Abstract
MicroRNA-221/222 (miRNA-221/222, miR-221/222) is a noncoding microRNA which is widely distributed in eukaryotic organisms and deeply involved in the posttranscriptional regulation of gene expressions. According to recent studies, abnormal expressions of miR-221/222 are closely related to the occurrence and development of various kinds of malignant tumors. The role of miR-221/222 in tumor development and their potential molecular mechanism in various cancers, including liver cancer, colorectal cancer, cervical cancer, ovarian cancer, and endometrial carcinoma, are summarized and reviewed in this paper. Moreover, the potential translational biomarker role of abnormal miR-221/222 level in tumor or blood circulation for tumor diagnosis is also discussed.
1. Introduction
MicroRNAs (miRNAs) are short, endogenous, noncoding RNAs with a length of 18–25 nucleotides. In the human genome, several thousands of miRNAs are encoded, which regulate more than 30% of the genes, thereby participating in the regulation of almost all cellular functions, such as cell differentiation, proliferation, growth, migration, and apoptosis. As illustrated, microRNA-221/222 (miRNA-221/222, miR-221/222) is a noncoding microRNA which is widely distributed in eukaryotic organisms and deeply involved in the posttranscriptional regulation of gene expressions [1–4].
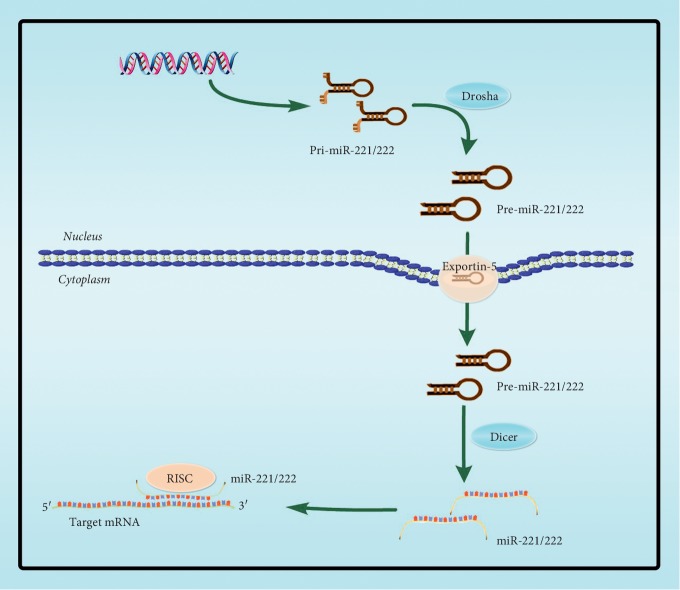
Hsa-miR-221 and hsa-miR-222 are encoded tandemly in chromosome Xp11.3, which are highly homologous miRNAs sharing the same “seed sequence.” The present study investigated the role of miR-221/222, which plays an important regulatory role in the development and progression of tumors, both in promoting or suppressing cancers [5]. Since there are different subtypes like miR-221/222 3p or 5p, whose heterogeneity may perform different biological functions, their new progresses are also reviewed in this paper. The biogenesis of miR-221/222 is similar to other miRNAs' biogenesis, which is initially transcribed principally by either RNA polymerase II or RNA polymerase III as long primary transcriptions and is further processed by the nuclear RNase Drosha and cytoplasmic RNase Dicer to generate precursor miRNAs and mature miRNAs, respectively (Figure 1) [6]. In the cytoplasm, miRNAs recognize and bind to partially complementary sites in the 3′ untranslated region (3′UTR) of target mRNAs, resulting in either translational repression or target degradation [7].
Figure 1.
Biogenesis process of miR-221/222. The gene of miR-221/222 is transcribed into primary miR-221/222 (pri-miR-221/222) by RNA polymerase II in nucleus and then processed to a stem-loop precursor miR-221/222 (pre-miR-221/222) by the nuclear RNase Drosha in the nucleus. Pre-miR-221/222 is transported from the nucleus to the cytoplasm by the exportin-5 transporter, in which the endoribonuclease Dicer cleaves it into a double-stranded miRNA, one of which binds to the RNA-induced silencing complex (RISC).
Recently, accumulated investigations showed that the abnormal expression of miR-221/222 involved in various tumor initiation and progressions. As noted, abnormal miR-221/222 expression promotes cancer cell proliferation, invasion and metastasis, stemness promotion, cell survival, chemoresistance, or the relapse of cancer cells [5, 8]. In this review, the role and its research progress of potential mechanism of miR-221/222 in tumor development are summarized and discussed.
2. Role of miR-221/222 in Tumorigenesis and Progression
2.1. Upstream Regulation of miR-221/222
In recent years, studies have found that in miRNA biogenesis (Figure 1), the pri-miRNA hairpins by Drosha and Dicer could result in a change of the length at the 5′- or 3′-end of miRNA [9]. Thus, the isoform of miRNA is longer or shorter than the consensus miRNA length [10]. The heterogeneity at the 5′-end could result in “seed shifts,” thereby changing the expected pool of target genes [11], while the occurrence of 5′ variation is relatively rare. In contrast, the 3′ heterogeneity is discovered as more common situations [12–14]. As an essential endonuclease to produce mature miRNAs, more researchers suggested Dicer is related to the regulation of miR-222. Ueda et al. reported [15] that Dicer-disrupted cells exhibited upregulated intercellular cell adhesion molecule-1 (ICAM-1) expression and enhanced susceptibility to CTL-mediated cytolysis and directly demonstrated that miR-222 functionally targeted the 3′ UTR of ICAM-1 mRNA according with luciferase reporter assays. After transfected with Dicer siRNA, or inhibitors of miR-222, the results of flow cytometric analyses and immunoblotting suggested the inhibition of Dicer or miR-222 led to an increase in ICAM-1 expression in human malignant glioma cells and promoted their susceptibility to cytotoxic T-lymphocytes (CTLs). Zhang et al. suggested that adenosine deaminases acting on RNA(ADAR) 1 could promote the expression of miRNA-222 by complexing with Dicer and the expression of phosphatase-and-tensin (PTEN) protein was significantly reduced by miRNA-222 target PTEN gene's 3′UTR [16]. Chen et al. found that Dicer expression was significantly increased in gefitinib-resistant non-small-cell lung cancer (NSCLC) PC9/GR cell line compared with the gefitinib-sensitive NSCLC PC9 cell line, while the silenced Dicer induced sensitivity to gefitinib in NSCLC cells through the downregulation of miR-221/222 to increase the protein level of caspase-3 [17]. Cochrane et al. found that miR-221/222 was higher in estrogen receptor 1 (ESR1) negative breast cancer cell lines than positive breast cancer cell lines and miR-221/222 directly targets on Dicer and ESR1 [18].
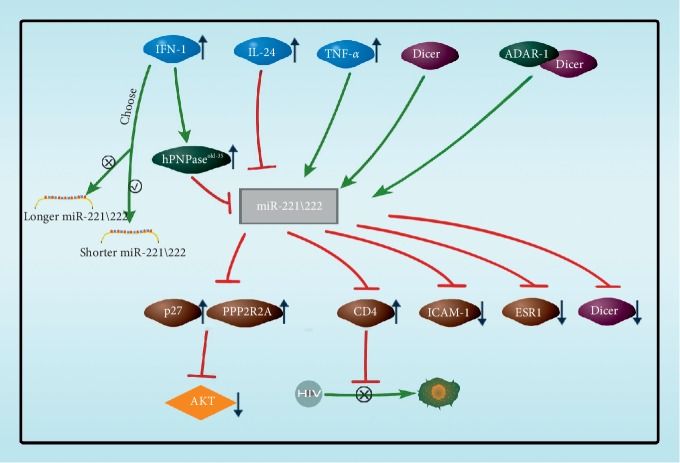
Cytokines play key regulatory roles in the expression of miR-221/222 subtypes (Figure 2). Human polynucleotide phosphorylase (hPNPaseold-35), a highly evolutionarily conserved gene that catalyzes 3′-to-5′ phosphorolysis as well as 5′-to-3′ polymerization of RNA, acts as a trimer in the expression of miR-221/222 subtypes. Das et al. [19] reported that the overexpression of hPNPaseold-35 by type-I IFN treatment resulted in robust and preferentially targeted downregulation of miRNA-221/222 with consequent upregulation of its suppressed target cyclin-dependent kinase inhibitor p27Kip1. Nejad et al. discovered that type-I IFN stimulation selectively decreases the levels of longer miR-221-3p and miR-222-3p isoforms, while sparing the shorter ones [20]. Yu et al. [21] analyzed the endogenous length variation across breast cancer tumor samples and observed the frequent occurrence of templated 3′ isomiRs of miRNA-221/222. In breast cancer cell line MCF10A, the longer miR-222 (extending 2–4 nucleotides at the 3′ end) reduced cell fusion and promotes cell apoptosis by downregulating PI3K-AKT, and the longer miR-222 displayed an increased proclivity for nuclear localization, which indicates that different subtypes of miRNA may have different functions. And when these subtypes are regulated, cellular activity is also affected. Panneerselvam et al. [22] found that IL-24 could decrease the levels of miR-222-3p and -5p, thus increasing the target PPP2R2A level of miR-222, inhibiting the activation of protein kinase B (PKB, AKT), and inhibiting the expression of AKT in lung cancer cells. Lodge reported [23] that the cytokine TNF-α produced by macrophages could enhance the expression of miR-221/222 and that miR-221/222 binds to CD4 mRNA. It could also be found that cytokines not only regulate the expression of miR-221/222 but also selectively reduce the level of allogeneic cells in order to realize the regulation of cell activity.
Figure 2.
Cytokine regulations on miR-221/222. IFN-1 decreased the levels of longer miR-221/222 isoforms and increased the levels of shorter miR-221/222 isoforms. The longer miR-222 reduces cell fusion and promotes cell apoptosis by downregulating PI3K-AKT. miR-221/222 downregulated by IFN-1 and IL-24 treatment, and downregulation of miR-221/222 upregulated the target p27 and protein phosphatase 2 regulatory subunit B alpha (PPP2R2A). But miR-221/222 was upregulated by TNF-α produced by macrophages and downregulation of miR-221/222 inhibits CD4 receptor expression in macrophages to inhibit HIV-1 entry into macrophages. Dicer generates mature miR-222, which suppresses ICAM-1 expression on tumor cells. ADAR-1 complexing with Dicer can promote the expression of miR-222. miR-221/222 was higher in ESR1-negative breast cancer cell line and directly targets Dicer and ESR1.
2.2. Regulation of Competing Endogenous RNA on miR-221/222
The proposition of competing endogenous RNA (ceRNA) was based on a series studies on the “talk” between RNAs [24]. They present a hypothesis about ceRNA as that the reversed RNAs influence each other's levels by competing for a limited pool of miRNAs, which is essentially referred to “RNA ⟶ miRNA” and also a supplement to the traditional theory of “miRNA⟶RNA.” In the context of cancer, pseudogenes and long noncoding RNAs (IncRNAs) could act as potential tumor suppressors and oncogenes through their ceRNA function by binding miRNA response elements (MREs). Lu et al. used molecular network-based identification of ceRNA (MNIceRNA) to construct miRNA-mRNA-IncRNA networks in thyroid carcinoma, while the hsa-miR-221/222 acted as key driver RNAs during the process [25]. Liu et al. verified that the expression of IncRNA GAS5 was significantly lower in the HCT116 and SW480 cell lines compared with that in the normal NCM460 cell line, whereas the expression of miR-222-3p was significantly higher [26]. The dual-luciferase reporter assays, RNA immunoprecipitations (IP), and RNA pull-down assays revealed that miR-222-3p could specifically bind to PTEN and IncRNA GAS5 and suggested that IncRNA GAS5 could regulate PTEN through competitively binding to miR-222-3p.
2.3. miR-221/222's Targets and Molecular Modulation Pathways
PTEN, TIMP3 (tissue inhibitor of metalloproteinases 3), p27, and p57 are well-known tumor suppressor genes, but their expressions are often dysregulated in human cancer [27]. The research showed that both miR-221 and miR-222 directly targeted the 3′UTR of p27 and p57 mRNA and decreased the protein level of p27 and p57, thus activating the AKT pathway [28]. On the other hand, miR-221 and miR-222 also targeted PTEN and to induce TRAIL tolerance and enhance cell migration [29]. PUMA is a member of Bcl-2 protein family and has a strong proapoptotic effect [30], while miR-221/222 could bind to 3′UTR of PUMA mRNA to inhibit the expression of PUMA and then resist normal apoptosis [31] (Table 1).
Table 1.
Role of miR-221/222 in various cancers and their direct target genes.
| Roles in cancer | Cancer types | miR-221/222 | Target genes | References |
|---|---|---|---|---|
| Onco-miRNA | Glioblastoma | Both | puma | [31] |
| Gastric cancer | miR-222 | reck, hipk2 | [32, 33] | |
| Both | pten | [34] | ||
| Bladder cancer | miR-222 | ppp2r2a | [35] | |
| Lung cancer | miR-222 | socs3, ppp2r2a | [36, 37] | |
| Liver cancer | miR-221 | p27, p57, ddit4, bmf, hdac6, adipor1, erα, caspase-3, socs3 | [38–46] | |
| miR-222 | ppp2r2a, gnai3, bbc3, pbx3 | [47–50] | ||
| Both | pten, timp3, p27 | [29, 51] | ||
| Colorectal cancer | miR-221 | p27, p57, pten, tp53inp1, reck | [49, 52, 53] | |
| miR-222 | pten, mst3, mia3 | [54–56] | ||
| Both | p27, p57, pten, pdlim2 | [57, 58] | ||
| Breast cancer | miR-221 | A20 | [59] | |
| Both | erα, esr1, mybl1, cdkn1b, bim, cdkn1c, pten, timp3, ddit4 | [60–63] | ||
| Cervical cancer | miR-221 | vash1, thbs | [64, 66] | |
| miR-222 | p27, pten | [67] | ||
| Both | timp3 | [68] | ||
| Ovarian cancer | miR-221 | apaf1, bmf, | [69, 70] | |
| miR-222 | p27, socs3, gnai2 | [71, 72] | ||
| Both | p57, pten | [66, 73] | ||
| Endometrial carcinoma | miR-222 | erα | [74] | |
| Melanoma | Both | p27, c-kit | [75, 76] | |
| Pancreatic cancer | miR-222 | p27 | [77] | |
| Thyroid cancer | Both | p27, pten | [78–79] | |
| Multiple myeloma | miR-221 | p27kip1 | [80] | |
| Chronic lymphocytic leukemia | Both | p27 | [81] | |
| Oral carcinoma | Both | p27 | [82] | |
| Retinoblastoma | Both | ap2α | [83] | |
| Nasopharyngeal carcinoma | miR-222 | pten | [84] | |
| Prostate carcinoma | Both | p27 | [85] | |
|
| ||||
| Tumor suppressor miRNA | Prostate cancer | Both | ecm29 | [61] |
| Erythroleukemia | Both | kit | [86] | |
| Gastrointestinal stromal tumors | Both | kit | [87] | |
| Tongue squamous cell carcinoma | miR-222 | mmp1, sod2 | [88] | |
| Cholangiocarcinoma | miR-221 | pik3r1 | [89] | |
| Ovarian cancer | miR-221 | arf 4 | [90] | |
| miR-222 | gnai 2 | [91] | ||
| Colorectal cancer | miR-222 | adam-17 | [92] | |
| Medulloblastoma | miR-221 | eif5a2 | [93] | |
2.4. miR-221/222 Dysregulation in Human Cancer
The abnormal high expression of miR-221/222 is involved in various cancers (Table 1), which promotes the malignant proliferation, immune escape, invasion, and metastasis of tumor cells [94]. Through the retrieval and statistics of the databases (including PubMed and Web of Science), the oncogene upregulated with overexpression of miR-221/222, including glioblastoma [31], gastric cancer [32, 33], bladder cancer [35], hepatocellular carcinoma [38, 39, 51, 59], lung cancer [36, 37], liver cancer [29, 51], breast cancer [60–63], cervical cancer [64, 68, 95], ovarian cancer [66, 69, 70], endometrial carcinoma [74], melanoma [75, 76], pancreatic cancer [77], thyroid cancer [78, 79], multiple myeloma [80], chronic lymphocytic leukemia [79], oral carcinoma [82], retinoblastoma [83, 96], nasopharyngeal carcinoma [84], and prostate carcinoma [85], was summarized. However, the upregulation of miR-221/222, also referred to as tumor suppressor miRNA, targeting overexpress oncogenes, leads to tumor suppressions. For example, miR-221/222 cluster directly targeted Ecm29 and attenuated migration and invasion in PCa cells in prostate cancer [61]. miRNA-221/222 also targets on kit+ in erythroleukemic cells [86] and gastrointestinal stromal tumor pathogenesis [87], to inhibit cell proliferation and induce apoptosis. Functional analysis and luciferase reporter gene assays indicated that has-miR-222 inhibits oral tongue squamous cell carcinoma cell invasion by targeting on matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) mRNAs [88]. Furthermore, in HuH28 human cholangiocarcinoma cells' research, Okamoto and colleagues found miR-221 could target phosphoinositide-3-kinase, regulatory subunit 1 (PIK3R1), to inhibit HuH28 cell proliferation and conferred Gem sensitivity [89].
3. Role of miR-221/222 in Various Kinds of Cancers
3.1. Role of miR-221/222 in Liver Cancer
Hepatocellular carcinoma (HCC) is one of the most high-mortality malignant tumors [97]. Recently, Song et al. [98] published a review on effect of overexpression of miR-221/222 on liver cancer several years ago, focusing on the relationship between overexpression of miR-221 and HCC, while more details between miR-221 and -222 and HCC are reported in recent papers [38, 79, 90, 91, 99]. Several papers demonstrated that the miR-221/222 level is related to the tumor TNM stages [39, 100, 101], among which Fu et al. found miR-221 is mainly located in the plasma firstly [102]. Li et al. [103] found the miR-221/222 levels were higher in HCC patients, and the overall survival rate of the high miR-221 expression group was significantly lower than that of the low miR-221 expression group according to the analysis of 46 HCC patients and 20 healthy normal controls by RT-PCR. Sohn et al. [104] investigated the levels of serum exosomal miRNAs in HCC patients and compared them with the levels observed in chronic hepatitis B (CHB) or liver cirrhosis (LC) patients. As a result, the levels of serum exosomal miR-221 and miR-222 were significantly higher in patients with HCC than in patients with CHB or LC.
It has been demonstrated that PTEN, TIMP3, and p27/CDKN1B are identified as the targets of miR-221/222 in HCC samples [29, 51]; thereby, the G1/S transition is loss of control in HCC. Gramantieri and colleagues [41] used three prediction algorithms (miRanda, TargetScould, and PicTar) predicted and the luciferase reporter assay verified bone marrow failure (BMF), a proapoptotic BH3-only member of the Bcl-2 family, is the target of miR-221 [105]. Recent reports showed that miR-222 targeted on 3′UTR of the mRNA of B-cell lymphoma-2 binding component 3 (BBC3) in HepG2 cell line [48]. Wong et al. [39] suggested that the AKT signaling was the major pathway influenced by miR-222 and that PPP2R2A was a miR-222 target in silico, thereby enhancing HCC cell invasion and motility. Pineau et al. [51] observed miR-221 expression could distinguish malignant from adjacent cirrhotic tissues and identify DNA damage-inducible transcript 4 (DDIT4) as a direct target of miR-221. Ectopic overexpression of histone deacetylase 6 (HDAC6) causes JNK/c-Jun activation, leading to autophagic cell death in HCC cells [106]. Bae et al. [42] found that the miR-221 increased according to the Met/JNK-activated c-Jun and cytokine-induced NF-κBp65 transcription factor activation, while the miR-221 suppressed HDAC6 and thus promoted hepatocellular malignant transformation and proliferation. Zhang et al. [47] found alpha inhibiting activity polypeptide 3 (GNAI3) inhibits HCC cell migration and invasion, but GNAI3 is downregulated in HCC at the protein level but not at the mRNA level. They also identify miR-222 directly binds to the 3′UTR of GNAI3 and posttranscriptionally regulates GNAI3 expression. Epithelial-mesenchymal transition (EMT) increases the migratory capacity of epithelial cells and plays a key role in the metastasis of cancer cells [105]; some reports show that the miR-221 promotes EMT in HCC cell lines by targeting on adiponectin receptor 1 (AdipoR1) and cytokine signaling 3 (SOCS3), which implies that the JAK/STAT3 signaling pathway may be involved [44, 45]. Hepatitis B virus X (HBx) protein plays an important role in the development of hepatocellular carcinoma (HCC). Chen et al. [46] identified that miR-221 promotes HBV-related HCC cancer cell proliferation by directly targeting on estrogen receptor 1 (ERα).
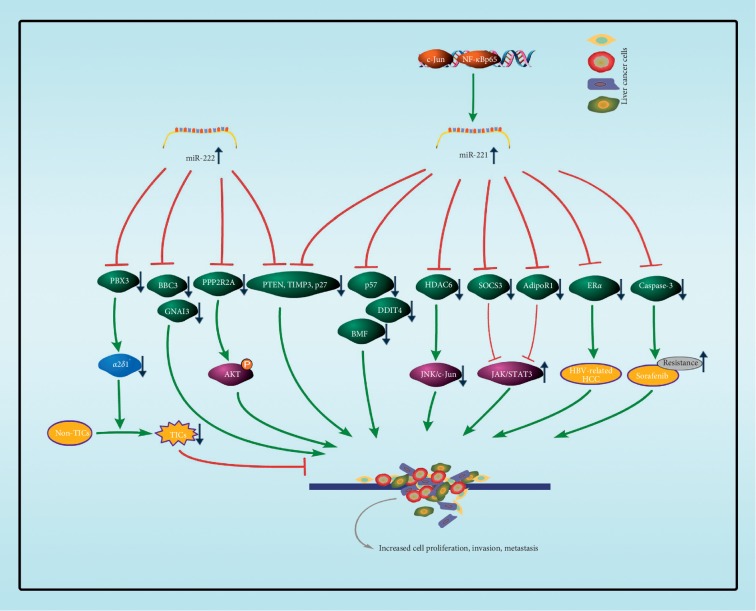
miR-221/222 is not only acting as a biomarker in patients with HCC but also directly targets on key targets in liver cancer (Figure 3). Fornari et al. [43] demonstrated that caspase-3 is a direct target of miR-221 in HCC, further suggesting miR-221 inhibition as a therapeutic approach aimed to contrast resistance to sorafenib. Moreover, the roles of miRNAs are cell context-dependent. For example, Han et al. [50] confirmed that α2δ1 (encoded by the gene CACNA2D1) is a surface marker that marks HCC tumor-initiating cells (TICs), and they identified miR-222 as a tumor suppressor in HCC by controlling the stemness and tumorigenicity of α2δ1+ TICs by directly targeting on pre-B-cell leukemia homeobox 3 (PBX3), which activated the expression of CACNA2D1.
Figure 3.
The main targets of miR-221/222 involved in liver cancer. miR-222 is a tumor suppressor by inhibiting PBX3 expression, which activates the α2δ1 expression. α2δ1 is a surface marker that marks HCC tumor initiating cells (TICs). However, miR-221/222 is often used as an onco-miRNA in liver cancer cells. BBC3, GNAI3, and PPP2R2A have been identified as a target for miR-222 and p57. DDIT4, BMF, HDAC6, AdioR1, and ERα have been identified as a target for miR-221. PTEN, TIMP3, and p27/CDKN1B have been identified as a target for miR-221/222. The downregulation of these targets promotes liver cell proliferation, invasion, and metastasis.
3.2. Role of miR-221/222 in Colorectal Cancer
Colorectal cancer (CRC) is one of the most common cancers and the fifth most common cause of cancer-related death world widely [97]. Several reports revealed miR-221/222 levels are elevated in human CRCs [58]. Early effective detection is critical for disease prevention; however, CRC is asymptomatic in the early stage and difficult to diagnose until advanced stages. Thus, there is a compelling need to identify molecular biomarkers for mass screening and early diagnosis of CRC. A report showed that miR-221 (p < 0.0001) was significantly higher in peripheral blood of 71 patients with colorectal cancer in comparison with 80 matched healthy control individuals [107]. Yau and colleagues evaluated the plasma miR-221 level [108] and the stool miR-221 level [109] and found that the plasma miR-221 had a high sensitivity (86%) but poor specificity (41%) and an AUC of only 0.61. In contrast, the stool miR-221 had an AUC of 0.73, a sensitivity rate of 62%, and a specificity rate of 74%; hence, the detection of miR-221 in stool is a better way. There are some reports that studied the changes of miR-221/222 in circulating tumor cells (CTCs) and miRNAs from CTCs during CRC treatment and found the changes in counts reflected the disease progression and/or response to chemotherapy; however, the miRNA levels (including miR-221/222) from CTCs showed transient expression and did not correlate with CTC counts [110].
The oncogene KRAS induces increased expression of miR-221/222 in human CRC cell lines [111]. It has been demonstrated that p27/CDKN1B, p57/CDKN1C, and PTEN have been identified as targets of miR-221/222 in CRC cells [49, 53, 54, 57]. Liao et al. [52] demonstrated that miR-221 promotes the cell proliferation of CRC via the inhibition of autophagy and targeted tumor protein 53-induced nuclear protein 1 (TP53INP1). Liu et al. [58] proposed a miR-221/222-NF-κB-STAT3 positive feedback loop in human CRC development and progression. They demonstrated that miR-221/222 regulates NF-κB and STAT3 signaling by directly binding to the rela coding region and stabilizing rela mRNA. Besides, miR-221/222 upregulates both RelA and STAT3 protein through binding to the 3′UTR of PDLIM2 as the E3 ligase for both RelA and STAT3.
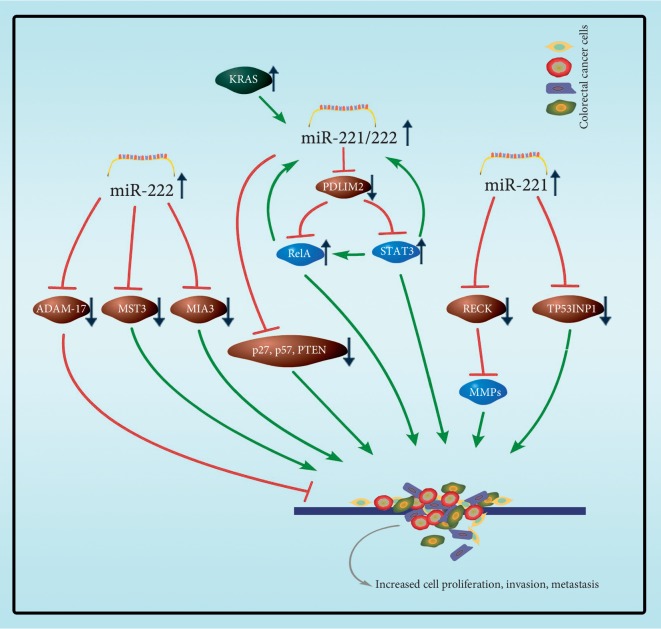
The overwhelming cause of death from CRC is metastasis and reversion (Figure 4) [112, 113]. Qin and Luo [114] found miR-221 modulates CRC cell migration and invasion in vitro and in vivo, and they demonstrated that miR-221 could directly bind to reversion-inducing-cysteine-rich protein with Kazal motifs (RECK) 3′UTR and promotes metastasis in CRC. More reports demonstrated that mammalian STE20-like protein kinase 3 (MST3) may be the miR-222 target, and therefore, overexpression of miR-222 plays a critical role in regulating CRC cell migration and invasion [56]. Gao et al. [55] demonstrated that miR-222 enhances the migration and invasion in CRC cells by specifically targeting on the 3′UTR of melanoma inhibitory activity member 3 (MIA3). However, miR-222 may be a tumor suppressor miRNA in CRC [92]. Scientists investigated the role of ADAM-17 (a disintegrin and metalloprotease 17) as a novel multidrug resistance (MDR) mechanism in multidrug-resistant CRC and found that miR-222 was directly targeting on ADAM-17, which was downregulated in multidrug-resistant CRC cells and increased cancer cells' apoptosis.
Figure 4.
The main targets of miR-221/222 involved in colorectal cancer. miR-222 directly targets ADAM-17, which was downregulated in multidrug-resistant CRC cells and increase cancer cells' apoptosis. Meanwhile, miR-221/222 is an onco-miRNA in colorectal cancer cells. MST3 and MIA3 have been identified as a target for miR-222. RECK and TP53INP1 have been identified as a target for miR-221. p27/CDKN1B, p57/CDKN1C, and PTEN have been identified as a target for miR-221/222. The downregulation of these targets promotes colorectal cell proliferation, invasion, and metastasis.
3.3. Role of miR-221/222 in Cervical Cancer
Although a great deal of scientific research has been made in recent years, the mortality rates of gynecological cancers continue to rise [97, 115]. Early diagnosis and limited options of treatment for advanced gynecological cancers are the factors to their high mortality. However, the expression level of miRNAs in gynecological cancers is closely related to its disease diagnosis, clinic pathological features, and prognostic markers, while it has been found that miR-221/222, as a common ectopic miRNA, is almost at the abnormal expression level in gynecological cancer [116].
Sun et al. [67] observed that the expression of miR-222 in cervical cancer tissue was 2.48 times higher than that in normal tissues adjacent to cancer according to the samples of 18 patients. The upregulation of miR-222 was correlated with the deterioration of cervical cancer, and the analysis showed that p27/Kip1 and PTEN were negatively correlated with miR-222, which affected the proliferation and migration of Hela cells and SiHa cells, suggesting that miR-222 might be a new target for cervical cancer therapy. Wilting et al. [95] analyzed different expression profiles of miRNAs in the development of cervical cancer, and they found that the high expression of miR-221 was a specific marker of HPV positive cervical cancer. Cervical squamous cell carcinoma (CSCC), the most common histological subtype of cervical cancer, spreads principally by migrating into the lymphatics or by invading adjacent soft tissue. Wei et al. [116] demonstrated that there was a higher level of miR-221-3p, in human primary cervical cancer tissues derived from cervical cancer patients with or without lymph node metastasis, and it enhanced the malignancy of cervical cancer cells. The calculation and experiment showed that twist homolog 2 (TWIST2) targeted on the promoter region of miR-221-3p.
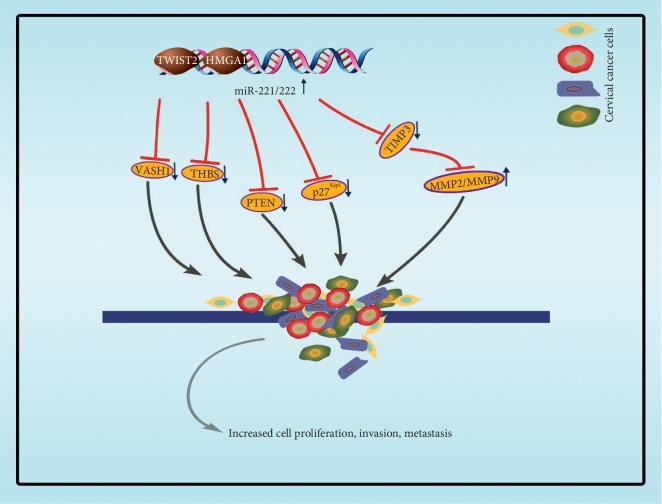
As a consequence of the metastasis of cervical cancer lymphocytes, the transcription and expression of miR-221-3p were stimulated, and miR-221-3p targeted on the 3′UTR of twist homolog 2, thus accelerating the metastasis of cervical cancer lymphocytes. Some latest reports confirms that the miR-221-3p is characteristically enriched in and transferred by CSCC-secreted exosomes into human lymphatic endothelial cells (HLECs) to promote HLECs migration and tube formation in vitro and facilitate lymph angiogenesis and lymph node (LN) metastasis in vivo according to both gain-of-function and loss-of-function experiments. Furthermore, they identify vasohibin-1 (VASH1) as a novel direct target of miR-221-3p through bioinformatics target prediction and luciferase reporter assays [64]. High-mobility group AT-hook1 (HMGA1, formerly HMG-I/Y), an architectural transcription factor, participates in a number of tumor biological processes. Fu et al. [68] have shown that HMGA1 is highly expressed in CSCC tissue, which promotes the migration and invasion of cervical cancer cells and showed that the mechanism of HMGA1 cancer promotion is targeting the promoter region of miR-221/222 to enhance the expression of miR-221/222. Among them, miR-221/222 targeted on the 3′UTR of tissue inhibitor of metalloproteinases 3 (TIMP3), while TIMP3 downregulated and MMP2/MMP9 upregulated, and miR-221/222-TIMP3-MMP2/MMP9 axis participates in the migration and invasion process (Figure 5).
Figure 5.
The main targets of miR-221/222 involved in cervical cancer. TWIST2 targets the promoter region of miR-221-3p and HMGA1 targets the promoter region of miR-221/222, both to enhance the expression of miR-221/222 in cervical cancer cells. P27/Kipq and PTEN have been identified as a target for miR-222. THBS2 and VASH1 have been identified as a target for miR-221. And miR-221/222-TIMP3-MMP2/MMP9 axis participates in the migration and invasion process.
3.4. Role of miR-221/222 in Ovarian Cancer
Epithelial ovarian cancer (EOC) is the most common type of ovarian cancer, accounting for 90% of the total ovarian cancer, and the 5-year survival rate is relatively low, which is the main cause of death in gynecological malignant tumors [117, 118]. Therefore, early detection and chemosensitivity are essential to control disease development and reduce mortality. Some research found that the expression of miR-221 and miR-222 in human ovarian cancer tissues and cell lines reaches the highest levels of miRNA relative to immortalized ovarian surface epithelial cultures [119, 120]. Studies have shown that miR-222 is overexpressed in epithelial ovarian cancer cases and promotes cell proliferation through downregulation of p27Kip1 [71]. Shang et al. indicted that patients with positive expressions of human epidermal growth factor receptor 2 (HER2), signal transducer, and activator of transcription 3 (STAT3) and p-STAT3 or with negative expressions of suppressors of cytokine signaling 3 (SOCS3) had shorter survival time [72]. Moreover, Ying et al. indicate miR-222-3p was enriched in exosomes released from EOC cells and it could be transferred to macrophages. Overexpression of miR-222-3p in macrophages induced polarization of the M2 phenotype. TargetScould prediction and the luciferase assay proved that the 3′ UTR of SOCS3 was targeted by miR-222-3p in these studies. Downregulation of SOCS3 correlated with an increased expression of STAT3 activation and induced activation of JAK/STAT pathway, which increased M2 macrophages and promoted angiogenesis and lymphangiogenesis in tumor microenvironment, which accelerated progression of EOC [121].
More studies showed that the miR-221 was upregulated in 63 samples of ovarian cancer tissues, and miR-221 level was upregulated with larger tumor size, deeper tumor invasion, and higher FIGO stage. There was a negative correlation between the expression of apoptosis protease activator 1 (APAF1) protein and miR-221 in 5 of 63 ovarian cancer tissues and 6 cell lines, including A2780, OVCAR3, SKOV3, and 3AO5. The APAF1 gene was confirmed to be a direct target of miR-221, which induced the proliferation of ovarian cancer cells and hindered the apoptosis of ovarian cancer cells in vitro [69]. MiR-221 also targets on BCL-2 modifying factor (BMF) promoting cell proliferation in ovarian cancer cell line SKOV3 [70]. Amini-Farsani et al. demonstrated that, in human epithelial ovarian cancer cell lines A2780 S and A2780/CP, miR-221/222 was expressed at a higher level in A2780/CP cells. An in vitro cell viability assay showed that downregulation of miR-221/222 sensitized A2780/CP cells to cisplatin-induced cytotoxicity. Bioinformatics analysis and luciferase reporter assays proved that miR-221/222 was found to directly target on PTEN, and miR-221/222 induced cisplatin resistance by targeting PTEN-mediated PI3K/Akt pathway [73].
Like the abovementioned cancers, other studies have also found that the high expression of miR-222-3p and miR-221-3p has an anticancer effect. For example, high expression of miR-222-3p could inhibit the deterioration of cancer cells, and the patients with high expression seem owning higher survival rate and longer survival time. Fu et al. analyzed miR-222-3p expression with qRT-PCR in 74 EOC patients, and the results suggest the higher the mean expression level of miR-222-3p, the longer the median overall survival time of EOC. The expression of miR-222-3p in six ovarian cancer cell lines (Tara R182, SKOV3, SKOV3DDP, SKOV3IP, HO8910, and HO8910-PM) was detected with qRT-PCR analysis, and then, low level of miR-222-3p was found in SKOV3/DDP and Tara R182 cells with high cell growth rate. SKOV3 and HO8910-PM cells with low cell growth rate expressed high level of miR-222-3p. The overexpression of miR-222-3p in the SKOV3/DDP cell line demonstrated that the migration of the tumor cells was blocked. When miR-222-3p was overexpressed in six cell lines, it was found that miR-222-3p targets on AKT phosphorylation protein regulator G protein alpha inhibiting activity polypeptide 2 (GNAI2), which inhibited AKT phosphorylation to reduce the proliferation of ovarian cancer cells [91]. Recently, there are also some investigations showing that higher expression of miR-221-3p is associated with better overall survival in EOC patients. Wu et al. examined the expression level of miR-221-3p in three EOC cell lines (SKOV3, SKOV3-IP, and SKOV3-R), which showed lower expression level in SKOV3-IP and SKOV3-R but showed more proliferations and migrations.
In vitro experiments indicated that miR-221-3p inhibited EOC cell proliferation and migration. By performing subsequent systematic molecular biological, bioinformatics analyses and luciferase reporter assay, scientists found and confirmed ADP-ribosylation factor (ARF) 4 is miR-221-3p′s targeting genes [90]. Interestingly, Wurz et al. [66] found that the expression of CDKN1C (p57) protein was negatively correlated with miR-221/222 in EOC, while the expression of CDKN1B (p27) protein was not correlated with miR-221/222 in EOC (Figure 6).
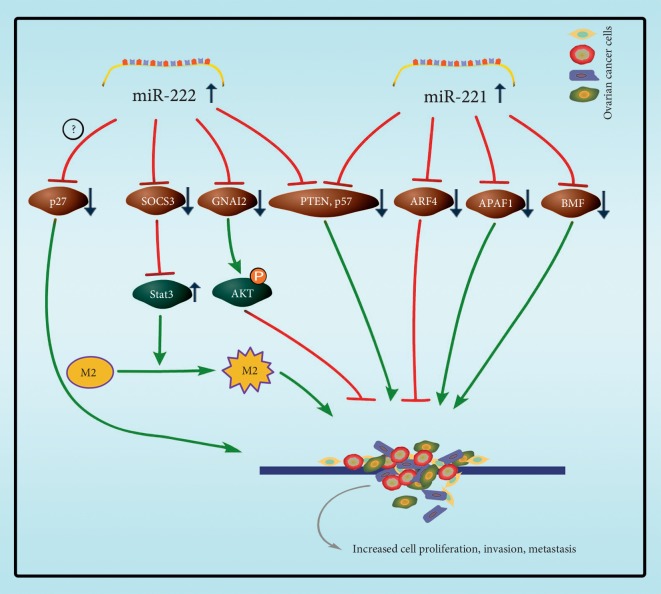
Figure 6.
The main targets of miR-221/222 involved in ovarian cancer. Overexpression of miR-222 inhibits SOCS3 in progression of EOC, and downregulation of SOCS3 increased expression of STAT3 activation, inducing activation of JAK/STAT pathway, which increased M2 macrophages and promoted angiogenesis in tumor microenvironment. Upregulated miR-222 inhibits GNAI2 expression, which inhibited AKT phosphorylation to reduce the proliferation of ovarian cancer cells.
3.5. Role of miR-221/222 in Endometrial Carcinoma
Endometrial carcinoma (EC) is the fourth most common malignant tumor in women in developed countries and the most common cancer in female genital tract; however, the incidence of endometrial carcinoma has been moving to the young population due to obesity and lifestyle factors in China [122]. Estrogen is a classic etiological factor for endometrial tumorigenesis [123]. In endometrial carcinoma, deregulated ERα caused by genomic or epigenetic aberrations was a prevalent phenomenon, which reduced the expression of ERα and associated with extensive invasion and high-stage and poor prognosis [124, 125].
Liu et al. found miR-222-3p expression was much lower in ERα-positive than in ERα-negative endometrial carcinoma tissue samples, and the level of miR-222-3p expression was lower in tumors of lower grades and earlier stage. TargetScould and luciferase reporter assays revealed that ERα was a target of miR-222-3p. Therefore, miR-222-3p was overexpressed in ERα-negative endometrial carcinoma tumors and was associated with high grade, late stage, and nodal metastasis, and high level of miR-222-3p was a mechanism for raloxifene resistance in endometrial carcinoma therapy. Moreover, a significant increase in serum miR-222 levels of endometrial carcinoma patients was found by qRT-PCR miRNA analysis in 46 EC patients and 28 women without cancer history. It may be used as a proper marker for the diagnosis of endometrial carcinoma in future [126].
4. Conclusion
As summarized above, microRNA-221/222 (miR-221/222) is a noncoding microRNA which is widely distributed in eukaryotic organisms and deeply involved in the posttranscriptional regulation of gene expressions. It is important that the miR-221/222 expression level is closely related to tumor stage and prognosis. It may be used as a biomarker for the diagnosis of premalignant tumors [104] and provide a new target for tumor therapy [8–33, 35, 38, 51, 94], as a therapeutic tool for drug resistance or sensitivity to anticancer drugs [127].
However, more questions also need to be answered as translated into clinical strategies. Firstly, given the complex networks in which cancer cells are located, the roles of miR-221/222 in various cells' background as well as its target genes need more micromesh clarifications. For example, a laboratory announced that miR-222 promotes the proliferation of human non-small cell lung cancer cell line H460 [128]; in contrast, at almost the same time, Yamashita and colleagues [129] demonstrated that miR-221/222 promoted the growth of H460 cell lines while miR-221 inhibited the growth of the other four NSCLC cell lines. Another example is that the low expression of miR-221/222 inhibited the malignant proliferation of vascular smooth muscle cells [130] but also inhibited the growth of myocardial cells and the increase of muscle cell proliferation markers induced by exercise [131]. It is worth noting that the model miRNA analyzed and transfected in tumor samples may have isomiR properties, which may cause some of the results to be contradictory.
Secondly, with the deepening of the research, the expression level of miR-221 and miR-222 is not proportional to the development of tumor, while its functions are abundant. Therefore, the ratio of miR-221 and miR-222 expression level is expected to become a new research field [66].
Finally, it is expected that a single miRNA could act as a therapeutic inhibitor towards the cancer cellular pathway by regulating the different genes involved in the network. For example, miR-221/222 could inhibit the expression of ER-α and FOXO3A transcription factors, which results in the overall change of gene expression through the expression inhibition of ER-α and FOXO3A at posttranscriptional stage. The suppressed ER-α has the ability to negatively regulate miR-221/222, and the suppressed FOXO3A blocked the transcriptional activation of p27 and Bim [63]. This shows that the regulation of individual miRNA may have great therapeutic potential towards various kinds of cancers. Back to the clinical strategy, for effectively suppressing onco-miRNA, locked nucleic acid- (LNA-) based oligonucleotides has been demonstrated to have great potential as inhibitors of small RNA targets [132].
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFC0310900), the Open Research Fund of State Key Laboratory of Bio-organic and Natural Products Chemistry (SIOC, CAS), and the Fudan University Outstanding Talent Plan (Fudan Excellence 2025).
Contributor Information
Xiaoling Lu, Email: luxiaoling80@126.com.
Ning Zhang, Email: zning818@163.com.
Xin Cao, Email: caox@fudan.edu.cn.
Data Availability
Some or all data, models, or code generated or used during the study are available from the corresponding author by request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Lewis B. P., Burge C. B., Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 3.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Farazi T. A., Spitzer J. I., Morozov P., Tuschl T. miRNAs in human cancer. The Journal of Pathology. 2011;223(2):102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garofalo M., Quintavalle C., Romano G., Croce C. M., Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Current Molecular Medicine. 2012;12(1):27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuchiya S., Okuno Y., Tsujimoto G. MicroRNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. Journal of Pharmacological Sciences. 2006;101(4):267–270. doi: 10.1254/jphs.cpj06013x. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Martino M. T., Rossi M., Caracciolo D., Gullà A., Tagliaferri P., Tassone P. mir-221/222 are promising targets for innovative anticancer therapy. Expert Opinion on Therapeutic Targets. 2016;20(9):1099–1108. doi: 10.1517/14728222.2016.1164693. [DOI] [PubMed] [Google Scholar]
- 9.Ha M., Kim V. N. Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 10.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42(D1):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan G. C., Chan E., Molnar A., et al. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Research. 2014;42(14):9424–9435. doi: 10.1093/nar/gku656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh T., Hojo H., Suzuki T. Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Research. 2015;43(15):7521–7534. doi: 10.1093/nar/gkv669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh T., Sakaguchi Y., Miyauchi K., et al. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & Development. 2009;23(4):433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcinowski L., Tanguy M., Krmpotic A., et al. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathogens. 2012;8(2) doi: 10.1371/journal.ppat.1002510.e1002510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda R., Kohanbash G., Sasaki K., et al. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proceedings of the National Academy of Sciences. 2009;106(26):10746–10751. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., Gao X., Hu J., et al. ADAR1p150 forms a complex with dicer to promote miRNA-222 activity and regulate PTEN expression in CVB3-induced viral myocarditis. International Journal of Molecular Sciences. 2019;20(2):p. 407. doi: 10.3390/ijms20020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J.-C., Su Y.-H., Chiu C.-F., et al. Suppression of dicer increases sensitivity to gefitinib in human lung cancer cells. Annals of Surgical Oncology. 2014;21(S4):555–563. doi: 10.1245/s10434-014-3673-y. [DOI] [PubMed] [Google Scholar]
- 18.Cochrane D. R., Cittelly D. M., Howe E. N, et al. MicroRNAs link estrogen receptor alpha status and Dicer levels in breast cancer. Hormones and Cancer. 2010;1(6):306–319. doi: 10.1007/s12672-010-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S. K., Sokhi U. K., Bhutia S. K., et al. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proceedings of the National Academy of Sciences. 2010;107(26):11948–11953. doi: 10.1073/pnas.0914143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nejad C., Pillman K. A., Siddle K. J., et al. miR-222 isoforms are differentially regulated by type-I interferon. RNA. 2018;24(3):332–341. doi: 10.1261/rna.064550.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu F., Pillman K. A., Neilsen C. T., et al. Naturally existing isoforms of miR-222 have distinct functions. Nucleic Acids Research. 2017;45(19):11371–11385. doi: 10.1093/nar/gkx788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panneerselvam J., Srivastava A., Muralidharan R., et al. IL-24 modulates the high mobility group (HMG) A1/miR222/AKT signaling in lung cancer cells. Oncotarget. 2016;7(43):70247–70263. doi: 10.18632/oncotarget.11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodge R., Ferreira Barbosa J. A., Lombard-Vadnais F., et al. Host microRNAs-221 and -222 inhibit HIV-1 entry in macrophages by targeting the CD4 viral receptor. Cell Reports. 2017;21(1):141–153. doi: 10.1016/j.celrep.2017.09.030. [DOI] [PubMed] [Google Scholar]
- 24.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu M., Xu X., Xi B., et al. Molecular network-based identification of competing endogenous RNAs in thyroid carcinoma. Genes. 2018;9(1):p. 44. doi: 10.3390/genes9010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L., Wang H.-J., Meng T., et al. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Molecular Therapy—Nucleic Acids. 2019;17:644–656. doi: 10.1016/j.omtn.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Sarkar S., Dubaybo H., Ali S., et al. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27kip1, p57kip2, and PUMA. American Journal of Cancer Research. 2013;3(5):p. 465. [PMC free article] [PubMed] [Google Scholar]
- 28.Medina R., Zaidi S. K., Liu C.-G., et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Research. 2008;68(8):2773–2780. doi: 10.1158/0008-5472.can-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garofalo M., Di Leva G., Romano G., et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Hikisz P., Kiliańska Z. M. Puma, a critical mediator of cell death—one decade on from its discovery. Cellular & Molecular Biology Letters. 2012;17(4):646–669. doi: 10.2478/s11658-012-0032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C.-Z., Zhang J.-X., Zhang A.-L., et al. miR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Molecular Cancer. 2010;9(1):p. 229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li N., Tang B., Zhu E.-D., et al. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS Letters. 2012;586(6):722–728. doi: 10.1016/j.febslet.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 33.Tan X., Tang H., Bi J., Li N., Jia Y. MicroRNA-222-3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion, and inhibits apoptosis in gastric cancer. Journal of Cellular Biochemistry. 2018;119(7):5153–5162. doi: 10.1002/jcb.26542. [DOI] [PubMed] [Google Scholar]
- 34.Chun-zhi Z., Lei H., An-ling Z., et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10(1):p. 367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng L.-P., Hu Z.-M., Li K., Xia K. miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR axis in bladder cancer cells. Journal of Cellular and Molecular Medicine. 2016;20(3):559–567. doi: 10.1111/jcmm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Ma T., Yang S., et al. High-mobility group A1 proteins enhance the expression of the oncogenic miR-222 in lung cancer cells. Molecular and Cellular Biochemistry. 2011;357(1-2):363–371. doi: 10.1007/s11010-011-0907-1. [DOI] [PubMed] [Google Scholar]
- 37.Wei F., Ma C., Zhou T., et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Molecular Cancer. 2017;16(1):p. 132. doi: 10.1186/s12943-017-0694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fornari F., Gramantieri L., Ferracin M., et al. miR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27(43):5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 39.Wong Q. W.-L., Ching A. K.-K., Chan A. W.-H., et al. miR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clinical Cancer Research. 2010;16(3):867–875. doi: 10.1158/1078-0432.ccr-09-1840. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Liao X., Huang K., et al. Clustered microRNAs hsa-miR-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for hepatocellular carcinoma. Journal of Cancer. 2019;10(11):2520–2533. doi: 10.7150/jca.29207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gramantieri L., Fornari F., Ferracin M., et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clinical Cancer Research. 2009;15(16):5073–5081. doi: 10.1158/1078-0432.ccr-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae H. J., Jung K. H., Eun J. W., et al. MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. Journal of Hepatology. 2015;63(2):408–419. doi: 10.1016/j.jhep.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 43.Fornari F., Pollutri D., Patrizi C., et al. In hepatocellular carcinoma miR-221 modulates sorafenib resistance through inhibition of caspase-3-mediated apoptosis. Clinical Cancer Research. 2017;23(14):3953–3965. doi: 10.1158/1078-0432.ccr-16-1464. [DOI] [PubMed] [Google Scholar]
- 44.Li T., Li M., Hu S., et al. MiR-221 mediates the epithelial-mesenchymal transition of hepatocellular carcinoma by targeting AdipoR1. International Journal of Biological Macromolecules. 2017;103:1054–1061. doi: 10.1016/j.ijbiomac.2017.05.108. [DOI] [PubMed] [Google Scholar]
- 45.Huang S., Zhou D., Li Y.-X., et al. In vivo and in vitro effects of microRNA-221 on hepatocellular carcinoma development and progression through the JAK-STAT3 signaling pathway by targeting SOCS3. Journal of Cellular Physiology. 2019;234(4):3500–3514. doi: 10.1002/jcp.26863. [DOI] [PubMed] [Google Scholar]
- 46.Chen J.-J., Tang Y.-S., Huang S.-F., Ai J.-G., Wang H.-X., Zhang L.-P. HBx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBV-related hepatocellular carcinoma by targeting estrogen receptor-α. Oncology Reports. 2015;33(2):792–798. doi: 10.3892/or.2014.3647. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y., Yao J., Huan L., et al. GNAI3 inhibits tumor cell migration and invasion and is post-transcriptionally regulated by miR-222 in hepatocellular carcinoma. Cancer Letters. 2015;356(2):978–984. doi: 10.1016/j.canlet.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z., Sun J., Liu B., Zhao M., Xing E., Dang C. miRNA-222 promotes liver cancer cell proliferation, migration and invasion and inhibits apoptosis by targeting BBC3. International Journal of Molecular Medicine. 2018;42(1):141–148. doi: 10.3892/ijmm.2018.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun K., Wang W., Zeng J.-J., Wu C.-T., Lei S.-T., Li G.-X. MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacologica Sinica. 2011;32(3):375–384. doi: 10.1038/aps.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han H., Du Y., Zhao W., et al. PBX3 is targeted by multiple miRNAs and is essential for liver tumour-initiating cells. Nature Communications. 2019;10(1):p. 2259. doi: 10.1038/s41467-019-10052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pineau P., Volinia S., McJunkin K., et al. miR-221 overexpression contributes to liver tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(1):264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao D., Li T., Ye C., et al. miR-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Experimental and Therapeutic Medicine. 2017;15(2):1712–1717. doi: 10.3892/etm.2017.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue Q., Sun K., Deng H.-J., Lei S.-T., Dong J. Q., Li G. X. Anti-miRNA-221 sensitizes human colorectal carcinoma cells to radiation by upregulating PTEN. World Journal of Gastroenterology. 2013;19(48):9307–9317. doi: 10.3748/wjg.v19.i48.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khoshinani H. M., Afshar S., Pashaki A. S., et al. Involvement of miR-155/FOXO3a and miR-222/PTEN in acquired radioresistance of colorectal cancer cell line. Japanese Journal of Radiology. 2017;35(11):664–672. doi: 10.1007/s11604-017-0679-y. [DOI] [PubMed] [Google Scholar]
- 55.Gao H., Cong X., Zhou J., Guan M. MicroRNA-222 influences migration and invasion through MIA3 in colorectal cancer. Cancer Cell International. 2017;17(1):p. 78. doi: 10.1186/s12935-017-0447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo F., Zhou J., Wang S., Sun Z., Han Q., Bai C. MicroRNA-222 promotes colorectal cancer cell migration and invasion by targeting MST3. FEBS Open Bio. 2019;9(5):901–913. doi: 10.1002/2211-5463.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.le Sage C., Nagel R., Egan D. A., et al. Regulation of the p27Kip1 tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. The EMBO Journal. 2007;26(15):3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu S., Sun X., Wang M., et al. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFκB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147(4):847–859. doi: 10.1053/j.gastro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santolla M. F., Lappano R., Cirillo F., et al. miR-221 stimulates breast cancer cells and cancer-associated fibroblasts (CAFs) through selective interference with the A20/c-Rel/CTGF signaling. Journal of Experimental & Clinical Cancer Research. 2018;37(1):p. 94. doi: 10.1186/s13046-018-0767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao J.-J., Lin J., Yang H., et al. MicroRNA-221/222 negatively regulates estrogen receptorα and is associated with tamoxifen resistance in breast cancer. Journal of Biological Chemistry. 2008;283(45):31079–31086. doi: 10.1074/jbc.m806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Goto Y., Kojima S., Nishikawa R., et al. MicroRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. British Journal of Cancer. 2015;113(7):1055–1065. doi: 10.1038/bjc.2015.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gorbatenko A., Sokilde R., Sorensen E. E., et al. HER2 and p95HER2 differentially regulate miRNA expression in MCF-7 breast cancer cells and downregulate MYB proteins through miR-221/222 and miR-503. Scientific Reports. 2019;9(1):p. 3352. doi: 10.1038/s41598-019-39733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Leva G., Gasparini P., Piovan C., et al. MicroRNA cluster 221-222 and estrogen receptor α interactions in breast cancer. JNCI: Journal of the National Cancer Institute. 2010;102(10):706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou C.-F., Ma J., Huang L., et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene. 2019;38(8):1256–1268. doi: 10.1038/s41388-018-0511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei W. F., Zhou C. F., Wu X. G., et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death & Disease. 2017;8(12):p. 3220. doi: 10.1038/s41419-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Wurz K., Garcia R. L., Goff B. A., et al. miR-221 and miR-222 alterations in sporadic ovarian carcinoma: relationship to CDKN1B, CDKNIC and overall survival. Genes Chromosomes & Cancer. 2010;49(7):577–584. doi: 10.1002/gcc.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun Y., Zhang B., Cheng J., et al. MicroRNA-222 promotes the proliferation and migration of cervical cancer cells. Clinical & Investigative Medicine. 2014;37(3):p. E131. doi: 10.25011/cim.v37i3.21380. [DOI] [PubMed] [Google Scholar]
- 68.Fu F. F., Wang T., Wu Z. Y., et al. HMGA1 exacerbates tumor growth through regulating the cell cycle and accelerates migration/invasion via targeting miR-221/222 in cervical cancer. Cell Death & Disease. 2018;9(6):p. 594. doi: 10.1038/s41419-018-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J., Li Q., Huang H., et al. Overexpression of miRNA-221 promotes cell proliferation by targeting the apoptotic protease activating factor-1 and indicates a poor prognosis in ovarian cancer. International Journal of Oncology. 2017;50(4):1087–1096. doi: 10.3892/ijo.2017.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie X., Huang Y., Chen L., Wang J. miR-221 regulates proliferation and apoptosis of ovarian cancer cells by targeting BMF. Oncology Letters. 2018;16(5):6697–6704. doi: 10.3892/ol.2018.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun C., Li N., Zhou B., et al. miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27kip1. Oncology Letters. 2013;6(2):507–512. doi: 10.3892/ol.2013.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shang A.-Q., Wu J., Bi F., et al. Relationship between HER2 and JAK/STAT-SOCS3 signaling pathway and clinicopathological features and prognosis of ovarian cancer. Cancer Biology & Therapy. 2017;18(5):314–322. doi: 10.1080/15384047.2017.1310343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amini-Farsani Z., Sangtarash M. H., Shamsara M., Teimori H. miR-221/222 promote chemoresistance to cisplatin in ovarian cancer cells by targeting PTEN/PI3K/AKT signaling pathway. Cytotechnology. 2018;70(1):203–213. doi: 10.1007/s10616-017-0134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu B., Che Q., Qiu H., et al. Elevated MiR-222-3p promotes proliferation and invasion of endometrial carcinoma via targeting ERα. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0087563.e87563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Felicetti F., Errico M. C., Bottero L., et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Research. 2008;68(8):2745–2754. doi: 10.1158/0008-5472.can-07-2538. [DOI] [PubMed] [Google Scholar]
- 76.Felli N., Errico M. C., Pedini F., et al. AP2α controls the dynamic balance between miR-126&126∗ and miR-221&222 during melanoma progression. Oncogene. 2016;35(23):3016–3026. doi: 10.1038/onc.2015.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee C., He H., Jiang Y., et al. Elevated expression of tumor miR-222 in pancreatic cancer is associated with Ki67 and poor prognosis. Medical Oncology. 2013;30(4):p. 700. doi: 10.1007/s12032-013-0700-y. [DOI] [PubMed] [Google Scholar]
- 78.Mardente S., Mari E., Consorti F., et al. HMGB1 induces the overexpression of miR-222 and miR-221 and increases growth and motility in papillary thyroid cancer cells. Oncology Reports. 2012;28(6):2285–2289. doi: 10.3892/or.2012.2058. [DOI] [PubMed] [Google Scholar]
- 79.Mardente S., Mari E., Massimi I., et al. HMGB1-Induced cross talk between PTEN and miRs 221/222 in thyroid cancer. Biomed Research International. 2015;2015:7. doi: 10.1155/2015/512027.512027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Di Martino M. T., Gullà A., Gallo Cantafio M. E., et al. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0089659.e89659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frenquelli M., Muzio M., Scielzo C., et al. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115(19):3949–3959. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- 82.Yang C.-J., Shen W. G., Liu C.-J., et al. miR-221 and miR-222 expression increased the growth and tumorigenesis of oral carcinoma cells. Journal of Oral Pathology & Medicine. 2011;40(7):560–566. doi: 10.1111/j.1600-0714.2010.01005.x. [DOI] [PubMed] [Google Scholar]
- 83.Shen F., Mo M.-H., Chen L., et al. MicroRNA-21 down-regulates Rb1 expression by targeting PDCD4 in retinoblastoma. Journal of Cancer. 2014;5(9):804–812. doi: 10.7150/jca.10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu W., Chen X., Yu S., Wang R., Zhao R., Du C. MicroRNA-222 promotes tumor growth and confers radioresistance in nasopharyngeal carcinoma by targeting PTEN. Molecular Medicine Reports. 2017;17(1):1305–1310. doi: 10.3892/mmr.2017.7931. [DOI] [PubMed] [Google Scholar]
- 85.Galardi S., Mercatelli N., Giorda E., et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. Journal of Biological Chemistry. 2007;282(32):23716–23724. doi: 10.1074/jbc.m701805200. [DOI] [PubMed] [Google Scholar]
- 86.Felli N., Fontana L., Pelosi E., et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proceedings of the National Academy of Sciences. 2005;102(50):18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gits C. M. M., van Kuijk P. F., Jonkers M. B. E., et al. MiR-17-92 and miR-221/222 cluster members target KIT and ETV1 in human gastrointestinal stromal tumours. British Journal of Cancer. 2013;109(6):1625–1635. doi: 10.1038/bjc.2013.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X., Yu J., Jiang L., et al. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics. 2009;6(3):131–139. [PMC free article] [PubMed] [Google Scholar]
- 89.Okamoto K., Miyoshi K., Murawaki Y. miR-29b, miR-205 and miR-221 enhance chemosensitivity to gemcitabine in HuH28 human cholangiocarcinoma cells. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077623.e77623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu Q., Ren X., Zhang Y., et al. miR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochemical and Biophysical Research Communications. 2018;497(4):1162–1170. doi: 10.1016/j.bbrc.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 91.Fu X., Li Y., Alvero A., et al. MicroRNA-222-3p/GNAI2/AKT axis inhibits epithelial ovarian cancer cell growth and associates with good overall survival. Oncotarget. 2016;7(49):p. 80633. doi: 10.18632/oncotarget.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu K., Liang X., Shen K., et al. MiR-222 modulates multidrug resistance in human colorectal carcinoma by down-regulating ADAM-17. Experimental Cell Research. 2012;318(17):2168–2177. doi: 10.1016/j.yexcr.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 93.Yang Y., Cui H., Wang X. Downregulation of EIF5A2 by miR-221-3p inhibits cell proliferation, promotes cell cycle arrest and apoptosis in medulloblastoma cells. Bioscience, Biotechnology, and Biochemistry. 2019;83(3):400–408. doi: 10.1080/09168451.2018.1553604. [DOI] [PubMed] [Google Scholar]
- 94.Dufresne S., Rébillard A., Muti P., Friedenreich C. M., Brenner D. R. A review of physical activity and circulating miRNA expression: implications in cancer risk and progression. Cancer Epidemiology Biomarkers & Prevention. 2018;27(1):11–24. doi: 10.1158/1055-9965.epi-16-0969. [DOI] [PubMed] [Google Scholar]
- 95.Wilting S. M., Snijders P. J. F., Verlaat W., et al. Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis. Oncogene. 2013;32(1):106–116. doi: 10.1038/onc.2012.20. [DOI] [PubMed] [Google Scholar]
- 96.Liu H., Cao B., Zhao Y., Liang H., Liu X. Upregulated miR-221/222 promotes cell proliferation and invasion and is associated with invasive features in retinoblastoma. Cancer Biomarkers. 2018;22(4):621–629. doi: 10.3233/cbm-170721. [DOI] [PubMed] [Google Scholar]
- 97.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 98.Song J., Ouyang Y., Che J., et al. Potential value of miR-221/222 as diagnostic, prognostic, and therapeutic biomarkers for diseases. Frontiers in Immunology. 2017;8:p. 56. doi: 10.3389/fimmu.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Di Martino M. T., Gullà A., Cantafio M. E., et al. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget. 2013;4(2):242–255. doi: 10.18632/oncotarget.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ladeiro Y., Couchy G., Balabaud C., et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47(6):1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 101.Murakami Y., Yasuda T., Saigo K., et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 102.Fu X., Wang Q., Chen J., et al. Clinical significance of miR-221 and its inverse correlation with p27Kip1 in hepatocellular carcinoma. Molecular Biology Reports. 2011;38(5):3029–3035. doi: 10.1007/s11033-010-9969-5. [DOI] [PubMed] [Google Scholar]
- 103.Li J., Wang Y., Yu W., Chen J., Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochemical and Biophysical Research Communications. 2011;406(1):70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 104.Sohn W., Kim J., Kang S. H., et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Experimental & Molecular Medicine. 2015;47(9):p. e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nakano K., Vousden K. H. PUMA, a novel proapoptotic gene, is induced by p53. Molecular Cell. 2001;7(3):683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 106.Jung K. H., Noh J. H., Kim J. K., et al. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology. 2012;56(2):644–657. doi: 10.1002/hep.25699. [DOI] [PubMed] [Google Scholar]
- 107.Sarlinova M., Halasa M., Mistuna D., et al. miR-21, miR-221 and miR-150 are deregulated in peripheral blood of patients with colorectal cancer. Anticancer Research. 2016;36(10):5449–5454. doi: 10.21873/anticanres.11124. [DOI] [PubMed] [Google Scholar]
- 108.Pu X.-X., Huang G.-L., Guo H.-Q., et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. Journal of Gastroenterology and Hepatology. 2010;25(10):1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 109.Yau T. O., Wu C. W., Dong Y., et al. MicroRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. British Journal of Cancer. 2014;111(9):1765–1771. doi: 10.1038/bjc.2014.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tan K., Leong S. M., Kee Z., et al. Longitudinal monitoring reveals dynamic changes in circulating tumor cells (CTCs) and CTC-associated miRNAs in response to chemotherapy in metastatic colorectal cancer patients. Cancer Letters. 2018;423(1):1–8. doi: 10.1016/j.canlet.2018.02.039. [DOI] [PubMed] [Google Scholar]
- 111.Tsunoda T., Takashima Y., Yoshida Y., et al. Oncogenic KRAS regulates miR-200c and miR-221/222 in a 3D-specific manner in colorectal cancer cells. Anticancer Research. 2011;31(7):2453–2459. [PubMed] [Google Scholar]
- 112.Chaffer C. L., Weinberg R. A. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 113.Simizu S., Takagi S., Tamura Y., Osada H. RECK-mediated suppression of tumor cell invasion is regulated by glycosylation in human tumor cell lines. Cancer Research. 2005;65(16):7455–7461. doi: 10.1158/0008-5472.can-04-4446. [DOI] [PubMed] [Google Scholar]
- 114.Qin J., Luo M. MicroRNA-221 promotes colorectal cancer cell invasion and metastasis by targeting RECK. FEBS Letters. 2014;588(1):99–104. doi: 10.1016/j.febslet.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 115.Bourla A. B., Zamarin D. Immunotherapy: new strategies for the treatment of gynecologic malignancies. Oncology (Williston Park) 2016;30(1):59–66. [PMC free article] [PubMed] [Google Scholar]
- 116.Srivastava S. K., Ahmad A., Zubair H., et al. MicroRNAs in gynecological cancers: small molecules with big implications. Cancer Letters. 2017;407:123–138. doi: 10.1016/j.canlet.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mutch D. G., Prat J. 2014 FIGO staging for ovarian, fallopian tube and peritoneal cancer. Gynecologic Oncology. 2014;133(3):401–404. doi: 10.1016/j.ygyno.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 118.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2018. CA: A Cancer Journal for Clinicians. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 119.Dahiya N., Sherman-Baust C. A., Wang T. L., et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3(6) doi: 10.1371/journal.pone.0002436.e2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang L., Volinia S., Bonome T., et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(19):7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ying X., Wu Q., Wu X., et al. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. 2016;7(28):43076–43087. doi: 10.18632/oncotarget.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Y., Liu Z., Yu X., et al. The association between metabolic abnormality and endometrial cancer: a large case-control study in China. Gynecologic Oncology. 2010;117(1):41–46. doi: 10.1016/j.ygyno.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 123.Shang Y. Molecular mechanisms of oestrogen and SERMs in endometrial carcinogenesis. Nature Reviews Cancer. 2006;6(5):360–368. doi: 10.1038/nrc1879. [DOI] [PubMed] [Google Scholar]
- 124.Srijaipracharoen S., Tangjitgamol S., Tanvanich S., et al. Expression of ER, PR, and Her-2/neu in endometrial cancer: a clinicopathological study. Asian Pacific Journal of Cancer Prevention. 2010;11(1):215–220. [PubMed] [Google Scholar]
- 125.Tan D., Lambros M., Marchiò C., Reis-Filho J. ESR1 amplification in endometrial carcinomas: hope or hyperbole? The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2008;216(3):271–274. doi: 10.1002/path.2432. [DOI] [PubMed] [Google Scholar]
- 126.Montagnana M., Benati M., Danese E., et al. Aberrant microRNA expression in patients with endometrial cancer. International Journal of Gynecological Cancer. 2017;27(3):459–466. doi: 10.1097/igc.0000000000000913. [DOI] [PubMed] [Google Scholar]
- 127.Mandraffino G., Aragona C. O., Cairo V., et al. Circulating progenitor cells in hypertensive subjects: effectiveness of a treatment with olmesartan in improving cell number and miR profile in addition to expected pharmacological effects. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0173030.e0173030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhong C., Ding S., Xu Y., Huang H. MicroRNA-222 promotes human non-small cell lung cancer H460 growth by targeting p27. International Journal of Clinical and Experimental Medicine. 2015;8(4):5534–5540. [PMC free article] [PubMed] [Google Scholar]
- 129.Yamashita R., Sato M., Kakumu T., et al. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Medicine. 2015;4(4):551–564. doi: 10.1002/cam4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doran A. C., Meller N., McNamara C. A. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28(5):812–819. doi: 10.1161/atvbaha.107.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu X., Xiao J., Zhu H., et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metabolism. 2015;21(4):584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Elmén J., Lindow M., Schütz S., et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data, models, or code generated or used during the study are available from the corresponding author by request.