We read with great interest the article by Ma et al. (1), which investigated the role of N6-methyladenosine (m6A) modification in hepatocellular carcinoma (HCC) and the mechanism of methyltransferase-like 14 (METTL14) involved in the progression of liver cancer. The authors found a decreased m6A level in HCC compared with peritumor and normal specimens. They discovered that depleted METTL14 regulated the recognition and binding of DGCR8 and pri-miR-126 via m6A modification, suppressing the miR-126 expression and causing HCC metastasis. In contrast, Chen et al. (2) reported a significant increase of m6A level in HCC (data not shown). It was confirmed that METTL14 was not significantly decreased in HCC. They also demonstrated the promoting effect of METTL14 in liver cancer cell proliferation and migration.
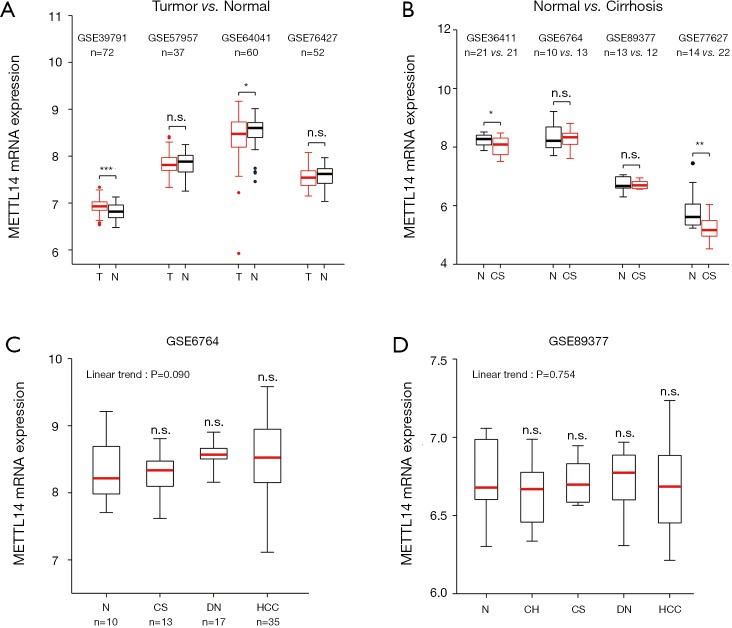
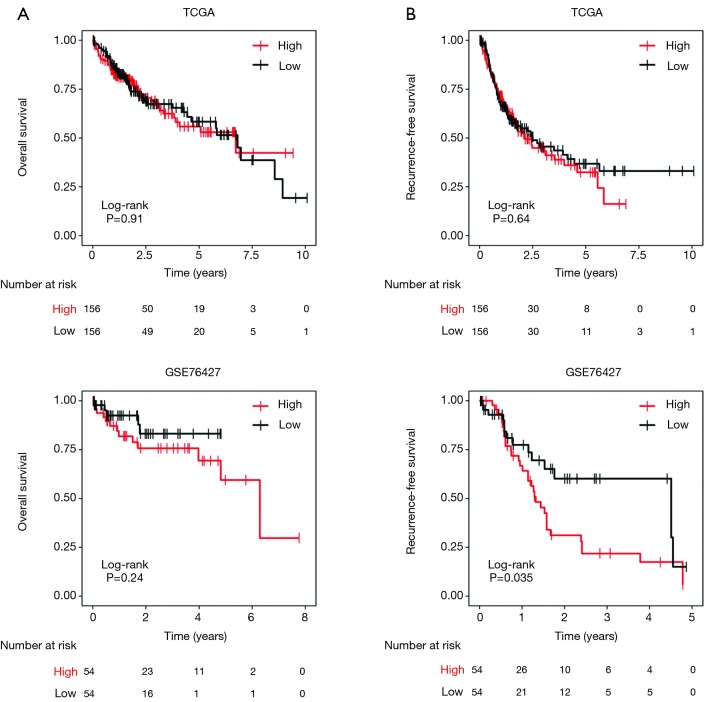
Considering the paradoxical expression pattern of METTL14 in HCC, we analysed paired HCC and normal samples in multiple microarray datasets. A differential expression of METTL14 mRNA was not observed in GSE57957 and GSE76427, while significantly high and low expressions of METTL14 in HCC were captured in GSE39791 and GSE64041 respectively (Figure 1A). Intriguingly, hepatic METTL14 expression was downregulated in cirrhotic patients compared with normal individuals (Figure 1B). Datasets containing a multistep carcinogenic process from healthy liver, to chronic hepatitis, to cirrhosis, to dysplastic nodule, and to HCC, could not corroborate the linear expression pattern of METTL14, nor its significant expression in HCC or normal liver tissues (Figure 1C,D). The survival analyses concerning overall survival showed no difference between high and low METTL14 expression groups (Figure S1A). As for recurrence-free survival, higher METTL14 expression implied a worse outcome in GSE76427 but not in TCGA (Figure S1B).
Figure 1.
METTL14 mRNA expression in human liver disease from microarray datasets. (A) Boxplots showing METTL14 mRNA expression in normal liver and HCC. *, P<0.05; ***, P<0.001, paired t-test; (B) METTL14 mRNA expression in normal liver and cirrhotic liver. *, P<0.05; **, P<0.01, unpaired t-test; (C,D) METTL14 mRNA expression in patients with various stages of liver disease leading to HCC. Statistical analysis was performed using one-way ANOVA comparing the mean of each column to normal liver and the posttest for the linear trend. n.s., no significance; T, tumor; N, normal; v., versus; CS, cirrhosis; DN, dysplastic nodules; HCC, hepatocellular carcinoma; CH, chronic hepatitis.
We ascribed the uncertainty of METTL14 expression pattern between HCC and normal samples to selection bias and different detection levels: Chen et al. (2) only detected the mRNA expression values of METTL14 while Ma et al. (1) measured the expression values of METTL14 at both mRNA and protein levels. However, the reduction of METTL14 mRNA in cirrhotic liver compared with normal liver suggests a destructive role of METTL14 in liver cirrhosis. Additionally, the association between METTL14 mRNA expression and recurrence-free survival indicates a tumor protective role of METTL14 in HCC. Contradiction between the impact of METTL14 on migration ability may be caused by the difference in cell lines, the versatility of methyltransferase-like 3 (METTL3)-METTL14 heterocomplex, and m6A-independent manner. For example, METTL3 directly associates with translation machinery and enhances the translation of target mRNAs independent of its methyltransferase activity (3). METTL14 is known to be primarily localized to the nucleus, thus experiments can be devised and completed at the nuclear and non-nuclear levels to ascertain the exact role of METTL14 in liver cancer. In sum, METTL14 may have a variety of roles as opposed to singularly being a tumor suppressor or promoter in hepatocarcinogenesis.
Figure S1.
Survival analysis using median expression value of METTL14 as the cut-off. (A) Correlation of METTL14 mRNA expression with overall survival in TCGA and GSE76427; (B) correlation of METTL14 mRNA expression with recurrence-free survival in TCGA and GSE76427.
Acknowledgments
None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6‐methyladenosine‐dependent primary MicroRNA processing. Hepatology 2017;65:529-43. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- 2.Chen M, Wei L, Law C, et al. RNA N6‐methyladenosine methyltransferase‐like 3 promotes liver cancer progression through YTHDF2‐dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. 10.1002/hep.29683 [DOI] [PubMed] [Google Scholar]
- 3.Lin S, Choe J, Du P, et al. The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Molecular Cell 2016;62:335-45. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]