]. These studies have been confirmed and extended to rodent models [100–102]. It was reported that mtDNA levels are also decreased in PD [103]. Moreover deficiency in maintaining the wild-type mtDNA pool, but not the deleted one, suggested that dysregulation of mtDNA homeostasis is an important process in the pathogenesis of neuronal loss in PD [100].
Other age-related neurodegenerative diseases linked to mitochondrial dysfunction and oxidative damage
In the case of other age-associated neurodegenerative diseases there are associations between disease progression and elevated ROS, suggesting mitochondrial dysfunction. However, there is no clear link between damaged or mutated mtDNA (particularly the common deletion and disease pathogenesis.
In Alzheimer’s disease (AD), the most common late-onset progressive neurodegenerative disorder, defects in cytochrome c oxidase have been implicated in the progression of the disease [3]. Amyloid beta (Aβ) fragments form cytotoxic plaques, which are more commonly found in the cortex and hippocampus [104,105]. Mitochondrial function is negatively affected by Aβ fragments, suggesting that the mitochondrial dysfunction is a consequence of Aβ toxicity [106–108]. The ‘mitochondrial cascade hypothesis for AD’ suggested that an individual’s genes for the proteins which make up the respiratory chain determines the inherent ROS production, which would then govern the severity of oxidative damage. Furthermore, it was suggested that with this accumulation of oxidative mtDNA damage, which would in turn lead to decreased ATP levels, increased oxidative stress, and finally Aβ toxicity, which would then cycle back to ultimately lead to neurodegeneration [109]. While studies in AD patients have not shown a causative mtDNA mutation that would link to the disease pathogenesis, an increased aggregate burden of individually rare point mutations has been seen in AD patients compared to young controls, but not age-matched controls [110]. As with PD, the concern in assessing for mtDNA damage in patients is the late-stage of AD disease progression, where the neurons with high levels of mtDNA damage may have already been lost. When assessing early-stage AD patients, an increase in mtDNA mutation frequency was seen in the hippocampus; however, these mutations were found to be due to replication errors, and not oxidative damage [111]. Numerous studies into the prevalence of the common deletion in AD patients and age-matched controls were inconclusive. Additionally, studies done looking at different mtDNA haplogroups as risk factors for AD are controversial [112].
Amyotrophic lateral sclerosis (ALS) is an adultonset neurodegenerative disorder characterized by the loss of motor neurons in the motor cortex, brainstem and spinal cord [113]. Mutations in the gene superoxide dismutase 1, SOD1, have been identified as an ALS gene, so there are potential implications for oxidative damage of mtDNA [114]. Numerous studies have shown increased oxidative damage to proteins in ALS postmortem samples compared to controls, as well as increased protein carbonyl levels in the motor cortex [115]. In a transgenic mouse model of ALS which expresses mutant human SOD1 (G93A), there was increased oxidative damaged to proteins, lipids and DNA; interestingly one of the most oxidized proteins was the mutant SOD1 itself [116,117]. Histology in ALS patient spinal cords showed a specific neuronal decrease in COX activity [118]. Patients with ALS were found to only have slightly higher levels of the common deletion in muscle and the motor cortex compared to age-matched controls, but not in other brain regions [113,119]. Because the levels of the deletion are still relatively low, there is no causative link between the common deletion and ALS. Studies on the association of mtDNA haplogroups and ALS are also controversial [120].
Multiple sclerosis (MS) is a chronic, inflammatory disease caused by the loss of myelin and gliosis [121,122]. Though the etiology of MS remains elusive, mitochondrial defects are increasingly recognized to play a role in disease pathogenesis. In patient brains, there is a reduction in the activities of Complexes I and III, and a decrease in COX/SDH staining in neurons compared to age-matched controls [121,122]. Multiple groups have reported on the presence of mtDNA deletions (not necessarily the common deletion) and mutations in patients in with MS [123]. There is not a single mtDNA mutation or deletion associated with MS, so the direct role of mtDNA in disease pathogenesis is still up for debate [124].
Mouse models of mitochondrial DNA damage in the central nervous system
As described previously, mtDNA changes have been implicated in several human neurodegenerative diseases as well as normal ageing. Mouse models serve as viable options to study the mechanisms behind these pathologies. Unfortunately, there are no robust techniques to manipulate the mtDNA. Moreover, even when an endogenous mutation is identified, it is a technically challenging endeavour to create a mouse model of specific mtDNA damage as the mutations or deletions may not pass down to the progeny, due to the ‘bottleneck effect’ during fertilization, and the segregation of the pathological mtDNA molecules in different tissues. One of the first mouse models of mtDNA dysfunction was the mitoMouse, reported in 2000, which contains a large ageing-derived mtDNA deletion [125]. However, due to mtDNA segregation during development, the predominant phenotype is renal failure, along with low body weight, lactic acidosis, ischaemia, myopathy, heart block, deafness and male infertility with no evidence of oxidative damage [126,127].
Double-strand breaks in mitochondrial DNA do not induce reactive oxygen species in the central nervous system
One of the first models of mitochondrial DNA damage is the mitoPstI mouse, which expresses a mitochondrial-targeted restriction endonuclease, PstI [128]. MitoPstI expression can be induced with doxycycline in a tissue-specific manner depending on the promoter that controls transgene expression [129]. In the mouse mtDNA, there are two recognition sites for PstI, so mitoPstI would create double-strand breaks in the mtDNA at these two specific sites, primarily leading to the depletion of mtDNA, and in rare cases the formation of a smaller deleted mtDNA (DmtDNA) molecule. Double-strand breaks could serve as the mechanism behind the generation of age-related mtDNA deletions.
When mitoPstI was expressed in neurons (under the control of the CamKIIα promoter) the phenotype was different depending on the age of induction, as well as the intensity of the induction. When mitoPstI was strongly expressed from birth, the mice developed a limb-clasping behaviour at 2 months, indicative of a neurodegenerative disorder [130]. When mitoPstI expression was repressed until P21, then induced for adulthood, the mice did not develop this limb-clasping behaviour, but did progressively become less active, and died before P100. These mice also showed decreased COX activity in the forebrain, but not in the cerebellum, consistent with CamKIIα expression patterns. Southern blot analyses of the different brain regions only showed mtDNA depletion, and did not show significant amounts of a ΔmtDNA molecule, so the depletion accompanied by the low levels of the ΔmtDNA molecule contributed to this neurodegenerative phenotype. When mitoPstI was not expressed as strongly, induced mice were able to survive up to 16 months (we now have evidence that these mice can survive up to 24+ months), with no obvious phenotype until 6–8 months of age and a specific, progressive neurodegeneration in the striatum and cortex [131].
The mitoPstI mouse has also been crossed with a mouse model of AD which shows Aβ plaques forming in the cortex and hippocampus. When mitoPstI was induced for 2 months, starting at 4 months of age, 6 month old mice showed a reduction in the amyloid plaques [56,132]. As discussed above, the amyloid plaques were associated with oxidative damage, and the decreased plaque formation with mitoPstI expression indicates the depletion of mtDNA was not associated with increased ROS. Because there is a depletion of mtDNA with mitoPstI expression, the steady-state level of the OXPHOS complexes decreases, leading to a decrease in ROS formation.
Increased reactive oxygen species in a mouse model of Leber’s hereditary optic neuropathy does not lead to decreased ATP production
A mouse model of LHON was created by Lin et al. in 2012 [133]. These mice are homoplasmic for the mtDNA mutation G13397A in ND6 which causes the amino acid substitution P25L. This is the same amino acid substitution seen in a family of patients with optic atrophy and LS. The ND6 G13397A P25L mouse exhibited reduced retinal response, axonal swelling of retinal ganglion cells, loss of the smallest axons in these RGCs, abnormal mitochondrial morphology and proliferation RGC axons, and reduced Complex I activity in isolated synaptosomes. The authors found that synaptosomes have increased ROS production, while energy levels remained constant. As discussed previously, there is also an increase in oxidative damage seen in patients with LHON that is not accompanied by a decrease in ATP, which this mouse model is able to readily recapitulate.
Increased reactive oxygen species production is not always detected in models of deficient mitochondrial DNA maintenance
Because Polg knockout mice are embryonic lethal, two different groups created exonuclease-deficient Polg mouse models, known as the ‘mutator’ mouse [50,51]. As the mutator mouse has impaired proofreading capability, these mice accumulate mtDNA point mutations and deletions in different tissues. While they did exhibit a premature ageing phenotype, reduced lifespan, kyphosis, alopecia, weight loss, decreased fertility, osteoporosis and reduced fat content, these mice did not show obvious signs of neurodegeneration [50]. Interestingly, when studying the mtDNA point mutation and deletion load in the mutator mouse, deletions correlate well with the premature ageing phenotype [52,53]. Later studies showed that point mutation in protein coding genes also correlated well with the phenotype [134]. Next generation sequencing done on brain mtDNA from the mutator mouse showed the accumulation of a mutant species of mtDNA with an abnormal D-loop structure, termed control region multimers (CRMs). These CRMs might reflect disrupted mtDNA replication, but the functional consequence is not known [135]. While, several studies have been done and concluded that there is no evidence of oxidative damage in the mutator mouse [134,136], the Suomalainen group has found the neural stem cell population was reduced in the mutator mouse [137]. This reduction could be rescued with the antioxidant NAC, suggesting that alterations in ROS affect the viability of neural stem cells [137].
The Twinkle mouse was made with A360T amino acid substitution or an in frame amino acid duplication at position 353–365, which are seen in patients with PEO [138]. These mutant Twinkle mice are known as the ‘deletor’ mice as they have multiple mtDNA deletions, along with progressive respiratory dysfunction and late-onset mitochondrial disease. The multiple deletions seen in the deletor mouse have been attributed to replication pausing or stalling, but this can also lead to mtDNA depletion, which is seen in PEO patients [10,139]. Furthermore, there was no evidence of oxidative damage or increased ROS in the deletor mouse [10,138].
From these studies, there is no evidence of increased ROS production, as a consequence of the increased mtDNA mutations in the mutator mouse and deletion loads in the deletor mouse, giving further strength to the mitohormesis theory (Fig. 2C) [10,134,136,138].
Mitochondrial DNA mutations and oxidative damage in the CNS: what is the relationship?
Mitochondria are essential organelles which provide most of the cellular energy through oxidative phosphorylation. Mutations and deletions in the essential genes that are encoded by the mtDNA can dramatically disrupt cellular function. When these pathogenic defects occur in the CNS, neurons are particularly affected due to their high energy quota. Mitochondrial ROS have been suggested to increase mtDNA mutations, but recent studies suggest that ROS have a more important signalling role rather than a harmful one. Therefore, increases in ROS are not necessarily associated with increases in mitochondrial dysfunction or increased mtDNA mutation load in the CNS.
Mitochondrial diseases stemming from mtDNA point mutations and deletions present a wide clinical spectrum of phenotypes. Patient-derived cell lines have been used to better understand the biochemical features associated with these defects. In many cases, the pathogenic mutations were associated with increases in ROS levels in cultured cells, while oxidative damage was not clearly documented in patients. Damage to mtDNA, including point mutations and deletions have also been reported to increase in ageing, but the correlation between accumulating mutated mtDNA as a cause of ageing has not been confirmed. In aged patients, the levels of damaged mtDNA molecules are very low, around 1%, but higher levels can be seen in specific brain regions, such as the substantia nigra, or in neurodegenerative diseases, like Parkinson’s. However, still, the increased frequency of mutated mtDNA is not associated with increased oxidative damage. Although there is strong evidence for oxidative stress in age-related neurodegenerative disorders, particularly PD and AD, it is unclear how this relates to mtDNA alterations.
The current knowledge in the field leads us to the conclusion that certain mtDNA alterations, such as the ones associated with reduced mitochondrial protein synthesis (e.g. tRNA mutations or deletions) are commonly associated with a decrease in the steady-state levels of OXPHOS complexes and reduced ROS (Fig. 2C). In contrast, mtDNA changes that result in normal levels of OXPHOS complexes with altered biochemistry (e.g. LHON or NARP mutations), are more likely to increase ROS beyond physiological concentrations, causing neuronal injury (Fig. 2C). In either case, the levels of ROS could have different consequences depending on the cell type and effective concentration, as stated by the mitohormesis theory.
The role of mtDNA damage and ROS in neurodegeneration is far from clear, but recent evidence changed our negative perception of ROS. At the same time, it shed light on the complexity of mitochondrial genetics and its relationship with ROS-induced signalling and damage. We expect that experiments with in vivo models will help resolve some of the remaining questions.
Acknowledgements
Our work was supported by the National Institutes of Health Grants 1R01AG036871, 5R01EY010804 and 1R01NS079965 (CTM); American Heart Association 16PRE30480009 and Lois Pope LIFE Fellows Program (NN). We acknowledge support from the NEI center grant P30-EY014801 from the National Institutes of Health (NIH).
Abbreviations
- AD
Alzheimer’s disease
- ALS
amyotrophic lateral sclerosis
- CNS
central nervous system
- CPEO
chronic progressive external ophthalmoplegia
- CRM
control region multimers
- KSS
Kearns-Sayre syndrome
- LHON
Leber’s hereditary optic neuropathy
- LS
Leigh syndrome
- MELAS
mitochondrial myopathy, encephalomyopathy, lactic acidosis and stroke-like episodes
- MERRF
myoclonic epilepsy with ragged red fibers
- MS
multiple sclerosis
- mtDNA
mitochondrial DNA
- NARP
neuropathy, ataxia, and retinitis pigmentosa
- nDNA
nuclear DNA
- OXPHOS
oxidative phosphorylation
- PD
Parkinson’s disease
- PS
Pearson syndrome
- RET
reverse electron transport
- ROS
reactive oxygen species
References
- 1.Mattiazzi M, Vijayvergiya C, Gajewski CD, DeVivo DC, Lenaz G, Wiedmann M and Manfredi G (2004) The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet 13, 869–879. [DOI] [PubMed] [Google Scholar]
- 2.Nijtmans LG, Henderson NS, Attardi G and Holt IJ (2001) Impaired ATP synthase assembly associated with a mutation in the human ATP synthase subunit 6 gene. J Biol Chem 276, 6755–6762. [DOI] [PubMed] [Google Scholar]
- 3.Coppede F and Migliore L (2015) DNA damage in neurodegenerative diseases. Mutat Res 776, 84–97. [DOI] [PubMed] [Google Scholar]
- 4.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F et al. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 5.Nass MM and Nass S (1963) Intramitochondrial fibers with DNA characteristics. I. Fixation and electron staining reactions. J Cell Biol 19, 593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V and Wallace DC (2013) mtDNA variation and analysis using Mitomap and Mitomaster. Curr Protoc Bioinformatics 44, 1.23.1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuels DC, Boys RJ, Henderson DA and Chinnery PF (2003) A compositional segmentation of the human mitochondrial genome is related to heterogeneities in the guanine mutation rate. Nucleic Acids Res 31, 6043–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiesner RJ, Ruegg JC and Morano I (1992) Counting target molecules by exponential polymerase chain reaction: copy number of mitochondrial DNA in rat tissues. Biochem Biophys Res Commun 183, 553–559. [DOI] [PubMed] [Google Scholar]
- 9.Bogenhagen D and Clayton DA (1977) Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell 11, 719–727. [DOI] [PubMed] [Google Scholar]
- 10.Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A and Spelbrink JN (2009) Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet 18, 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidzon G, Greene P, Mancuso M, Klos KJ, Ahlskog JE, Hirano M and DiMauro S (2006) Earlyonset familial parkinsonism due to POLG mutations. Ann Neurol 59, 859–862. [DOI] [PubMed] [Google Scholar]
- 12.Luoma PT, Eerola J, Ahola S, Hakonen AH, Hellstrom O, Kivisto KT, Tienari PJ and Suomalainen A (2007) Mitochondrial DNA polymerase gamma variants in idiopathic sporadic Parkinson disease. Neurology 69, 1152–1159. [DOI] [PubMed] [Google Scholar]
- 13.Young MJ and Copeland WC (2016) Human mitochondrial DNA replication machinery and disease. Curr Opin Genet Dev 38, 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorman GS, Schaefer AM, Ng Y, Gomez N, Blakely EL, Alston CL, Feeney C, Horvath R, Yu-Wai-Man P, Chinnery PF et al. (2015) Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol 77, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP and Letellier T (2003) Mitochondrial threshold effects. Biochem J 370, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt IJ, Harding AE, Petty RK and Morgan-Hughes JA (1990) A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet 46, 428–433. [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy SR, Salk JJ, Schmitt MW and Loeb LA (2013) Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet 9, e1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinnery PF, Elliott HR, Hudson G, Samuels DC and Relton CL (2012) Epigenetics, epidemiology and mitochondrial DNA diseases. Int J Epidemiol 41, 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falkenberg M, Larsson NG and Gustafsson CM (2007) DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem 76, 679–699. [DOI] [PubMed] [Google Scholar]
- 20.Holt IJ, Harding AE and Morgan-Hughes JA (1988) Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719. [DOI] [PubMed] [Google Scholar]
- 21.Reeve AK, Krishnan KJ, Taylor G, Elson JL, Bender A, Taylor RW, Morris CM and Turnbull DM (2009) The low abundance of clonally expanded mitochondrial DNA point mutations in aged substantia nigra neurons. Aging Cell 8, 496–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T et al. (2006) High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet 38, 515–517. [DOI] [PubMed] [Google Scholar]
- 23.Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S and Aiken JM (2006) Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am J Hum Genet 79, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW and Khrapko K (2006) Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet 38, 518–520. [DOI] [PubMed] [Google Scholar]
- 25.Yu-Wai-Man P, Lai-Cheong J, Borthwick GM, He L, Taylor GA, Greaves LC, Taylor RW, Griffiths PG and Turnbull DM (2010) Somatic mitochondrial DNA deletions accumulate to high levels in aging human extraocular muscles. Invest Ophthalmol Vis Sci 51, 3347–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balaban RS, Nemoto S and Finkel T (2005) Mitochondria, oxidants, and aging. Cell 120, 483–495. [DOI] [PubMed] [Google Scholar]
- 27.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL and Lesnefsky EJ (2003) Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem 278, 36027–36031. [DOI] [PubMed] [Google Scholar]
- 28.Gyulkhandanyan AV and Pennefather PS (2004) Shift in the localization of sites of hydrogen peroxide production in brain mitochondria by mitochondrial stress. J Neurochem 90, 405–421. [DOI] [PubMed] [Google Scholar]
- 29.Andreyev AY, Kushnareva YE and Starkov AA (2005) Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 70, 200–214. [DOI] [PubMed] [Google Scholar]
- 30.Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, Pelliccia G, Luzi L, Minucci S, Marcaccio M et al. (2005) Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell 122, 221–233. [DOI] [PubMed] [Google Scholar]
- 31.Murphy MP (2009) How mitochondria produce reactive oxygen species. Biochem J 417, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brand MD (2010) The sites and topology of mitochondrial superoxide production. Exp Gerontol 45, 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller FL, Liu Y and Van Remmen H (2004) Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem 279, 49064–49073. [DOI] [PubMed] [Google Scholar]
- 34.Zorov DB, Juhaszova M and Sollott SJ (2014) Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev 94, 909–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao VK, Carlson EA and Yan SS (2014) Mitochondrial permeability transition pore is a potential drug target for neurodegeneration. Biochim Biophys Acta 1842, 1267–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massaad CA and Klann E (2011) Reactive oxygen species in the regulation of synaptic plasticity and memory. Antioxid Redox Signal 14, 2013–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattson MP and Magnus T (2006) Ageing and neuronal vulnerability. Nat Rev Neurosci 7, 278–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X and Michaelis EK (2010) Selective neuronal vulnerability to oxidative stress in the brain. Front Aging Neurosci 2, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halliwell B (1992) Reactive oxygen species and the central nervous system. J Neurochem 59, 1609–1623. [DOI] [PubMed] [Google Scholar]
- 40.Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT and Surmeier DJ (2010) Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 468, 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finkel T (2011) Signal transduction by reactive oxygen species. J Cell Biol 194, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finkel T (2001) Reactive oxygen species and signal transduction. IUBMB Life 52, 3–6. [DOI] [PubMed] [Google Scholar]
- 43.Simon AR, Rai U, Fanburg BL and Cochran BH (1998) Activation of the JAK-STAT pathway by reactive oxygen species. Am J Physiol 275, C1640–C1652. [DOI] [PubMed] [Google Scholar]
- 44.Lubos E, Kelly NJ, Oldebeken SR, Leopold JA, Zhang YY, Loscalzo J and Handy DE (2011) Glutathione peroxidase-1 deficiency augments proinflammatory cytokine-induced redox signaling and human endothelial cell activation. J Biol Chem 286, 35407–35417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CT, Shih YR, Kuo TK, Lee OK and Wei YH (2008) Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 26, 960–968. [DOI] [PubMed] [Google Scholar]
- 46.Owusu-Ansah E and Banerjee U (2009) Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tormos KV, Anso E, Hamanaka RB, Eisenbart J, Joseph J, Kalyanaraman B and Chandel NS (2011) Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab 14, 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11, 298–300. [DOI] [PubMed] [Google Scholar]
- 49.Harman D (1972) The biologic clock: the mitochondria? J Am Geriatr Soc 20, 145–147. [DOI] [PubMed] [Google Scholar]
- 50.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA et al. (2005) Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481–484. [DOI] [PubMed] [Google Scholar]
- 51.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R et al. (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423. [DOI] [PubMed] [Google Scholar]
- 52.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA and Loeb LA (2007) Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet 39, 540–543. [DOI] [PubMed] [Google Scholar]
- 53.Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA and Loeb LA (2008) DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet 40, 392–394. [DOI] [PubMed] [Google Scholar]
- 54.Lapointe J, Wang Y, Bigras E and Hekimi S (2012) The submitochondrial distribution of ubiquinone affects respiration in long-lived Mclk1+/− mice. J Cell Biol 199, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang W and Hekimi S (2010) A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol 8, e1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fukui H, Diaz F, Garcia S and Moraes CT (2007) Cytochrome c oxidase deficiency in neurons decreases both oxidative stress and amyloid formation in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA 104, 14163–14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz F, Garcia S, Padgett KR and Moraes CT (2012) A defect in the mitochondrial complex III, but not complex IV, triggers early ROS-dependent damage in defined brain regions. Hum Mol Genet 21, 5066–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hekimi S, Lapointe J and Wen Y (2011) Taking a “good” look at free radicals in the aging process. Trends Cell Biol 21, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan Y, Schroeder EA, Ocampo A, Barrientos A and Shadel GS (2011) Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab 13, 668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ II and Nikoskelainen EK (1988) Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science 242, 1427–1430. [DOI] [PubMed] [Google Scholar]
- 61.Huoponen K, Vilkki J, Aula P, Nikoskelainen EK and Savontaus ML (1991) A new mtDNA mutation associated with Leber hereditary optic neuroretinopathy. Am J Hum Genet 48, 1147–1153. [PMC free article] [PubMed] [Google Scholar]
- 62.Johns DR, Neufeld MJ and Park RD (1992) An ND-6 mitochondrial DNA mutation associated with Leber hereditary optic neuropathy. Biochem Biophys Res Commun 187, 1551–1557. [DOI] [PubMed] [Google Scholar]
- 63.Jun AS, Brown MD and Wallace DC (1994) A mitochondrial DNA mutation at nucleotide pair 14459 of the NADH dehydrogenase subunit 6 gene associated with maternally inherited Leber hereditary optic neuropathy and dystonia. Proc Natl Acad Sci USA 91, 6206–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malfatti E, Bugiani M, Invernizzi F, de Souza CF, Farina L, Carrara F, Lamantea E, Antozzi C, Confalonieri P, Sanseverino MT et al. (2007) Novel mutations of ND genes in complex I deficiency associated with mitochondrial encephalopathy. Brain 130, 1894–1904. [DOI] [PubMed] [Google Scholar]
- 65.Trounce IA, Kim YL, Jun AS and Wallace DC (1996) Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol 264, 484–509. [DOI] [PubMed] [Google Scholar]
- 66.Vergani L, Rossi R, Brierley CH, Hanna M and Holt IJ (1999) Introduction of heteroplasmic mitochondrial DNA (mtDNA) from a patient with NARP into two human rho degrees cell lines is associated either with selection and maintenance of NARP mutant mtDNA or failure to maintain mtDNA. Hum Mol Genet 8, 1751–1755. [DOI] [PubMed] [Google Scholar]
- 67.Rahman S, Blok RB, Dahl HH, Danks DM, Kirby DM, Chow CW, Christodoulou J and Thorburn DR (1996) Leigh syndrome: clinical features and biochemical and DNA abnormalities. Ann Neurol 39, 343–351. [DOI] [PubMed] [Google Scholar]
- 68.Manfredi G, Gupta N, Vazquez-Memije ME, Sadlock JE, Spinazzola A, De Vivo DC and Schon EA (1999) Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. J Biol Chem 274, 9386–9391. [DOI] [PubMed] [Google Scholar]
- 69.Goto Y, Horai S, Matsuoka T, Koga Y, Nihei K, Kobayashi M and Nonaka I (1992) Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS): a correlative study of the clinical features and mitochondrial DNA mutation. Neurology 42, 545–550. [DOI] [PubMed] [Google Scholar]
- 70.Manwaring N, Jones MM, Wang JJ, Rochtchina E, Howard C, Mitchell P and Sue CM (2007) Population prevalence of the MELAS A3243G mutation. Mitochondrion 7, 230–233. [DOI] [PubMed] [Google Scholar]
- 71.Garrido-Maraver J, Cordero MD, Moñino ID, Pereira-Arenas S, Lechuga-Vieco AV, Cotán D, De la Mata M, Oropesa-Ávila M, De Miguel M, Bautista Lorite J et al. (2012) Screening of effective pharmacological treatments for MELAS syndrome using yeasts, fibroblasts and cybrid models of the disease. Br J Pharmacol 167, 1311–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marin SE, Mesterman R, Robinson B, Rodenburg RJ, Smeitink J and Tarnopolsky MA (2013) Leigh syndrome associated with mitochondrial complex I deficiency due to novel mutations In NDUFV1 and NDUFS2. Gene 516, 162–167. [DOI] [PubMed] [Google Scholar]
- 73.Martín MA, Blázquez A, Gutierrez-Solana LG, Fern andez-Moreira D, Briones P, Andreu AL, Garesse R, Campos Y and Arenas J (2005) Leigh syndrome associated with mitochondrial complex I deficiency due to a novel mutation in the NDUFS1 gene. Arch Neurol 62, 659–661. [DOI] [PubMed] [Google Scholar]
- 74.Sarzi E, Brown MD, Lebon S, Chretien D, Munnich A, Rotig A and Procaccio V (2007) A novel recurrent mitochondrial DNA mutation in ND3 gene is associated with isolated complex I deficiency causing Leigh syndrome and dystonia. Am J Med Genet A 143A, 33–41. [DOI] [PubMed] [Google Scholar]
- 75.Kirby DM, Boneh A, Chow CW, Ohtake A, Ryan MT, Thyagarajan D and Thorburn DR (2003) Low mutant load of mitochondrial DNA G13513A mutation can cause Leigh’s disease. Ann Neurol 54, 473–478. [DOI] [PubMed] [Google Scholar]
- 76.Lake NJ, Compton AG, Rahman S and Thorburn DR (2016) Leigh syndrome: one disorder, more than 75 monogenic causes. Ann Neurol 79, 190–203. [DOI] [PubMed] [Google Scholar]
- 77.Makino M, Horai S, Goto Y and Nonaka I (2000) Mitochondrial DNA mutations in Leigh syndrome their phylogenetic implications. J Hum Genet 45, 69–75. [DOI] [PubMed] [Google Scholar]
- 78.Taylor RW, Singh-Kler R, Hayes CM, Smith PE and Turnbull DM (2001) Progressive mitochondrial disease resulting from a novel missense mutation in the mitochondrial DNA ND3 gene. Ann Neurol 50, 104–107. [DOI] [PubMed] [Google Scholar]
- 79.Nesbitt V, Morrison PJ, Crushell E, Donnelly DE, Alston CL, He L, McFarland R and Taylor RW (2012) The clinical spectrum of the m.10191T>C mutation in complex I-deficient Leigh syndrome. Dev Med Child Neurol 54, 500–506. [DOI] [PubMed] [Google Scholar]
- 80.Wojtala A, Karkucinska-Wieckowska A, Sardao VA, Szczepanowska J, Kowalski P, Pronicki M, Duszynski J and Wieckowski MR (2017) Modulation of mitochondrial dysfunction-related oxidative stress in fibroblasts of patients with Leigh syndrome by inhibition of prooxidative p66Shc pathway. Mitochondrion 37, 62–79. [DOI] [PubMed] [Google Scholar]
- 81.Tanji K, Kunimatsu T, Vu TH and Bonilla E (2001) Neuropathological features of mitochondrial disorders. Semin Cell Dev Biol 12, 429–439. [DOI] [PubMed] [Google Scholar]
- 82.Sparaco M, Bonilla E, DiMauro S and Powers JM (1993) Neuropathology of mitochondrial encephalomyopathies due to mitochondrial DNA defects. J Neuropathol Exp Neurol 52, 1–10. [DOI] [PubMed] [Google Scholar]
- 83.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF and Wallace DC (1992) Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet 2, 324–329. [DOI] [PubMed] [Google Scholar]
- 84.Indo HP, Davidson M, Yen HC, Suenaga S, Tomita K, Nishii T, Higuchi M, Koga Y, Ozawa T and Majima HJ (2007) Evidence of ROS generation by mitochondria in cells with impaired electron transport chain and mitochondrial DNA damage. Mitochondrion 7, 106–118. [DOI] [PubMed] [Google Scholar]
- 85.Lax NZ, Gorman GS and Turnbull DM (2017) Review: central nervous system involvement in mitochondrial disease. Neuropathol Appl Neurobiol 43, 102–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shigenaga MK, Hagen TM and Ames BN (1994) Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA 91, 10771–10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meissner C, Bruse P, Mohamed SA, Schulz A, Warnk H, Storm T and Oehmichen M (2008) The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol 43, 645–652. [DOI] [PubMed] [Google Scholar]
- 88.Keogh MJ and Chinnery PF (2015) Mitochondrial DNA mutations in neurodegeneration. Biochim Biophys Acta 1847, 1401–1411. [DOI] [PubMed] [Google Scholar]
- 89.Sims-Robinson C, Hur J, Hayes JM, Dauch JR, Keller PJ, Brooks SV and Feldman EL (2013) The role of oxidative stress in nervous system aging. PLoS One 8, e68011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J and Yankner BA (2004) Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891. [DOI] [PubMed] [Google Scholar]
- 91.Reeve A, Simcox E and Turnbull D (2014) Ageing and Parkinson’s disease: why is advancing age the biggest risk factor? Ageing Res Rev 14, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gibb WR and Lees AJ (1991) Anatomy, pigmentation, ventral and dorsal subpopulations of the substantianigra, and differential cell death in Parkinson’s disease. J Neurol Neurosurg Psychiatry 54, 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wakabayashi K, Tanji K, Mori F and Takahashi H (2007) The Lewy body in Parkinson’s disease: molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology 27, 494–506. [DOI] [PubMed] [Google Scholar]
- 94.Lotharius J, Barg S, Wiekop P, Lundberg C, Raymon HK and Brundin P (2002) Effect of mutant alpha-synuclein on dopamine homeostasis in a new human mesencephalic cell line. J Biol Chem 277, 38884–38894. [DOI] [PubMed] [Google Scholar]
- 95.Lotharius J and Brundin P (2002) Impaired dopamine storage resulting from alpha-synuclein mutations may contribute to the pathogenesis of Parkinson’s disease. Hum Mol Genet 11, 2395–2407. [DOI] [PubMed] [Google Scholar]
- 96.Exner N, Lutz AK, Haass C and Winklhofer KF (2012) Mitochondrial dysfunction in Parkinson’s disease: molecular mechanisms and pathophysiological consequences. EMBO J 31, 3038–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meredith GE and Rademacher DJ (2011) MPTP mouse models of Parkinson’s disease: an update. J Parkinsons Dis 1, 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin MT, Cantuti-Castelvetri I, Zheng K, Jackson KE, Tan YB, Arzberger T, Lees AJ, Betensky RA, Beal MF and Simon DK (2012) Somatic mitochondrial DNA mutations in early Parkinson and incidental Lewy body disease. Ann Neurol 71, 850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Franco-Iborra S, Vila M and Perier C (2016) The Parkinson disease mitochondrial hypothesis: where are we at? Neuroscientist 22, 266–277. [DOI] [PubMed] [Google Scholar]
- 100.Dolleö C, Flønes I, Nido GS, Miletic H, Osuagwu N, Kristoffersen S, Lilleng PK, Larsen JP, Tysnes OB, Haugarvoll K et al. (2016) Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat Commun 7, 13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tzoulis C, Schwarzlmuller T, Biermann M, Haugarvoll K and Bindoff LA (2016) Mitochondrial DNA homeostasis is essential for nigrostriatal integrity. Mitochondrion 28, 33–37. [DOI] [PubMed] [Google Scholar]
- 102.Parkinson GM, Dayas CV and Smith DW (2014) Increased mitochondrial DNA deletions in substantia nigra dopamine neurons of the aged rat. Curr Aging Sci 7, 155–160. [DOI] [PubMed] [Google Scholar]
- 103.Grunewald A, Rygiel KA, Hepplewhite PD, Morris CM, Picard M and Turnbull DM (2016) Mitochondrial DNA depletion in respiratory chain-deficient Parkinson disease neurons. Ann Neurol 79, 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hyman BT, Van Hoesen GW, Damasio AR and Barnes CL (1984) Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science 225, 1168–1170. [DOI] [PubMed] [Google Scholar]
- 105.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL and Beyreuther K (1985) Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA 82, 4245–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sheng B, Wang X, Su B, Lee HG, Casadesus G, Perry G and Zhu X (2012) Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J Neurochem 120, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Du H, Guo L, Yan S, Sosunov AA, McKhann GM and Yan SS (2010) Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci USA 107, 18670–18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mossmann D, Vögtle FN, Taskin AA, Teixeira PF, Ring J, Burkhart JM, Burger N, Pinho CM, Tadic J, Loreth D et al. (2014) Amyloid-beta peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab 20, 662–669. [DOI] [PubMed] [Google Scholar]
- 109.Swerdlow RH, Burns JM and Khan SM (2014) The Alzheimer’s disease mitochondrial cascade hypothesis: progress and perspectives. Biochim Biophys Acta 1842, 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin MT, Simon DK, Ahn CH, Kim LM and Beal MF (2002) High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum Mol Genet 11, 133–145. [DOI] [PubMed] [Google Scholar]
- 111.Hoekstra JG, Hipp MJ, Montine TJ and Kennedy SR (2016) Mitochondrial DNA mutations increase in early stage Alzheimer disease and are inconsistent with oxidative damage. Ann Neurol 80, 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mancuso M, Filosto M, Orsucci D and Siciliano G (2008) Mitochondrial DNA sequence variation and neurodegeneration. Hum Genomics 3, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dhaliwal GK and Grewal RP (2000) Mitochondrial DNA deletion mutation levels are elevated in ALS brains. NeuroReport 11, 2507–2509. [DOI] [PubMed] [Google Scholar]
- 114.Sau D, De Biasi S, Vitellaro-Zuccarello L, Riso P, Guarnieri S, Porrini M, Simeoni S, Crippa V, Onesto E, Palazzolo I et al. (2007) Mutation of SOD1 in ALS: a gain of a loss of function. Hum Mol Genet 16, 1604–1618. [DOI] [PubMed] [Google Scholar]
- 115.Ferrante RJ, Browne SE, Shinobu LA, Bowling AC, Baik MJ, MacGarvey U, Kowall NW, Brown RH Jr and Beal MF (1997) Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem 69, 2064–2074. [DOI] [PubMed] [Google Scholar]
- 116.Andrus PK, Fleck TJ, Gurney ME and Hall ED (1998) Protein oxidative damage in a transgenic mouse model of familial amyotrophic lateral sclerosis. J Neurochem 71, 2041–2048. [DOI] [PubMed] [Google Scholar]
- 117.Liu R, Althaus JS, Ellerbrock BR, Becker DA and Gurney ME (1998) Enhanced oxygen radical production in a transgenic mouse model of familial amyotrophic lateral sclerosis. Ann Neurol 44, 763–770. [DOI] [PubMed] [Google Scholar]
- 118.Borthwick GM, Johnson MA, Ince PG, Shaw PJ and Turnbull DM (1999) Mitochondrial enzyme activity in amyotrophic lateral sclerosis: implications for the role of mitochondria in neuronal cell death. Ann Neurol 46, 787–790. [DOI] [PubMed] [Google Scholar]
- 119.Mawrin C, Kirches E, Krause G, Wiedemann FR, Vorwerk CK, Bogerts B, Schildhaus HU, Dietzmann K and Schneider-Stock R (2004) Single-cell analysis of mtDNA deletion levels in sporadic amyotrophic lateral sclerosis. NeuroReport 15, 939–943. [DOI] [PubMed] [Google Scholar]
- 120.Ingram CJ, Weale ME, Plaster CA, Morrison KE, Goodall EF, Pall HS, Beck M, Jablonka S, Sendtner M, Fisher EM et al. (2012) Analysis of European case-control studies suggests that common inherited variation in mitochondrial DNA is not involved in susceptibility to amyotrophic lateral sclerosis. Amyotroph Lateral Scler 13, 341–346. [DOI] [PubMed] [Google Scholar]
- 121.Campbell GR, Ziabreva I, Reeve AK, Krishnan KJ, Reynolds R, Howell O, Lassmann H, Turnbull DM and Mahad DJ (2011) Mitochondrial DNA deletions and neurodegeneration in multiple sclerosis. Ann Neurol 69, 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dutta R, McDonough J, Yin X, Peterson J, Chang A, Torres T, Gudz T, Macklin WB, Lewis DA, Fox RJ et al. (2006) Mitochondrial dysfunction as a cause of axonal degeneration in multiple sclerosis patients. Ann Neurol 59, 478–489. [DOI] [PubMed] [Google Scholar]
- 123.Andalib S, Talebi M, Sakhinia E, Farhoudi M, Sadeghi-Bazargani H, Motavallian A and Pilehvar-Soltanahmadi Y (2013) Multiple sclerosis and mitochondrial gene variations: a review. J Neurol Sci 330, 10–15. [DOI] [PubMed] [Google Scholar]
- 124.Campbell GR, Reeve A, Ziabreva I, Polvikoski TM, Taylor RW, Reynolds R, Turnbull DM and Mahad DJ (2013) Mitochondrial DNA deletions and depletion within paraspinal muscles. Neuropathol Appl Neurobiol 39, 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Inoue K, Nakada K, Ogura A, Isobe K, Goto Y, Nonaka I and Hayashi JI (2000) Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat Genet 26, 176–181. [DOI] [PubMed] [Google Scholar]
- 126.Nakada K, Inoue K, Chen CS, Nonaka I, Goto Y, Ogura A and Hayashi JI (2001) Correlation of functional and ultrastructural abnormalities of mitochondria in mouse heart carrying a pathogenic mutant mtDNA with a 4696-bp deletion. Biochem Biophys Res Commun 288, 901–907. [DOI] [PubMed] [Google Scholar]
- 127.Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I and Hayashi JI (2001) Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat Med 7, 934–940. [DOI] [PubMed] [Google Scholar]
- 128.Srivastava S and Moraes CT (2001) Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum Mol Genet 10, 3093–3099. [DOI] [PubMed] [Google Scholar]
- 129.Srivastava S and Moraes CT (2005) Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet 14, 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fukui H and Moraes CT (2009) Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet 18, 1028–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pickrell AM, Fukui H, Wang X, Pinto M and Moraes CT (2011) The striatum is highly susceptible to mitochondrial oxidative phosphorylation dysfunctions. J Neurosci 31, 9895–9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pinto M, Pickrell AM, Fukui H and Moraes CT (2013) Mitochondrial DNA damage in a mouse model of Alzheimer’s disease decreases amyloid beta plaque formation. Neurobiol Aging 34, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin CS, Sharpley MS, Fan W, Waymire KG, Sadun AA, Carelli V, Ross-Cisneros FN, Baciu P, Sung E, McManus MJ et al. (2012) Mouse mtDNA mutant model of Leber hereditary optic neuropathy. Proc Natl Acad Sci USA 109, 20065–20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Edgar D, Shabalina I, Camara Y, Wredenberg A, Calvaruso MA, Nijtmans L, Nedergaard J, Cannon B, Larsson NG and Trifunovic A (2009) Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab 10, 131–138. [DOI] [PubMed] [Google Scholar]
- 135.Williams SL, Huang J, Edwards YJ, Ulloa RH, Dillon LM, Prolla TA, Vance JM, Moraes CT and Züchner S (2010) The mtDNA mutation spectrum of the progeroid Polg mutator mouse includes abundant control region multimers. Cell Metab 12, 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT and Larsson NG (2005) Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci USA 102, 17993–17998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ahlqvist KJ, Hämäläinen RH, Yatsuga S, Uutela M, Terzioglu M, Götz A, Forsström S, Salven P, Angers-Loustau A, Kopra OH et al. (2012) Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab 15, 100–109. [DOI] [PubMed] [Google Scholar]
- 138.Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A and Suomalainen A (2005) Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci USA 102, 17687–17692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tyynismaa H and Suomalainen A (2009) Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep 10, 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]