Abstract
Osteosarcoma is one of the most common malignant tumors in children and adolescents and is characterized by early metastasis. High-mobility group N (HMGN) domains are involved in the development of several tumors. Our previous study found that HMGN5 is highly expressed in osteosarcoma tissues and knockdown of HMGN5 inhibits migration and invasion of U-2 OS and Saos-2 cells. A hypoxic environment is commonly found in solid tumors such as osteosarcoma and is likely to be associated with tumor metastasis, so we further explored the relationship between HMGN5 and the hypoxic environment. Hypoxia-inducible factor 1A (HIF1A) is an adaptive factor in the hypoxic environment. We found that HIF1A and HMGN5 were upregulated in osteosarcoma (OS) cells cultured in the hypoxic environment, and the results of overexpression and knockdown experiments showed that HIF1A upregulated the transcription factor GATA1 and further promoted the expression of HMGN5. In addition, MMP2 and MMP9 were subsequently upregulated through the c-jun pathway, and finally, this promoted the migration and invasion of OS cells. It is suggested that HMGN5 may be an important downstream factor for HIF1A to promote osteosarcoma metastasis. It has an important clinical significance for the selection of therapeutic targets for osteosarcoma.
1. Introduction
Osteosarcoma is a highly malignant bone tumor disease derived from mesenchymal cells and is one of the most common malignant tumors in children and adolescents [1, 2]. It tends to metastasize at an early stage. When osteosarcoma is diagnosed, about 15–20% of patients already have macroscopic evidence of metastases, most commonly in the lungs (85–90%), sometimes in the bone (8–10%), and occasionally in the lymph nodes [3–5]. The main treatments are still tumor resection and nonspecific combination chemotherapy [6–9]. With the development of treatment of primary tumor, the long-term survival rate of osteosarcoma patients has increased [10], but the five-year survival rate of patients with distant metastases is still low (0–29%) [11, 12]. In addition, fast-growing solid tumors such as osteosarcoma often have a hypoxic microenvironment inside, which further promotes the metastasis of osteosarcoma [10].
HIF1A is a key factor in the hypoxic microenvironment and is closely related to the expression of various tumor-associated factors. Meanwhile, HMGN5 has been shown to be associated with metastasis of tumors [12]. Our previous study found that HMGN5 is highly expressed in osteosarcoma tissues and knockdown of HMGN5 inhibits migration and invasion of U-2 OS and Saos-2 cells [13].
We speculated that HMGN5 may be a downstream factor of HIF1A which plays an important role in osteosarcoma metastasis under hypoxic conditions. Therefore, we plan to explore the relationship between HIF1A and HMGN5 in a hypoxic environment and its effect on osteosarcoma metastasis.
2. Materials and Methods
Our research team has established standardized experimental procedures for cell culture, viral transfection, RT-qPCR, western blot, etc. The process of these assays in this article is similar to that of our previous paper [14].
2.1. Cell Culture
DMEM (Hyclone, Utah, USA) medium contains 10% FBS (FBS, Gibco, NY, USA), penicillin (100 U/L, Invitrogen, NY, USA), and streptomycin (100 mg/L, Invitrogen, NY, USA). McCoy's 5a (Invitrogen, NY, USA) medium contains 15% FBS (FBS, Gibco, NY, USA), penicillin (100 U/L, Invitrogen, NY, USA), and streptomycin (100 mg/L, Invitrogen, NY, USA). U-2 OS osteosarcoma cells were cultured in DMEM and placed in a humidified atmosphere of 5% CO2 at 37°C. Saos-2 osteosarcoma cells were cultured in McCoy's 5a medium and placed in a humidified atmosphere of 5% CO2 at 37°C. We divided the cells into two groups: a normoxic group and a hypoxic group. The normoxic group was cultured in 20% O2, and the hypoxic group was cultured in 1% O2. All experimental results from U-2 OS cells were repeated in Saos-2 cells.
2.2. Overexpression of HIF1A and Knockdown of HMGN5 and GATA1
Recombinant lentiviruses overexpressing HIF1A and blank lentivirus were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China), and OS cells were transfected with overexpression lentiviruses (HIF1AOE) and blank lentivirus (Cherry).
Recombinant lentiviruses knocking down HMGN5 and GATA1 were also purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China), and HIF1A-overexpressed OS cells were transfected with knockdown lentiviruses (HMGN5sh or GATA1sh and blank lentivirus (GFP, Table 1)).
Table 1.
Cellular virus infection.
| Group | Overexpression | Knockdown |
|---|---|---|
| Cherry-GFP | — | — |
| HIF1AOE-GFP | HIF1A | — |
| HIF1AOE-HMGN5sh | HIF1A | HMGN5 |
| HIF1AOE-GATA1sh | HIF1A | GATA1 |
We determined that the optimal multiplicity of infection (MOI) for U-2 OS virus infection was 10 by preexperiment and MOI for Saos-2 was 15. We conducted the formal experiments based on these MOI values. In order to increase the proportion of infected cells, we performed flow sorting after 3 days of cell infection and continued to culture the cells with cherry or green fluorescence. We extracted RNA and proteins when the cells reached 70–80% confluence. Overexpression and knockdown efficiency were confirmed by RT-qPCR and western blot.
2.3. RT-qPCR
RT-qPCR was applied to quantitatively determine the expression level of HIF1A, HMGN5, and GATA1 using the SYBR-Premix Ex Taq (Takara, Japan) and ABI Prism 7900HT sequence detection system (Applied Biosystems, Carlsbad, CA). Total RNA of each group was extracted with TRIzol according to the manufacturer's instructions. The mRNA was transcribed into cDNA using a Reverse Transcription kit (Applied Biosystems, Carlsbad, CA). The genes were amplified using specific primers, and human β-actin gene was used as an endogenous control. The PCR primer sequences used were as follows: HIF1A: forward: 5′-GAACGTCGAAAAGAAAAGTCTCG-3′, reverse: 5′-CCTTATCAAGATGCGAACTCACA-3′; HMGN5: forward: 5′-CAGGTCAAGGTGATATGAGGCA-3′, reverse: 5′-GCTTGGGCACTTGTATCTATGT-3′; GATA1: forward: 5′-CTGTCCCCAATAGTGCTTATG-3′, reverse: 5′-GAATAGGCTGCTGAATTGAGGG-3′; β-actin: forward: 5′-ACCGAGCGCGGCTACAG-3′, reverse: 5′-CTTAATGTCACGCACGATTTCC-3′. Data were analyzed using the comparative Ct method (2−ΔΔCt). We repeated the RT-qPCR experiment three times with the same samples and the same primers.
2.4. Western Blot Assay
HMGN5 and GATA1 are mainly expressed in the nucleus, and HIF1A is mainly expressed in the nucleus and cytosol (https://compartments.jensenlab.org). Nuclear proteins were extracted using a NE-PER Nuclear Extraction Reagents (Thermo Fisher Scientific, MA, USA) and whole proteins were extracted using a protein extract kit (Beyotime, Shanghai, China), and the concentration was determined by using a BCA Protein Assay Kit (Pierce). Proteins were run on SDS-PAGE gels and transferred onto polyvinylidene fluoride (PVDF, Millipore, MA, USA) membranes and blocked in 5% bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO, USA). Membranes were probed overnight at 4°C with the appropriate primary antibody. Antibody were used as follows: HIF1A (1 : 1000 dilution, ab51608, Abcam, Cambridge, UK), HMGN5 (1 : 2000 dilution, ab186001, Abcam), GATA1 (1 : 500 dilution, ab28839, Abcam), c-jun (1 : 1000 dilution, ab40766, Abcam), MMP2 (1 : 500 dilution, ab97779, Abcam), MMP9 (1 : 1000 dilution, ab76003, Abcam), MMP16 (1 : 500 dilution, ab73877, Abcam), LMNB1 (1 : 10000 dilution, ab16048, Abcam), and β-actin (1 : 10000 dilution, ab8226, Abcam). Membranes were then probed with a horseradishperoxidase-conjugated secondary antibody for 1 hour at room temperature. The immunoreactive bands were visualized using an enhanced chemiluminescence (ECL) detection system (No. 32106, Pierce). We chose loading control according to Abcam recommendation (http://www.abcam.com/primary-antibodies/loading-control-guide). The relative protein level in different cell lines was normalized to LMNB1 (Lamin B1) or β-actin concentration. Three separate experiments were performed for each group.
2.5. Wound-Healing Assay
OS cells were plated in 6-well culture plates and grown to 90% confluence. Wounds were created using a 200 μl micropipette tip. The migration of cells towards the wound was monitored and photographed. Calculated cell migration rate = (0 h width − 24 h or 48 h width)/0 h width × 100%.
2.6. Transwell Invasion Assay
The invasive ability of cells was determined in a 6.5 mm transwell (Coring, NY, USA). The filter of the top chamber was matrigel-coated with 50 μl of diluted matrigel, and the lower chamber was filled with 500 μl of DMEM containing 10% FBS. Cells were resuspended in serum-free medium and added to each top chamber. The cells were cultured for 24 h, and the noninvading cells were removed. Invading cells were fixed with 4% paraformaldehyde for 30 minutes and then stained with crystal violet staining solution (Sigma-Aldrich). The number of invading cells were counted and analyzed.
2.7. Statistical Analysis
Three separate experiments were performed for each experiment. Analysis of the data was performed using SPSS version 23.0, with P value <0.05 considered statistically significant. All data were presented as the mean ± SD. The Mann–Whitney U test was used to compare the difference between two groups (Figures 1(a) and 1(b)). The Kruskal–Wallis H test was used to analyze the differences between three groups (Figures 2(a), 2(b), 3(b), 3(c), 3(e), and 4(a)∼4(c)).
Figure 1.
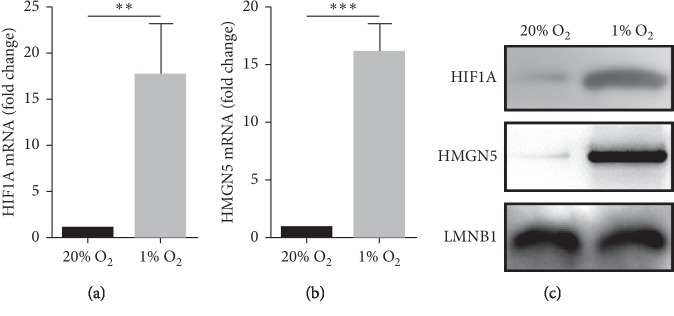
Expression levels of HIF1A and HMGN5 in hypoxic cells were higher than in normoxic cells. (a, b) RT-qPCR showed that the expression levels of HIF1A mRNA and HMGN5 mRNA were higher in hypoxic OS cells. (c) Western blot showed that the expression levels of HIF1A protein and HMGN5 protein were higher in hypoxic OS cells. Data were presented as the mean ± SD. ∗∗P < 0.01; ∗∗∗P < 0.001.
Figure 2.
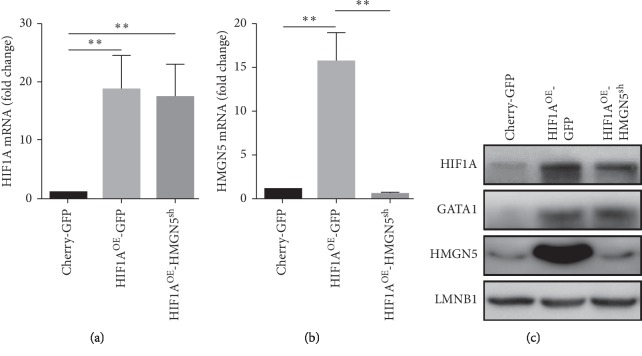
HIF1A upregualtes HMGN5. (a) The expression levels of HIF1A mRNA were higher in the HIF1AOE-GFP group and HIF1AOE-HMGN5sh group than in the Cherry-GFP group. (b) The expression level of HMGN5 mRNA in the HIF1AOE-GFP group was significantly higher than in the Cherry-GFP group and was knocked down in the HIF1AOE-HMGN5sh group. (c) The expression levels of HIF1A and GATA1 were higher in HIF1AOE-GFP and HIF1AOE-HMGN5sh groups than in the Cherry-GFP group. The expression level of HMGN5 in the HIF1AOE-GFP group was higher than in the Cherry-GFP group and was knocked down in the HIF1AOE-HMGN5sh group. Data were presented as the mean ± SD. ∗∗P < 0.01.
Figure 3.
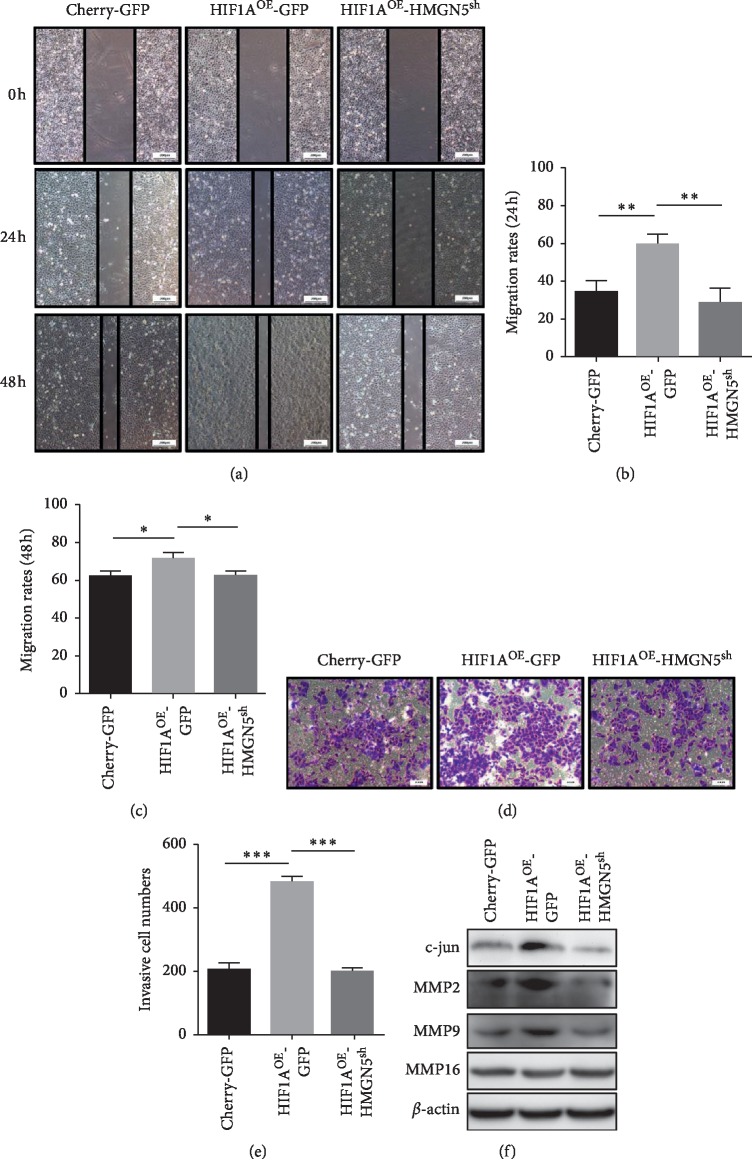
Reversal of enhanced migration and invasion of osteosarcoma cells under hypoxic conditions by knocking down HMGN5. (a) Wound-healing assay in 24 hours and 48 hours for each group. (b, c) Statistical analysis of cell migration rate of each group in 24 hours and 48 hours; the results showed that the migration ability of the HIF1AOE-GFP group was significantly enhanced compared with the Cherry-GFP group, while the migration ability of the HIF1AOE-HMGN5sh group was significantly lower than that of HIF1AOE-GFP group. (d) Transwell invasion assay for each group. (e) Statistical analysis of invading cells of each group; the results showed that the invasion ability of the HIF1AOE-GFP group was significantly enhanced compared with the Cherry-GFP group, while the invasion ability of the HIF1AOE-HMGN5sh group was significantly lower than that of the HIF1AOE-GFP group. (f) The expression levels of c-jun, MMP2, and MMP9 in the HIF1AOE-GFP group were higher than in Cherry-GFP group. The expression levels of c-jun, MMP2, and MMP9 in the HIF1AOE-HMGN5sh group were lower than in the HIF1AOE-GFP group. Data were presented as the mean ± SD. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Figure 4.
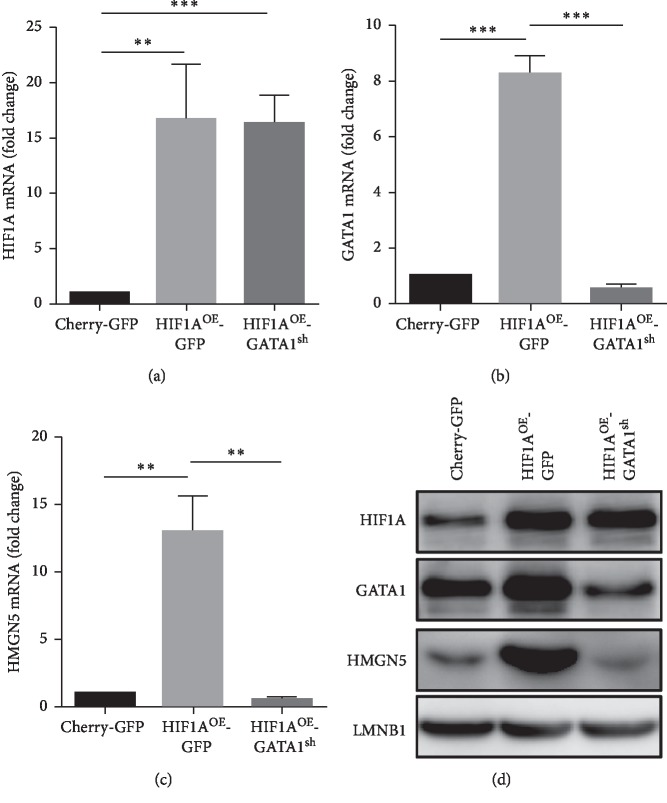
HIF1A upregulates GATA1 to promote the expression of HMGN5. (a) The expression levels of HIF1A mRNA were higher in HIF1AOE-GFP and HIF1AOE-GATA1sh groups than in the Cherry-GFP group. (b) The expression level of GATA1 mRNA in the HIF1AOE-GFP group was significantly higher than in the Cherry-GFP group and was knocked down in the HIF1AOE-GATA1sh group. (c) The expression level of HMGN5 mRNA in the HIF1AOE-GFP group was significantly higher than in the Cherry-GFP group, and the expression level of HMGN5 mRNA in HIF1AOE-GATA1sh group was significantly lower than in the HIF1AOE-GFP group. (d) GATA1 and HMGN5 had higher expression levels in the HIF1AOE-GFP group (compared with Cherry-GFP group) but lower expression in the HIF1AOE-GATA1sh group (compared with HIF1AOE-GFP group). Data were presented as the mean ± SD. ∗∗P < 0.01; ∗∗∗P < 0.001.
3. Results
3.1. High Expression of HIF1A and HMGN5 in Hypoxic Environment-Cultured OS Cells
We cultured OS cells in 20% O2 (normoxic group) and 1% O2 (hypoxic group). The results of RT-qPCR and western blot showed that the expression level of HMGN5 was higher in hypoxic OS cells (Figures 1(b) and 1(c)). It suggests that some hypoxia-related factors may upregulate the expression of HMGN5. HIF1A is an adaptive factor that is highly expressed in the hypoxic environment (Figures 1(a) and 1(c)), which is closely related to the expression of various tumor-associated factors. We subsequently did some work to clarify the relationship between HIF1A and HMGN5.
3.2. HIF1A Was Overexpressed and HMGN5 Was Knocked Down Successfully in OS Cells
We overexpressed HIF1A and knocked down HMGN5 in OS cells, and finally, we have 3 groups of cells to explore the relationship between HMGN5 and HIF1A: Cherry-GFP group (control), HIF1AOE-GFP group (HIF1A overexpression), and HIF1AOE-HMGN5sh group (HIF1A overexpression and HMGN5 knockdown) (Table 1). RT-qPCR results showed that HIF1A was overexpressed in the HIF1AOE-GFP group and HIF1AOE-HMGN5sh group, and HMGN5 was knocked down in the HIF1A-HMGN5sh group (Figures 2(a) and 2(b)). The results of western blot further confirmed the overexpression and knockdown (Figure 2(c)).
3.3. Knockdown of HMGN5 Inhibits Migration and Invasion of OS Cells under Hypoxic Conditions
The role of HIF1A and HMGN5 in the migration and invasion of OS cells was determined using wound-healing and transwell invasion assay. The results showed that the migration and invasion of the HIF1AOE-GFP group were significantly enhanced compared with the Cherry-GFP group, while the migration and invasion of the HIF1AOE-HMGN5sh group were significantly lower than those of the HIF1A-GFP group. The results showed that overexpression of HIF1A in OS cells significantly increased cell migration and invasion, while knockdown of HMGN5 reversed these effects (Figures 3(a)–3(e)), suggesting that HIF1A enhances cell migration and invasion partially through HMGN5 in OS cells.
Previous studies have confirmed that HMGN5 regulates the expression of MMP2 and MMP9 via c-jun and then contributed to the migration and invasion of clear cell renal cell carcinoma (ccRCC) cells [15]. Our results showed that upregulated HMGN5 in a hypoxic environment increased the expression of c-jun and finally increased the expression of MMP2 and MMP9, which was associated with migration and invasion of osteosarcoma (Figure 3(f)).
3.4. HIF1A Regulates the Expression of HMGN5 through the Transcription Factor GATA1
HIF1A upregulated the expression of HMGN5 mRNA (Figure 2(b)). We predicted transcription factors binding to the HMGN5 promoter by PROMO transcription factor prediction software and found that GATA1 is probably an important transcription factor of HMGN5. Western blot confirmed that HIF1A upregulates GATA1 expression (Figure 2(c)). We obtained a new group through lentiviral infection named “HIF1AOE-GATA1sh group” (HIF1A overexpression and GATA1 knockdown, Figures 4(a) and 4(b)) to confirm that HIF1A regulates HMGN5 through GATA1. The results of RT-qPCR and western blot showed that overexpression of HIF1A upregulated GATA1 and HMGN5, while knockdown of GATA1 reduced the expression of HMGN5 (Figures 4(a)–4(d)).
4. Discussion
It is estimated that 20% of patients with osteosarcoma have metastases at the time of initial diagnosis [16], and more than 80% of patients with osteosarcoma develop pulmonary metastases within two years and, if left untreated, die of disease within a few months [17]. The main treatments for osteosarcoma are chemotherapy and surgical intervention. However, a significant proportion of these patients eventually develop pulmonary metastases and succumb to their disease even after chemotherapy and surgical excision [18–20]. Despite complete tumor resection and intensive chemotherapy, approximately 30% to 40% of patients with osteosarcoma relapse [21–25], and recurrence of osteosarcoma is the most common in the lungs [26]. Therefore, there is a need to develop new and safe approaches to the treatment of osteosarcoma metastasis.
Osteosarcoma is a solid malignant tumor characterized by rapid growth and a high rate of metastasis [27]. It increases in a short period of time and leads to an oxygen-deficient environment inside the tumor [10]. Hypoxia has been shown to play important roles in the progression and metastasis of many cancers such as breast cancer [28], colon cancer [29], and melanoma [30]. HIF is a key regulator of local and systemic responses to hypoxia that occur during normal development and pathological/physiological processes [31]. HIF1A stability is regulated at the posttranslational level by oxygen concentration, which is continuously degraded under normoxia conditions. The activity of prolyl hydroxylases is diminished under hypoxic conditions since they require ferrous ions and O2 for their function, which leads to HIF1A stabilization [32]. Overexpression of HIF1A has been reported to indicate poor prognosis in osteosarcoma patients [33]. In our study, we found that HIF1A upregulated HMGN5 and further promoted migration and invasion of OS cells.
PROMO transcription factor prediction software was used to find that GATA1 is probably a transcription factor of HMGN5. Our results showed that overexpression of HIF1A promoted the expression of HMGN5 by upregulating GATA1. GATA1 was originally identified in 1989 as a transcription factor that binds to the β-globin regulatory region, and it was later revealed to be a member of the GATA transcription factor family [34–36]. GATA1 is widely expressed in human and mammalian cells [37, 38]. Researchers found that GATA1 plays an important role in blood cell differentiation [39–41]. Zhang et al. found that the upregulation of GATA1 during hypoxia is directly mediated by HIF1. The mRNA expression of some erythroid differentiation markers were increased under hypoxic conditions but decreased with RNA interference of HIF1A or GATA1 [42].
The high-mobility group (HMG) includes HMGA, HMGB, and HMGN. Previous studies have confirmed that HMGA and HMGB are related to tumor metastasis. In recent years, the function of HMGN in tumor invasion and metastasis has been extensively studied [12]. Studies have shown that HMGN5 plays an important role in the metastasis of prostate cancer [43], bladder cancer [44, 45], kidney cancer [46], and breast cancer [47]. Our previous study found that HMGN5 is highly expressed in osteosarcoma tissues, and knockdown of HMGN5 inhibits migration and invasion of U-2 OS and Saos-2 cells [13]. HMGN5 plays a role in PI3K/AKT signaling pathway or MAPK/ERK signaling pathway [12]. In addition, Ji et al. found that the expression level of HMGA1 mRNA and protein were significantly higher in the hypoxic environment [48]. In our study, we found that overexpression of HIF1A upregulated the expression of transcription factor GATA1 and further promoted the expression of HMGN5. The higher expression of HMGN5 upregulated MMP2 and MMP9 through the c-jun pathway and finally promoted the migration and invasion of OS cells. It is suggested that HMGN5 is an important downstream factor for HIF1A to promote osteosarcoma metastasis. It has an important clinical significance for the selection of therapeutic targets for osteosarcoma.
This is a limited research because we did not repeat the experiment in animal models. In vivo assays are necessary to confirm our in vitro results, but it is very difficult to raise animals for a long time under hypoxic conditions due to the limitations of research facilities. In the next step, we plan to explore the effects of HIF1A and HMGN5 on osteosarcoma cell metastasis by injecting three groups of cells (Cherry-GFP, HIF1AOE-GFP, and HIF1AOE-HMGN5sh) into the tail vein of nude mice.
5. Conclusion
We found that HIF1A upregulated the transcription factor GATA1 and further promoted the expression of HMGN5. In addition, MMP2 and MMP9 were subsequently upregulated through the c-jun pathway, and this finally promoted the migration and invasion ability of osteosarcoma cells. It is suggested that HMGN5 may be an important downstream factor for HIF1A to promote osteosarcoma metastasis.
Acknowledgments
This work was supported by the Shanghai Science and Technology Committee of China (No. 16140901800) and National Natural Science Foundation of China (No. U1603118).
Abbreviations
- HIF1A:
Hypoxia-inducible factor 1A
- HMGN5:
High-mobility group nucleosome binding domain 5
- GATA1:
GATA binding protein 1
- OS:
Osteosarcoma
- MMP2:
Matrix metallopeptidase 2
- MMP9:
Matrix metallopeptidase 9.
Contributor Information
Jun Ma, Email: jmaspine@163.com.
Xuhui Zhou, Email: xhzhouspine@163.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Enjie Xu, Zhe Ji, and Heng Jiang contributed equally to this work.
References
- 1.Group E. E. S. N. W. Bone sarcomas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2012;23(7):vii100–vii109. doi: 10.1093/annonc/mds254. [DOI] [PubMed] [Google Scholar]
- 2.Jo V. Y., Fletcher C. D. M. WHO classification of soft tissue tumours: an update based on the 2013 (4th) edition. Pathology. 2014;46(2):95–104. doi: 10.1097/pat.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 3.Marina N., Gebhardt M., Teot L., Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. The Oncologist. 2004;9(4):422–441. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 4.Chou A. J., Geller D. S., Gorlick R. Therapy for Osteosarcoma. Pediatric Drugs. 2008;10(5):315–327. doi: 10.2165/00148581-200810050-00005. [DOI] [PubMed] [Google Scholar]
- 5.Bacci G., Longhi A., Bertoni F., et al. Bone metastases in osteosarcoma patients treated with neoadjuvant or adjuvant chemotherapy the Rizzoli experience in 52 patients. Acta Orthopaedica. 2006;77(6):938–943. doi: 10.1080/17453670610013268. [DOI] [PubMed] [Google Scholar]
- 6.Palmerini E., Jones R. L., Marchesi E., et al. Gemcitabine and docetaxel in relapsed and unresectable high-grade osteosarcoma and spindle cell sarcoma of bone. BMC Cancer. 2016;16(1):p. 280. doi: 10.1186/s12885-016-2312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner M. J., Livingston J. A., Patel S. R., Benjamin R. S. Chemotherapy for bone sarcoma in adults. Journal of Oncology Practice. 2016;12(3):208–216. doi: 10.1200/jop.2015.009944. [DOI] [PubMed] [Google Scholar]
- 8.Reed D. R., Hayashi M., Wagner L., et al. Treatment pathway of bone sarcoma in children, adolescents, and young adults. Cancer. 2017;123(12):2206–2218. doi: 10.1002/cncr.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakano K., Takahashi S. Current molecular targeted therapies for bone and soft tissue sarcomas. International Journal of Molecular Sciences. 2018;19(3) doi: 10.3390/ijms19030739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wittig J. C., Bickels J., Priebat D., et al. Osteosarcoma: a multidisciplinary approach to diagnosis and treatment. American Family Physician. 2002;65(6):1123–1132. [PubMed] [Google Scholar]
- 11.Tsuchiya H., Kanazawa Y., Abdel-Wanis M. E., et al. Effect of timing of pulmonary metastases identification on prognosis of patients with osteosarcoma: the Japanese Musculoskeletal Oncology Group study. Journal of Clinical Oncology. 2002;20(16):3470–3477. doi: 10.1200/jco.2002.11.028. [DOI] [PubMed] [Google Scholar]
- 12.Shi Z., Tang R., Wu D., Sun X. Research advances in HMGN5 and cancer. Tumor Biology. 2016;37(2):1531–1539. doi: 10.1007/s13277-015-4693-3. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X., Yuan B., Yuan W., Wang C., Gao R., Wang J. The expression and clinical significance of high mobility group nucleosome binding domain 5 in human osteosarcoma. Tumor Biology. 2014;35(7):6539–6547. doi: 10.1007/s13277-014-1825-0. [DOI] [PubMed] [Google Scholar]
- 14.Xu E., Lin T., Jiang H., et al. Asymmetric expression of GPR126 in the convex/concave side of the spine is associated with spinal skeletal malformation in adolescent idiopathic scoliosis population. European Spine Journal. 2019;28(9):1977–1986. doi: 10.1007/s00586-019-06001-5. [DOI] [PubMed] [Google Scholar]
- 15.Ji S.-Q., Yao L., Zhang X.-Y., Li X.-S., Zhou L.-Q. Knockdown of the nucleosome binding protein 1 inhibits the growth and invasion of clear cell renal cell carcinoma cells in vitro and in vivo. Journal of Experimental & Clinical Cancer Research. 2012;31(1):p. 22. doi: 10.1186/1756-9966-31-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longhi A., Errani C., De Paolis M., Mercuri M., Bacci G. Primary bone osteosarcoma in the pediatric age: state of the art. Cancer Treatment Reviews. 2006;32(6):423–436. doi: 10.1016/j.ctrv.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Martini N., Huvos A. G., Miké V., Marcove R. C., Beattie E. J., Jr. Multiple pulmonary resections in the treatment of osteogenic sarcoma. The Annals of Thoracic Surgery. 1971;12(3):271–280. doi: 10.1016/s0003-4975(10)65124-7. [DOI] [PubMed] [Google Scholar]
- 18.Yang J., Zhang W. New molecular insights into osteosarcoma targeted therapy. Current Opinion in Oncology. 2013;25(4):398–406. doi: 10.1097/cco.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 19.Botter S. M., Neri D., Fuchs B. Recent advances in osteosarcoma. Current Opinion in Pharmacology. 2014;16:15–23. doi: 10.1016/j.coph.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 20.He J.-P., Hao Y., Wang X.-L., et al. Review of the molecular pathogenesis of osteosarcoma. Asian Pacific Journal of Cancer Prevention. 2014;15(15):5967–5976. doi: 10.7314/apjcp.2014.15.15.5967. [DOI] [PubMed] [Google Scholar]
- 21.Bramwell V. H., Burgers M., Sneath R., et al. A comparison of two short intensive adjuvant chemotherapy regimens in operable osteosarcoma of limbs in children and young adults: the first study of the European Osteosarcoma Intergroup. Journal of Clinical Oncology. 1992;10(10):1579–1591. doi: 10.1200/jco.1992.10.10.1579. [DOI] [PubMed] [Google Scholar]
- 22.Bacci G., Picci P., Ferrari S., et al. Primary chemotherapy and delayed surgery for nonmetastatic osteosarcoma of the extremities. Results in 164 patients preoperatively treated with high doses of methotrexate followed by cisplatin and doxorubicin. Cancer. 1993;72(11):3227–3238. doi: 10.1002/1097-0142(19931201)72:11<3227::aid-cncr2820721116>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 23.Provisor A. J., Ettinger L. J., Nachman J. B., et al. Treatment of nonmetastatic osteosarcoma of the extremity with preoperative and postoperative chemotherapy: a report from the Children’s Cancer Group. Journal of Clinical Oncology. 1997;15(1):76–84. doi: 10.1200/jco.1997.15.1.76. [DOI] [PubMed] [Google Scholar]
- 24.Meyers P. A., Gorlick R., Heller G., et al. Intensification of preoperative chemotherapy for osteogenic sarcoma: results of the Memorial Sloan-Kettering (T12) protocol. Journal of Clinical Oncology. 1998;16(7):2452–2458. doi: 10.1200/jco.1998.16.7.2452. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs N., Bielack S. S., Epler D., et al. Long-term results of the co-operative German-Austrian-Swiss osteosarcoma study group’s protocol COSS-86 of intensive multidrug chemotherapy and surgery for osteosarcoma of the limbs. Annals of Oncology. 1998;9(8):893–899. doi: 10.1023/a:1008391103132. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari S., Briccoli A., Mercuri M., et al. Postrelapse survival in osteosarcoma of the extremities: prognostic factors for long-term survival. Journal of Clinical Oncology. 2003;21(4):710–715. doi: 10.1200/jco.2003.03.141. [DOI] [PubMed] [Google Scholar]
- 27.Legare M. E., Bush J., Ashley A. K., Kato T., Hanneman W. H. Cellular and phenotypic characterization of canine osteosarcoma cell lines. Journal of Cancer. 2011;2:262–270. doi: 10.7150/jca.2.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilkes D. M., Semenza G. L. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncology. 2013;9(11):1623–1636. doi: 10.2217/fon.13.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romain B., Hachet-Haas M., Rohr S., et al. Hypoxia differentially regulated CXCR4 and CXCR7 signaling in colon cancer. Molecular Cancer. 2014;13(1):p. 58. doi: 10.1186/1476-4598-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nys K., Maes H., Dudek A. M., Agostinis P. Uncovering the role of hypoxia inducible factor-1α in skin carcinogenesis. Biochimica et Biophysica Acta (BBA)—Reviews on Cancer. 2011;1816(1):1–12. doi: 10.1016/j.bbcan.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Semenza G. L. Targeting HIF-1 for cancer therapy. Nature Reviews Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 32.Maxwell P. H., Pugh C. W., Ratcliffe P. J. The pVHL-HIF-1 system. Advances in Experimental Medicine and Biology. 2001;502:365–376. doi: 10.1007/978-1-4757-3401-0_24. [DOI] [PubMed] [Google Scholar]
- 33.Yang Q.-C., Zeng B.-F., Dong Y., Shi Z.-M., Jiang Z.-M., Huang J. Overexpression of hypoxia-inducible factor-1α in human osteosarcoma: correlation with clinicopathological parameters and survival outcome. Japanese Journal of Clinical Oncology. 2007;37(2):127–134. doi: 10.1093/jjco/hyl137. [DOI] [PubMed] [Google Scholar]
- 34.Tsai S.-F., Martin D. I. K., Zon L. I., D’Andrea A. D., Wong G. G., Orkin S. H. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339(6224):446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto M., Ko L. J., Leonard M. W., Beug H., Orkin S. H., Engel J. D. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes & Development. 1990;4(10):1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu R., Yamamoto M. Gene expression regulation and domain function of hematopoietic GATA factors. Seminars in Cell & Developmental Biology. 2005;16(1):129–136. doi: 10.1016/j.semcdb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Fagerberg L., Hallström B. M., Oksvold P., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Molecular & Cellular Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.m113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yue F., Cheng Y., Breschi A., et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García P., Berlanga O., Vegiopoulos A., Vyas P., Frampton J. c-Myb and GATA-1 alternate dominant roles during megakaryocyte differentiation. Journal of Thrombosis and Haemostasis. 2011;9(8):1572–1581. doi: 10.1111/j.1538-7836.2011.04396.x. [DOI] [PubMed] [Google Scholar]
- 40.Nei Y., Obata-Ninomiya K., Tsutsui H., et al. GATA-1 regulates the generation and function of basophils. Proceedings of the National Academy of Sciences. 2013;110(46):18620–18625. doi: 10.1073/pnas.1311668110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolff L., Humeniuk R. Concise review: erythroid versus myeloid lineage commitment: regulating the master regulators. Stem Cells. 2013;31(7):1237–1244. doi: 10.1002/stem.1379. [DOI] [PubMed] [Google Scholar]
- 42.Zhang F.-L., Shen G.-M., Liu X.-L., Wang F., Zhao Y.-Z., Zhang J.-W. Hypoxia-inducible factor 1-mediated human GATA1 induction promotes erythroid differentiation under hypoxic conditions. Journal of Cellular and Molecular Medicine. 2012;16(8):1889–1899. doi: 10.1111/j.1582-4934.2011.01484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei P., Qiao B., Li Q., et al. microRNA-340 suppresses tumorigenic potential of prostate cancer cells by targeting high-mobility group nucleosome-binding domain 5. DNA and Cell Biology. 2016;35(1):33–43. doi: 10.1089/dna.2015.3021. [DOI] [PubMed] [Google Scholar]
- 44.Gan Y., Tan J., Yang J., et al. Knockdown of HMGN5 suppresses the viability and invasion of human urothelial bladder cancer 5637 cells in vitro and in vivo. Medical Oncology. 2015;32:p. 136. doi: 10.1007/s12032-015-0611-1. [DOI] [PubMed] [Google Scholar]
- 45.Wu J., Wang J. HMGN5 expression in bladder cancer tissue and its role on prognosis. European Review for Medical and Pharmacological Sciences. 2018;22:970–975. doi: 10.26355/eurrev_201802_14378. [DOI] [PubMed] [Google Scholar]
- 46.Wei X., Yu L., Kong X. miR-488 inhibits cell growth and metastasis in renal cell carcinoma by targeting HMGN5. OncoTargets and Therapy. 2018;11:2205–2216. doi: 10.2147/ott.s156361. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Weng M., Song F., Chen J., et al. The high-mobility group nucleosome-binding domain 5 is highly expressed in breast cancer and promotes the proliferation and invasion of breast cancer cells. Tumor Biology. 2015;36(2):959–966. doi: 10.1007/s13277-014-2715-1. [DOI] [PubMed] [Google Scholar]
- 48.Ji Y.-S., Xu Q., Schmedtje J. F., Jr. Hypoxia induces high-mobility-group protein I(Y) and transcription of the cyclooxygenase-2 gene in human vascular endothelium. Circulation Research. 1998;83(3):295–304. doi: 10.1161/01.res.83.3.295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.