Abstract
Background
Gallbladder cancer (GBC) is often diagnosed at an advanced stage with limited therapeutic options and poor prognosis. The five-year survival rate of this cancer when diagnosed at an advanced stage is below 5%, and the median survival time is less than a year with standard gemcitabine-based chemotherapy. Survival benefit with second-line treatment is unknown. Thus, there is an urgent need for novel treatment strategies and targeted therapy based on next generation sequencing (NGS) may be of value.
Methods
Comprehensive genomic profiling (CGP) was performed with NGS panel on paraffin-embedded tumors from a cohort of 108 Chinese and 107 US GBC patients. Clinical data were collected using an IRB approved protocol from a single-center in US and from China.
Results
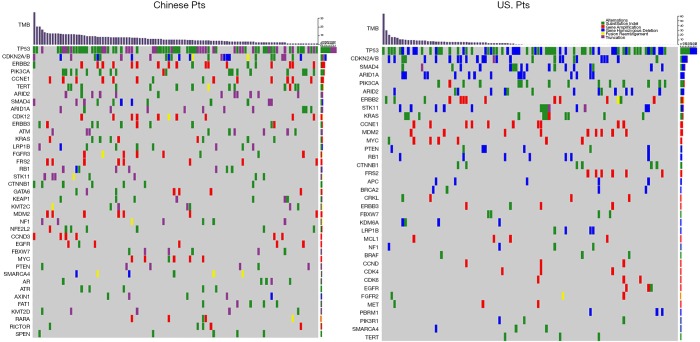
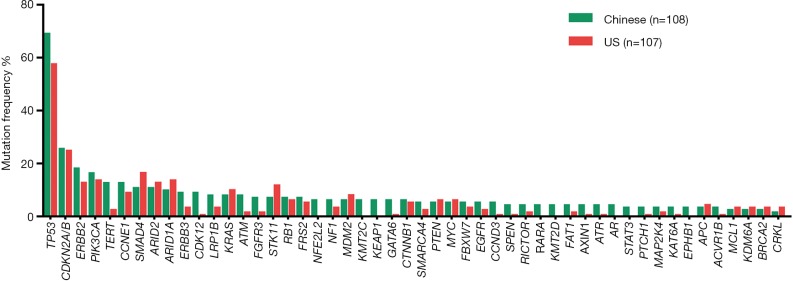
In Chinese and US GBC cohorts, an average of 6.4 vs. 3.8 genomic alterations (GAs) were identified per patient. The most frequent alterations were TP53 (69.4%), CDKN2A/B (26%), ERBB2 (18.5%), PIK3CA (17%) and CCNE1 (13%) in Chinese cohort, TP53 (57.9%), CDKN2A/B (25%), SMAD4 (17%), ARID1A (14%), PIK3CA (14%) and ERBB2 (13.1%) in US patients. NFE2L2 mutations were present in 6.5% of Chinese patients and not observed in the US cohort. Interestingly, ERBB2 genetic aberrations were significantly associated with better pathological tumor differentiation and tended to co-occurrence with CDKN2A/B mutations in both the Chinese and US GBC cases. Out of the top 9 dysregulated genetic pathways in cancer, Chinese patients harbored more frequent mutations in ERBB genes (30.6% vs. 19.0%, P=0.04). High frequency of PI3K/mTOR pathway variations was observed in both Chinese (37%) and US cohort (33%) (P=0.5). Additionally, both Chinese and US GBC patients exhibited a relatively high tumor mutational burden (TMB) (17.6% and 17.0%, respectively). In the Chinese cohort, a significant association was seen between direct repair gene alterations and TMB ≥10 muts/Mb (P=0.004).
Conclusions
In our study, over 83% Chinese and 68% US GBC patients had actionable alterations that could potentially guide and influence personalized treatment options. The identification of high TMB, ERBB2, CDKN2A/B, PI3K/mTOR pathway and DNA repair mutations indicated that both Chinese and US GBC patients may benefit from targeted or immune checkpoint inhibitors.
Keywords: Gallbladder cancer (GBC), comprehensive genomic profiling (CGP), ERBB2, CDKN2A/B, PI3K/mTOR
Introduction
Gallbladder cancer (GBC) is the most common malignancy of the biliary tract cancer. It often presents at an advanced, unresectable stage, and has an aggressive course with a 5-year survival rate of less than 5% (1). This disease is considered rare in the US, while it is more common in geographic pockets, including the West coast of Latin America, particularly Chile, Eastern Europe, Northern India and East Asia.
While, surgery can offer potential cure, GBC is often diagnosed at an advanced stage with limited treatment options. Systemic therapy comprising of gemcitabine and cisplatin is the first line therapy for locally advanced and metastatic and the benefit of second line treatment is unknown at this time (2,3). This is therefore an area of unmet need and novel targeted therapy or immunotherapy approaches are needed.
In recent times, next generation sequencing (NGS) has been incorporated in the management of various cancers. Prior studies have documented important differences in mutation spectra between the Asian and Western patients within the same cancer type. For example, the EGFR L858R mutation rate in Chinese non-small cell lung cancer population was higher (39%), while KRAS mutation rate was lower (11%) as compared with the Western patients (4). Also, RHPN2, GLI3 and MRC2 have higher mutation rates in the Chinese population (5). These results are of clinical value both for targeted therapy as well as to understand the underlying basis for clinical and epidemiologic differences. Data regarding genetic heterogeneity of GBC in different populations are very limited and need further investigation given the varied geographic distribution of this cancer.
Methods
Comprehensive genomic profiling (CGP)
Comprehensive profiling of genomic alterations (GAs) was performed for formalin-fixed, paraffin-embedded (FFPE) tissue samples obtained from 108 Chinese and 107 US GBC patients using NGS based cancer gene panel as described below.
In all cases, 4 µm thick paraffin sections were stained by hematoxylin and eosin (H&E), tumor cell content was assessed by the pathologist; 20% (tumor cellularity) were considered to be of acceptable quality. DNA extraction was performed, and 50–250 ng double-stranded DNA was interrupted ultrasonically.
Hybrid selection and sequencing
A custom hybridization capture panel including over 23,660 individually synthesized 5'-biotinylated DNA 120 bp oligonucleotides, and the library construction used the APA Hyper Prep Kit (KAPA Biosystems) were used, and the amount of sample DNA extracted for subsequent sequencing needed to be greater than 40 ng. Customized panel for approximately 2.6 Mb (7,029 exons of 450 cancer-associated genes and selected introns of 38 genes which commonly rearranged in cancer) was used. The captured libraries sequenced on an Illumina NextSeq 500 (sequencing mean coverage ≥700×). CGP was performed using the Yuansu assay (Origimed, China) Paired end sequencing (2×75 bp) followed the manufacturer’s protocol. For the US cohort, CGP was performed using the FoundationOne assay (Cambridge, USA), as previously reported (6,7). Following DNA extraction of ≥50 ng, hybridization capture of exons of 322 genes and introns of 31 genes that are commonly rearranged in cancer was performed. These libraries were sequenced with the mean coverage sequencing ≥700× (6). For the purpose of estimation of sequencing error rate, a PhiX spike-in was added as an external control to measure the percentage of reads with 0−4 mismatches, following the method described by Manley et al. (8). The gene list of the Yuansu (OrigiMed) and FoundationOne assay (Cambridge) between the two cohorts were presented in http://fp.amegroups.cn/cms/hbsn.2019.04.11-1.pdf.
Bioinformatics pipeline for single nucleotide variation (SNV)and short indels, long indel, copy number alternations (CNA) and gene rearrangement
Alignment of raw reads to the human genome reference sequence (hg19) was done with the Burrows-Wheeler Aligner (BWA, v0.6.2), followed by PCR duplicates removal using MarkDuplicates algorithm from Picard (version 1.47). Local realignment and base quality recalibration for SNV was performed using GATK (v3.1-1) and subsequently called by MUTECT (v1.7). Short insertion/deletions were calibrated for alignment using ABRA (v0.97) and then called by PINDEL (v0.2.5a8). Common single nucleotide polymorphism (SNPs) defined as those from dbSNP database (version 147).
For detection of long indel (deletion size 100 bp−3 kbp, insertions 25 bp−3 kbp), adaptor sequences were first removed and the resulted reads having more than 1% bases with quality score less than 15 were excluded, error bases were then corrected. All high-quality reads were collected and potential error was corrected for each base to achieve matched k-mer (k =33) with high confidence, the cleaned reads were then assembled into multiple Unitigs by FM-index algorithm (9), the resulted Unitigs were aligned to the hg19 reference genome and L-indels were inferred from breakpoint information. The matched normal reference DNA was needed for making this L-indel calls.
To identify CNA, aligned reads were first normalized by EXCAVATOR (version v2.2). Log2 ratio of the read depths between tumor tissue and matched normal blood was calculated. Segmentation was performed based on Bayesian information criterion (10). Tumor cellularity was estimated by allele frequencies of over 4,000 sequenced SNPs, following the method in ASCAT (11).
To validate gene rearrangement, paired-end reads with abnormal insert size of over 2,000 bp aligned to the same chromosome or aligned to different ones were collected and used as discordant reads. The group consisting of discordant reads with the distance less than 500 bp formed a cluster and paired clusters were obtained according to the pairing relationship. Consistent breakpoints from the paired-end discordant reads within a cluster were identified and the corresponding reads were further assembled by fermi-lite to establish potential rearrangement breakpoints.The candidates reads supporting gene rearrangement were genomic annotated.
Tumor mutational burden (TMB)
For Chinese cohort, TMB was estimated for each sample by counting its somatic mutations including coding SNVs and indels per megabase. Driver mutations and germline alterations in dbSNP database were not counted, following the method of Chalmers et al. (12). Innovatively, TMB of each sample were turned into adjusted panel TMB with a linear predictive model.
For US cohort, all base substitutions and indels in the coding region of targeted gene were counted (12), Alterations listed as known somatic alterations in COSMIC and truncations of tumor suppressor genes, germline mutation were not counted for TMB calculation (13,14).
Microsatellite instability (MSI)
MSI status was determined in all cases with detectable TMB status. According to the MSI score, we classified the samples as MSI high, defined as instability in 2 or more microsatellite loci; MSI low, defined as instability in only 1 locus; and microsatellite stable (MSS), defined as absence of any evidence of microsatellite loci instability. If the results for a sample were ambiguous, the analysis would be performed again. Polymerase chain reaction (PCR) validation confirms the diagnosis of MSI high.
Immunohistochemistry (IHC)
FFPE Sections (4 µm thick) were incubated with antibody against programmed death-ligand 1 (PD-L1) using 28-8 pharmDx assay and the Dako PD-L1 IHC 22C3 pharmDx assay IHC assays were performed on a Gene Stainer automated staining instrument according to the manufacturer’s instructions. IHC was carried out on FFPE sample using a well-established method (15). All immunohistochemical stained sections were independently analyzed by two pathologists, and the positive criteria used the standards method (16-18).
Statistical analysis
Alignment of raw reads to the human genome reference sequence (hg19) was done. Comparison between the Chinese and US cohort in all classes of GAs, TMB, MSI was done. χ2 or Fisher exact test was used to assess associations between clinical characteristics and high frequency mutation between the two cohorts. Statistical analysis was performed using IBM SPSS version 23.0 (Armonk, USA). P values greater than 0.05 were considered significant.
Results
Patient population
One hundred and eight Chinese and 107 US GBC patients were included. The Chinese cohort comprised of 70 females and 38 males with male-to-female ratio 0.5:1, median age at diagnosis was 61 years and most of them (92.6%) had adenocarcinoma. Other histologies were squamous, mixed adenosquamous, or neuroendocrine. The US GBC patients comprised of 69 females and 38 males with median age at diagnosis of 64 years. In this cohort, 98 (91.6%) had adenocarcinoma (Table 1).
Table 1. Patients’ demographics and clinical characteristics.
| Parameters | Chinese cohort, N=108 (%) | US cohort, N=107 (%) | P value |
|---|---|---|---|
| Age (years): median [range] | 61 [31−85] | 64 [38−86] | 0.5 |
| Gender | 1 | ||
| Female | 70 (64.8) | 69 (64.5) | |
| Male | 38 (35.2) | 38 (35.5) | |
| Subtype | 0.1 | ||
| Gallbladder adenocarcinoma | 100 (92.6) | 98 (91.6) | |
| Mixed adenocarcinoma | 2 (1.9) | 3 (2.8) | |
| Others | 6 (5.6) | 6 (5.6) | |
| Histological grade | 0.3 | ||
| Well/moderately differentiated | 38 (35.2) | 48 (44.9) | |
| Poorly/undifferentiated | 59 (54.6) | 51 (47.7) | |
| Not available | 11 (10.2) | 8 (7.5) |
Mutational landscape of GBC patients
Among Chinese patients, the most frequently altered genes were TP53 (69.4%), CDKN2A/B (26%), ERBB2 (18.5%), PIK3CA (17%) and CCNE1 (13%) with 83.3% of patients having at least one actionable genetic aberration. Actionable genetic aberration was based on databases such as oncokb (www.oncokb.org), cosmic (cancer.sanger.ac.uk/cosmic), clinicaltrials (cancer.sanger.ac.uk/cosmic/) and pubmed (www.ncbi.nlm.nih.gov/pubmed). While, in US patients the most commonly reported genetic aberrations were TP53 (57.9%), CDKN2A/B (25%), SMAD4 (17%), PIK3CA (14%) ARID1A (14%) and ERBB2 (13.1%) with 68.2% of patients having at least one actionable genetic aberration (Figures 1,2). Out of the most commonly dysregulated genetic pathways in cancer, genetic aberrations in ERBB pathway were more frequent in Chinese cohort as compared to US cohort (30.6% vs. 19.0%, P=0.04). Additionally, genetic aberrations in DNA repair pathway were more frequent in Chinese cohort, in particular ATM (8.3% vs. 1.9%, P=0.03) and CDK12 (9.3% vs. 0.9%, P=0.006). DNA repair gene mutation status for these cohorts is depicted in Table 2. Moreover, high frequency of PI3K/mTOR pathway genetic aberrations was noticed in both Chinese (37%) and US cohort (33%) (37% vs. 33%, P=0.5).
Figure 1.
Mutational landscape of GBC patients. Top panel indicates the distribution of TMB value in Chinese and US GBC patients. Bottom panel shows mutational landscape among the two cohorts. Mutations are colored according to mutation types of substitution/indel, gene amplification, gene homozygous deletion, truncation and fusion/rearrangement. Alteration types are presented in the legend at the top right. Pts, patients; TMB, tumor mutational burden; GBC, gallbladder cancer.
Figure 2.
The most common alterations in both cohorts, each gene is separately stated in China (108 patients) and US (107 patients).
Table 2. Direct repair genes genetic aberrations in Chinese and US gallbladder cancer patients.
| Category | Gene | Chinese cohort N=108 (%) | US cohort N=107 (%) | P value |
|---|---|---|---|---|
| Care taker | TP53 | 75 (69.4) | 62 (57.9) | 0.08 |
| CDK12 | 10 (9.3) | 1 (0.9) | 0.006 | |
| BAP1 | 3 (2.8) | 0 (0.0) | 0.2 | |
| Direct repair gene | ATM | 9 (8.3) | 2 (1.9) | 0.03 |
| ATR | 5 (4.6) | 1 (0.9) | 0.2 | |
| RAD50 | 3 (2.8) | 0 (0.0) | 0.2 | |
| BRCA2 | 3 (2.8) | 4 (3.7) | 1 | |
| BRCA1 | 3 (2.8) | 0 (0.0) | 0.2 | |
| PALB2 | 2 (1.9) | 1 (0.9) | 1 | |
| MSH2 | 2 (1.9) | 2 (1.9) | 0.6 | |
| PRKDC | 1 (0.9) | 0 (0.0) | NA | |
| POLE | 1 (0.9) | 0 (0.0) | NA | |
| POLD1 | 1 (0.9) | 0 (0.0) | NA | |
| MSH6 | 1 (0.9) | 1 (0.9) | NA | |
| FANCA | 1 (0.9) | 1 (0.9) | NA |
NA, not available.
TMB was analyzed in 108 Chinese patients and 53 US patients. Notably, both Chinese and US GBC patients exhibited high TMB ≥10 muts/Mb in 17.6% and 17.0%, respectively. While, in Chinese cohort the median TMB (range) was 5.4 muts/Mb (0.8–36.7), it was 5 mut/Mb (0–65) in US cohort. All samples in Chinese cohort were MSS. For US Cohort, microsatellite instability-high (MSI-H) accounted for 3.3% (1/30) of GBC, with average TMB of 30 muts/Mb.
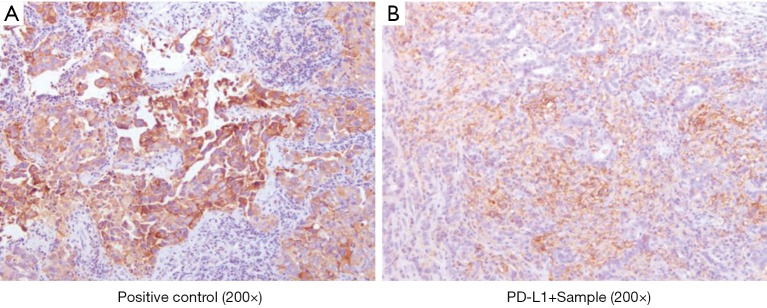
PD-L1 expression was assessed in 21 out of 108 Chinese patients with no available data about PD-L1 expression in US GBC patients. At cut off point 1%, 6/21 (29%) had PD-L1 overexpression; and at cut-off point 50%, 3/21 patients (14%) had PD-L1 tumor sample expression. Representative picture is shown in Figure 3.
Figure 3.
PD-L1 expression status of a Chinese GBC patient. PD-L1 expression was assessed in 21 out of 108 Chinese patients. (A) Positive control (200× magnification); (B) representative example of PD-L1 positive sample (200× magnification). GBC, gallbladder cancer; PD-L1, programmed death-ligand 1.
Actionable mutations
ErbB family mutation status
In our cohorts, mutations in ErbB family including EGFR, ERBB2, ERBB3, and ERBB4 were identified. However, ERBB4 mutation occurred only in Chinese patients (Table 3, Figure 4). Chinese patients harbored more frequent mutations in ERbB family (30.6% vs. 18.7%, P=0.04). ERBB2 variation was significantly associated with better tumor differentiation in both cohorts (Table 4). In our study, 5% (1/20) of ERBB2 mutated cases in Chinese cohort had coexisting KRAS mutation while this association was not observed in US patients. Interestingly, ERBB2 genetic aberrations were tended to co-occurrence with CDKN2A/B mutations in Chinese GBC cases [95% confidence interval (CI): 1.7−17.6, odds ratio 5.4, P=0.0014], and were strong tended to co-occurrence with CDKN2A/B variations in US GBC cases (95% CI: 2.7−53.2; odds ratio 10.8, P=0.0001). Eleven (55%) out of 20 Chinese patients with ERBB2 genetic aberrations had CDKN2A/B mutations whereas 10 (71.4%) out of 14 ERBB2 US patients had CDKN2A/B mutations (Table 5). For Chinese patients with ERBB2 genetic aberrations, 80.0% (16/20) patients had ERBB2 gene amplification, 20.0% (4/20) patients had substitution. In US cohort, 64.3% (9/14) patients with ERBB2 gene amplification, 28.6% (4/14) patients had ERBB2 gene substitution and 7.1% (1/14) patients had ERBB2 gene fusion/rearrangement. ERBB2 S310Y and S310F were the most common substitution types in both cohorts.
Table 3. ErbB family gene mutation frequency in Chinese and US gallbladder cancer patients.
| Gene variation | Chinese patients, N=108 (%) | US patients, N=107 (%) | P value |
|---|---|---|---|
| EGFR | 6 (5.6) | 3 (2.8) | 0.5 |
| ERBB2 | 20 (18.5) | 14 (13.1) | 0.4 |
| Amplification | 16/20 (80.0) | 9/14 (64.3) | |
| Mutation | 4/20 (20.0) | 4/14 (28.6) | |
| Fusion/rearrangement | 0 (0.0) | 1/14 (7.1) | |
| ERBB3 | 10 (9.3) | 4 (3.7) | 0.1 |
| ERBB4 | 1 (0.9) | 0 (0.0) | 1 |
| ErbB family | 33 (30.6) | 20 (18.7) | 0.04 |
Figure 4.
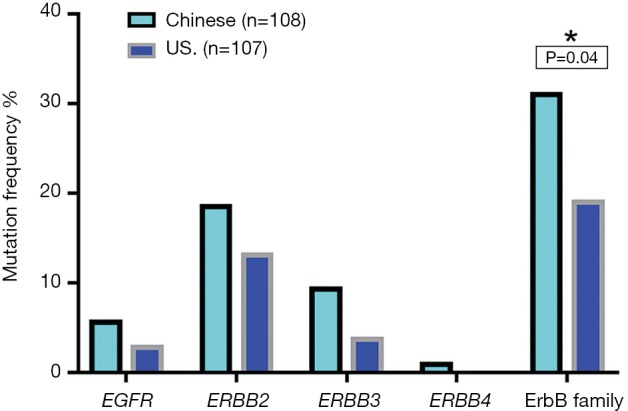
Chinese patients harbored more frequent mutations in ErbB family. Mutations in ErbB family including EGFR, ERBB2, ERBB3, and ERBB4 has been identified except for ERBB4 which was found only in Chinese patients, Chinese patients harbored more frequent mutations in ErbB family (30.6% vs. 19.0%, P=0.04).
Table 4. Correlation between ERBB2 and histological grade.
| Cohort | ERBB2 | Poorly/undifferentiated (%) | Well/moderately differentiated (%) | P value |
|---|---|---|---|---|
| Chinese cohort | MT* | 6 (33.3) | 12 (66.7) | 0.008 |
| WT** | 53 (67.1) | 26 (32.9) | ||
| US cohort | MT*** | 3 (23.1) | 10 (76.9) | 0.03 |
| WT**** | 25 (58.1) | 18 (41.9) |
*, data is not available in 2 patients; **, data is not available in 9 patients; ***, data is not available in 1 patient; ****, data is not available in 50 patients. MT, mutant type; WT, wild type.
Table 5. Correlation between ERBB2 and CDKN2A/B mutation status.
| Cohort | ERBB2 | CDKN2A/B MT (%) | CDKN2A/B WT (%) | Odds ratio | P value |
|---|---|---|---|---|---|
| Chinese cohort | MT* | 11 (55.0) | 9 (45.0) | 5.4 | 0.0014 |
| WT** | 16 (18.2) | 72 (81.8) | |||
| US cohort | MT*** | 10 (71.4) | 4 (28.6) | 10.8 | 0.0001 |
| WT**** | 17 (18.3) | 76 (81.7) |
*, data is not available in 2 patients; **, data is not available in 9 patients; ***, data is not available in 1 patient; ****, data is not available in 50 patients. NA, not available; MT, mutant type; WT, wild type.
DNA repair pathway
DNA repair genetic aberrations were classified into direct, which include ATM, ATR, BRCA1, BRCA2, FANCA, FANCD2, MLH1, MSH2, MSH6, PALB2, POLD1, POLE, PRKDC, RAD50, and SLX4 and caretaker genes comprising of BAP1, CDK12, MLL3, TP53, BLM. Significant association was seen between direct repair genetic aberrations and TMB ≥10 in Chinese cohort (P=0.004).
PI3K/mTOR pathway
High frequency PI3K/mTOR pathway variation was observed in both Chinese (37%) and US cohort (33%) (P=0.5), including PIK3CA, FBXW7, TSC1/2, STK11, and PTEN (Figure 5). There was no significant difference in the variation of these genes between two cohorts.
Figure 5.
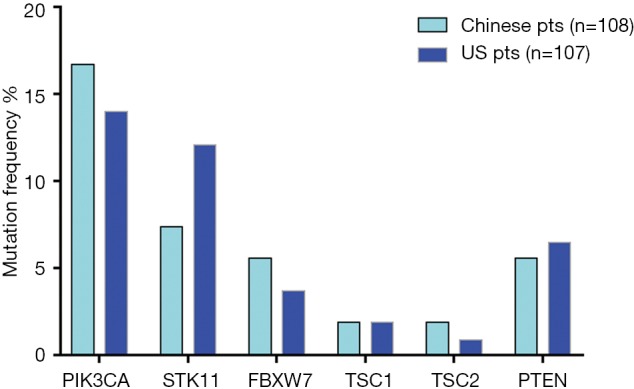
PI3K/mTOR pathway gene mutation status in Chinese and US cohorts.
Discussion
To successful treatment of GBC, the important strategy is to understand the pathogenesis of it. Gallstones, gallbladder polyps, chronic cholecystitis and primary sclerosing cholangitis are etiologic factors in the western countries while chronic Salmonella and H. Pylori infections predispose to this cancer in the developing world (19-21). Additionally, although only a small proportion of patients with gallstone disease develop GBC, the presence of gallstones has been consistently linked with increased risk of GBC (22). Molecular alterations playing a role in the pathogenesis of GBC: from chronic inflammation (TP53); hyperplasia, dysplasia, in situ carcinoma (loss of p16/gain of CDK4/cyclin D1); invasive adenocarcinoma and finally metastatic disease (pERK1/2, Hedehog pathway/VHL overexpression) (23). In our study, genetic aberrations: TP53, (69.4% vs. 57.9%), CDKN2A/B (26% vs. 25%), ERBB2 (18.5% vs. 13.1%) PIK3CA (17% vs. 14%) was observed in Chines and US cohort respectively, The evidence available is supporting the involvement of some genetic aberrations in the development of GBC.
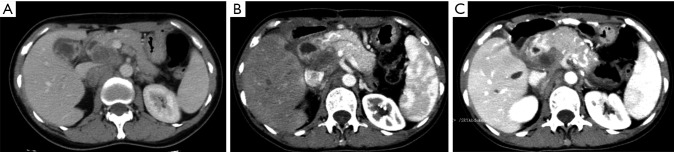
Our study examines the somatic genomic landscape of GBCs in Asia and the West and has made several interesting observations. Both Chinese and US patients with GBC have a relatively high TMB. Genetic aberrations in ERBB, PI3K/mTOR and DNA repair pathway occur in both Chinese and US GBC patients and targeted therapies or immune checkpoint inhibitors are worthy of further investigation in this cancer. Currently, none of genomically matched targeted therapy or immune checkpoint inhibitors are FDA-approved for treatment of GBCs.ERBB2 genetic aberration was one of the most common actionable genetic aberrations in both the Chinese and US cohort. In our study, ERBB2 genetic aberrations account for 18.5% in Chinese patients and 13.1% in US patients, and ERBB2 gene amplification was the most common variation type. ERBB2 genetic aberrations, particularly amplification may represent an important opportunity for targeted therapy. The use of HER2/neu-directed therapy including trastuzumab, lapatinib, pertuzumab and ado-trastuzumab emtansine have been under investigation. In a retrospective study of eight GBC patients with ERBB2 gene amplification treated with trastuzumab, lapatinib, or pertuzumab, over 50% of patients achieved partial response or complete response (24). Similar results were reported when ERBB2-directed therapy was combined with taxanes (25). In our Chinese cohort, a 45-year-old female patient diagnosed with locally advanced T4N2M0 GBC underwent cholecystectomy. CGP showed ERBB2 amplification, CDK12 amplification, ARID2 exon 4-exon 18 deletion (108 kb), ARID2 c.5148-2A>G (splicing site mutation), TP53 M237I, and TMB 4.6 muts/Mb. Post-operative computerized tomography showed multiple enlarged hepatic hilar and retroperitoneal lymph node. Patient was treated with trastuzumab plus nab-paclitaxel and remained stable for 5 months (Figure 6). Additionally, we observed that ERBB2 alteration is positively correlated with differentiation both in Chinese and US cohort. Similarly, Yoshikawa et al. reported a higher incidence of ERBB2 gene amplification in well differentiated cholangiocarcinoma (26). ERBB2 acts as an oncogene in several human cancers (24,27). Overexpression of ErbB-2 in the basal layer of biliary tract epithelium led to the development of gallbladder adenocarcinoma in 100% of transgenic mice by 3 months of age (28). This suggests that ERBB2 overexpression can potentially drive gallbladder carcinogenesis.
Figure 6.
A 45-year-old Chinese T4N2M0 gallbladder papillary adenocarcinoma patient. CT images demonstrate (A) before cholecystectomy; (B) after 1.5 months of trastuzumab; (C) after 3 months (3 cycles) of trastuzumab with nab-paclitaxel.
KRAS genetic aberrations have been previously reported in 3−30% of GBC patients (29). Previous studies identified coexisting KRAS mutations with ERBB3 genetic aberrations in primary GBC tumor tissue and GBC cell lines (30). In our study, we observed concurrent KRAS G12A mutation with ERBB2 in 5% of Chinese GBC patients but not in US cohort. The presence of KRAS mutation may be a negative predictive factor for HER2-directed therapy (30,31). The current study is the first to report co-occurrence between ERBB2 and CDKN2A/B genetic aberration. Based on The cBioPortal for Cancer Genomics (32) (http://cbioportal.org), 2< odds ratio <10 indicated the tendency toward co-occurrence, strong tendency towards co-occurrence (Odds Ratio >10). ERBB2 genetic aberrations were tended to co-occurrence with CDKN2A/B mutations in Chinese GBC cohort (odds ratio 5.4, P=0.0014), and were strong tended to co-occurrence with CDKN2A/B variations in US GBC cohort (odds ratio 10.8, P=0.0001). On another hand, CDKN2A/2B alterations were significantly associated with distant metastases in our study. The biological interrelationships between these mutations need further study.
In our study, high tumor mutational burden (TMB-H) was defined as TMB ≥10, based on the recently published results of the CheckMate 568 that showed 44% overall response rate in patients with non-small cell lung cancer treated with nivolumab and ipilimumab regardless of the PD-1 expression. We identified that 17.6% of the Chinese GBC patients and 17.0% of US GBC patients have TMB-H. Additionally, a significant association between direct DNA repair GAs and TMB-H has been observed in Chinese patients. This emphasizes the importance of using PD-1 inhibitors alone and combined with PARP inhibitors in this subpopulation.
Our study has some important limitations. We have used two different NGS platforms that have two distinct targeted gene panels. However, in the analysis we included 320 overlapping genes (http://fp.amegroups.cn/cms/hbsn.2019.04.11-1.pdf). Furthermore, due to lack of epidemiological and clinicopathological data, we did not assess genomic profiling variation in regards to disease risk factors and staging. The data regarding PD-L1 expression was only available in 19% of Chinese patients, PD-L1 data was not available in US cohort. Therefore, we could not identify variations in immune biomarker expression between the two cohorts in this study.
In conclusion, GBC is highly mutated cancer with over 83% Chinese and 68% US gallbladder patients had at least 1 actionable alteration with high ERBB2 mutation rates and high TMB. We identified some actionable alterations between Chinese and US gallbladder patients, some of which may have therapeutic implications. The findings presented here highlight the need for prospective genomic characterization of patients with GBC, and enrich drug target in the design of future clinical trials, development of model systems, and the assessment of patient outcomes. Future clinical trials are also required to gauge the efficacy and safety of targeted and immunotherapy in this cancer.
Acknowledgments
None.
Ethical Statement: The study was approved by Medical ethics committee, Hunan Provincial People’s Hospital/The First Affiliated Hospital of Hunan Normal University (IEC number: 2017-15).
Footnotes
Conflicts of Interest: F Pang, C Zou, S Mu, Y Xing and K Wang are employees of Origimed. The other authors have no conflicts of interest to declare.
References
- 1.Rakić M, Patrlj L, Kopljar M, et al. Gallbladder cancer. Hepatobiliary Surg Nutr 2014;3:221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kabbach G, Assi HA, Bolotin G, et al. Hepatobiliary Tumors: Update on Diagnosis and Management. J Clin Transl Hepatol 2015;3:169-81. 10.14218/JCTH.2015.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamarca A, Barriuso J, McNamara MG, et al. Biliary Tract Cancer: State of the Art and potential role of DNA Damage Repair. Cancer Treat Rev 2018;70:168-77. 10.1016/j.ctrv.2018.09.002 [DOI] [PubMed] [Google Scholar]
- 4.Wu K, Zhang X, Li F, et al. Frequent alterations in cytoskeleton remodelling genes in primary and metastatic lung adenocarcinomas. Nat Commun 2015;6:10131. 10.1038/ncomms10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He J, Abdel-Wahab O, Nahas MK, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 2016;127:3004-14. 10.1182/blood-2015-08-664649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manley LJ, Ma D, Levine SS. Monitoring Error Rates In Illumina Sequencing. J Biomol Tech 2016;27:125-8. 10.7171/jbt.16-2704-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H. Fast construction of FM-index for long sequence reads. Bioinformatics 2014;30:3274-5. 10.1093/bioinformatics/btu541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xi R, Lee S, Xia Y, et al. Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants. Nucleic Acids Res 2016;44:6274-86. 10.1093/nar/gkw491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Loo P, Nordgard SH, Lingjaerde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA 2010;107:16910-5. 10.1073/pnas.1009843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 2004;91:355-8. 10.1038/sj.bjc.6601894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285-91. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCourt CM, Boyle D, James J, et al. Immunohistochemistry in the era of personalised medicine. J Clin Pathol 2013;66:58-61. 10.1136/jclinpath-2012-201140 [DOI] [PubMed] [Google Scholar]
- 16.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 17.Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol 2011;6:466-72. 10.1097/JTO.0b013e31820b82e8 [DOI] [PubMed] [Google Scholar]
- 18.Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 2014;138:241-56. 10.5858/arpa.2013-0953-SA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma A, Sharma KL, Gupta A, et al. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol 2017;23:3978-98. 10.3748/wjg.v23.i22.3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLeon TT, Ahn DH, Bogenberger JM, et al. Novel targeted therapy strategies for biliary tract cancers and hepatocellular carcinoma. Future Oncol 2018;14:553-66. 10.2217/fon-2017-0451 [DOI] [PubMed] [Google Scholar]
- 21.Wistuba II, Gazdar AF. Gallbladder cancer: lessons from a rare tumour. Nat Rev Cancer 2004;4:695-706. 10.1038/nrc1429 [DOI] [PubMed] [Google Scholar]
- 22.Mhatre S, Wang Z, Nagrani R, et al. Common genetic variation and risk of gallbladder cancer in India: a case-control genome-wide association study. Lancet Oncol 2017;18:535-44. 10.1016/S1470-2045(17)30167-5 [DOI] [PubMed] [Google Scholar]
- 23.Barreto SG, Dutt A, Chaudhary A. A genetic model for gallbladder carcinogenesis and its dissemination. Ann Oncol 2014;25:1086-97. 10.1093/annonc/mdu006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javle M, Churi C, Kang HC, et al. HER2/neu-directed therapy for biliary tract cancer. J Hematol Oncol 2015;8:58. 10.1186/s13045-015-0155-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czink E, Heining C, Weber TF, et al. Durable remission under dual HER2 blockade with Trastuzumab and Pertuzumab in a patient with metastatic gallbladder cancer. Z Gastroenterol 2016;54:426-30. 10.1055/s-0042-103498 [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa D, Ojima H, Iwasaki M, et al. Clinicopathological and prognostic significance of EGFR, VEGF, and HER2 expression in cholangiocarcinoma. Br J Cancer 2008;98:418-25. 10.1038/sj.bjc.6604129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutierrez C, Schiff R. HER2: biology, detection, and clinical implications. Arch Pathol Lab Med 2011;135:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiguchi K, Carbajal S, Chan K, et al. Constitutive expression of ErbB-2 in gallbladder epithelium results in development of adenocarcinoma. Cancer Res 2001;61:6971-6. [PubMed] [Google Scholar]
- 29.Müller BG, De Aretxabala X, González Domingo M. A review of recent data in the treatment of gallbladder cancer: what we know, what we do, and what should be done. Am Soc Clin Oncol Educ Book 2014:e165-70. 10.14694/EdBook_AM.2014.34.e165 [DOI] [PubMed] [Google Scholar]
- 30.Iyer P, Shrikhande SV, Ranjan M, et al. ERBB2 and KRAS alterations mediate response to EGFR inhibitors in early stage gallbladder cancer. Int J Cancer 2019;144:2008-19. 10.1002/ijc.31916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A phase 2b of eryaspase in combination with gemcitabine or FOLFOX as second-line therapy in patients with metastatic pancreatic adenocarcinoma (NCT02195180). Ann Oncol 2017;28:v209-68. [Google Scholar]
- 32.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]