Abstract
Objective
To assess the association between MUC expression levels in colorectal cancer (CRC) tissues and prognosis and investigate the associations between MUC expression levels and CRC clinicopathological characteristics.
Methods
The PubMed, Embase, Cochrane Library, and Web of Science databases were searched from inception through September 13, 2019, to identify studies investigating the association between MUC expression levels in CRC tissues and prognosis. Pooled hazard ratios (HRs) or odds ratio (ORs) with 95% confidence intervals (CIs) were used to evaluate associations between MUC expression levels and prognosis or clinicopathological characteristics, respectively. The heterogeneity between studies was assessed by the I2 values, whereas the likelihood of publication bias was assessed by Egger's linear regression and Begg's rank correlation test.
Results
Among 33 included studies (n = 6032 patients), there were no associations between combined MUC phenotype expression levels and overall survival (OS) or disease-free survival (DFS)/relapse-free survival (RFS) in patients with CRC. In subgroup analyses, the upregulated MUC1 expression (HR = 1.50; 95% CI, 1.29–1.74; P < 0.00001) was associated with poor OS. However, the upregulated MUC2 expression (HR = 0.64; 95% CI, 0.52–0.79; P < 0.00001) was associated with better OS. Furthermore, a high level of MUC1 expression (HR = 1.99; 95% CI, 0.99–3.99; P = 0.05) was associated with shorter DFS/RFS. However, patients with a low level of MUC2 tumors showed better DFS/RFS than patients with a high level of MUC2 tumors (HR = 0.71; 95% CI, 0.49–1.04; P = 0.08; P = 0.0.009, I2 = 67%) and MUC5AC expression (HR = 0.56; 95% CI, 0.38–0.82; P = 0.003) was associated with longer DFS/RFS. In addition, a high level of MUC1 expression was associated with CRC in the rectum, deeper invasion, lymph node metastasis, distant metastasis, advanced tumor stage, and lymphatic invasion. A high level of MUC2 expression had a protective effect. High secretion of MUC5AC is associated with colon cancer compared with rectal cancer.
Conclusion
The protein expression of MUC1 might be a poor biomarker in colorectal cancer and might play a role in tumor transformation and metastasis. However, the protein expression of MUC2 expression might have a protective effect. Furthermore, randomized controlled trials (RCTs) of large patients are needed to confirm the results.
1. Introduction
Colorectal cancer (CRC) is among the most frequently diagnosed cancers in the United States (US) [1]. In 2018, an estimated 140,250 Americans will be diagnosed with CRC and 50,630 individuals will die from the disease [2]. Although morbidity and mortality in CRC are reduced by high-quality healthcare and healthy lifestyles, the 5-year overall survival (OS) rates after initial diagnosis remain at 67% for patients with rectal cancer and 64% for patients with colon cancer [1]. Furthermore, CRC survivors have a high risk of cancer recurrence [3, 4] and secondary tumors, particularly in the digestive system [5].
The classic tumor, node, and metastasis (TNM) staging system is regarded as the standard prognostic parameter and forms the basis for treatment decisions in CRC [6]. However, since the TNM system fails to reflect the intrinsic biological heterogeneity of CRC, especially in patients with atypical early or occult metastases, only 40% of CRCs are diagnosed at an early stage and approximately 50% of recently diagnosed cases will progress to metastatic cancer [7]. In addition, the prognostic value of TNM in patients with CRC is suboptimal [8]. Currently, there is an unmet need for biomarkers that accurately predict CRC progression, metastasis, and treatment outcomes [9].
In recent years, increasing attention has been given to the role of mucins (MUC) in the pathogenesis of cancer. MUC are a family of high molecular weight glycosylated proteins [10], which have a highly polymorphic tandem repeat in the central region [11]. At present, approximately 20 MUC have been identified. These can be divided into two major subfamilies, secreting gel-type mucins and transmembrane mucins, according to their structure and function [12]. MUC are usually expressed on the apical surfaces of normal glandular epithelial cells and luminal epithelial cells and have key functions in immunity, cell adhesion, and intracellular signaling [13]. Studies on the subcellular distribution of MUC and biochemical characteristics of malignant transformation and progression implicate MUC in tumorigenesis and metastasis [14–18], suggesting that abnormal MUC expression may be a predictive biomarker of CRC.
Evidence suggests that MUC expression is involved in the invasion and metastasis of various malignancies, including gallbladder cancer [19], breast cancer [20], ovarian cancer [21], gastric carcinoma [22, 23], pancreatic carcinoma [24–26], ampullary cancer [27, 28], lung cancer [16, 29], prostate cancer [30], renal cell carcinoma [31], and appendiceal carcinoma [32]. However, the prognostic value of MUC expression in CRC remains controversial [33–37]. To clarify the inconsistent findings from previously published studies investigating the role of MUC in CRC, this meta-analysis was conducted to assess the association between MUC expression levels and prognosis in CRC and investigate the associations between MUC expression levels and several CRC clinicopathological characteristics.
2. Materials and Methods
This systematic review and meta-analysis is reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline [38]. Basing on previously published studies, our study does not include any research with humans or animals, so ethical recognition and patient consent are not required.
2.1. Search Strategy
Two review authors independently searched the PubMed, Embase, Cochrane Library, and Web of Science databases from inception through September 13, 2019. Keywords included (“mucins” OR “mucin” OR “MUC”) AND (“colorectal cancer” OR “colorectal neoplasm” OR “colorectal tumor” OR “colonic cancer” OR “colon cancer” OR “rectal cancer” OR “CRC”) AND (“prognostic “OR “prognosis” OR “outcome” OR “survival”). A manual search of the reference lists of relevant articles was performed. Searches were limited to articles published in English or Chinese language.
2.2. Inclusion and Exclusion Criteria
The inclusion criteria were (1) study design: cohort study; (2) population: patients with CRC; (3) parameter: MUC expression levels in CRC tissues; and (4) outcome: association between MUC expression levels in CRC tissues and prognosis.
The exclusion criteria were as follows: (1) duplicate publications; (2) in vitro or animal studies; (3) reviews, conference reports, meta-analyses, books, case reports, or letters; or (4) studies that reported insufficient data. When articles reported data from the same study, the most recent article was included.
2.3. Data Extraction
Two review authors independently extracted data from the eligible studies, including the surname of the first author, year, country, sample size, patients' mean age, MUC phenotype, antibody for MUC, cut-off value for MUC, frequency of high MUC expression, detection method, TNM stage, histologic type, mean tumor dimensions, median follow-up, and outcomes. Disagreements about data extraction were resolved by discussion with a third reviewer until consensus was reached.
2.4. Quality Assessment
Two review authors independently conducted an assessment of the methodological quality of included studies using the Newcastle Ottawa Scale (NOS) [39]. The NOS assessed the quality of the enrolled groups, the comparability and outcomes of the study populations, and study quality on a scale from 0 to 9 points, with ≥7 considered high-quality research.
Publication bias was evaluated using Egger's linear regression and Begg's rank correlation test [40].
2.5. Statistical Analysis
Statistical analyses were performed using Review Manager, version 5.3 (Cochrane Collaboration, Copenhagen, Denmark) and STATA, version 12.0 (Stata Corporation, College Station, TX, USA). Survival analysis was performed according to Moher et al. [38]. Hazard ratios (HRs) were directly extracted from included studies, or digitized and extracted using Engauge Digitizer version 4.1 (http://markummitchell.github.io/engauge-digitizer/) software when prognostic information was plotted as a Kaplan-Meier curve [41]. Pooled HRs with corresponding 95% confidence intervals (CIs) were used to assess the association between MUC expression levels (low vs. high) in CRC tissues and OS or disease-free survival (DFS)/relapse-free survival (RFS). Odds ratios (ORs) with 95% CIs were used to assess the impact of MUC expression levels on clinicopathological characteristics.
Studies with significant heterogeneity were identified with the chi-squared test (P ≤ 0.10) and the inconsistency index (I2 ≥ 50%) [42]. When significant heterogeneity was found, a random effects model was adopted. Otherwise, a fixed effects model is used. Subgroup analyses stratified by MUC phenotype and metaregression analysis were performed to explore sources of heterogeneity. The likelihood of publication bias was assessed by Egger's linear regression and Begg's rank correlation test. Sensitivity analysis evaluated the robustness of the data by omitting one study at a time. P < 0.05 was considered statistically significant.
3. Results
3.1. Search Results
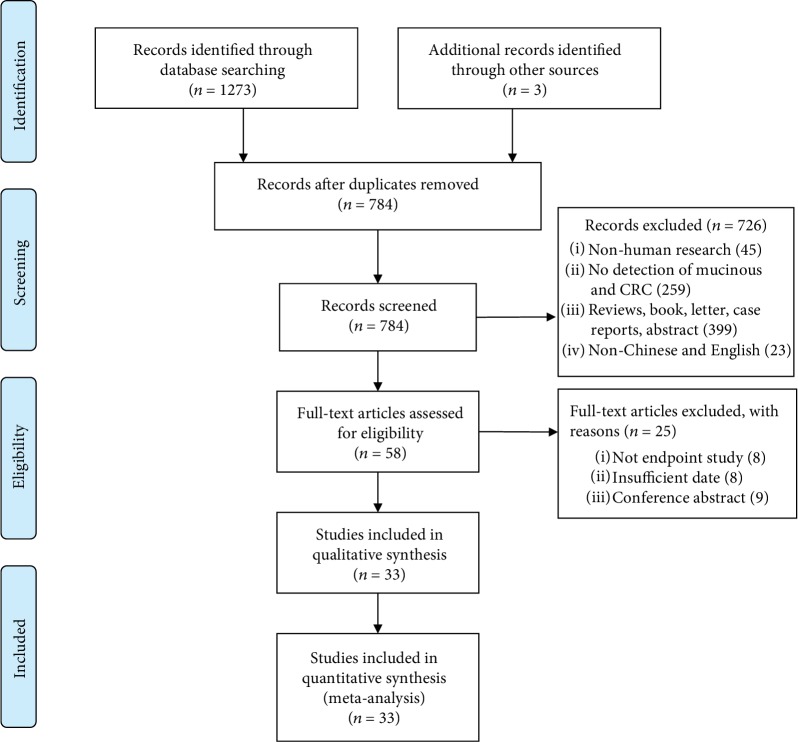
A total of 1273 articles were identified from the electronic search of the databases, and 3 additional studies were obtained from the manual search of the reference lists of relevant articles. After excluding 492 duplicates, titles and abstracts were screened, and 726 studies that did not meet the inclusion criteria were excluded. The full text of 58 studies was retrieved for further review, and 8 articles that did not report an endpoint, 8 articles with insufficient data, and 9 conference abstracts were excluded. Finally, 33 observational studies [33–37, 43–70] were found eligible for inclusion in our review (Figure 1).
Figure 1.
Flow diagram of included studies.
3.2. Characteristics of the Included Studies
The characteristics of the included studies are shown in Table 1. The 33 eligible studies were published between 1987 and 2019. The studies included a total of 6032 cases. The mean age of patients ranged from 54.3 to 72.0 years, and the median follow-up ranged from 18.0 to 116.0 months. All included studies evaluated the correlation between MUC expression levels in CRC tissues and prognosis. 31 studies evaluated MUC expression using immunohistochemistry (IHC), and 2 studies used reverse transcriptase polymerase chain reaction (qRT-PCR). Nine MUC phenotypes, determined by the expression of MUC1, MUC2, MUC3, MUC4, MUC5AC, MUC12, MUC16, MUC20, and sialomucin, were associated with prognosis in CRC. Various anti-MUC monoclonal antibodies were utilized to identify the MUC phenotypes, and each study applied a different cut-off point (low/high level) to assess MUC expression.
Table 1.
Characteristics of the included studies.
| First author | Year | Country | Patient number | Detection method | Mean age (years) | Media follow-up (mounts) | Outcome | Mucins phenotype | Antibody | Cut-off value (high level) | High MUC expression |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adams | 2009 | Switzerland | 938 | IHC | 70.5 | 128.0 | OS | MUC2 | NR | PP > 5% | NR |
| Al-Maghrabi | 2019 | Saudi Arabia | 128 | IHC | NR | NR | OS/DFS | MUC2 | MRQ-18 | PP ≥ 25% | 36.7% |
| Baldus | 2000 | Germany | 264 | IHC | 64.8 | NR | OS | MUC1 | NCL-MUC1 | PP > 5% | 58.0% |
| Baldus | 2004 | Germany | 205 | IHC | 65.0 | NR | OS | MUC1 | HMFG-2 | PP > 35% | 49.8% |
| Betge | 2016 | Germany | 381 | IHC | 68.5 | NR | OS/DFS | MUC1 | Ma695 | PP > 0% | 64.0% |
| MUC2 | Ccp-58 | PP > 0% | 77.0% | ||||||||
| MUC5AC | 45M1 | PP > 0% | 48.9% | ||||||||
| MUC6 | MCN6.01 | PP > 0% | 28.7% | ||||||||
| Dawson | 1987 | UK | 358 | IHC | 65.7 | 18.0 | OS | Sialomucin | High iron diamine-alcian blue | Blue staining | 29.6% |
| Diaz | 2018 | Spain | 96 | IHC | 65.9 | NR | DFS | MUC1 | Clone E29 | PP ≥ 50% | 46.0% |
| Duncan | 2007 | UK | 403 | IHC | 72.0 | 116.0 | OS | MUC1 | Ma695 | PP ≥ 30% | 31.5% |
| MUC3 | 1143/B7 | PP ≥ 30% | 73.9% | ||||||||
| Elzagheid | 2013 | Libya | 141 | IHC | NR | 77.0 | OS/DFS | MUC2 | MRQ-18 | PP > 0% | 50.0% |
| Hiraga | 1998 | Japan | 100 | IHC | 62.7 | 80.0 | OS | MUC1 | KL-6 | PP > 30% | 71.0% |
| Imai | 2013 | Japan | 250 | IHC | 66.9 | NR | OS/RFS | MUC2 | Ccp-58 | PP ≥ 25% | 49.4% |
| MUC5AC | CLH2 | PP ≥ 1% | 46.8% | ||||||||
| Ionescu | 2014 | Romania | 39 | qRT-PCR | 66.0 | NR | OS | MUC12 | NR | NR | NR |
| Kang | 2011 | Korea | 229 | IHC | NR | 108 | OS | MUC2 | NR | Score ≥ 6 | 24.2% |
| Kasprzak | 2018 | Poland | 34 | IHC | NR | NR | OS | MUC1 | Ma552 | PP ≥ 2.57% | 100% |
| MUC2 | Ccp-58 | PP ≥ 4.97% | 100% | ||||||||
| Khanh | 2013 | Japan | 206 | IHC | NR | NR | OS/RFS | MUC1 | Ma695 | PP ≥ 25% | 62.6% |
| MUC2 | Ccp-58 | PP ≥ 50% | 32.5% | ||||||||
| MUC4 | 1G8 | PP ≥ 50% | 33.0% | ||||||||
| MUC5AC | CLH2 | PP ≥ 5% | 33.5% | ||||||||
| Kimura | 2000 | Japan | 110 | IHC | 63.1 | 68.5 | OS | MUC1 | KL-6 | PP ≥ 30% | 69.1% |
| Kocer | 2002 | Turkey | 41 | IHC | 56.3 | NR | DFS | MUC5AC | 45M1 | ISS > 0.1 | 34.1% |
| Kocer | 2006 | USA | 30 | IHC | 59.0 | 39.0 | OS | MUC5AC | 45M1 | PP > 10% | 60.0% |
| Lennerz | 2016 | USA | 33 | IHC | 58.0 | 51.2 | OS | MUC2 | Ccp58 | PP ≥ 10% | 84.0% |
| MUC5AC | CLH2 | PP ≥ 10% | 45.0% | ||||||||
| MUC6 | CLH5 | PP ≥ 10% | 0.0% | ||||||||
| Manne | 2000 | USA | 166 | IHC | 65.3 | NR | OS | MUC1 | DF3 | SI ≥ 0.5 | 39.8% |
| MUC2 | Ccp58 | SI ≥ 0.5 | 80.7% | ||||||||
| Matsuda | 2010 | Japan | 569 | IHC | 68.0 | NR | OS | MUC2 | Anti-MUC2 | PP ≥ 10% | 65.0% |
| MUC5AC | Anti-MUC5 | PP ≥ 10% | 15.1% | ||||||||
| MUC6 | Anti-MUC6 | PP ≥ 10% | 1.9% | ||||||||
| Matsuyama | 2010 | Japan | 100 | qRT-PCR | 65.1 | 27.0 | DFS | MUC12 | Rabbit polyclonal antibody | NR | NR |
| Perez | 2008 | Brazil | 35 | IHC | 62.2 | NR | OS/DFS | MUC1 | Ma695 | PP > 10% | 20.0% |
| MUC2 | Ccp-58 | PP > 10% | 65.7% | ||||||||
| MUC5AC | CLH2 | PP > 10% | 22.9% | ||||||||
| Shanmugam | 2010 | USA | 132 | IHC | 65.0 | NR | OS | MUC4 | Clone 8G7 | ISS > 2 | 24.2% |
| Sun | 2018 | China | 118 | IHC | 54.3 | 57.0 | OS/DFS | MUC1 | MXB Biotechnologies | PP ≥ 10% | 14.4% |
| Streppel | 2012 | USA | 39 | IHC | 63.6 | NR | OS | MUC16 | Monoclonal antibody | PP > 0% | 64.1% |
| Wang | 2016 | China | 81 | IHC | 63.5 | NR | OS | MUC1 | ZM-0391 | ISS > 1 | 53.1% |
| Wang | 2017 | China | 139 | IHC | NR | NR | OS | MUC2 | NCL-MUC2 | PP > 20% | 48.2% |
| MUC5AC | NCL-MUC5 | PP > 20% | 28.1% | ||||||||
| Xiao | 2013 | China | 150 | IHC | 55.0 | NR | OS/DFS | MUC20 | Mouse antihuman polyclonal antibody | ISS > 2 | 60.7% |
| You | 2006 | China | 203 | IHC | NR | 111.9 | OS | MUC1 | Ma695 | IRS ≥ 2 | 40.7% |
| Yu | 2007 | China | 150 | IHC | 57.5 | NR | OS | MUC1 | Ma695 | ISS ≥ 2 | 45.3% |
| MUC2 | Ccp-58 | ISS ≥ 2 | 52.6% | ||||||||
| MUC5AC | 45M1 | ISS ≥ 2 | 44.0% | ||||||||
| Zhang | 2008 | Japan | 77 | IHC | 64.9 | NR | OS | MUC1 | KL-6 | SI (positive) | 55.8% |
| Zwenger | 2014 | Argentina | 90 | IHC | NR | NR | OS | MUC1 | HMFG1 | Score > 0 | 94.0% |
| MUC2 | H300 | Score > 0 | 52.4% |
NR: not reported; RT-PCR: reverse transcriptase polymerase chain reaction; IHC: immunohistochemistry; SI: staining intensity; PP: positive cell percentage; immunostaining score (ISS): PP∗SI (while groups I and II (absent and low) were considered negative expression).
3.3. Methodological Quality
According to the NOS, all included studies were of high methodological quality (score ≥ 7) ().
3.4. MUC Expression and Overall Survival in CRC
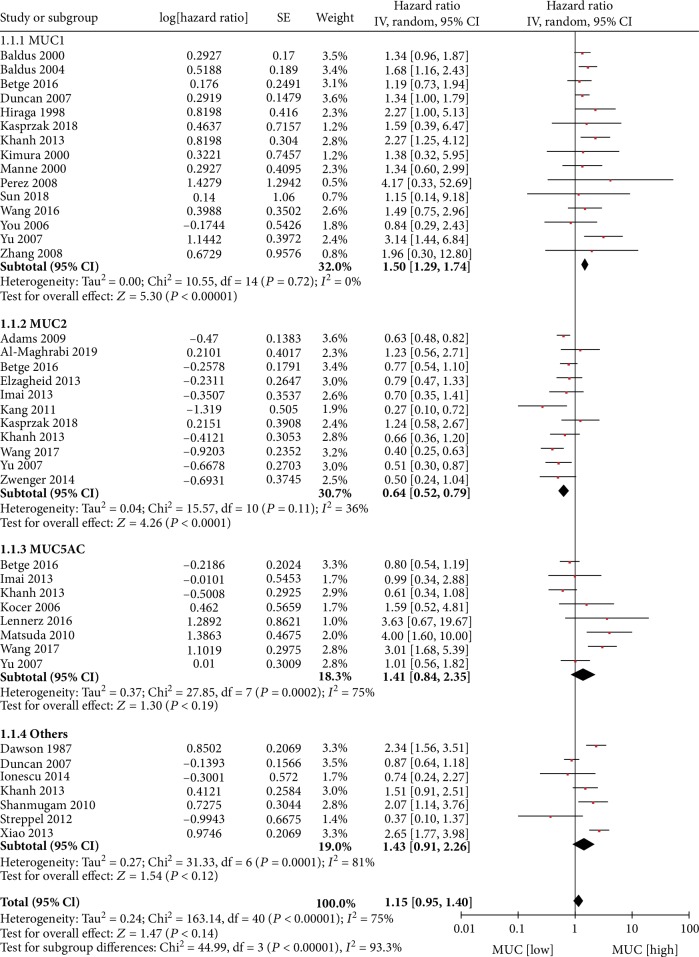
The association between MUC expression levels in CRC tissues and OS was investigated in 41 datasets from 30 articles; each dataset represented various MUC phenotypes. The meta-analysis demonstrated no association between combined MUC phenotype expression levels and OS (HR = 1.15; 95% CI, 0.95–1.40; P = 0.14). There was evidence of significant heterogeneity between studies (P < 0.00001, I2 = 75%). The source of the heterogeneity was investigated in a subgroup analysis stratified by specific MUC phenotype. The subgroup analysis demonstrated that a high level vs. a low level of MUC1 expression (HR = 1.50; 95% CI, 1.29–1.74; P < 0.00001; P = 0.72, I2 = 0%) or a low level vs. a high level of MUC2 expression (HR = 1.56; 95% CI, 1.27–1.92; P < 0.00001; P = 0.11, I2 = 36%) was associated with poor OS in patients with CRC. However, associations between the levels of MUC5AC (HR = 1.41; 95% CI, 0.84–2.35; P = 0.19; P = 0.0002, I2 = 75%), other MUC phenotypes (HR = 1.43; 95% CI, 0.91–2.26; P = 0.12; P < 0.00001, I2 = 81%), and OS were not significant (Figure 2).
Figure 2.
MUC expression and OS.
3.5. MUC Expression and Disease-Free Survival/Recurrence-Free Survival in CRC
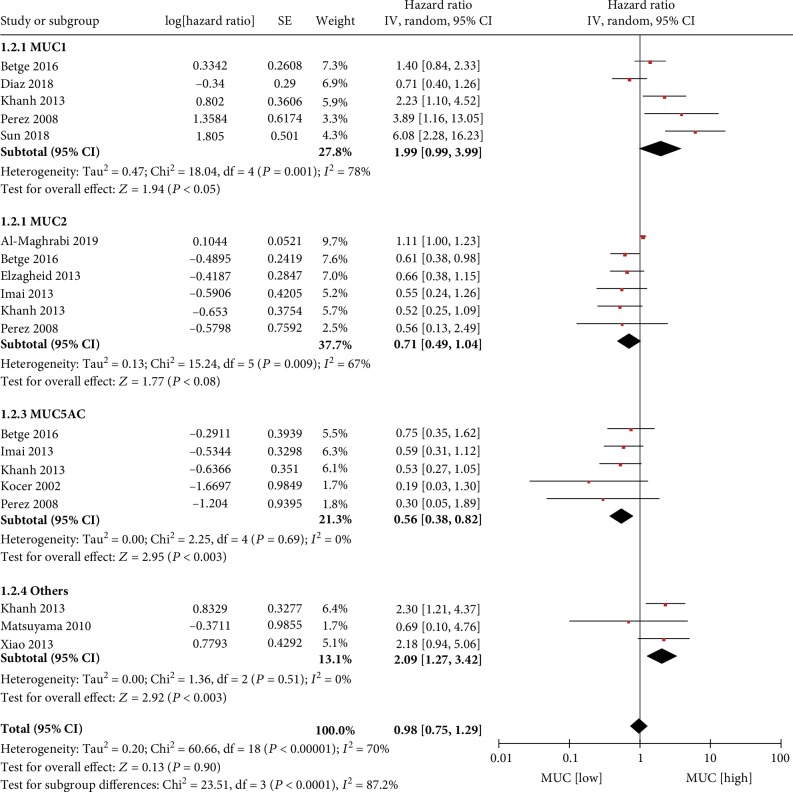
The association between MUC expression level in CRC tissues and DFS/RFS was investigated in 19 datasets from 11 articles. The meta-analysis demonstrated no association between combined MUC phenotype expression levels and DFS/RFS (HR = 0.98; 95% CI, 0.75–1.29; P = 0.90). There was evidence of significant heterogeneity between studies (P < 0.00001, I2 = 70%). The source of the heterogeneity was investigated in a subgroup analysis stratified by specific MUC phenotype. The subgroup analysis demonstrated that a high level vs. a low level of MUC1 expression (HR = 1.99; 95% CI, 0.99–3.99; P = 0.05; P = 0.0001, I2 = 78%) or other MUC expression (HR = 2.09; 95% CI, 1.27–3.42; P = 0.003; P = 0.51, I2 = 0%) was associated with shorter DFS/RFS in patients with CRC. However, a high level vs. a low level of MUC5AC expression (HR = 0.56; 95% CI, 0.38–0.82; P = 0.003; P = 0.69, I2 = 0%) was associated with longer DFS/RFS and patients with a low level of MUC2 tumors showed better DFS/RFS than patients with a high level of MUC2 tumors (HR = 0.71; 95% CI, 0.49–1.04; P = 0.08; P = 0.0.009, I2 = 67%).(Figure 3).
Figure 3.
MUC expression and DFSRFS.
3.6. MUC Expression and CRC Clinicopathological Characteristics
The meta-analysis demonstrated no association between combined MUC phenotype expression levels and CRC clinicopathological characteristics. In all analyses, there was evidence of significant heterogeneity between studies. The source of the heterogeneity was investigated in subgroup analyses stratified by specific MUC phenotype (Table 2).
Table 2.
Meta-analysis of the correlation between MUC expression and clinicopathological factors of colorectal cancer.
| Clinicopathological parameter | Mucins phenotype | No. of studies | OR (95% CI) | Analysis model | Test for overall effect | Heterogeneity | ||
|---|---|---|---|---|---|---|---|---|
| Z test | P value | I 2 (%) | P value | |||||
| TNM stage (III/IV vs. I/II) | MUC1 | 11 | 2.17 (1.31-3.59) | Random | 3.03 | 0.002 | 83 | <0.00001 |
| MUC2 | 7 | 0.52 (0.36-0.76) | Random | 3.35 | 0.0008 | 52 | 0.05 | |
| MUC5AC | 8 | 1.00 (0.67-1.49) | Random | 0.01 | 0.99 | 55 | 0.03 | |
|
| ||||||||
| Depth of invasion (T3/T4 vs. T1/T2) | MUC1 | 11 | 1.79 (1.41-2.26) | Fixed | 4.86 | <0.00001 | 40 | 0.08 |
| MUC2 | 6 | 0.65 (0.37-1.13) | Random | 1.53 | 0.13 | 63 | 0.02 | |
| MUC5AC | 4 | 0.64 (0.35-1.18) | Random | 1.42 | 0.15 | 61 | 0.05 | |
|
| ||||||||
| Lymph node metastasis (+ vs. -) | MUC1 | 10 | 2.45 (1.38-4.35) | Random | 3.07 | 0.002 | 81 | <0.00001 |
| MUC2 | 8 | 0.59 (0.47-0.73) | Fixed | 4.64 | <0.00001 | 48 | 0.06 | |
| MUC5AC | 7 | 1.07 (0.67-1.72) | Random | 0.29 | 0.77 | 67 | 0.006 | |
|
| ||||||||
| Tumor site (colon vs. rectum) | MUC1 | 7 | 0.79 (0.63-0.98) | Fixed | 2.12 | 0.03 | 0 | 0.63 |
| MUC2 | 5 | 1.64 (1.01-2.67) | Random | 2.02 | 0.04 | 55 | 0.06 | |
| MUC5AC | 6 | 1.97 (1.48-2.62) | Fixed | 4.63 | <0.00001 | 49 | 0.08 | |
|
| ||||||||
| Distant metastasis (+ vs. -) | MUC1 | 3 | 2.47 (1.47-4.13) | Fixed | 3.43 | 0.0006 | 49 | 0.14 |
| MUC2 | 3 | 0.83 (0.48-1.41) | Fixed | 0.70 | 0.49 | 0 | 0.61 | |
| MUC5AC | 2 | 0.86 (0.15-4.87) | Random | 0.17 | 0.87 | 73 | 0.06 | |
|
| ||||||||
| Lymphatic invasion (+ vs. -) | MUC1 | 5 | 3.39 (1.69-9.14) | Random | 3.19 | 0.001 | 72 | 0.007 |
| MUC2 | 3 | 0.53 (0.27-1.03) | Random | 1.88 | 0.06 | 60 | 0.08 | |
| MUC5AC | 4 | 0.76 (0.55-1.05) | Fixed | 1.64 | 0.10 | 20 | 0.29 | |
|
| ||||||||
| Mucinous component (high vs. low) | MUC1 | 7 | 0.71 (0.42-1.19) | Random | 1.31 | 0.19 | 59 | 0.02 |
| MUC2 | 2 | 14.46 (1.71-121.97) | Random | 2.46 | 0.01 | 59 | 0.12 | |
| MUC5AC | 3 | 1.41 (0.85-2.34) | Fixed | 1.32 | 0.19 | 0 | 0.62 | |
|
| ||||||||
| Gender (male vs. female) | MUC1 | 7 | 1.10 (0.86-1.41) | Fixed | 0.77 | 0.44 | 0 | 0.75 |
| MUC2 | 7 | 0.87 (0.68-1.12) | Fixed | 1.07 | 0.29 | 8 | 0.29 | |
| MUC5AC | 6 | 0.93 (0.69-1.24) | Random | <0.00001 | 1.00 | 55 | 0.005 | |
|
| ||||||||
| Tumor size (large vs. small) | MUC1 | 4 | 0.77 (0.53-1.12) | Fixed | 1.38 | 0.17 | 19 | 0.30 |
| MUC2 | 2 | 0.70 (0.47-1.05) | Fixed | 1.73 | 0.08 | 0 | 0.39 | |
| MUC5AC | 2 | 0.80 (0.48-1.32) | Fixed | 0.87 | 0.38 | 0 | 0.41 | |
|
| ||||||||
| Histological grade (3 vs. 1 and 2) | MUC1 | 12 | 1.39 (0.87-2.21) | Random | 1.39 | 0.16 | 66 | 0.0007 |
| MUC2 | 7 | 0.75 (0.56-0.99) | Fixed | 2.02 | 0.04 | 44 | 0.10 | |
| MUC5AC | 5 | 1.44 (0.70-2.97) | Random | 0.99 | 0.32 | 79 | 0.0007 | |
RR: risk ratio; Random: random effects model; Fixed: fixed.
A high level of MUC1 expression (III/IV vs. I/II: OR = 2.17, 95% CI = 1.31–3.59, P = 0.002) was associated with advanced tumor stage in patients with CRC than MUC2 expression (III/IV vs. I/II: OR = 0.52, 95% CI = 0.36–0.76, P = 0.0008), but the association between MUC5AC expression and tumor stage was not significant.
A high level of MUC1 expression (T3/T4 vs. T1/T2: OR = 1.79, 95% CI = 1.41–2.26, P < 0.00001) was associated with deeper invasion in patients with CRC, but the association between MUC5AC and MUC2 expression and depth of invasion was not significant.
A high level of MUC1 expression (positive vs. negative: OR = 2.45, 95% CI = 1.38–4.35, P = 0.002) was associated with lymph node metastasis in patients with CRC than MUC2 expression (positive vs. negative: OR = 0.59, 95% CI = 0.47–0.73, P < 0.00001), but the association between MUC5AC expression and lymph node metastasis was not significant.
A high level of MUC1 expression (positive vs. negative: OR = 0.79, 95% CI = 0.63–0.98, P = 0.03) was associated with rectum cancer. However, the elevated MUC2 expression (positive vs. negative: OR = 1.64, 95% CI = 1.01–2.67, P = 0.04) and MUC5AC expression (positive vs. negative: OR = 1.97, 95% CI = 1.48–2.62, P < 0.00001) were associated with colon cancer.
A high level of MUC1 expression was associated with distant metastasis (positive vs. negative: OR = 2.47, 95% CI = 1.47–4.13, P = 0.0006) and lymphatic invasion (positive vs. negative: OR = 3.39, 95% CI = 1.69–9.14, P = 0.001) in patients with CRC. A high level of MUC2 expression was associated mucinous cancer (high vs. low: OR = 14.46, 95% CI = 1.71–121.97, P = 0.01) and low histological grade (3 vs. 1 and 2: OR = 0.75, 95% CI = 0.56–0.99, P = 0.04).
There were no associations between the expression levels of any MUC phenotypes and other clinicopathological characteristics, including gender or tumor size.
3.7. Sensitivity Analysis and Publication Bias
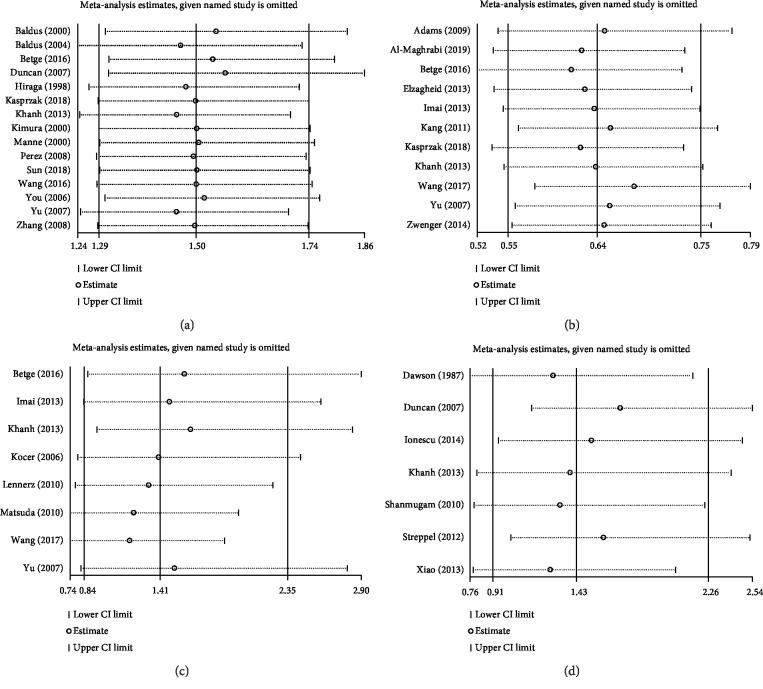
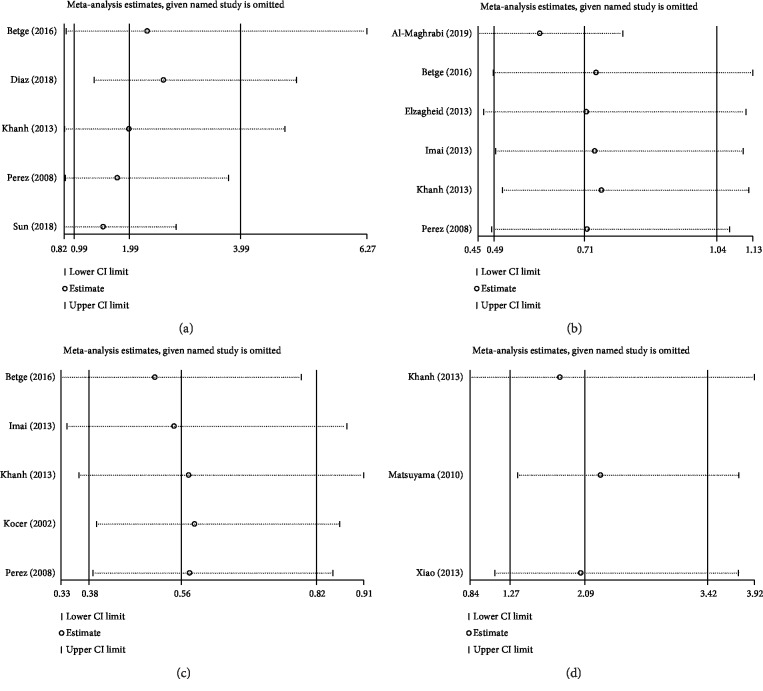
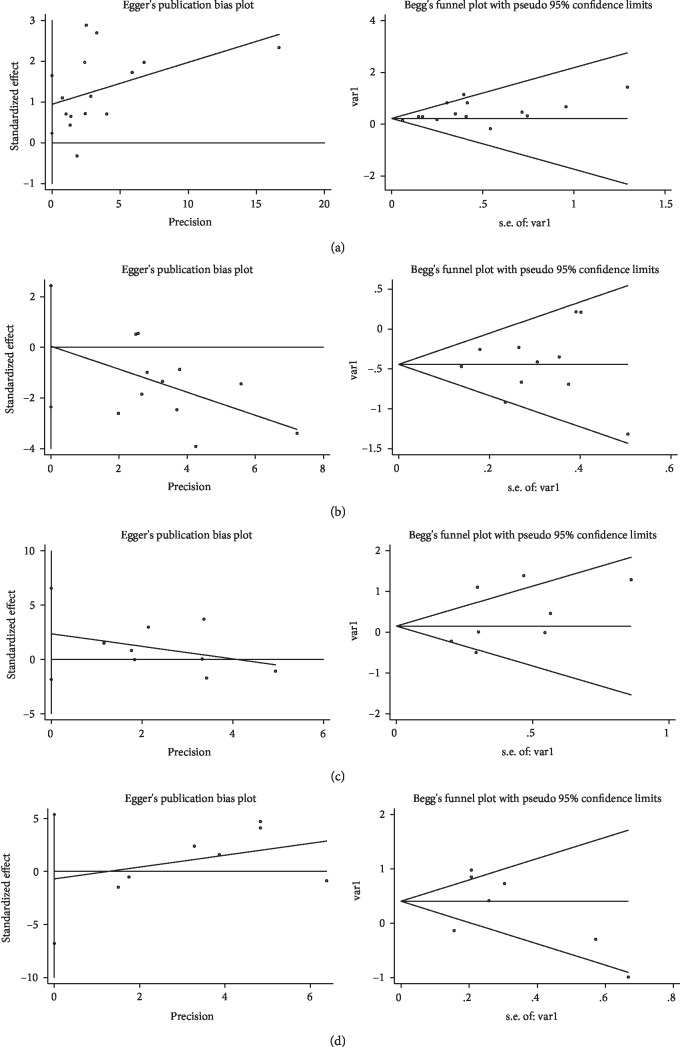
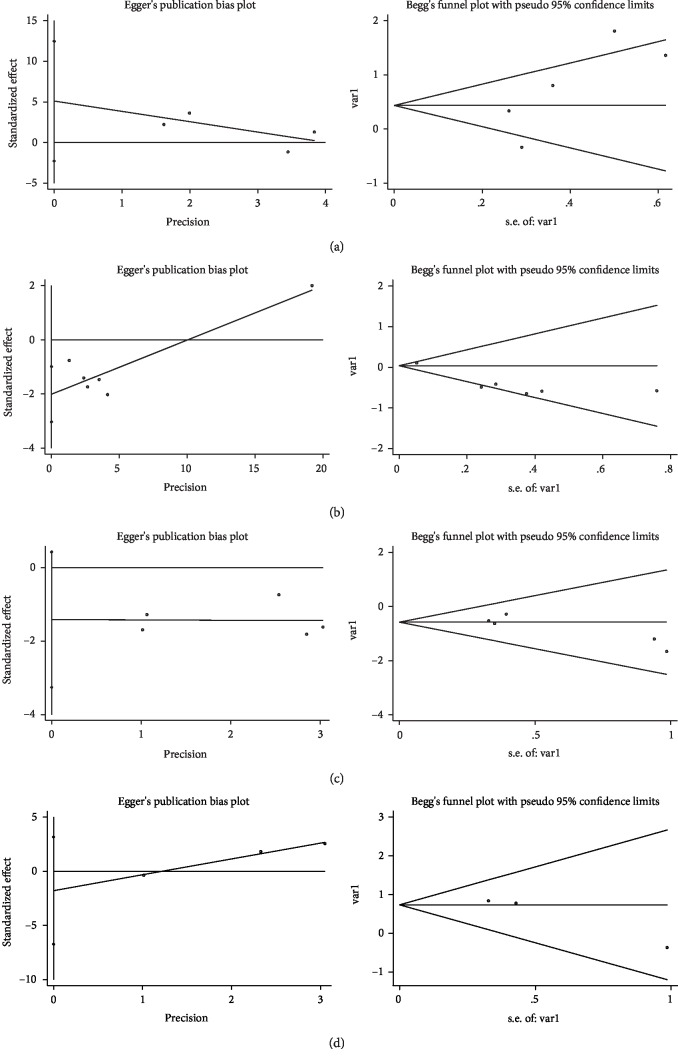
Sensitivity analysis omitting one study at a time demonstrated the associations of MUC family members' expression with OS (Figure 4) and DFS/RFS (Figure 5) in CRC were robust. Begg's rank correlation test and Egger's linear regression showed no publication bias among studies investigating OS (Figure 6) and DFS/RFS (Figure 7).
Figure 4.
Sensitivity analysis for MUC expression ((a): MUC1, (b): MUC2, (c): MUC5AC, (d): Others MUC) and OS.
Figure 5.
Sensitivity analysis for MUC expression ((a): MUC1, (b): MUC2, (c): MUC5AC, (d): Others MUC) and DFS/RFS.
Figure 6.
Publication bias for MUC expression ((a): MUC1, (b): M UC2, (c): MUC5AC, (d): Others MUC) and OS.
Figure 7.
Publication bias for MUC expression ((a): MUC1, (b): MUC2, (c): MUC5AC, (d): Others MUC) and DFS/RFS.
3.8. Metaregression
Metaregression was performed to explore the factors influencing the association of MUC expression with OS and DFS/RFS in CRC. None of the covariates (cut-off value, antibody, TNM stage, country, and years) analyzed were identified as potential sources of heterogeneity (Table 3).
Table 3.
Results of meta-regression analysis exploring the source of heterogeneity with OS and DFS/RFS.
| Mucins phenotype | Covariates | Univariate analysis (OS) | Univariate analysis (DFS) | ||||
|---|---|---|---|---|---|---|---|
| Coefficient | SE | P value | Coefficient | SE | P value | ||
| MUC1 | Antibody | 0.055 | 0.087 | 0.538 | -0.142 | 0.882 | 0.883 |
| Cut-off value | 0.0297 | 0.032 | 0.369 | 0.155 | 0.295 | 0.635 | |
| TNM stage | 0.365 | 0.324 | 0.281 | 0.773 | 1.106 | 0.535 | |
| Country | 0.048 | 0.462 | 0.323 | 0.155 | 0.295 | 0.635 | |
| Year | -0.001 | 0.014 | 0.964 | -0.077 | 0.115 | 0.552 | |
|
| |||||||
| MUC2 | Antibody | -0.204 | 0.215 | 0.367 | 0.550 | 0.252 | 0.094 |
| Cut-off value | -0.027 | 0.043 | 0.552 | -0.030 | 0.221 | 0.898 | |
| TNM stage | -0.309 | 0.124 | 0.054 | -0.270 | 0.838 | 0.763 | |
| Country | 0.007 | 0.048 | 0.891 | 0.180 | 0.050 | 0.023 | |
| Year | 0.036 | 0.030 | 0.264 | 0.108 | 0.030 | 0.022 | |
|
| |||||||
| MUC5AC | Antibody | 0.464 | 0.269 | 0.135 | -0.139 | 0.434 | 0.769 |
| Cut-off value | 0.187 | 0.158 | 0.282 | -0.248 | 0.193 | 0.288 | |
| TNM stage | 0.923 | 0.211 | 0.055 | -0.652 | 0.961 | 0.546 | |
| Country | 0.250 | 0.240 | 0.339 | -0.379 | 0.293 | 0.287 | |
| Year | 0.135 | 0.073 | 0.859 | 0.102 | 0.069 | 0.236 | |
4. Discussion
In this meta-analysis, we assessed the association between MUC expression levels in CRC tissues and prognosis and investigate the associations between MUC expression levels and several CRC clinicopathological characteristics. Interestingly, findings demonstrated no association between combined MUC phenotype expression levels in CRC tissues and prognosis. However, in subgroup analyses stratified by MUC phenotype, a high level of MUC1 expression was associated with poor OS and DFS/RFS, a high level of MUC2 expression was associated with improved OS and DFS/RFS, and a high level of MUC5AC was associated with improved DFS/RFS. Generally, heterogeneity between studies was significantly reduced in the subgroup analyses stratified by MUC phenotype. Meanwhile, meta-regression analysis revealed that antibody for MUC, cut-off value for MUC, TNM stage, and histologic type were not significant sources of heterogeneity.
However, importantly, several studies have shown a correlation between MUC expression and patient with various cancers. For example, a meta-analysis reported that MUC expression was significantly higher in patients with esophageal adenocarcinoma than in normal squamous esophageal mucosa [71]. The study by Lu et al. [72] also indicated that increased MUC expression was associated with worse OS and more detrimental clinicopathological outcomes in head and neck cancer patients. Overall, it is reasonable that the expression of MUC was associated with variable clinical outcomes in different tumors. These differences may be due to different mechanisms, pathways, and treatment options. An earlier meta-analysis have shown that abnormal expression of MUC in CRC tissues compared with healthy mucosa plays an important role in the pathogenesis and progression of CRC [73]. Several meta-analyses have explored the association between MUC expression and CRC clinicopathological characteristics [74–76]. Furthermore, compared with two earlier meta-analyses for various types of cancer by Xu et al. [77] and Huang et al. [78], the present analysis not only added additional 26 and 27 studies in colorectal cancer subtype but also examined the correlation between MUC expression and the clinicopathological factors of colorectal cancer.
The current study explored the association between MUC expression levels in CRC tissues and CRC clinicopathological characteristics. A high level of MUC1 expression was associated with CRC in the rectum, deeper invasion, lymph node metastasis, distant metastasis, advanced tumor stage, and lymphatic invasion. Elevated MUC2 expression was associated with CRC in the colon, shallower lesions, negative lymph node metastasis, early stage of tumor, mucinous carcinoma, and larger tumor size. MUC5AC was more easily expressed in colon cancer. These findings implicate MUC1 in mechanisms that promote tumor invasion, lymph node metastasis, high stage, lymphatic invasion, and poor survival in CRC, while MUC2 may have a protective role. A number of studies have demonstrated a unique role for MUC in proliferation, survival, metastasis, epithelial-mesenchymal transition, and antiapoptosis in tumors [13, 17, 79–82]. As a ligand of cell adhesion molecules, MUC 1 induces circulating tumor cells (CTCs) to adhere to endothelial cells or transport to distant sites, establishing secondary tumors [81]. MUC2 is major structural component of the inner mucus layer in the colon, which is impervious to bacteria and protects the colon epithelium. Decreased MUC2 expression allows bacteria to contact the epithelial surface, triggering inflammatory bowel disease, which can lead to colon cancer [83]. Studies characterizing the function of MUC5AC are scarce. Hoshi et al. [84] showed that MUC5AC protects pancreatic cancer cells from TRAIL-induced apoptosis, while other reports suggest that MUC5AC has no effect on cell growth, cell survival, proliferation, or morphology in vitro [85].
Findings from the current meta-analysis indicate MUC1 may be a biomarker of poor prognosis in CRC and suggest that combined detection of MUC1 and MUC2 should be used to accurately predict CRC progression, metastasis, and treatment outcomes. Understanding the association between MUC expression levels and metastasis in CRC may help clarify the risk of metastasis at the time of diagnosis in patients with CRC, especially in those patients without symptoms or signs of metastasis. Clinically, MUC detection is simple and easy to implement.
This study was associated with several limitations. First, HRs from some of the included studies were calculated from Kaplan-Meier curves, which may have influenced the robustness of our findings. Second, the lack of a standardized detection methods and antibodies to detect MUC status may have affected the accuracy of our results. Third, despite the use of subgroup analysis and meta-regression to identify potential sources of heterogeneity between studies, they may have been additional sources of heterogeneity that impacted our findings. Finally, the sample size was small, and results should be considered preliminary.
In conclusion, findings from the current study suggest that MUC1 and MUC2 expression levels in CRC tissues are associated with OS, DFS/RFS, tumor site, depth of invasion, lymph node metastasis, distant metastasis, tumor stage, histologic type, and lymphatic invasion. These results indicate that MUC status can be used to differentiate between normal cells and CRC cells and predict a patient's clinicopathological characteristics and prognosis. The clinical relevance of MUC regulation in CRC tissues remains to be elucidated in large well-designed cohort studies.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Chao Li is assigned to the data curation, investigation, methodology, resources, validation, and writing of the original draft. Didi Zuo is also assigned to the data curation, formal analysis, and investigation. Tao Liu is responsible for the formal analysis, investigation, and validation. Libin Yin is also responsible for the formal analysis and software. Chenyao Li is also assigned to software. Lei Wang is in charge of the conceptualization, funding acquisition, project administration, supervision, visualization, and writing of the review and editing.
Supplementary Materials
Table S1: quality assessment of the included studies.
References
- 1.Siegel R. L., Miller K. D., Fedewa S. A., et al. Colorectal cancer statistics, 2017. CA: a Cancer Journal for Clinicians. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.National Cancer Institute Surveillance. Epidemiology, and End Results Program. Cancer Stat Facts: Colorectal Cancer 2018. May 2018, https://seer.cancer.gov/statfacts/html/colorect.html.
- 3.Cole B. F., Baron J. A., Sandler R. S., et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297(21):2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 4.Vollset S. E., Clarke R., Lewington S., et al. Effects of folic acid supplementation on overall and site-specific cancer incidence during the randomised trials: meta-analyses of data on 50 000 individuals. The Lancet. 2013;381(9871):1029–1036. doi: 10.1016/S0140-6736(12)62001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doubeni C. A. The impact of colorectal cancer screening on the US population: is it time to celebrate? Cancer. 2014;120(18):2810–2813. doi: 10.1002/cncr.28789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compton C. C. Optimal pathologic staging: defining stage II disease. Clinical Cancer Research. 2007;13(22):6862s–6870s. doi: 10.1158/1078-0432.CCR-07-1398. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Pons M., Cruz-Correa M. Colorectal Cancer biomarkers: where are we now? BioMed Research International. 2015;2015:14. doi: 10.1155/2015/149014.149014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueno H., Mochizuki H., Akagi Y., et al. Optimal colorectal cancer staging criteria in TNM classification. Journal of Clinical Oncology. 2012;30(13):1519–1526. doi: 10.1200/JCO.2011.39.4692. [DOI] [PubMed] [Google Scholar]
- 9.Paul D., Kumar A., Gajbhiye A., Santra M. K., Srikanth R. Mass spectrometry-based proteomics in molecular diagnostics: discovery of cancer biomarkers using tissue culture. BioMed Research International. 2013;2013:16. doi: 10.1155/2013/783131.783131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrianifahanana M., Moniaux N., Batra S. K. Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2006;1765(2):189–222. doi: 10.1016/j.bbcan.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Hollingsworth M. A., Swanson B. J. Mucins in cancer: protection and control of the cell surface. Nature Reviews Cancer. 2004;4(1):45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 12.Itoh Y., Kamata-Sakurai M., Denda-Nagai K., et al. Identification and expression of human epiglycanin/MUC21: a novel transmembrane mucin. Glycobiology. 2008;18(1):74–83. doi: 10.1093/glycob/cwm118. [DOI] [PubMed] [Google Scholar]
- 13.Hattrup C. L., Gendler S. J. Structure and function of the cell surface (tethered) mucins. Annual Review of Physiology. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 14.Singh P. K., Hollingsworth M. A. Cell surface-associated mucins in signal transduction. Trends in Cell Biology. 2006;16(9):467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Krishn S. R., Kaur S., Smith L. M., et al. Mucins and associated glycan signatures in colon adenoma-carcinoma sequence: prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Letters. 2016;374(2):304–314. doi: 10.1016/j.canlet.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awaya H., Takeshima Y., Yamasaki M., Inai K. Expression of MUC1, MUC2, MUC5AC, and MUC6 in atypical adenomatous hyperplasia, bronchioloalveolar carcinoma, adenocarcinoma with mixed subtypes, and mucinous bronchioloalveolar carcinoma of the lung. American Journal of Clinical Pathology. 2004;121(5):644–653. doi: 10.1309/U4WG-E9EB-FJN6-CM8R. [DOI] [PubMed] [Google Scholar]
- 17.Kimura T., Finn O. J. MUC1 immunotherapy is here to stay. Expert Opinion on Biological Therapy. 2013;13(1):35–49. doi: 10.1517/14712598.2012.725719. [DOI] [PubMed] [Google Scholar]
- 18.Rakha E. A., Boyce R. W. G., Abd el-Rehim D., et al. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Modern Pathology. 2005;18(10):1295–1304. doi: 10.1038/modpathol.3800445. [DOI] [PubMed] [Google Scholar]
- 19.Hiraki T., Yamada S., Higashi M., et al. Immunohistochemical expression of mucin antigens in gallbladder adenocarcinoma: MUC1-positive and MUC2-negative expression is associated with vessel invasion and shortened survival. Histology and Histopathology. 2017;32(6):585–596. doi: 10.14670/HH-11-824. [DOI] [PubMed] [Google Scholar]
- 20.Patel D. S., Khandeparkar S. G. S., Joshi A. R., et al. Immunohistochemical study of MUC1, MUC2 and MUC5AC expression in primary breast carcinoma. Journal of Clinical and Diagnostic Research. 2017;11(4):EC30–EC34. doi: 10.7860/jcdr/2017/26533.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y. F., Zhang M. Y., Wu X., et al. High MUC2 expression in ovarian cancer is inversely associated with the M1/M2 ratio of tumor-associated macrophages and patient survival time. PLoS One. 2013;8(12, article e79769) doi: 10.1371/journal.pone.0079769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao L. J., Zhao S., Zhao E. H., et al. Clinicopathological and prognostic significance of MUC-2, MUC-4 and MUC-5AC expression in Japanese gastric carcinomas. Asian Pacific Journal of Cancer Prevention. 2012;13(12):6447–6453. doi: 10.7314/apjcp.2012.13.12.6447. [DOI] [PubMed] [Google Scholar]
- 23.Javanbakht M., Akhavanmoghadam J., Talaei A. J., et al. Differential expression of two genes Oct-4 and MUC5AC associates with poor outcome in patients with gastric cancer. Clinical and Experimental Pharmacology & Physiology. 2017;44(11):1099–1105. doi: 10.1111/1440-1681.12840. [DOI] [PubMed] [Google Scholar]
- 24.Yang H. S., Tamayo R., Almonte M., et al. Clinical significance of MUC1, MUC2 and CK17 expression patterns for diagnosis of pancreatobiliary arcinoma. Biotechnic & Histochemistry. 2012;87(2):126–132. doi: 10.3109/10520295.2011.570276. [DOI] [PubMed] [Google Scholar]
- 25.Dotan E., Alpaugh R. K., Ruth K., et al. Prognostic significance of MUC-1 in circulating tumor cells in patients with metastatic pancreatic adenocarcinoma. Pancreas. 2016;45(8):1131–1135. doi: 10.1097/MPA.0000000000000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chauhan S. C., Ebeling M. C., Maher D. M., et al. MUC13 mucin augments pancreatic tumorigenesis. Molecular Cancer Therapeutics. 2012;11(1):24–33. doi: 10.1158/1535-7163.MCT-11-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriya T., Kimura W., Hirai I., Takasu N., Mizutani M. Expression of MUC1 and MUC2 in ampullary cancer. International Journal of Surgical Pathology. 2011;19(4):441–447. doi: 10.1177/1066896911405654. [DOI] [PubMed] [Google Scholar]
- 28.Xue Y., Reid M. D., Balci S., et al. Immunohistochemical classification of ampullary carcinomas: critical reappraisal fails to confirm prognostic relevance for recently proposed panels, and highlights MUC5AC as a strong prognosticator. The American Journal of Surgical Pathology. 2017;41(7):865–876. doi: 10.1097/PAS.0000000000000863. [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa N., Hattori N., Yokoyama A., et al. Usefulness of monitoring the circulating Krebs von den Lungen-6 levels to predict the clinical outcome of patients with advanced nonsmall cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors. International Journal of Cancer. 2008;122(11):2612–2620. doi: 10.1002/ijc.23411. [DOI] [PubMed] [Google Scholar]
- 30.Cozzi P. J., Wang J., Delprado W., et al. MUC1, MUC2, MUC4, MUC5AC and MUC6 expression in the progression of prostate cancer. Clinical & Experimental Metastasis. 2005;22(7):565–573. doi: 10.1007/s10585-005-5376-z. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H., Liu Y., Xie H., et al. High mucin 5AC expression predicts adverse postoperative recurrence and survival of patients with clear-cell renal cell carcinoma. Oncotarget. 2017;8(35):59777–59790. doi: 10.18632/oncotarget.15894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shibahara H., Higashi M., Yokoyama S., et al. A comprehensive expression analysis of mucins in appendiceal carcinoma in a multicenter study: MUC3 is a novel prognostic factor. PLoS One. 2014;9(12, article e115613) doi: 10.1371/journal.pone.0115613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ionescu C., Braicu C., Chiorean R., et al. TIMP-1 expression in human colorectal cancer is associated with SMAD3 gene expression levels: a pilot study. Journal of Gastrointestinal and Liver Diseases. 2014;23(4):413–418. doi: 10.15403/jgld.2014.1121.234.smad. [DOI] [PubMed] [Google Scholar]
- 34.Khanh D. T., Mekata E., Mukaisho K. I., et al. Transmembrane mucin MUC1 overexpression and its association with CD10+ myeloid cells, transforming growth factor-β1 expression, and tumor budding grade in colorectal cancer. Cancer Science. 2013;104(7):958–964. doi: 10.1111/cas.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Streppel M. M., Vincent A., Mukherjee R., et al. Mucin 16 (cancer antigen 125) expression in human tissues and cell lines and correlation with clinical outcome in adenocarcinomas of the pancreas, esophagus, stomach, and colon. Human Pathology. 2012;43(10):1755–1763. doi: 10.1016/j.humpath.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H., Jin S., Lu H., et al. Expression of survivin, MUC2 and MUC5 in colorectal cancer and their association with clinicopathological characteristics. Oncology Letters. 2017;14(1):1011–1016. doi: 10.3892/ol.2017.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lennerz J. K., van der Sloot K. W. J., le L. P., et al. Colorectal cancer in Crohn’s colitis is comparable to sporadic colorectal cancer. International Journal of Colorectal Disease. 2016;31(5):973–982. doi: 10.1007/s00384-016-2574-x. [DOI] [PubMed] [Google Scholar]
- 38.Moher D., Liberati A., Tetzlaff J., Altman D. G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Medicine. 2009;6(7, article e1000097) doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 40.Begg C. B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 41.Tierney J. F., Stewart L. A., Ghersi D., Burdett S., Sydes M. R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8(1):p. 16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgins J. P., Thompson S. G., Deeks J. J., Altman D. G. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams H., Tzankov A., Lugli A., Zlobec I. New time-dependent approach to analyse the prognostic significance of immunohistochemical biomarkers in colon cancer and diffuse large B-cell lymphoma. Journal of Clinical Pathology. 2009;62(11):986–997. doi: 10.1136/jcp.2008.059063. [DOI] [PubMed] [Google Scholar]
- 44.Baldus S. E., Mönig S. P., Huxel S., et al. MUC1 and nuclear β-catenin are coexpressed at the invasion front of colorectal carcinomas and are both correlated with tumor prognosis. Clinical Cancer Research. 2004;10(8):2790–2796. doi: 10.1158/1078-0432.ccr-03-0163. [DOI] [PubMed] [Google Scholar]
- 45.Baldus S. E., Zirbes T. K., Hanisch F. G., et al. Thomsen-Friedenreich antigen presents as a prognostic factor in colorectal carcinoma - a clinicopathologic study of 264 patients. Cancer. 2000;88(7):1536–1543. [PubMed] [Google Scholar]
- 46.Betge J., Schneider N. I., Harbaum L., et al. MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: expression profiles and clinical significance. Virchows Archiv. 2016;469(3):255–265. doi: 10.1007/s00428-016-1970-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dawson P. M., Habib N. A., Rees H. C., Williamson R. C., Wood C. B. Influence of sialomucin at the resection margin on local tumour recurrence and survival of patients with colorectal cancer: a multivariate analysis. The British Journal of Surgery. 1987;74(5):366–369. doi: 10.1002/bjs.1800740514. [DOI] [PubMed] [Google Scholar]
- 48.Duncan T. J., Watson N. F. S., al-Attar A. H., Scholefield J. H., Durrant L. G. The role of MUC1 and MUC3 in the biology and prognosis of colorectal cancer. World Journal of Surgical Oncology. 2007;5(1):p. 31. doi: 10.1186/1477-7819-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elzagheid A., Emaetig F., Buhmeida A., et al. Loss of MUC2 expression predicts disease recurrence and poor outcome in colorectal carcinoma. Tumor Biology. 2013;34(2):621–628. doi: 10.1007/s13277-012-0588-8. [DOI] [PubMed] [Google Scholar]
- 50.Hiraga Y., Tanaka S., Haruma K., et al. Immunoreactive MUC1 expression at the deepest invasive portion correlates with prognosis of colorectal cancer. Oncology. 1998;55(4):307–319. doi: 10.1159/000011868. [DOI] [PubMed] [Google Scholar]
- 51.Imai Y., Yamagishi H., Fukuda K., Ono Y., Inoue T., Ueda Y. Differential mucin phenotypes and their significance in a variation of colorectal carcinoma. World Journal of Gastroenterology. 2013;19(25):3957–3968. doi: 10.3748/wjg.v19.i25.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang H., Min B. S., Lee K. Y., et al. Loss of E-cadherin and MUC2 expressions correlated with poor survival in patients with stages II and III colorectal carcinoma. Annals of Surgical Oncology. 2011;18(3):711–719. doi: 10.1245/s10434-010-1338-z. [DOI] [PubMed] [Google Scholar]
- 53.Kimura T., Tanaka S., Haruma K., et al. Clinical significance of MUC1 and E-cadherin expression, cellular proliferation, and angiogenesis at the deepest invasive portion of colorectal cancer. International Journal of Oncology. 2000;16(1):55–64. doi: 10.3892/ijo.16.1.55. [DOI] [PubMed] [Google Scholar]
- 54.Kocer B., McKolanis J., Soran A. Humoral immune response to MUC5AC in patients with colorectal polyps and colorectal carcinoma. BMC Gastroenterology. 2006;6(1) doi: 10.1186/1471-230X-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kocer B., Soran A., Erdogan S., et al. Expression of MUC5AC in colorectal carcinoma and relationship with prognosis. Pathology International. 2002;52(7):470–477. doi: 10.1046/j.1440-1827.2002.01369.x. [DOI] [PubMed] [Google Scholar]
- 56.Manne U., Weiss H. L., Grizzle W. E. Racial differences in the prognostic usefulness of MUC1 and MUC2 in colorectal adenocarcinomas. Clinical Cancer Research. 2000;6(10):4017–4025. [PubMed] [Google Scholar]
- 57.Matsuda M., Sentani K., Noguchi T., et al. Immunohistochemical analysis of colorectal cancer with gastric phenotype: Claudin-18 is associated with poor prognosis. Pathology International. 2010;60(10):673–680. doi: 10.1111/j.1440-1827.2010.02587.x. [DOI] [PubMed] [Google Scholar]
- 58.Matsuyama T., Ishikawa T., Mogushi K., et al. MUC12 mRNA expression is an independent marker of prognosis in stage II and stage III colorectal cancer. International Journal of Cancer. 2010;127(10):2292–2299. doi: 10.1002/ijc.25256. [DOI] [PubMed] [Google Scholar]
- 59.Perez R. O., Bresciani B. H., Bresciani C., et al. Mucinous colorectal adenocarcinoma: influence of mucin expression (Muc 1, 2 and 5) on clinico-pathological features and prognosis. International Journal of Colorectal Disease. 2008;23(8):757–765. doi: 10.1007/s00384-008-0486-0. [DOI] [PubMed] [Google Scholar]
- 60.Shanmugam C., Jhala N. C., Katkoori V. R., et al. Prognostic value of mucin 4 expression in colorectal adenocarcinomas. Cancer. 2010;116(15):3577–3586. doi: 10.1002/cncr.25095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X., Zhan P., Guo W. Expression of MUC1 and Gal-3 in colorectal carcinoma and its relation to prognosis. Journal of Practical Oncology. 2016;31(3):250–254. [Google Scholar]
- 62.Xiao X., Wang L., Wei P., et al. Role of MUC20 overexpression as a predictor of recurrence and poor outcome in colorectal cancer. Journal of Translational Medicine. 2013;11(1):p. 151. doi: 10.1186/1479-5876-11-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.You J. F., Hsieh L. L., Changchien C. R., et al. Inverse effects of mucin on survival of matched hereditary nonpolyposis colorectal cancer and sporadic colorectal cancer patients. Clinical Cancer Research. 2006;12(14 Part 1):4244–4250. doi: 10.1158/1078-0432.CCR-06-0202. [DOI] [PubMed] [Google Scholar]
- 64.Yu X. W., Rong W., Xu F. L., Xu G. Y., Sun Y. R., Feng M. Y. Expression and clinical significance of Mucin and E-cadherin in colorectal tumors. Chinese Journal of Cancer. 2007;26(11):1204–1210. [PubMed] [Google Scholar]
- 65.Zhang W., Tang W., Inagaki Y., et al. Positive KL-6 mucin expression combined with decreased membranous beta-catenin expression indicates worse prognosis in colorectal carcinoma. Oncology Reports. 2008;20(5):1013–1019. [PubMed] [Google Scholar]
- 66.Zwenger A., Rabassa M., Demichelis S., Grossman G., Segal-Eiras A., Croce M. V. High expression of sLex associated with poor survival in Argentinian colorectal cancer patients. International Journal of Biological Markers. 2014;29(1):E30–E39. doi: 10.5301/jbm.5000060. [DOI] [PubMed] [Google Scholar]
- 67.Sun Y., Wu X., Zhang Y., et al. Pathological complete response may underestimate distant metastasis in locally advanced rectal cancer following neoadjuvant chemoradiotherapy and radical surgery: incidence, metastatic pattern, and risk factors. European Journal of Surgical Oncology. 2019;45(7):1225–1231. doi: 10.1016/j.ejso.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 68.Al-Maghrabi J., Sultana S., Gomaa W. Low expression of MUC2 is associated with longer disease-free survival in patients with colorectal carcinoma. Saudi Journal of Gastroenterology. 2019;25(1):61–66. doi: 10.4103/sjg.SJG_199_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kasprzak A., Siodła E., Andrzejewska M., et al. Differential expression of mucin 1 and mucin 2 in colorectal cancer. World Journal of Gastroenterology. 2018;24(36):4164–4177. doi: 10.3748/wjg.v24.i36.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Díaz del Arco C., Garré P., Molina Roldán E., Lorca V., Cerón Nieto M. Á., Fernández Aceñero M. J. MUC1 expression in colorectal carcinoma: clinicopathological correlation and prognostic significance. Revista Española de Patología. 2018;51(4):204–209. doi: 10.1016/j.patol.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 71.Niv Y., Ho S. B., Fass R., Rokkas T. Mucin expression in the esophageal malignant and pre-malignant states: a systematic review and meta-analysis. Journal of Clinical Gastroenterology. 2018;52(2):91–96. doi: 10.1097/MCG.0000000000000863. [DOI] [PubMed] [Google Scholar]
- 72.Lu H., Liang D., Zhu Y., et al. Prognostic and clinicopathological significance of MUC expression in head and neck cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(56):96359–96372. doi: 10.18632/oncotarget.19648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niv Y., Rokkas T. Mucin expression in colorectal cancer (CRC): systematic review and meta-analysis. Journal of Clinical Gastroenterology. 2019;53(6):434–440. doi: 10.1097/MCG.0000000000001050. [DOI] [PubMed] [Google Scholar]
- 74.Zeng Y., Zhang Q., Zhang Y., et al. MUC1 predicts colorectal cancer metastasis: a systematic review and meta-analysis of case controlled studies. PLoS One. 2015;10(9, article e0138049) doi: 10.1371/journal.pone.0138049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li L., Huang P. L., Yu X. J., Bu X. D. Clinicopathological significance of mucin 2 immuno-histochemical expression in colorectal cancer: a meta-analysis. Chinese Journal of Cancer Research. 2012;24(3):190–195. doi: 10.1007/s11670-012-0190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X., Yan F., Shi R., et al. Hyper expression of mucin 5ac indicates poor cancer prognoses: a meta-analysis. Medicine. 2016;95(1, article e2396) doi: 10.1097/MD.0000000000002396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu F., Liu F., Zhao H., An G., Feng G. Prognostic significance of mucin antigen MUC1 in various human epithelial cancers: a meta-analysis. Medicine. 2015;94(50, article e2286) doi: 10.1097/MD.0000000000002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang X., Wang X., Lu S.-M., et al. Clinicopathological and prognostic significance of MUC4 expression in cancers: evidence from meta-analysis. International Journal of Clinical and Experimental Medicine. 2015;8(7):10274–10283. [PMC free article] [PubMed] [Google Scholar]
- 79.Roy L. D., Sahraei M., Subramani D. B., et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30(12):1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mohr A. M., Bailey J. M., Lewallen M. E., et al. MUC1 regulates expression of multiple microRNAs involved in pancreatic tumor progression, including the miR-200c/141 cluster. PLoS One. 2013;8(10, article e73306) doi: 10.1371/journal.pone.0073306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayashi T., Takahashi T., Motoya S., et al. MUC1 mucin core protein binds to the domain 1 of ICAM-1. Digestion. 2001;63(1):87–92. doi: 10.1159/000051917. [DOI] [PubMed] [Google Scholar]
- 82.Jepson S., Komatsu M., Haq B., et al. Muc4/sialomucin complex, the intramembrane ErbB2 ligand, induces specific phosphorylation of ErbB2 and enhances expression of p27kip, but does not activate mitogen-activated kinase or protein kinaseB/Akt pathways. Oncogene. 2002;21(49):7524–7532. doi: 10.1038/sj.onc.1205970. [DOI] [PubMed] [Google Scholar]
- 83.Hansson G. C., Johansson M. E. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes. 2010;1(1):51–54. doi: 10.4161/gmic.1.1.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoshi H., Sawada T., Uchida M., et al. MUC5AC protects pancreatic cancer cells from TRAIL-induced death pathways. International Journal of Oncology. 2013;42(3):887–893. doi: 10.3892/ijo.2013.1760. [DOI] [PubMed] [Google Scholar]
- 85.Hoshi H., Sawada T., Uchida M., et al. Tumor-associated MUC5AC stimulates in vivo tumorigenicity of human pancreatic cancer. International Journal of Oncology. 2011;38(3):619–627. doi: 10.3892/ijo.2011.911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: quality assessment of the included studies.