Abstract
AIM
To compare the effectiveness of two well described machine learning modalities, ocular coherence tomography (OCT) and fundal photography, in terms of diagnostic accuracy in the screening and diagnosis of glaucoma.
METHODS
A systematic search of Embase and PubMed databases was undertaken up to 1st of February 2019. Articles were identified alongside their reference lists and relevant studies were aggregated. A Meta-analysis of diagnostic accuracy in terms of area under the receiver operating curve (AUROC) was performed. For the studies which did not report an AUROC, reported sensitivity and specificity values were combined to create a summary ROC curve which was included in the Meta-analysis.
RESULTS
A total of 23 studies were deemed suitable for inclusion in the Meta-analysis. This included 10 papers from the OCT cohort and 13 from the fundal photos cohort. Random effects Meta-analysis gave a pooled AUROC of 0.957 (95%CI=0.917 to 0.997) for fundal photos and 0.923 (95%CI=0.889 to 0.957) for the OCT cohort. The slightly higher accuracy of fundal photos methods is likely attributable to the much larger database of images used to train the models (59 788 vs 1743).
CONCLUSION
No demonstrable difference is shown between the diagnostic accuracy of the two modalities. The ease of access and lower cost associated with fundal photo acquisition make that the more appealing option in terms of screening on a global scale, however further studies need to be undertaken, owing largely to the poor study quality associated with the fundal photography cohort.
Keywords: machine learning, glaucoma, ocular coherence tomography, fundal photography, diagnosis, Meta-analysis
INTRODUCTION
Glaucoma is a term used to describe a group of optic neuropathies which cause damage to retinal ganglion cells, and is the second leading cause of permanent blindness in developed countries[1]. The damage caused by glaucoma is irreversible and therefore early detection and treatment is vital to halt visual damage[2]. From a global perspective, the number of people diagnosed with the disease is expected to almost double from 76 million in 2020 to 112 million in 2040[3]. Glaucoma is usually related to an increase in intraocular pressure which leads to stress induced damage on the retinal ganglion cells resulting in a characteristic appearance of the optic nerve head and associated visual field defects[4]. Pressure lowering medications and/or surgery can be used to halt its progression, especially if it is detected at an early stage. However, the disease has an insidious onset and so therefore patients can remain asymptomatic for many years before they attend for investigation and/or treatment. Early detection is essential to ensure patients continue to have an adequate quality of life and allow them to retain their independence and the ability to drive[5]. The economic and social impact that glaucomatous optic neuropathy can have on society has been well described[6]. The pathogenesis and progression of glaucoma is still poorly understood[7].
Currently there are numerous methods used to diagnose and screen for glaucoma, however these techniques are expensive, time consuming, require skilled operators and are manual[8]. Four modalities are routinely used; perimetry to detect a visual field defect, pachymetry to detect corneal thickness, tonometry to measure intraocular pressure and fundoscopy to examine the optic nerve head. Glaucoma is a disease that is seen to increase significantly with advancing age[9] and the projected increase in the population over the age of 50 is expected to double over the next 20y[10]. It is therefore imperative that an efficient screening and/or diagnosing system is established to halt both the disease burden and the burden on ophthalmic departments.
Glaucomatous optic neuropathy can result in a thinning of the retinal nerve fibre layer (RNFL) and an associated enlargement of the cup-to-disc ratio (CDR). Peripapillary atrophy is also a well-known sign associated with glaucoma[11], however this can be seen in other ocular pathologies such as high myopia[12]. Confusingly, individuals with myopia have an increased risk of developing glaucoma[13].
The Anderson Patella criteria is the gold standard criteria for manifest glaucoma diagnosis. It is stated for a diagnosis to be made, the following must be seen on a 30-2 Humphrey visual field test (Humphrey Field Analyser, Carl Zeiss Meditec, Dublin, California): 1) abnormal glaucoma hemi-field test, 2) three or more non-edge points, which are contiguous, must be depressed with a P<5% with at least one of these having a P<1%, 3) this must be demonstrated on two or more field tests[14].
Ocular coherence tomography (OCT) is a non-invasive imaging technique which provides micrometre resolution cross sectional views of the retina[15]. It can be utilised to assess RNFL thinning around the optic nerve head and macular area. Fundal photography is another imaging technique which takes photographs of the inner retina mainly using a widefield fundus camera. Parameters including the size of the optic nerve head and the CDR, alongside peripapillary atrophy, vessel branching and tortuosity can be examined using fundal photos[16]. OCT scanners can interpret these parameters alongside RNFL thickness. OCT scans are accurate, reproducible and are not patient dependant. They can supply us with information about change in thickness of the RNFL and can be used in differentiate glaucomatous from non-glaucomatous eyes[17]. RNFL thickness as determined by OCT scans has shown a high correlation with the functional status of the optic nerve[18].
Fundal photos[19] and OCT scanning techniques[20] have proven to be useful screening and diagnosing modalities for glaucoma. A contrast between the two modalities exist relating to the ease of access and speed of acquisition of fundal photos in comparison to cost and user expertise associated with OCT scanning. The use of a fundal camera negates the price associated with more expensive diagnostic equipment or in setting where one is just not available (e.g., lower income countries). The average cost of an OCT scanner is approximately $40 000 whereas one can augment their smartphone with a lens to aid in the acquisition of basic fundal photos[21]. Fundal photos can be used concomitantly to diagnose other ocular pathologies such as age related macular degeneration (ARMD) or diabetic retinopathy (DR)[22].
Artificial intelligence (AI), in particular “Machine learning”, has seen a recent upsurge, particularly its use in medicine, namely ophthalmology, and is currently being developed as a screening and diagnostic tool in many ophthalmic conditions. Machine learning refers to any process in which an algorithm is iteratively improved or “trained” in performing a task, usually a classification or identification task, by repeated exposure to many examples, known as the training data or training set. The trained algorithm can then be tested by measuring its performance in classifying novel unseen data (the test set).
In particular, “supervised” learning algorithms have proven highly successful in automating binary classification tasks, such as determining the presence or absence of a pathology. “Supervised learning” refers to training regimes in which the algorithm is given both the input (e.g., an OCT or fundus image) and the correct output (e.g., the correct diagnosis) for each element in the training set. In this way, the algorithm implicitly learns a mathematical function which maps each input to the correct output. If new data is applied to this function, the machine learning algorithm should be able to classify it correctly. Machine learning can be utilised when we can't directly express how a problem should be solved using an algorithm but we can illustrate to the machine examples that are both positive and negative and allow it to identify a function for itself. As a result, the validity of a machine learning algorithm depends heavily on the size and quality of the training data, and so validation of algorithms is highly important to ensure that the results will generalise. Steps for constructing an AI model include pre-processing the raw data, training the model, validating it and then testing it[23].
Machine Learning Algorithms
Artificial neural networks
Artificial neural network (ANN) models are inspired by the structure of the brain, in particular the human visual system, making them highly useful in automated image analysis. ANNs consists of many simple simulated processing units (“neurons”) connected in one or more layers. Neurons receive input from preceding layers, combine the inputs according to simple summation rules, and generate an output which is fed forward to the next layer. The lowest layer of the network represents the input (e.g., image pixel values), while the final layer represents the output or classification. Inputs from one neuron to another are “weighted”, with values analogous to synaptic weights in neural connections. As the algorithm is trained, these weights are updated according to simple feedback rules to improve the accuracy of the classification.
Support vector machine
Support vector machines (SVMs) apply a multi-dimensional transform to the input data (image pixels). The algorithm then attempts to identify the hyperplane in this higher-dimensional space which best separates the training data into the desired categories (e.g., glaucomatous and non-glaucomatous). The further away the data points lie form the plane, the more confident the model is that it identified them correctly[24]. The algorithm's objective is to find the plane with the greatest margin, i.e., the greatest distance away from points and the plane so that it can achieve the greatest accuracy.
Random forest
Random forest (RAN) uses multiple non-correlated decision trees. Each decision tree predicts an output and the output with the highest prediction rate is the one which has the greatest likelihood to be correct. In essence, the outcome which gets the greatest number of “votes” from multiple non-correlated prediction models is the answer which is presumed to be the most accurate[25].
K-nearest neighbour
This is an algorithm that works on the principal that data with similar characteristics will lie in close proximity to each other. For a new piece of data, the algorithm determines how close it lies in relation to another piece of predesignated data and will then make an assumption on whether the new data has a positive or negative value[26].
Validation
Validation is a process in which the trained model is evaluated with a testing data set. It is used to determine how well the algorithm can classify images that it has never seen before. Cross validation is a commonly utilised method of testing the validity of machine learning algorithms to reduce the risk of overfitting. Overfitting is when a machine learning algorithm learns the details and noise in a testing set too well and subsequently impacts negatively on the classification of future data[27]. The most commonly applied cross-validation method is “K-fold cross-validation”. In this method, the dataset is randomly split into k subsets of equal size. Common choices of k are 5 or 10 (5-fold or 10-fold cross validation). The training is then performed using k-1 of the subsets as the training set, and the remaining 1 subset as the test set. This process is repeated k times, leaving a different subset out of the training each time. The final estimate of the model's accuracy is given by pooling the results of the validation in each of the k subsets. Although k-fold cross-validation uses the same data for both training and testing, an individual data point is never included in both the training and test sets for a particular iteration, reducing the likelihood of overfitting. This model has proven to be effective to avoid the overfitting or under fitting of data[28]. It has been stated that cross validation is a better method for testing and training than random allocation[29]. Random allocation is when the training and validation set are randomly split, with one section used for training and another used for validation.
Ophthalmology and machine learning
Machine learning is a technology that is still in its embryonic stage. Deep learning (a subset of machine learning focusing on ANN), which only found its feet in the 2000s, is a technology with widespread use in modern society including speech recognition, real time language translation and, most notably, image recognition[30]. Its transition to medical imaging analysis was an obvious step.
Ophthalmology is an ideal specialty for the implantation of machine learning due to the ability to obtain high resolution images of the posterior of the eye in the form of fundal photos or OCT scans. These are non-invasive techniques with no radiation or potential for harm. A recent study[31] examining the ability of a machine learning algorithm to identify referable retinal diseases from OCT scans revealed that its success was comparable with clinical retinal specialists. It demonstrated that it could work in a real-world setting, with the benefit of being able to diagnose multiple pathologies.
Multiple machine learning parameters are currently being assessed by numerous authors to aid in glaucoma diagnosis. There is debate over which screening frame work is the most sensitive and/or specific. On review of the literature, the most utilised screening modalities include either fundal photographs (which incorporates optic nerve head assessment and retinal vascular geometry) or OCT imaging. As previously stated, techniques offer the advantage of quick assessment time, however the OCT scanner is a significantly more expensive than the fundal camera and requires a skilled operator.
The literature has indicated that there are many studies examining the efficacy of these modalities in the screening/diagnosing of glaucoma, but none have compared the sensitivity and specificity of these tests in comparison to one another. To facilitate the mass screening of glaucoma, it would be beneficial to identify the most appropriate diagnostic test and to date the literature has failed to examine this.
Study aims and objectives
To determine the diagnostic accuracy, in terms of sensitivity and specificity and/or area under the receiver operating curve (AUROC), of machine learning (including, but not restricted to, SVM, ANN, convolutional neural network (CNN), K-nearest neighbour, least square SVM (LS-SVM), naïve Bayes and sequential minimal optimisation in diagnosing glaucoma and identifying those at risk. The two imaging modalities to be examined are fundal photographs of the optic disc, retinal vessels and OCT imaging.
These will be compared to the current reference test which is defined by the Anderson Patella criteria[14] for glaucoma diagnosis. Referable glaucomatous optic neuropathy is a term used when there is an increased CDR and therefore an associated suspicion of glaucoma. Not all suspicious discs are glaucomatous and a proportion of the normal population will have an increased CDR of greater than 0.7[32]. Functional damage, as represented by specific loss of peripheral visual field, has been demonstrated to be the most crucial component in the diagnosis of glaucoma[33].
In essence, we hope to elucidate from the general population (Population) which method of machine learning screening for glaucoma (Experimental test) is most accurate when we compare it to the gold standard test which is perimetry as defined by the Anderson Patella criteria (Reference test). This will allow us to determine the most appropriate imaging modality to utilise in terms of the automation of the mass screening of glaucoma.
MATERIALS AND METHODS
Search Strategy
A search of Pubmed and Embase was undertaken up to the first of February 2019. The search terms used in PubMed included (“glaucoma”[MeSH Terms] OR “glaucoma”[All Fields] OR “glaucomatous”[All Fields] OR “glaucomatous”[All Fields]) AND (“machine learning”[All Fields] OR “deep learning”[All Fields] OR “Computer Aided”[All]) AND (“diagnosis”[All Fields] OR “detection”[All Fields] OR “Screening”[All fields]). The search terms in Embase included (‘glaucoma’/exp OR glaucoma) AND (‘machine learning’ OR ‘deep learning’ OR ‘computer aided diagnosis’) AND (‘diagnosis’ OR ‘detection’ OR ‘screening’).
The retrieved studies were imported into RevMan 5 (version 5.3. Copenhagen: The Nordic Cochrane Center, the Cochrane Collaboration, 2014). All duplicates were deleted. The titles and abstracts of the remaining articles were reviewed by two authors (Murtagh P and Greene G) and those that did not meet the inclusion criteria were removed. For completion, the reference lists from the selected studies were also examined.
Inclusion and Exclusion Criteria
Machine learning in diagnostic imaging is a field which is still in its infancy and so therefore there were a limited number of robust papers on the subject. Inclusion criteria consisted of all observational studies examining machine learning in the diagnosis and/or screening of glaucoma involving fundal photographs and OCT imaging. Exclusion criteria included studies which used human interpretation of fundal images, those whose machine learning was based on perimetry, those only associated with diabetic macular oedema or ARMD, participants under 18 years old and those with neurological or other disorders which may confound visual field results. Some of these studies did not define their diagnostic criteria but stated that the diagnosis was of glaucoma was made by an ophthalmologist.
Data Extraction
For each study we recorded the name of the principal author, year of publication, the number of eyes involved in the study (both glaucomatous and healthy), the machine learning classifier used (if multiple were utilised, the classifier with the most favourable result was taken), how the classifier was trained and tested, their definition of glaucoma diagnosis, make of OCT scanner in the cohort that used OCT and their results.
Measurements of Diagnostic Accuracy
Results were recorded in the papers as either the AUROC or in terms of sensitivity and specificity. A receiver operating characteristic (ROC) curve is a statistical representation which demonstrates the diagnostic ability of a binary classifier at varying discrimination thresholds[34]. A ROC curve is generated by plotting true positive rates against false positive rates or by (1-specificty) on the x-axis and sensitivity on the y-axis. The AUROC informs us about the ability of the model to distinguish between different classes. It is the outcome measure most used to assess the reliability of a machine learning diagnosis. The results range from 0.5-1, the closer the result is to one, the better the performance of the machine leaning model[35]. Sensitivity is the proportion of true positives that are correctly identified by the test. Specificity is the proportion of true negatives that are correctly identified by the test[36]. Although these two outcome parameters are not directly comparable, we used an algorithm to calculate an average of the sensitivity and specificity values in terms of AUROC in the papers that failed to define one. ROC curves illustrate sensitivity and specificity at different cut-off values. If only sensitivity and specificity are stated in the studies, then there must be a single cut-off value being used, but this is not always stated (and may not be known, since it is sometimes a hidden parameter of the machine learning model).
Assessment of Study Quality
All studies available were observational studies and therefore there was no defined standard evaluation of bias. We consequently established an adapted scoring system based on the Newcastle-Ottawa Scale (NOS)[37]. Each study was assessed on the following criteria: 1) sample size (greater than 100, 1 point; less than 100, 0 points); 2) validation technique (cross validation, 1 point; other, 0 points); 3) unique database (unique database, 1 point; previously utilised database, 0 points); 4) their definition of glaucoma and diagnostic criteria (Anderson Patella criteria, 1 point; other, 0 points); 5) inclusion of a confidence intervals (CIs) around reported outcomes (yes, 1 point; no, 0 points), and 6) their interpretation and reporting of results (AUROC, 1 point; other, 0 points). Studies which scored four points or greater were deemed to be of a higher methodological standard.
Statistical Analysis
Results of studies employing fundal images and OCT were extracted using Cochrane RevMan software (version 5.3. Copenhagen: The Nordic Cochrane Center, the Cochrane Collaboration, 2014) and Meta-analysis was performed to compare the accuracy of diagnosis. In the majority of studies, diagnostic accuracy was summarised by the AUROC. Summary estimates of the combined AUROC for each imaging methodology were estimated by inverse-variance weighted Meta-analysis following the method of Zhou et al[38].
However, several studies employing fundal images reported only a single sensitivity and specificity point. A single summary AUROC value for these studies was derived by estimating a Heirarchical Summary ROC curve (HSROC)[39]. The HSROC was estimated by hierarchical logistic regression using the Metandi and Midas[40] packages in Stata version 15.0 (StataCorp, College Station, TX, USA). This summary AUROC value and associated standard error (SE) were included in the Meta-analysis of fundal image studies.
Comparison of accuracy of fundal image and OCT studies was performed by comparing pooled estimates and SE obtained through Meta-analysis of each cohort. Significant difference was defined by a P value less than 0.05.
Potential Confounders
Potential Confounders include the same data set being used by different studies, training and testing on the same data set, discrepancy in glaucoma diagnosis, concomitant neurological disorders which may confound results, use of crowdsourcing platforms and focus on computer methods as opposed to clinical outcomes.
RESULTS
Selection Process and Search Results
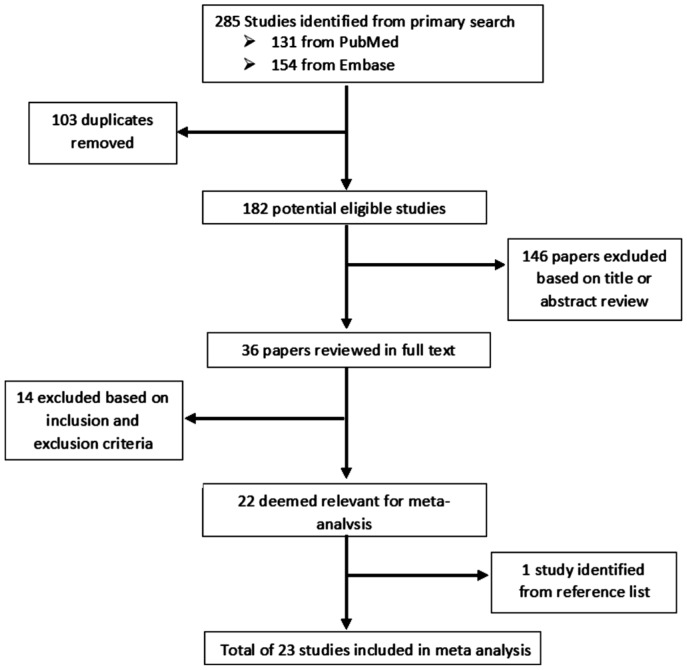
Our search parameters returned a total of 131 papers from PubMed and 154 from Embase giving us a total of 285 studies. The titles and abstracts of these studies were reviewed, duplicates were removed, alongside the papers that did not fulfil the inclusion or exclusion criteria, and 36 papers were deemed suitable for revision in full text. Following comprehensive appraisal, a total of 23 papers were deemed suitable for inclusion in this Meta-analysis. This consisted of 13 papers which examined machine learning in the diagnosis of glaucoma using fundal photos and 10 using OCT technology. All studies were population based observational studies. Figure 1 outlines the selection process.
Figure 1. Flow diagram depicting the selection process for inclusion in the Meta-analysis.
Table 1 tabulates the data with regards machine learning and fundal images. Ten of the thirteen studies were from Asia, nine of which were from India[41]–[49] and one from South Korea[50]. Of the remaining three studies, one utilised a dataset from Germany[51], one used fundal photos from two previous American studies[52] (the African Descent and Glaucoma Evaluation Study and the Diagnostic Innovations in Glaucoma Study) and the comprehensive study by Li et al[53] used the large online dataset Labelme (a crowdsourcing platform for labelling fundal photographs). Of the studies undertaken in India, six utilised the Kasturba Medical College dataset[42]–[44],[46],[48]–[49] and two used the Venu Eye Research Centre dataset[45],[47] but using different machine learning algorithms. The studies were published between 2009 and 2018. A total of 59 788 eyes were included in the studies, 39 745 coming from a single study[53].
Table 1. A summary of studies depicting automated diagnosis of glaucoma using fundal images.
| Paper | Classifier | Number, age | Training and testing | Results | Glaucoma diagnosis | Database |
| Nayak et al 2009[42] | ANN | 61, 37 G, 24 H, 25 to 60 | 46 images used for training, 15 images used for testing | AUROC 0.984 (sensitivity 100%, specificity 80%), no CI | Ill-defined but by an ophthalmologist | Kasturba Medical College, Manipal, India |
| Bock et al 2010[51] | SVM | 575, 239 G, 336 N, 56.1±11.4 | 5 fold cross validation | AUROC 0.88 P<0.07, sensitivity 73%, specificity 85% | Ill defined, stated gold standard | Erlangen Glaucoma Registry, Germany |
| Acharya et al2011[43] | SVM | 60, 30 G, 30 N, 20-70 | 5 fold cross validation | 91% accuracy, no CI, stated P significant is <0.05 | Ill defined | Kasturba Medical College, Manipal, India |
| Mookiah et al 2012[44] | SVM | 60, 30 G, 30 N, 20-70 | 3 fold stratified cross validation | Accuracy 93.33%, sensitivity 86.67%, specificity 93.33%, AUROC 0.984, no CI, stated P significant is <0.05 | Ill-defined but by an ophthalmologist | Kasturba Medical College, Manipal, India |
| Chakrabarty et al 2016[41] | CNN | 314, 169 G, 145 N | 1926 to train, 314 to test | AUROC 0.792 | Gold standard. Diagnosed by 4 glaucoma specialist | Aravind Eye Hospital, Madurai and Coimbatore, India |
| Issac et al 2015[45] | SVM | 67, 32 G, 35 N, 18-75 | Leave one out cross validation | Accuracy 94.11%, sensitivity 100%, specificity 90%, no CI, P significant if less than 0.05 | Ill-defined but by an ophthalmologist | Venu Eye Research Centre, New Delhi, India |
| Maheshwari et al 2017[46] | SVM | Two databases, 60, 30 G, 30 N, 505, 250 G, 255 N, no age range | Three fold and tenfold cross validation | Accuracy 98.33%, sensitivity 100%, specificity 96.67%, no CI, P significant if less than 0.05 | Ill-defined but by an ophthalmologist | Medical Images analysis Group Kasturba Medical College, Manipal, India |
| Singh et al 2016[47] | SVM | 63, 33 G, 30 N, 18-75 | Leave one out cross validation, 44 to train 19 to check | Accuracy 95.24%, sensitivity 96.97%, specificity 93.33%, no CI, P significant if less than 0.05 | Ill-defined but by an ophthalmologist | Venu Eye Research Centre, New Delhi, India |
| Maheshwari et al 2017[48] | LS-SVM | 488, 244 G, 244 N, no age range | Three fold and tenfold, cross validation | Accuracy 94.79%, sensitivity 93.62%, specificity 95.88% | Ill-defined but by an ophthalmologist | Kasturba Medical College, Manipal, India |
| Raghavendra et al 2018[49] | SVM | 1426, 837 G, 589 N | 70% raining, 30% testing, repeated 50 times, random training and testing partitions | Accuracy 98.13%, sensitivity 98%, specificity 98.3%, no CI, P significant if less than 0.05 | Ill-defined but by an ophthalmologist | Kasturba Medical College, Manipal, India |
| Ahn et al 2018[63] | CNN | 1542, 756 G, 786 N, no age range | Randomly partitioned into 754 training, 324 validation and 464 test datasets | AUROC 0.94, accuracy 87.9%, no CI | Ill-defined but likely Anderson Patella Criteria | Kim's Eye Hospital, Seoul, South Korea |
| Christopher et al 2018[52] | CNN | 14822, 5633 G, 9189 N | 10 fold cross validation | AUROC 0.91 (0.9-0.91 CI) | Independent masked graders | The ADAGES study, New and Alabama DIGS Study, California |
| Li et al 2018[53] | CNN | 39745, 9279 G, 30466 N | 8000 images as the validation set, and 31745 images as training set | AUROC 0.986 (95%CI, 0.984-0.988) | Grading by trained ophthalmologists | Label me Data Set |
G: Glaucoma; N: Normal; AUROC: Area under the receiver operating characteristics curve; CI: Confidence interval; CNN: Convolutional neural networks; ANN: Artificial neural network; SVM: Support vector machine; LS-SVM: Least squares support vectors machine; ADAGES: African descent and glaucoma evaluation study; DIGS: Diagnostic innovations in glaucoma study.
Table 2 illustrates the data with regards machine learning and OCT imaging techniques. There is a total of ten studies published between 2005 and 2019. Five of the studies are from the USA[54]–[58], two are from Japan[50],[59], two are from Brazil[60]–[61] and the remaining study is from Sweden[62]. There was no overlap between the between the studies as regards data sets. Three studies used the Stratus OCT[54]–[55],[62], three studies used the Cirrus OCT (one standard definition[60] and two high definition[56],[61]), Topcon OCT was used in two[50],[57] and the RS 3000[50] and Spectralis[58] was used in one study each. The studies were published between 2005 and 2019. A total of 1743 eyes were included in the OCT studies.
Table 2. A summary of studies depicting automated diagnosis of glaucoma using OCT.
| Paper | Classifier | Number | Training and testing | Results | OCT | Glaucoma diagnosis | Database |
| Burgansky-Eliash et al 2005[54] | Multiple-take SVM | 89, 47 G, 42 N | Six fold validation, leave one out | AUROC 0.981, no CI | Stratus OCT | Anderson Patella Criteria | Recruitment of Subjects, Pennsylvania |
| Bowd et al 2008[55] | RVM | 225, 156 G, 69 N | Tenfold cross validation | AUROC 0.809, no CI | Stratus OCT | Anderson Patella Criteria | Observational Cross Sectional Study, California |
| Bizios et al 2010[62] | SVM | 152, 62 G, 90 N | Tenfold cross validation | AUROC 0.977, CI 0.959-0.999 | Stratus OCT | Anderson Patella Criteria | Observational Cross Sectional Study, Citizens of Malmo Sweden |
| Barella et al 2013[60] | RAN | 103, 57 G, 46 N | Tenfold cross validation resampling | AUROC 0.877, CI 0.810-0.944 | Cirrus SD OCT | Anderson Patella Criteria | Glaucoma Service UNICAMP, Brazil, prospective, observational cross sectional |
| Silva et al 2013[61] | RAN | 110, 62 G, 48 N | Tenfold cross validation | AUROC 0.807, CI 0.721-0.876 | Cirrus HD OCT | Anderson Patella Criteria | Glaucoma Service UNICAMP, Brazil, observational cross sectional |
| Xu et al 2013[56] | Boosted logistic regression | 192, 148 G, 44N | Normative database, Tenfold cross validation | AUROC 0.903, no CI | Cirrus HD OCT | Anderson Patella Criteria | PITT trial, Pennsylvania |
| Muhammad et al 2017[57] | CNN | 102, 57 G, 45 N | Pretrained, leave one out cross validation | AUROC 0.945, CI 0.955-0.947 | Topcon OCT | Anderson Patella Criteria | From previous study for OCT and early glaucoma diagnosis, New York |
| Asaoka et al 2019[59] | SVM | 178, 94 G, 84 N | Pre training, glaucoma OCT database | AUROC 0.937, CI 0.906-0.968 | RS 3000 | Anderson Patella Criteria | Japanese Archives of Multicentral Images of Glaucomatous OCT database, Japan |
| Christopher et al 2018[58] | PCA | 235, 179 G, 56 N | Leave one out approach | AUROC 0.95, CI 0.92-0.98 | Spectralis OCT | Ill defined | DIGS dataset, California |
| An et al 2019[50] | CNN | 357, 208 G, 149 N | Tenfold cross validation | AUROC 0.963, Mean±SD 0.029 | Topcon OCT | Anderson Patella Criteria | Observational Cross Sectional Study, Japan |
G: Glaucoma; N: Normal; AUROC: Area under the receiver operating characteristics curve; CI: Confidence interval; CNN: Convolutional neural networks; ANN: Artificial neural network; SVM: Support vector machine; LS-SVM: Least squares support vectors machine; ADAGES: African descent and glaucoma evaluation study; DIGS: Diagnostic innovations in glaucoma study; PCA: Principal component analysis; RVM: Relevance vector machine.
Assessment of Study Quality
An assessment of study quality can be seen in Tables 3 and 4 with regards to the fundal photo and OCT groups respectively. We defined a superior methodology as a score of four or greater. It can be observed that the OCT group have a superior methodological standard than the fundal photo group. All of the OCT group have a score of four point of greater, while only 5 of the 13 (38.46%) studies in the fundal photo group achieved this score.
Table 3. Assessment of study quality using modified NOS with respect to fundal images.
| Paper | Sample size | Validation technique | Unique database | Definition of glaucoma | CI | AUROC | Total |
| Nayak et al 2009[42] | X | 1 | |||||
| Bock et al 2010[51] | X | X | X | X | X | 5 | |
| Acharya et al 2011[43] | X | 1 | |||||
| Mookiah et al 2012[44] | X | X | 2 | ||||
| Chakrabarty et al 2016[41] | X | X | X | X | 4 | ||
| Issac et al 2015[45] | X | 1 | |||||
| Maheshwari et al 2017[46] | X | X | 2 | ||||
| Singh et al 2016[47] | X | 1 | |||||
| Maheshwari et al 2017[48] | X | X | 2 | ||||
| Raghavendra et al 2018[49] | X | 1 | |||||
| Ahn et al 2018[63] | X | X | X | X | 4 | ||
| Christopher et al 2018[52] | X | X | X | X | X | 5 | |
| Li et al 2018[53] | X | X | X | X | 4 |
CI: Confidence interval; AUROC: Area under the receiver operating characteristics curve; NOS: Newcastle-Ottawa Scale; OCT: Ocular coherence tomography.
Table 4. Assessment of study quality using modified NOS with respect to OCT scans.
| Paper | Sample size | Validation technique | Unique database | Definition of glaucoma | CI | AUROC | Total |
| Burgansky-Eliash et al 2005[54] | X | X | X | X | 4 | ||
| Bowd et al 2008[55] | X | X | X | X | X | 5 | |
| Bizios et al 2010[62] | X | X | X | X | X | X | 6 |
| Barella et al 2013[60] | X | X | X | X | X | 4 | |
| Silva et al 2013[61] | X | X | X | X | X | 5 | |
| Xu et al 2013[56] | X | X | X | X | X | 5 | |
| Muhammad et al 2017[57] | X | X | X | X | X | X | 6 |
| Asaoka et al 2019[59] | X | X | X | X | X | 5 | |
| Christopher et al 2018[58] | X | X | X | X | 4 | ||
| An et al 2019[50] | X | X | X | X | X | X | 6 |
CI: Confidence interval, AUROC: Area under the receiver operating characteristics curve; NOS: Newcastle-Ottawa Scale; OCT: ocular coherence tomography.
Definition of Glaucoma
Definition of glaucoma diagnosis varied between the studies. In the fundal photo study cohort, the majority[42]–[49] were ill defined but stated that they were diagnosed by an ophthalmologist, three[41],[51],[63] stated that the diagnosis was gold standard and was likely the Anderson Patella Criteria and the remaining 2 studies[52]–[53] were diagnosed using trained independent masked graders. In the OCT studies group, nine of the ten studies[50],[54]–[57],[59]–[62] had their glaucoma diagnosis defined by the Anderson Patella criteria. In the remaining paper[58], the diagnosis was ill-defined but stated to be by two independent masked graders.
Machine Learning Classifier
As regards the machine learning classifier, SVM[43]–[47],[49]–[51] was used in seven of the fundal photo group. Neural networks were used in five[41]–[42],[52]–[53],[63] and LS-SVM[48] was used in one each. In the OCT cohort, the classifier used was more varied. Three studies utilised SVM[54],[59],[62]. RAN[60]–[61] and CNN[50],[57] was utilised by two studies each. Principal Component Analysis[58] and Relevance Vector Machine[55] were used in one study and the final study[56] employed boosted logistic regression.
Validation
Training and testing protocols are outlined in Tables 1 and 2. Cross validation was the most common method with tenfold, fivefold, threefold and leave one out cross validation accounting for the practices in nine[46],[48],[50],[52],[55]–[56],[60]–[62], two[43],[51], one[44] and five[45],[47],[54],[57]–[58] of the studies respectively. A random partitioning of training and testing occurred in four of the studies[49],[53],[59],[63].
Meta-Analysis
Since AUROC was the most widely reported and informative measure of diagnostic precision employed in the included studies, Meta-analysis was performed based on pooling AUROC estimates for each cohort.
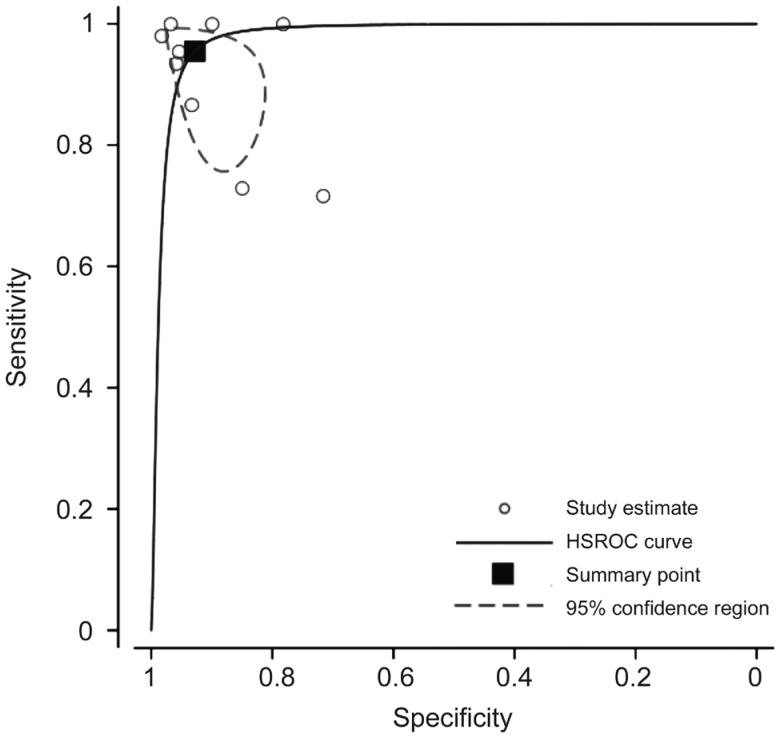
Nine of the fundal image studies did not report an AUROC, but gave only a single value of sensitivity and specificity[42]–[49],[51]. In order to include these studies in the larger Meta-analysis, we obtained a single pooled AUROC value by estimating a HSROC. This is shown in Figure 2. AUROC curve was calculated to be 0.979; 95%CI: 0.887-0.996. Studies which only reported an AUROC but did not include an estimate of variance or uncertainty (i.e., SE or CI) could not be included in the Meta-analysis.
Figure 2. An HSROC estimated by pooling results of nine fundal photo studies who did not report a AUROC value (sensitivity and specificity only).
Area under the summary ROC curve: 0.979; 95%CI: 0.887-0.996.
Tables 5 and 6 outline the results of the Meta-analysis in terms of the fundal photo cohort (with the HSROC addition) and the OCT cohort respectively.
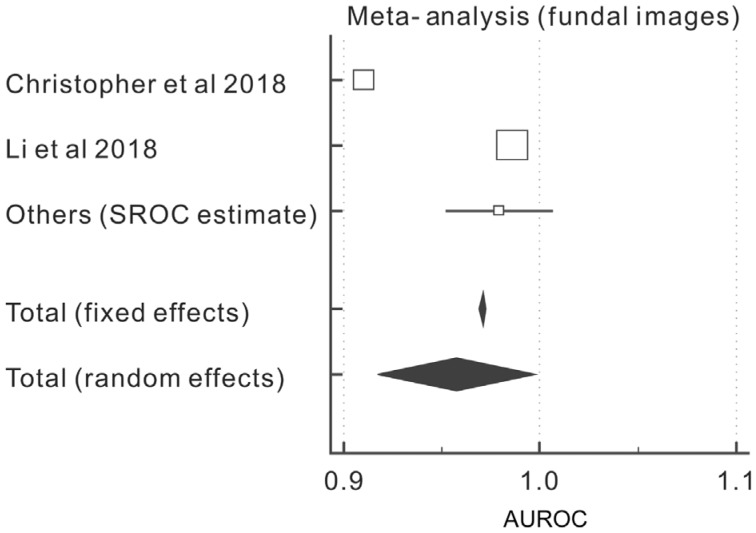
Table 5. Meta-analysis, AUROC and estimated HSROC of studies relating to fundal photos.
| Study | AUROC | SE | 95%CI | z | P | Weight (%) |
|
| Fixed | Random | ||||||
| Christopher et al 2018[52] | 0.910 | 0.00200 | 0.906 to 0.914 | 19.92 | 34.92 | ||
| Li et al 2018[53] | 0.986 | 0.00100 | 0.984 to 0.988 | 79.67 | 35.01 | ||
| Others (HSROC estimate) | 0.979 | 0.0140 | 0.952 to 1.000 | 0.41 | 30.08 | ||
| Total (random effects) | 0.957 | 0.0204 | 0.917 to 0.997 | 46.9 | <0.001 | 100.00 | 100.00 |
AUROC: Area under the receiver operating characteristic curve; CI: Confidence interval; HSROC: Hierarchical summary receiver operating characteristic curve; SE: Standard error.
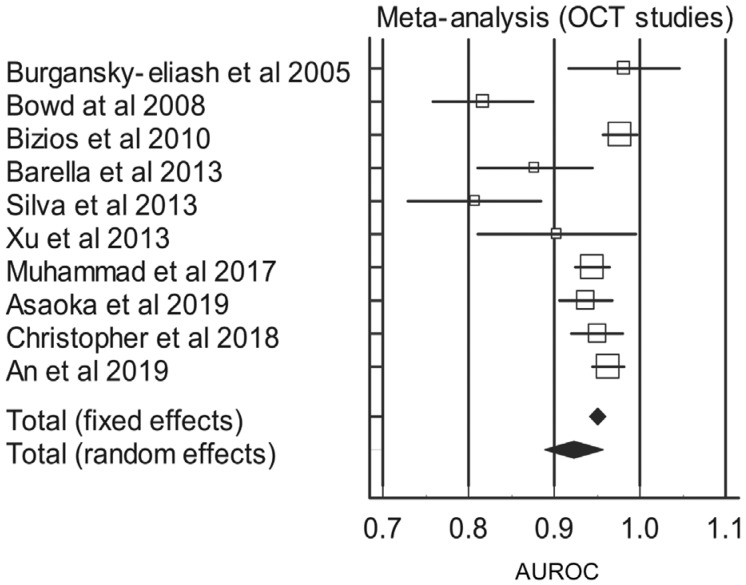
Table 6. Meta-analysis and AUROC of studies relating to OCT studies.
| Study | AUROC | SE | 95%CI | z | P | Weight (%) |
|
| Fixed | Random | ||||||
| Burgansky-Eliash et al 2005[54] | 0.981 | 0.0330 | 0.916 to 1.000 | 2.12 | 8.69 | ||
| Bowd at al 2008[55] | 0.817 | 0.0300 | 0.758 to 0.876 | 2.56 | 9.19 | ||
| Bizios et al 2010[62] | 0.977 | 0.0102 | 0.957 to 0.997 | 22.16 | 12.12 | ||
| Barella et al 2013[60] | 0.877 | 0.0342 | 0.810 to 0.944 | 1.97 | 8.49 | ||
| Silva et al 2013[61] | 0.807 | 0.0395 | 0.730 to 0.884 | 1.48 | 7.64 | ||
| Xu et al 2013[56] | 0.903 | 0.0472 | 0.810 to 0.996 | 1.04 | 6.54 | ||
| Muhammad et al 2017[57] | 0.945 | 0.0102 | 0.925 to 0.965 | 22.16 | 12.12 | ||
| Asaoka et al 2019[59] | 0.937 | 0.0158 | 0.906 to 0.968 | 9.22 | 11.45 | ||
| Christopher et al 2018[58] | 0.950 | 0.0153 | 0.920 to 0.980 | 9.85 | 11.52 | ||
| An et al 2019[50] | 0.963 | 0.00917 | 0.945 to 0.981 | 27.44 | 12.22 | ||
| Total (random effects) | 0.923 | 0.0174 | 0.889 to 0.957 | 53.1 | <0.001 | 100.00 | 100.00 |
OCT: Ocular coherence tomography; AUROC: Area under the receiver operating characteristic curve; CI: Confidence interval; SE: Standard error.
It can be seen that there is no statistically significant difference with respect to machine learning between fundal photos and OCT images in diagnosing or screening for glaucoma. The total AUROC, in terms of random effects, for the fundal photo cohort was calculated to be 0.957 (CI: 0.917 to 0.997, P<0.001) and 0.923 (CI: 0.889 to 0.957, P<0.001) for the OCT cohort.
Figures 3 and 4 are Forest plots depicting a graphical representation of weight and AUROC of both the fundal image cohort and the OCT cohort respectively.
Figure 3. Forest plot of the AUROC of the fundal images cohort.
Figure 4. Forest plot of the AUROC of the OCT cohort.
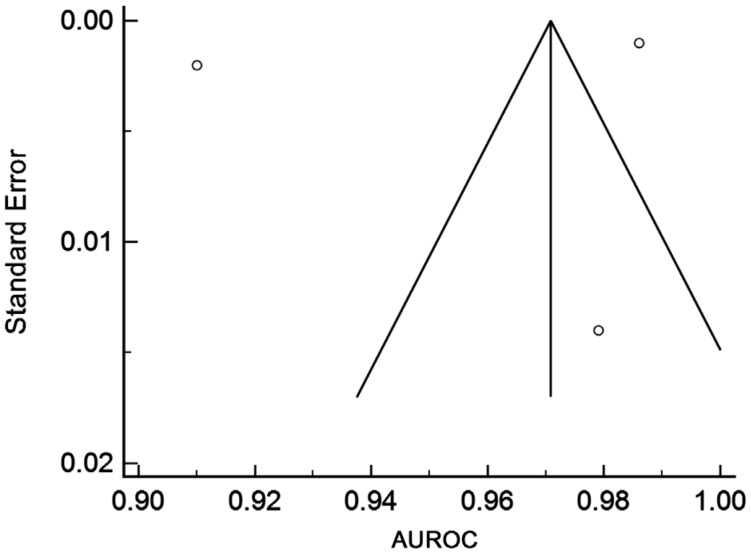
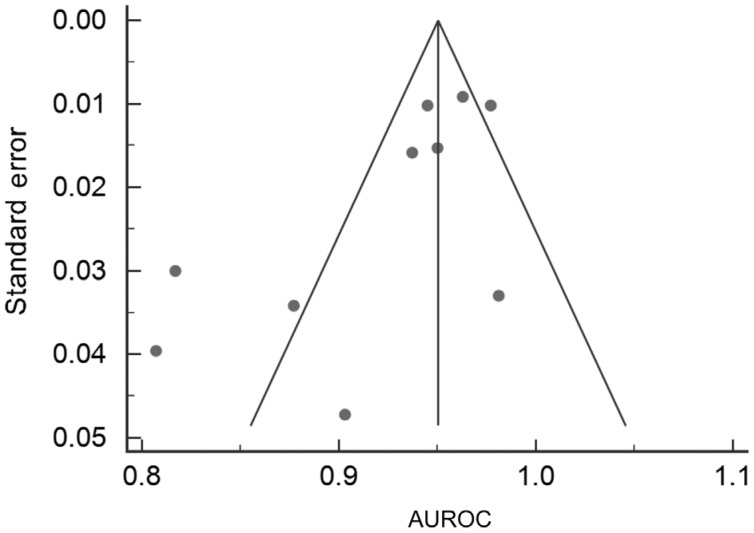
Funnel Plots for risk of bias were also performed for the two cohorts and are outlined in Figures 5 and 6. The fundal image group only has three points and therefore it has an ill-fitting funnel plot.
Figure 5. Funnel plot for fundal image studies.
Figure 6. Funnel plot for OCT studies.
The OCT funnel has every study include and is a greater assessment of study bias in comparison to the fundal image plot. Due to the fact that, unlike with standard diagnostic tests, diagnostic accuracy is expected to increase with sample size in machine learning studies, one would expect funnel plots in machine learning Meta-analysis to be asymmetric, with the majority of studies falling in the lower left quadrant. A large number of studies falling to the bottom-right would be suggestive of publication bias or perhaps overfitting of machine learning models.
Tests for heterogeneity were also performed and these are outlined in Table 7. The I2 value for the fundal image cohort and the OCT cohort is 99.83% and 81.66% respectively which is indicative of a high level of heterogeneity.
Table 7. Test for heterogeneity for fundal image studies and OCT studies.
| Parameters | Q | Significance level | I2 (inconsistency) | 95%CI for I2 |
| Fundal image studies | 1155.5417 | P<0.0001 | 99.83% | 99.77 to 99.87 |
| OCT studies | 49.0860 | P<0.0001 | 81.66% | 67.42 to 89.68 |
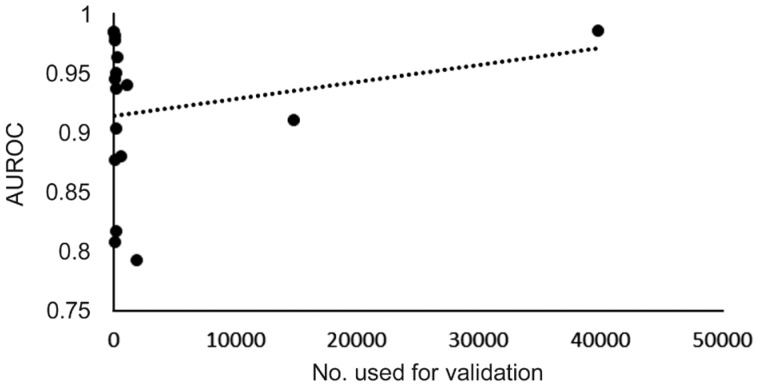
A comparison of sample size in terms of numbers used for validation versus diagnostic accuracy (given as AUROC) was performed to examine if any correlation existed.
Table 8 outlines the result of a Meta-analysis of the fundal image group without the Li et al's[53] study. The study was excluded due to the very high numbers, and therefore effect it may have on the outcome of the analysis. It can be seen that the total AUROC in terms of random effects has decreased from 0.957 to 0.942, a difference of 0.015.
Table 8. Meta-analysis, AUROC and estimated HSROC of studies relating to fundal photos with the exclusion of the Li study.
| Study | ROC area | SE | 95%CI | z | P | Weight (%) |
|
| Fixed | Random | ||||||
| Christopher et al 2018[52] | 0.910 | 0.00200 | 0.906 to 0.914 | 98.00 | 54.06 | ||
| Others (SROC estimate) | 0.979 | 0.0140 | 0.952 to 1.000 | 2.00 | 45.94 | ||
| Total (random effects) | 0.942 | 0.0242 | 0.894 to 0.989 | 38.858 | <0.001 | 100.0 | 100.00 |
AUROC: Area under the receiver operating characteristic curve; CI: Confidence interval; HSROC: Hierarchical summary receiver operating characteristic curve; SE: Standard error.
DISCUSSION
Meta-Analysis
The findings of this Meta-analysis have indicated that there is no statistically significant difference with respect to machine learning between fundal photos and OCT images in diagnosing or screening for glaucoma.
The total AUROC, in terms of random effects, for the fundal photo cohort was calculated to be 0.957 (95%CI: 0.917 to 0.997, P<0.001) and 0.923 (95%CI: 0.889 to 0.957, P<0.001) for the OCT cohort. Although there is a difference of 0.034 between the two results, the CIs of both groups overlap and there is no significant difference in diagnostic accuracy between the two cohorts (P=0.34; t-test based on pooled AUROC values and SE).
Sample Size
There is a notable discrepancy between the sample sizes of the OCT group (n=1743) and the fundal images group (n=59 788). Although the number of studies is approximately on par (10 studies for OCT and 13 for fundal photos), there is over a 30-fold increase in the numbers of eyes participating in the fundal photo group in comparison to the OCT group. However, the majority (39 745) of these eyes come from a single study[53]. If we remove this study from our Meta-analysis, as in seen in Table 8, the AUROC in terms of random effects is 0.942, leaving a difference of just 0.019 between the two groups. It is known that machine learning and deep learning are techniques that benefit from large databases for training hence the bigger the training database, the more accurate the model[64]. This can be illustrated with our data. Figure 7 depicts a scatter plot of validation numbers versus AUROC. A linear trend line shows that for increasing validation numbers there is an associated rise in diagnostic accuracy. A similar trend can be seen in the funnel plot with regards to the OCT cohort. There are multiple study sizes with low sample size and low accuracy. Accuracy improves as the sample size gets bigger (smaller SE and hence higher accuracy).
Figure 7. Graph of number used for validation versus AUROC.
One reason for the variance in numbers is the ease of acquisition of fundal photos in comparison to OCT scans. Large data sets such as the “Labelme” dataset (http://www.labelme.org/ assessed on 21/04/2019) is a crowdsourcing platform for fundal images which contains thousands of retinal fundal images from diverse populations and is available online. There is no comparable online database of OCT scans. Crowdsourcing is a method of obtaining information about data from a large number of people, usually through the internet[65].
Data Sets
There is substantial overlap between the datasets used in the fundal imaging cohort. Six studies in this group used the Kasturba Medical College dataset[42]–[44],[46],[48]–[49] and two used the Venu Eye Research Centre dataset[45],[47]. They used different machine learning models, however, three of the studies[43]–[44],[48] had the same number of participants, both glaucomatous and healthy, and it is possible that they used exactly the same fundal photos as it does not state how many images the dataset contains. This does not skew the findings but potentially hampers the power of these studies. It is also seen that there is a heavy Asian majority with regards number of papers in the fundal photo cohort. Ten of the thirteen papers came from Asia, nine of them from India. There is an obvious population based bias in this group and the same machine learning technique may not be comparable to glaucoma diagnosis in a different ethnic cohort[66]. There is a greater ethnic diversity observed in the OCT study groups. These are mainly population based studies and by their design should limit the effect of selection bias.
Validation
As stated in the results section above, cross validation was the most utilised method of training and testing with some form of it being used in seventeen out of the twenty-three studies. It has been stated that cross validation is a better method for testing and training than random allocation which occurred in four of the studies[29]. This is due to the fact that when random sampling is used, there is a chance that the sampling set does not contain the disease or features associated with the disease process. Four of our studies[49],[53],[59],[63] used random sampling and although initially may appear as the better teaching process, the more robust technique of cross validation may make their models more accurate.
Machine Classifier
The studies used a range of different classifiers or machine learning algorithms. The most commonly used algorithm was SVM being utilised in 10 of the studies. This is useful in classifying linear features and as such is the option of choice in classifying fundal images[67]. Problems arise with this classifier when non-linear features are extracted such as are employed in OCT scanning techniques. Hence more convoluted classifiers may be more appropriate when interpreting these scans, e.g., CNN and ANN, and this is reflected in the data.
The algorithms used in these studies were solely constructed to aid in the diagnosis of glaucoma. This is usually a binary classification as outlined with the abundant use of the SVM classifier. However, in the clinical setting many patients can suffer from a multitude of eye pathologies. Cataract, for instance, is an extremely common finding especially in the elderly. Significant cataract can hamper the acquisition of fundal photos and OCT images. It can also increase the amount of “noise” in the attained scans making it more difficult for the algorithm to interpret it[64]. Patients may also have features of DR and/or ARMD, pathologies which are very common in the aging population. A deep learning algorithm was previously developed[68] and tested on a small cohort (n=60) which aimed to detect a range of retinal disease from fundal images. Accuracy dropped from 87.4% in the cohort which had DR alone to 30.5% when multiple aetiologies were included. However, the small dataset used for 10 identifiable diseases is likely to inflict bias on the results.
Glaucoma Diagnosis
Glaucoma diagnosis is a multifactorial process. It is a significant proportion of the workload of general ophthalmologists. In order to make a definitive diagnosis perimetry, fundoscopy, gonioscopy and tonometry must all be undertaken. There is significant variance in the agreed diagnosis of glaucoma in the above studies. Many have the diagnostic criteria ill-defined but state that is was established by an ophthalmologist or multiple masked graders. The reason for this inconsistency is that a significant proportion of the reviewed papers were published in journals with an interest in computer methods as opposed to clinical and ophthalmological findings. Their definitions are vaguer than those published in the clinical journals. The variation in robust diagnoses can also be observed between the fundal images and OCT cohort with all but one[58] of the OCT group having their glaucoma diagnosis defined by the Anderson Patella criteria.
Incorporation of other patient parameters into the model process, e.g., age, smoking status, intraocular pressure, visual field testing has been shown to increase diagnostic accuracy[67], although the incorporation of perimetry is likely to prove to be a time and resource heavy inclusion parameter.
Methodological Quality
The methodological quality of the studies was assessed using a modified NOS. It can be witnessed that the OCT studies group have a greater standard of quality as opposed to the fundal image group. Only 5 of the 13 (38.46%) studies in the fundal image group received a methodological score of 4 or greater on our modified scale. This could be a potential confounder with respect to the results of the analysis but due to the fact the studies with a low score amount to 3.82% (2285 of 59 788) of the total number of eyes in the fundal photos group, its effect is unlikely to be statistically significant in the pooled analysis.
Publication Bias
The funnel plots, outlined in Figures 5 and 6, indicate that there is low risk of publication bias especially in OCT group. A total of three points are on the fundal image funnel plot and so therefore it is difficult to make an assumption about the bias of these studies. It can be seen that a larger sample size (and as such a smaller SE) will give a large degree of accuracy. Due to the fact that, unlike with standard diagnostic tests, diagnostic accuracy is expected to increase with sample size in machine learning studies, one would expect funnel plots in machine learning Meta-analysis to be asymmetric, with the majority of studies falling in the lower left quadrant. A large number of studies falling to the bottom-right would be suggestive of publication bias or perhaps overfitting of machine learning models.
Heterogeneity
There is a large degree of heterogeneity as outlined in Table 7. The I2 value for the fundal image cohort and the OCT cohort is 99.83% and 81.66% respectively. This is not surprising given the different methods, sample sizes and algorithms used in each.
Glaucoma Prevalence
The prevalence of primary open angle glaucoma in the general population is approximately 2% over the age of 40 and increases with age to affect 4% of those over 80[69]. On review of our data sets, it is seen that that the proportional of glaucomatous eyes in our cohorts ranged from 23.35%[53] to 77.08%[56], with an average of 53.05%±11.66% SD. This indicates that, during the training and testing process, the algorithm is more likely to observe glaucomatous eye on average thirteen times more frequently than it would in the general population. Validation on such datasets may lead to an increase in false positives either by the algorithm “expecting” to have more positive results than it has or secondary to overfitting of potential disease characteristics.
Unsupervised Machine Learning
Although our studies solely examined supervised machine learning, unsupervised machine learning in the form of deep learning will generate decisions based on high dimensional interpretation and neural networks that humans cannot interpret and is likely to be the next step in evolution of computer aided diagnosis. We fundamentally do not know how they make their judgments[70]. Areas of the image can be highlighted, but often they are not associated with the pathological process from our understanding. This can aid us to look for new aetiologies for retinal disease process. However, this is termed a “black box” as we don't fully understand how the algorithms are coming to their conclusions.
Challenges
In a recent review by Ting et al[71], a number of potential challenges for AI implementation into clinical practice were identified. The algorithm requires a large number of pathological images to train. The sharing of images between centres is currently an ethical grey area but to ensure adequate classification by the algorithm, data must be shared between centres. This includes a range of date from diverse populations. Rare ocular disease may also prove an issue, as there may not be enough images to adequately train an algorithm to recognise these diseases.
Limitations
Limitations of our studies include the high prevalence of articles in computer method journals as opposed to clinical journals especially in the fundal photo group. They were more preoccupied with how the algorithm and classifier functioned from a computer science point of view and their definitions of glaucoma were not as robust as they were in the clinical journals. Although our studies scored low on our assessment of quality, these studies had to be included due to the paucity of papers on the subject.
The use of the same database and crowdsourcing material has the potential to bias the results of the Meta-analysis. Ideally all studies would have the same definition of glaucoma, use a separate training, validation and testing set and use cross validation.
In conclusion, OCT scanning provides micrometre resolution of the RNFL and one would assume that it should be related to a more accurate screening and diagnostic tool. However, we have demonstrated that the literature to date has failed to corroborate this with respect to machine learning. The ease of access and lower cost associated with fundal photo acquisition make that the more appealing option in terms of screening on a global scale, however further studies need to be undertaken on both groups owing largely to the poor study quality associated with the fundal photography cohort.
The prospect of machine learning in the screening and diagnosing of ocular disease is a very appealing prospect. It can take pressure off ophthalmology departments and allow a greater throughput of the population to enjoy a better vision related quality of life. However, care should be taken in interpreting these findings. During an ocular assessment, ophthalmologists take in a holistic view of the patient, including past medical history and medications, and may undertake multimodal imaging e.g. angiograms and/or perimetry to come to a complete diagnosis. We know that patients greatly value their interaction with their doctor[72] and the “human-touch” associated with it, something which could be become extinct with the advent of AI. Another cause of concern is the “black box” nature of decision making in unsupervised machine learning. With so much emphasis nowadays on evidence-based medicine, it may not yet be time to place all blind trust in machines.
Acknowledgments
Conflicts of Interest: Murtagh P, None; Greene G, None; O'Brien C, None.
REFERENCES
- 1.Fingert JH, Alward WL, Kwon YH, Shankar SP, Andorf JL, Mackey DA, Sheffield VC, Stone EM. No association between variations in the WDR36 gene and primary open-angle glaucoma. Arch Ophthalmol. 2007;125(3):434–436. doi: 10.1001/archopht.125.3.434-b. [DOI] [PubMed] [Google Scholar]
- 2.Greco A, Rizzo MI, De Virgilio A, Gallo A, Fusconi M, de Vincentiis M. Emerging concepts in glaucoma and review of the literature. Am J Med. 2016;129(9):1000.e7–1001000.e13. doi: 10.1016/j.amjmed.2016.03.038. [DOI] [PubMed] [Google Scholar]
- 3.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Alhadeff PA, De Moraes CG, Chen M, Raza AS, Ritch R, Hood DC. The association between clinical features seen on fundus photographs and glaucomatous damage detected on visual fields and optical coherence tomography scans. J Glaucoma. 2017;26(5):498–504. doi: 10.1097/IJG.0000000000000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chauhan BC, Garway-Heath DF, Goñi FJ, Rossetti L, Bengtsson B, Viswanathan AC, Heijl A. Practical recommendations for measuring rates of visual field change in glaucoma. Br J Ophthalmol. 2008;92(4):569–573. doi: 10.1136/bjo.2007.135012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varma R, Lee PP, Goldberg I, Kotak S. An assessment of the health and economic burdens of glaucoma. Am J Ophthalmol. 2011;152(4):515–522. doi: 10.1016/j.ajo.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. JAMA. 2014;311(18):1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim TC, Chattopadhyay S, Acharya UR. A survey and comparative study on the instruments for glaucoma detection. Med Eng Phys. 2012;34(2):129–139. doi: 10.1016/j.medengphy.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Tuck MW, Crick RP. The age distribution of primary open angle glaucoma. Ophthalmic Epidemiol. 1998;5(4):173–183. doi: 10.1076/opep.5.4.173.4192. [DOI] [PubMed] [Google Scholar]
- 10.Dodds MK, Codd MB, Looney A, Mulhall KJ. Incidence of hip fracture in the Republic of Ireland and future projections: a population-based study. Osteoporos Int. 2009;20(12):2105–2110. doi: 10.1007/s00198-009-0922-1. [DOI] [PubMed] [Google Scholar]
- 11.Mataki N, Tomidokoro A, Araie M, Iwase A. Beta-peripapillary atrophy of the optic disc and its determinants in Japanese eyes: a population-based study. Acta Ophthalmol. 2018;96(6):e701–e706. doi: 10.1111/aos.13702. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, Gong L, Li Y, Zhu X, Stewart JM, Wang C. Peripapillary atrophy in high myopia. Curr Eye Res. 2017;42(9):1308–1312. doi: 10.1080/02713683.2017.1307992. [DOI] [PubMed] [Google Scholar]
- 13.Marcus MW, de Vries MM, Junoy Montolio FG, Jansonius NM. Myopia as a risk factor for open-angle glaucoma: a systematic review and meta-analysis. Ophthalmology. 2011;118(10):1989–1994.e2. doi: 10.1016/j.ophtha.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Anderson DR, Chauhan B, Johnson C, Katz J, Patella VM, Drance SM. Criteria for progression of glaucoma in clinical management and in outcome studies. Am J Ophthalmol. 2000;130(6):827–829. doi: 10.1016/s0002-9394(00)00665-6. [DOI] [PubMed] [Google Scholar]
- 15.Huang D, Swanson EA, Lin CP, et al. Optical coherence tomography. Science. 1991;254(5035):1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagiwara Y, Koh JEW, Tan JH, Bhandary SV, Laude A, Ciaccio EJ, Tong L, Acharya UR. Computer-aided diagnosis of glaucoma using fundus images: a review. Comput Methods Programs Biomed. 2018;165:1–12. doi: 10.1016/j.cmpb.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Grewal DS, Tanna AP. Diagnosis of glaucoma and detection of glaucoma progression using spectral domain optical coherence tomography. Curr Opin Ophthalmol. 2013;24(2):150–161. doi: 10.1097/ICU.0b013e32835d9e27. [DOI] [PubMed] [Google Scholar]
- 18.Schuman JS, Hee MR, Puliafito CA, Wong C, Pedut-Kloizman T, Lin CP, Hertzmark E, Izatt JA, Swanson EA, Fujimoto JG. Quantification of nerve fiber layer thickness in normal and glaucomatous eyes using optical coherence tomography. Arch Ophthalmol. 1995;113(5):586–596. doi: 10.1001/archopht.1995.01100050054031. [DOI] [PubMed] [Google Scholar]
- 19.Myers JS, Fudemberg SJ, Lee D. Evolution of optic nerve photography for glaucoma screening: a review. Clin Exp Ophthalmol. 2018;46(2):169–176. doi: 10.1111/ceo.13138. [DOI] [PubMed] [Google Scholar]
- 20.Bussel II, Wollstein G, Schuman JS. OCT for glaucoma diagnosis, screening and detection of glaucoma progression. Br J Ophthalmol. 2014;98(Suppl 2):ii15–19. doi: 10.1136/bjophthalmol-2013-304326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katherine M, Michael W. Direct ophthalmoscopy… soon to be forgotten? Ulster Med J. 2019;88(2):115–117. [PMC free article] [PubMed] [Google Scholar]
- 22.Agurto C, Barriga ES, Murray V, Nemeth S, Crammer R, Bauman W, Zamora G, Pattichis MS, Soliz P. Automatic detection of diabetic retinopathy and age-related macular degeneration in digital fundus images. Invest Ophthalmol Vis Sci. 2011;52(8):5862–5871. doi: 10.1167/iovs.10-7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 24.Rau CS, Wu SC, Chuang JF, Huang CY, Liu HT, Chien PC, Hsieh CH. Machine learning models of survival prediction in trauma patients. J Clin Med. 2019;8(6):E799. doi: 10.3390/jcm8060799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigatti SJ. Random Forest. J Insur Med. 2017;47(1):31–39. doi: 10.17849/insm-47-01-31-39.1. [DOI] [PubMed] [Google Scholar]
- 26.Ghaneei M, Ekyalimpa R, Westover L, Parent EC, Adeeb S. Customized k-nearest neighbourhood analysis in the management of adolescent idiopathic scoliosis using 3D markerless asymmetry analysis. Comput Methods Biomech Biomed Engin. 2019;22(7):696–705. doi: 10.1080/10255842.2019.1584795. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian J, Simon R. Overfitting in prediction models-is it a problem only in high dimensions? Contemp Clin Trials. 2013;36(2):636–641. doi: 10.1016/j.cct.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Zhang YC, Kagen AC. Machine learning interface for medical image analysis. J Digit Imaging. 2017;30(5):615–621. doi: 10.1007/s10278-016-9910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pérez-Guaita D, Kuligowski J, Lendl B, Wood BR, Quintás G. Assessment of discriminant models in infrared imaging using constrained repeated random sampling-cross validation. Anal Chim Acta. 2018;1033:156–164. doi: 10.1016/j.aca.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Lu W, Tong Y, Yu Y, Xing Y, Chen C, Shen Y. Applications of artificial intelligence in ophthalmology: general overview. J Ophthalmol. 2018;2018:5278196. doi: 10.1155/2018/5278196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 32.Ulas F, Dogan Ü, Kaymaz A, Çelik F, Çelebi S. Evaluation of subjects with a moderate cup to disc ratio using optical coherence tomography and Heidelberg retina tomograph 3: impact of the disc area. Indian J Ophthalmol. 2015;63(1):3–8. doi: 10.4103/0301-4738.151454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma P, Sample PA, Zangwill LM, Schuman JS. Diagnostic tools for glaucoma detection and management. Surv Ophthalmol. 2008;53(Suppl1):S17–32. doi: 10.1016/j.survophthal.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park SH, Goo JM, Jo CH. Receiver operating characteristic (ROC) curve: practical review for radiologists. Korean J Radiol. 2004;5(1):11–18. doi: 10.3348/kjr.2004.5.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4(2):627–635. [PMC free article] [PubMed] [Google Scholar]
- 36.Altman DG, Bland JM. Diagnostic tests. 1: Sensitivity and specificity. BMJ. 1994;308(6943):1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in Meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 38.Zhou XH, McClish DK, Obuchowski NA. Statistical methods in diagnostic medicine. John Wiley & Sons; 2009. [Google Scholar]
- 39.Takwoingi Y, Guo B, Riley RD, Deeks JJ. Performance of methods for meta-analysis of diagnostic test accuracy with few studies or sparse data. Stat Methods Med Res. 2017;26(4):1896–1911. doi: 10.1177/0962280215592269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harbord RM, Whiting P. Metandi: Meta-analysis of diagnostic accuracy using hierarchical logistic regression. Stata Jornal. 2009;9(2):211–229. [Google Scholar]
- 41.Chakrabarty L, Joshi GD, Chakravarty A, Raman GV, Krishnadas SR, Sivaswamy J. Automated detection of glaucoma from topographic features of the optic nerve head in color fundus photographs. J Glaucoma. 2016;25(7):590–597. doi: 10.1097/IJG.0000000000000354. [DOI] [PubMed] [Google Scholar]
- 42.Nayak J, Acharya UR, Bhat PS, Shetty N, Lim TC. Automated diagnosis of glaucoma using digital fundus images. J Med Syst. 2009;33(5):337–346. doi: 10.1007/s10916-008-9195-z. [DOI] [PubMed] [Google Scholar]
- 43.Acharya UR, Dua S, Du X, Sree SV, Chua CK. Automated diagnosis of glaucoma using texture and higher order spectra features. IEEE Trans Inf Technol Biomed. 2011;15(3):449–455. doi: 10.1109/TITB.2011.2119322. [DOI] [PubMed] [Google Scholar]
- 44.Mookiah MRK, Acharya UR, Lim C, Petznick A, Suri J. Data mining technique for automated diagnosis of glaucoma using higher order spectra and wavelet energy features. Knowl Based Syst. 2012;33:73–82. [Google Scholar]
- 45.Issac A, Partha Sarathi M, Dutta MK. An adaptive threshold based image processing technique for improved glaucoma detection and classification. Comput Methods Programs Biomed. 2015;122(2):229–244. doi: 10.1016/j.cmpb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Maheshwari S, Pachori RB, Acharya UR. Automated diagnosis of glaucoma using empirical wavelet transform and correntropy features extracted from fundus images. IEEE J Biomed Health Inform. 2017;21(3):803–813. doi: 10.1109/JBHI.2016.2544961. [DOI] [PubMed] [Google Scholar]
- 47.Singh A, Dutta MK, ParthaSarathi M, Uher V, Burget R. Image processing based automatic diagnosis of glaucoma using wavelet features of segmented optic disc from fundus image. Comput Methods Programs Biomed. 2016;124:108–120. doi: 10.1016/j.cmpb.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Maheshwari S, Pachori RB, Kanhangad V, Bhandary SV, Acharya UR. Iterative variational mode decomposition based automated detection of glaucoma using fundus images. Comput Biol Med. 2017;88:142–149. doi: 10.1016/j.compbiomed.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 49.Raghavendra U, Fujita H, Bhandary SV, Gudigar A, Tan JH, Acharya UR. Deep convolution neural network for accurate diagnosis of glaucoma using digital fundus images. Inf Sci. 2018;441:41–49. [Google Scholar]
- 50.An G, Omodaka K, Hashimoto K, Tsuda S, Shiga Y, Takada N, Kikawa T, Yokota H, Akiba M, Nakazawa T. Glaucoma diagnosis with machine learning based on optical coherence tomography and color fundus images. J Healthc Eng. 2019;2019:4061313. doi: 10.1155/2019/4061313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bock R, Meier J, Nyúl LG, Hornegger J, Michelson G. Glaucoma risk index: automated glaucoma detection from color fundus images. Med Image Anal. 2010;14(3):471–481. doi: 10.1016/j.media.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Christopher M, Belghith A, Bowd C, Proudfoot JA, Goldbaum MH, Weinreb RN, Girkin CA, Liebmann JM, Zangwill LM. Performance of deep learning architectures and transfer learning for detecting glaucomatous optic neuropathy in fundus photographs. Sci Rep. 2018;8(1):16685. doi: 10.1038/s41598-018-35044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, He Y, Keel S, Meng W, Chang RT, He M. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology. 2018;125(8):1199–1206. doi: 10.1016/j.ophtha.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 54.Burgansky-Eliash Z, Wollstein G, Chu TJ, Ramsey JD, Glymour C, Noecker RJ, Ishikawa H, Schuman JS. Optical coherence tomography machine learning classifiers for glaucoma detection: a preliminary study. Invest Ophthalmol Vis Sci. 2005;46(11):4147–4152. doi: 10.1167/iovs.05-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowd C, Hao J, Tavares IM, Medeiros FA, Zangwill LM, Lee TW, Sample PA, Weinreb RN, Goldbaum MH. Bayesian machine learning classifiers for combining structural and functional measurements to classify healthy and glaucomatous eyes. Invest Ophthalmol Vis Sci. 2008;49(3):945–953. doi: 10.1167/iovs.07-1083. [DOI] [PubMed] [Google Scholar]
- 56.Xu J, Ishikawa H, Wollstein G, Bilonick RA, Folio LS, Nadler Z, Kagemann L, Schuman JS. Three-dimensional spectral-domain optical coherence tomography data analysis for glaucoma detection. PLoS One. 2013;8(2):e55476. doi: 10.1371/journal.pone.0055476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muhammad H, Fuchs TJ, De Cuir N, De Moraes CG, Blumberg DM, Liebmann JM, Ritch R, Hood DC. Hybrid deep learning on single wide-field optical coherence tomography scans accurately classifies glaucoma suspects. J Glaucoma. 2017;26(12):1086–1094. doi: 10.1097/IJG.0000000000000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christopher M, Belghith A, Weinreb RN, Bowd C, Goldbaum MH, Saunders LJ, Medeiros FA, Zangwill LM. Retinal nerve fiber layer features identified by unsupervised machine learning on optical coherence tomography scans predict glaucoma progression. Invest Ophthalmol Vis Sci. 2018;59(7):2748–2756. doi: 10.1167/iovs.17-23387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asaoka R, Murata H, Hirasawa K, Fujino Y, Matsuura M, Miki A, Kanamoto T, Ikeda Y, Mori K, Iwase A, Shoji N, Inoue K, Yamagami J, Araie M. Using deep learning and transfer learning to accurately diagnose early-onset glaucoma from macular optical coherence tomography images. Am J Ophthalmol. 2019;198:136–145. doi: 10.1016/j.ajo.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Barella KA, Costa VP, Gonçalves Vidotti V, Silva FR, Dias M, Gomi ES. Glaucoma diagnostic accuracy of machine learning classifiers using retinal nerve fiber layer and optic nerve data from SD-OCT. J Ophthalmol. 2013;2013:789129. doi: 10.1155/2013/789129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silva FR, Vidotti VG, Cremasco F, Dias M, Gomi ES, Costa VP. Sensitivity and specificity of machine learning classifiers for glaucoma diagnosis using spectral domain OCT and standard automated perimetry. Arq Bras Oftalmol. 2013;76(3):170–174. doi: 10.1590/s0004-27492013000300008. [DOI] [PubMed] [Google Scholar]
- 62.Bizios D, Heijl A, Hougaard JL, Bengtsson B. Machine learning classifiers for glaucoma diagnosis based on classification of retinal nerve fibre layer thickness parameters measured by Stratus OCT. Acta Ophthalmol. 2010;88(1):44–52. doi: 10.1111/j.1755-3768.2009.01784.x. [DOI] [PubMed] [Google Scholar]
- 63.Ahn JM, Kim S, Ahn KS, Cho SH, Lee KB, Kim US. A deep learning model for the detection of both advanced and early glaucoma using fundus photography. PLoS One. 2018;13(11):e0207982. doi: 10.1371/journal.pone.0207982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee JG, Jun S, Cho YW, Lee H, Kim GB, Seo JB, Kim N. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18(4):570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wazny K. Applications of crowdsourcing in health: an overview. J Glob Health. 2018;8(1):010502. doi: 10.7189/jogh.08.010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kosoko-Lasaki O, Gong G, Haynatzki G, Wilson MR. Race, ethnicity and prevalence of primary open-angle glaucoma. J Natl Med Assoc. 2006;98(10):1626–1629. [PMC free article] [PubMed] [Google Scholar]
- 67.Kim SJ, Cho KJ, Oh S. Development of machine learning models for diagnosis of glaucoma. PLoS One. 2017;12(5):e0177726. doi: 10.1371/journal.pone.0177726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi JY, Yoo TK, Seo JG, Kwak J, Um TT, Rim TH. Multi-categorical deep learning neural network to classify retinal images: a pilot study employing small database. PLoS One. 2017;12(11):e0187336. doi: 10.1371/journal.pone.0187336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90(3):262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quellec G, Charrière K, Boudi Y, Cochener B, Lamard M. Deep image mining for diabetic retinopathy screening. Med Image Anal. 2017;39:178–193. doi: 10.1016/j.media.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 71.Ting DSW, Peng L, Varadarajan AV, Keane PA, Burlina PM, Chiang MF, Schmetterer L, Pasquale LR, Bressler NM, Webster DR, Abramoff M, Wong TY. Deep learning in ophthalmology: the technical and clinical considerations. Prog Retin Eye Res. 2019;72:100759. doi: 10.1016/j.preteyeres.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 72.Pilnick A, Dingwall R. On the remarkable persistence of asymmetry in doctor/patient interaction: a critical review. Soc Sci Med. 2011;72(8):1374–1382. doi: 10.1016/j.socscimed.2011.02.033. [DOI] [PubMed] [Google Scholar]