Abstract
MetaCyc (MetaCyc.org) is a comprehensive reference database of metabolic pathways and enzymes from all domains of life. It contains 2749 pathways derived from more than 60 000 publications, making it the largest curated collection of metabolic pathways. The data in MetaCyc are evidence-based and richly curated, resulting in an encyclopedic reference tool for metabolism. MetaCyc is also used as a knowledge base for generating thousands of organism-specific Pathway/Genome Databases (PGDBs), which are available in BioCyc.org and other genomic portals. This article provides an update on the developments in MetaCyc during September 2017 to August 2019, up to version 23.1. Some of the topics that received intensive curation during this period include cobamides biosynthesis, sterol metabolism, fatty acid biosynthesis, lipid metabolism, carotenoid metabolism, protein glycosylation, antibiotics and cytotoxins biosynthesis, siderophore biosynthesis, bioluminescence, vitamin K metabolism, brominated compound metabolism, plant secondary metabolism and human metabolism. Other additions include modifications to the GlycanBuilder software that enable displaying glycans using symbolic representation, improved graphics and fonts for web displays, improvements in the PathoLogic component of Pathway Tools, and the optional addition of regulatory information to pathway diagrams.
INTRODUCTION
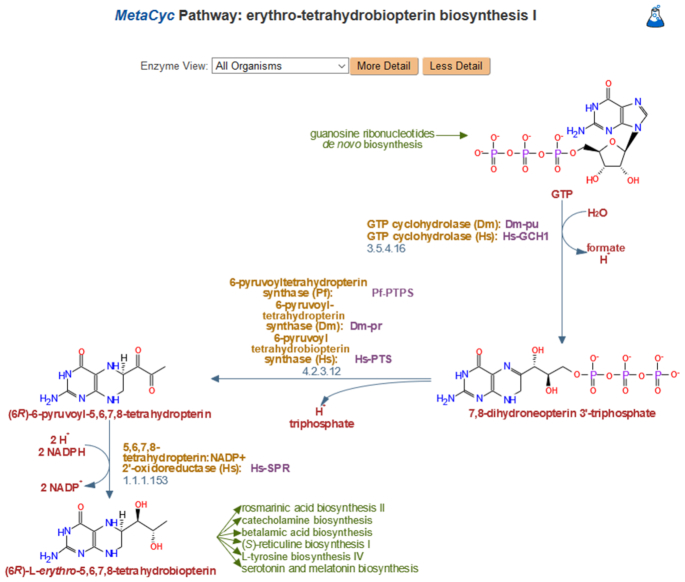
MetaCyc (MetaCyc.org) (pronounced ‘met-a-sike’, as in ‘encyclopedia’) is a highly curated reference database of metabolism from all domains of life that has been curated for over 20 years (1). It contains data about chemical compounds, reactions, enzymes and metabolic pathways that have been experimentally validated and reported in the scientific literature (2), and cover both small molecule metabolism and macromolecular metabolism (e.g. protein modification). Figure 1 shows an example of a MetaCyc pathway diagram. Due to its exclusively experimentally determined data, intensive curation, extensive referencing, and user-friendly and highly integrated interface, MetaCyc is commonly used in various fields, including genome annotation, biochemistry, enzymology, metabolomics, genome and metagenome analysis, and metabolic engineering.
Figure 1.
A typical MetaCyc pathway. A short pathway was selected for this figure; the average number of metabolites in a MetaCyc pathway is 13, with the largest pathway containing 244 metabolites. The green captions are links to the upstream and downstream pathways that produce the inputs and outputs for this pathway. Enzymes are labeled in ochre, genes are labeled in purple and compounds are labeled in red. When available, EC numbers are shown next to reactions. Each item in a pathway diagram is a clickable hyperlink to a dedicated page for that item. Below each pathway is a summary and additional information such as the pathway’s taxonomic range, some taxa known to possess it, the pathway’s position within the pathway ontology, and potential pathway variants (not shown).
In addition to its role as a general reference on metabolism, MetaCyc is used by the PathoLogic component of the Pathway Tools software (3,4) as a reference database, enabling the computational prediction of the metabolic reactions and pathways of any organism that has a sequenced and annotated genome (5). During this mostly automated process, the predicted metabolic network is captured in the form of a Pathway/Genome Database (PGDB). Pathway Tools also provides browsing and searching capabilities to explore the databases as well as editing tools for improving and updating the computationally generated PGDBs by manual curation. SRI has used MetaCyc to create 14 730 PGDBs (as of August 2019), which are available through the BioCyc (BioCyc.org) website (6). In addition, many groups outside SRI have generated thousands of additional PGDBs (7–11). Interested scientists may adopt any of the SRI PGDBs through the BioCyc website for further curation (https://biocyc.org/adopted.shtml).
EXPANSION OF MetaCyc DATA
Since the 2018 Nucleic Acids Research publication (2), we added 184 new base pathways (pathways composed of reactions only, where no portion of the pathway is designated as a subpathway) and 5 superpathways (pathways composed of at least one base pathway plus additional reactions or pathways), and updated 109 existing pathways, for a total of 298 new and revised pathways. The total number of base pathways grew by 7%, from 2572 (version 21.1) to 2749 (version 23.1) (the total increase is slightly <184 pathways, because some existing pathways were deleted from the database during this period). We have also added 910 new enzymes (an increase of 8%), 1687 reactions (12%) and 1806 chemical compounds (13%). The number of referenced organisms increased by 6% to 3045. The data in MetaCyc have been gathered from 60 935 scientific publications. Table 1 lists species with >20 experimentally elucidated pathways in MetaCyc, and Table 2 shows the taxonomic distribution of all MetaCyc pathways.
Table 1.
List of species with 20 or more experimentally elucidated pathways represented in MetaCyc (meaning experimental evidence exists for the occurrence of these pathways in the organism)
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Escherichia coli | 358 | Arabidopsis thaliana | 345 | Methanocaldococcus jannaschii | 31 |
| Pseudomonas aeruginosa | 80 | Homo sapiens | 318 | Methanosarcina barkeri | 25 |
| Bacillus subtilis | 64 | Saccharomyces cerevisiae | 204 | Sulfolobus solfataricus | 24 |
| Pseudomonas putida | 51 | Rattus norvegicus | 88 | Methanosarcina thermophila | 20 |
| Mycobacterium tuberculosis | 49 | Solanum lycopersicum | 67 | ||
| Salmonella Typhimurium | 48 | Glycine max | 65 | ||
| Synechocystis sp. PCC 6803 | 36 | Nicotiana tabacum | 58 | ||
| Pseudomonas fluorescens | 33 | Mus musculus | 56 | ||
| Klebsiella pneumoniae | 27 | Pisum sativum | 52 | ||
| Enterobacter aerogenes | 26 | Zea mays | 51 | ||
| Agrobacterium tumefaciens | 25 | Oryza sativa | 47 | ||
| Mycolicibacterium smegmatis | 23 | Solanum tuberosum | 47 | ||
| Corynebacterium glutamicum | 21 | Catharanthus roseus | 30 | ||
| Staphylococcus aureus | 21 | Spinacia oleraca | 29 | ||
| Hordeum vulgare | 28 | ||||
| Triticum aestivum | 26 | ||||
| Bos taurus | 24 | ||||
| Petunia x hybrida | 21 | ||||
| Sus scrofa | 20 | ||||
| Chlamydomonas reinhardtii | 20 |
The species are grouped by taxonomic domain and are ordered within each domain based on the number of pathways (number following species name) to which the given species was assigned.
Table 2.
The distribution of pathways in MetaCyc based on the taxonomic classification of associated species
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Proteobacteria | 1231 | Viridiplantae | 1021 | Thermoprotei | 50 |
| Actinobacteria | 412 | Fungi | 459 | Methanomicrobia | 50 |
| Firmicutes | 393 | Metazoa | 424 | Methanococci | 41 |
| Cyanobacteria | 101 | Euglenozoa | 28 | Halobacteria | 40 |
| Bacteroidetes/Chlorobi | 92 | Alveolata | 23 | Thermococci | 40 |
| Deinococcus–Thermus | 31 | Stramenopiles | 14 | Methanobacteria | 37 |
| Thermotogae | 26 | Amoebozoa | 11 | Archaeoglobi | 16 |
| Tenericutes | 18 | Rhodophyta | 9 | Thermoplasmata | 9 |
| Aquificae | 18 | Haptophyceae | 6 | Methanopyri | 4 |
| Spirochaetes | 16 | Fornicata | 4 | ||
| Chlamydiae–Verrucomicrobia | 9 | Parabasalia | 4 | ||
| Chloroflexi | 8 | ||||
| Planctomycetes | 8 | ||||
| Fusobacteria | 7 | ||||
| Nitrospirae | 2 | ||||
| Thermodesulfobacteria | 2 | ||||
| Chrysiogenetes | 1 | ||||
| Nitrospinae | 1 |
For example, the statement ‘Tenericutes 18’ means that experimental evidence exists for the occurrence of at least 18 MetaCyc pathways in members of this taxonomic group. Major taxonomic groups are grouped by domain and are ordered within each domain based on the number of pathways (number following taxon name) associated with the taxon. A pathway may be associated with multiple organisms.
Expansion of pathways
This section provides a partial description of pathways that were added or significantly revised over the past 2 years, illustrating the breadth of topics that have been covered during this time. When data are available, MetaCyc pathways include curated representative enzymes for each enzymatic step.
Cobamides biosynthesis
The cobamides are complex cobalt-containing cofactors that are required for the catalytic activity of many enzymes. The cobalt ion in cobamides is attached to the rest of the molecules by six coordination bonds: four planar and two located above and below the plane. The four planar coordination sites are provided by a corrin ring, while the other two are provided by upper (Coβ) and lower (Coα) ligands. Most readers are likely familiar with adenosylcobalamin, the cofactor used by eukaryotic organisms in which the two ligands are 5′-deoxyadenosyl and 5,6-dimethylbenzimidazole, respectively. However, most bacteria utilize different forms of cobamides in which the upper ligand is often a methyl group and the lower ligand is one of 16 different compounds. We have added a large number of pathways that describe the biosynthesis of these bacterial cobamides, rearranged our current cobamide biosynthesis pathways, and added a few new cobamide salvage pathways.
Sterol metabolism
Sterols such as ergosterol, sitosterol and cholesterol are isoprenoid-derived molecules that have essential functions in eukaryotes. Some bacterial species also produce sterols, although the function of the sterols in these organisms is still not well understood. We added a new pathway describing sterol biosynthesis by a group of aerobic methylotrophs and pathways describing the biosynthesis of cycloartenol, a precursor for all the steroids produced by plants, and of parkeol, a rare isomer of lanosterol that is produced by plants and some bacteria. We also revised the plant cholesterol biosynthesis pathway.
Fatty acid biosynthesis
Of the two basic types of fatty acid biosynthesis systems, type I systems are found in lower eukaryotes and animals, while type II systems are found in bacteria, plants, parasites of the Apicomplexa phylum and mitochondria. Thus, both systems operate in animals—the type I system in the cytosol and the type II system in the mitochondria. We added new pathways that describe fatty acid biosynthesis initiation in the cytosol and the mitochondria, respectively. While the type I system continues to extend the fatty acids to palmitate and stearate, the type II system in mitochondria mostly forms octanoate (for synthesis of (R)-lipoate) and possibly myristate. We added pathways to MetaCyc to describe both processes. Very-long-chain fatty acids (VLCFAs), components of eukaryotic cells, are involved in many different physiological functions in different organisms. VLCFAs are synthesized in eukaryotes in the endoplasmic reticulum (ER) by a membrane-bound enzyme complex known as the elongase. We have revised significantly the pathway describing this process. We also added a pathway that describes the biosynthesis of ultra-long-chain fatty acids (ULCFAs), which are fatty acids with aliphatic tails of 28 carbons or longer. ULCFAs are common in acylceramides, which are epidermis-specific ceramide species consisting of a ceramide ester linked to linoleate. Acylceramides are very important for skin barrier formation.
Lipid metabolism
Sphingolipids are a class of lipids derived from the unsaturated aliphatic amino alcohol sphingosine. They are essential components of the plasma membrane in all eukaryotic cells. Sphingolipids concentrate with sterols (cholesterol in animals and ergosterol in yeast) to form lipid rafts, specialized membrane microdomains implicated in a variety of cellular processes, including sorting of membrane proteins and lipids, as well as organizing and regulating signaling cascades. We have updated our yeast sphingolipid biosynthesis pathway to incorporate newly characterized enzymes. We added a pathway that describes the biosynthesis of acylceramides (described above), which are some of the most hydrophobic lipids in mammalian bodies, and a pathway for the biosynthesis of glycine lipids, acylated amino acid lipids that are found in bacteria. We also added several pathways describing the biosynthesis of (Kdo)2-lipid A in Pseudomonas putida.
Carotenoid metabolism
Carotenoids are isoprenoid pigments in the yellow to red color range. They are commonly produced by photosynthetic organisms, but also by some non-photosynthetic fungi and bacteria. In photosynthetic organisms, carotenoids participate in light harvesting and provide protection against photooxidative stress through energy-dissipation of excess light absorbed by the antenna pigments. In non-photosynthetic organisms, carotenoids serve multiple roles as antioxidants, virulence factors and modulators of membrane function. Animals, including humans, cannot synthesize carotenoids, but require them for the synthesis of retinoids and vitamin A. In September 2017, we reorganized and updated our coverage of carotenoid biosynthesis by adding all pathways known at the time. MetaCyc currently contains two pathways for the biosynthesis of C30 carotenoids, 37 pathways for the biosynthesis of C40 carotenoids and 4 pathways for the biosynthesis of C50 carotenoids.
Protein glycosylation
N-linked glycosylation is an important protein post-translational modification in eukaryotes and archaea, and very rarely, in bacteria. During this process, certain oligosaccharides are attached to an L-asparagine residue in the polypeptide chain of target proteins. Oligosaccharide composition and structure vary greatly among kingdoms and to a lesser degree among species. N-linked glycosylation in eukaryotes consists of three stages: the initial synthesis of a high-mannose, dolichol phosphate-linked precursor tetradecasaccharide and its transfer from the dolichol phosphate anchor to a newly synthesized polypeptide; the trimming (processing) of these high-mannose structures by α-glucosidases and α-mannosidases, which occurs in the ER and Golgi complex; and the synthesis of complex branched oligosaccharide chains, carried out by Golgi glycosyltransferases. In the past, MetaCyc contained a pathway describing only the first stage. During our November 2017 release, we added pathways describing all stages of the process in multiple kingdoms and revised an existing archaeal pathway. MetaCyc now contains N-glycosylation pathways for bacteria, yeast, plants, vertebrates and the archaeons Haloferax volcanii and Methanococcus voltae. In addition, we added pathways that describe the process of protein O-mannosylation in mammals and yeast. All of these pathways use the symbol nomenclature for graphical representations of glycans (12), which makes them much easier to understand and uses the GlycoCT format for the import/export of such structures (13).
Antibiotics and cytotoxins biosynthesis
Since the release of version 21.1, we added or significantly revised pathways for the biosynthesis of the antibiotics mupirocin, fosfomycin, chloramphenicol, aureothin, spectinabilin, rifamycin B, 3-[(E)-2-isocyanoethenyl]-1H-indole, and the ansamycin family members ansatrienin, chaxamycin, mitomycin, naphthomycin, saliniketal A and streptovaricin. We also added pathways for the biosynthesis of the cytotoxins sodorifen, rhabduscin, bryostatin, cylindrospermopsin, tabtoxinine-β-lactam (wild fire toxin), toxoflavin, ginkgotoxin and pederin (the latter is one of the most potent non-proteinaceous substances ever isolated).
Siderophore biosynthesis
Iron is an essential trace element. In the presence of oxygen, ferrous iron is rapidly oxidized to ferric iron, which tends to form insoluble compounds and becomes unavailable to organisms. As a result, the level of physiologically available iron can drop far below 1 μM, making it a growth-limiting nutrient. To survive, many bacteria and fungi evolved specialized transport systems called siderophores, which are low molecular mass compounds that complex and retract iron ions. MetaCyc now contains pathways for the biosynthesis of 28 different siderophores, namely 2′-deoxymugineic acid, achromobactin, acinetobactin, acinetoferrin, aerobactin, alcaligin, anguibactin, bacillibactin, baumannoferrin, bisucaberin, desferrioxamine B, desferrioxamine E, enterobactin, ferrichrome, ferrichrome A, hydroxylated mugineic acid, petrobactin, pseudomonine, putrebactin, pyochelin, pyoverdine I, rhizobactin 1021, salmochelin, staphyloferrin A, staphyloferrin B, vanchrobactin, vibriobactin and yersiniabactin.
Bioluminescence
Throughout evolution, bioluminescence has been reinvented many times; some 30 different independent systems are still extant. The responsible enzymes are unrelated in bacteria, unicellular algae, coelenterates, beetles, fishes and other organisms, although all systems involve exergonic reactions of molecular oxygen with different substrates (luciferins) and enzymes (luciferases), resulting in photons of visible light. MetaCyc now contains pathways that describe bioluminescence in bacteria, fungi, dinoflagellates, corals, jellyfish, squid and fireflies.
Vitamin K metabolism
Vitamin K-dependent proteins are modified in metazoans by carboxylation of clusters of glutamate residues to carboxylated glutamate as they transit through the ER. The carboxylation is catalyzed by EC 4.1.1.90, peptidyl-glutamate 4-carboxylase, an enzyme that uses various vitamin-K hydroquinones, including menaquinol, as co-substrates that are epoxidated during the reaction, generating vitamin K 2,3-epoxide. This post-translational protein modification is the only firmly established biochemical function of vitamin K in metazoa. MetaCyc now contains pathways that describe the biosynthesis of several forms of the vitamin as well as the recycling of the epoxide back to the quinol active form.
Brominated compound metabolism
Marine organisms, including bacteria, algae and invertebrates, produce many brominated compounds. In some marine sponges within the Dysideidae family, polybrominated diphenyl ethers can exceed 10% of the sponge tissue by dry weight. In many cases, the origin of the compounds was shown to be symbiotic bacteria. We have added several pathways that describe bacterial biosynthesis of organobromine compounds.
Degradation pathways
We have added or significantly revised pathways that describe the bacterial degradation of the plant hormone indole-3-acetate, the siderophore salmochelin, cyanuric acid (a common intermediate in the degradation of hundreds of s-triazine compounds), levulinate, N-methylpyrrolidone, pinoresinol, ellagic acid, thiocyanate, rubber, nylon-6 oligomer, (S)-propane-1,2-diol, toluene, 3,4,6-trichlorocatechol, 4,5-dichlorocatechol, 2,4,6-trichlorophenol, 3,5,6-trichloro-2-pyridinol, benzoyl-CoA, chlorpyrifos, methylphosphonate, glutarate, uracil, cyclohexane-1-carboxylate, limonene, sulfoacetaldehyde, androstenedione, picolinate, 4-coumarate, trans-caffeate, fructosyllysine and glucosyllysine, L-threitol and D-xylose. We also added pathways for the bacterial degradation of the amino acids glycine, L-phenylalanine, L-tryptophan and L-tyrosine by the Stickland reaction. Several new pathways, all drawn using the symbolic representation of glycans mentioned above, describe the degradation of the large plant-produced polymers glucuronoarabinoxylan, homogalacturonan, rhamnogalacturonan type I and plant arabinogalactan type II. New mammalian degradation pathways were added for heme and vitamin K, while new plant degradation pathways were added for the steroidal glycoalkaloid α-tomatine, the volatile methylsalicylate and for oxalate (the most oxidized two-carbon compound).
Plant secondary metabolism
MetaCyc contains a vast number of plant-specific secondary metabolism pathways. Among the pathways added or significantly revised since version 21.1 are those describing the biosynthesis of the indole alkaloids secologanin, strictosidine, ajmaline and camptothecin; the bisindole alkaloids vinblastine, vindorosine and vincristine; the sesquiterpene lactones artemisinin and arteannuin B; the diterpenoids cis-abienol, cembratrienediol, labdenediol, sclareol and the members of the dolabralexins branch; the triterpenoid marneral; the isoquinoline alkaloid noscapine; the flavonoid tricin; the ginsenosides; the arylnaphthalene lignan justicidin B (which has been used traditionally by native Taiwanese as a fish-killing agent); the steroidal alkaloids α-tomatine, solasodine and soladulcidine; the isoquinoline alkaloids (S)-reticuline, coptisine and epiberberine; the volatile organic compounds (VOCs) guaiacol and methylsalicylate and various acylsugars.
Human metabolism
Among the human pathways added are those for the biosynthesis of ophthalmate (a tripeptide analog of glutathione in which L-cysteine is replaced by (S)-2-aminobutanoate) and the endocannabinoids anandamide (N-arachidonoyl ethanolamide) and 2-arachidonoylglycerol. The endocannabinoids are the endogenous agonists of the cannabinoid receptors, CB1 and CB2. We revised the pathways for the biosynthesis of the anti-inflammatory mediators lipoxin, 15-epi-lipoxin and resolvin E, and a pathway for the biosynthesis of itaconate, which inhibits the growth of pathogenic bacteria. Two new protein glycosylation pathways describe the formation of the core M3 glycan on mammalian proteins, and its extension to the complex glycan found on the α-dystroglycan protein. Other new pathways describe the various ways in which hydrogen sulfide is produced and oxidized in mammalian tissues, as well as the metabolism of 5-oxo-L-proline (also known as L-pyroglutamate), which is formed via the spontaneous cyclization of L-glutamine. Finally, a new pathway describing pheomelanin biosynthesis has been added, complementing the existing pigment biosynthesis pathways for L-dopachrome and eumelanin.
Expansion of other objects in the database
Polypeptides
The total number of polypeptides in the database is 14 427 in version 23.1. In addition, MetaCyc contains 4190 protein complexes. Out of these 12 457 are associated with an enzymatic activity.
Compounds
The total number of compounds grew by 12.9% from 14 003 (version 21.1) to 15 809 (version 23.1). Of the compounds, 15 592 have structures and 10 194 participate in reactions. All compounds in MetaCyc are protonated to the state most prevalent at pH 7.3. Most compounds also contain standard Gibbs free energy of formation (ΔfG′°) values, most of which are computed by Pathway Tools using an algorithm developed internally that is based on techniques by Jankowski et al. (14) and Alberty (15). As of August 2019, a total of 15 368 compounds include Gibbs free energy values.
Reactions
As of August 2019, the total number of reactions in MetaCyc is 17 486, out of which 9980 participate in pathways. The number of enzymatic reactions (reactions associated with an enzyme) is 16 034, an increase of 11.8% from version 21.1. MetaCyc uses a reaction-balance-checking algorithm that checks for elemental composition and electric charge. Unlike many reaction resources available online, the vast majority of MetaCyc reactions are balanced taking into account the protonation state of the compounds. As of August 2019, a total of 16 161 reactions (92.4% of the total reactions) are balanced. The remaining reactions cannot be balanced due to assorted reasons (for example, a reaction may describe a polymeric process such as the hydrolysis of a polymer of an undefined length, may involve an ‘n’ coefficient, or may involve a substrate that lacks a defined structure, such as ‘an aldose’). Balanced reactions are also processed by an atom mapping algorithm, and currently 14 403 reactions have atom mapping data.
MetaCyc reactions contain standard change in Gibbs free energy (ΔrG′°) values that are computed based on the ΔfG′° values computed for reactants and products. As of August 2019, a total of 15 337 reactions include these Gibbs free energy values.
Linking to other databases
Objects in MetaCyc are extensively linked to other leading databases in the field, with a total of 264 375 links. MetaCyc proteins have a total of 144 347 links to a number of protein databases that include (only databases with more than 1000 links are listed) InterPro, PDB, Pfam, UniProt, Protein Model Portal, PROSITE, SMR, PRIDE, PID, PANTHER, PRINTS, MODBASE, SMART, RefSeq, EcoliWiki, PortEco, DIP, MINT, ProDB, SwissModel and PhylomeDB. MetaCyc genes have a total of 41 282 links to NCBI-Entrez, NCBI-Gene, STRING, RegulonDB, EchoBASE, ASAP, OU Microarray, RefSeq, MIM, CGSC and ArrayExpress. MetaCyc compounds have a total of 40 241 links to PubChem, ChEBI, KEGG, ChemSpider, HMDB, MetaboLights, RefMet and CAS. MetaCyc reactions have a total of 37 679 links to UniProt, Rhea and KEGG.
Enzyme commission numbers
The curation of MetaCyc is conducted in close collaboration with the Enzyme Commission (EC) (16). During the curation process, MetaCyc curators come across thousands of enzymes that have not yet been classified by the EC. In addition, curation exposes errors in older existing EC entries. While curating MetaCyc content, curators prepare and submit new and revised entries to the EC, leading to the creation of hundreds of new and modified EC entries over the past 2 years. Many enzymes that have not been classified in the EC system are assigned ‘M-numbers’ in MetaCyc, which are temporary numbers that indicate a well-characterized enzymatic activity that has not yet been classified by the EC (17). Our aim is to have as many M-numbers as possible eventually replaced by official EC numbers. MetaCyc currently contains 6349 official EC numbers, 139 provisional EC numbers (entries in internal review by the Enzyme Commission) and 327 M-numbers.
SOFTWARE AND WEBSITE ENHANCEMENTS
The following sections describe significant enhancements to Pathway Tools (the software that powers the BioCyc website) during the past 2 years that affect the MetaCyc user experience.
Glycan pathways
In 2012, we integrated the GlycanBuilder software with Pathways Tools (18,19), which enabled the drawing of glycan structures using colored symbols to represent the different monosaccharide building blocks of the glycan molecule, thus making it easier for users to comprehend the structures of complex polysaccharides (12). Utilizing these structures in glycan biosynthetic pathways enables users to quickly grasp how a sugar residue is added to the growing polymer. A year later, we introduced a new type of pathway diagram based on these structures to describe the complex process of glycan degradation. In these diagrams, arrows indicate where different enzymes cleave the glycan macro molecule, enabling the description of complex processes, in which multiple enzymes act in parallel with no particular order.
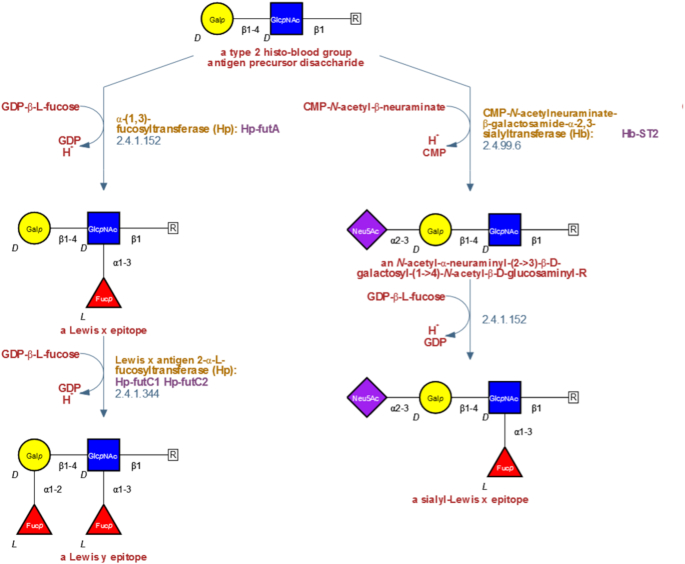
To make these structures more useful for BioCyc users, we next introduced several new features to the original GlycanBuilder code. Perhaps, the most important such feature is the ability to display text strings or other, non-glycan, compounds as part of the glycan structure. This allows, for example, to depict how an oligosaccharide is connected to a lipid membrane anchor or to a glycoprotein (see Figure 2). Unfortunately, a myriad of technical complications prevented us from using this useful addition for several years. However, for version 22.0 of Pathway Tools, these issues were resolved, allowing us to convert a large number of pathways to the symbolic representation format. MetaCyc currently contains 46 glycan biosynthesis pathways, 11 glycan degradation pathways and 3 O-antigen biosynthesis pathways that are drawn using symbolic representation.
Figure 2.
A section from a pathway that describes the biosynthesis of Lewis-type epitopes by the bacterium Helicobacter pylori. This pathway was drawn using a modified version of the GlycanBuilder software (18). The modifications, introduced by the SRI International, allow the software to display text strings or other, non-glycan, compounds as part of the structure (shown as R in this example). As in non-glycan type pathways, each item in the diagram is a clickable hyperlink to a dedicated page for that item.
Improved graphics and fonts
We have upgraded many web visualizations produced by Pathway Tools in its web server mode to use modern graphics and fonts, including pathways, enzymatic reactions and transport reactions. Many other diagrams that are available only when browsing organism-specific PGDBs (and thus not applicable to MetaCyc) have also been enhanced.
Changes in PathoLogic
PathoLogic is a component of the Pathway Tools software that allows the user to create a new PGDB using MetaCyc and an annotated genome. Starting with version 22.0, PathoLogic can accept input genomes in GFF format in addition to Genbank and PathoLogic format files. In addition, PathoLogic now extracts organism metadata from its input files for inclusion in a PGDB. Example metadata include the geographic location from which the organism was sampled, its aerobicity, and its human microbiome body site.
Pathway regulation
MetaCyc contains regulatory data for many enzymes, primarily substrate-level enzyme activation and inhibition and required cofactors (additional types of regulation, such as transcriptional and translational regulation, are available in some other BioCyc PGDBs). Starting with version 22.0, pathway diagrams can include detailed regulatory information for each step, provided such data are present. When regulatory data are available, a button labeled ‘Show Regulation Details’ appears above the pathway diagram and users can add or remove the regulatory information to/from the diagram by clicking on it. The button will be present only if regulation data are available for the pathway and if the pathway is displayed at a detail level that includes enzyme names. Users can mouse over any regulator icon for further information.
MetaCyc versus KEGG
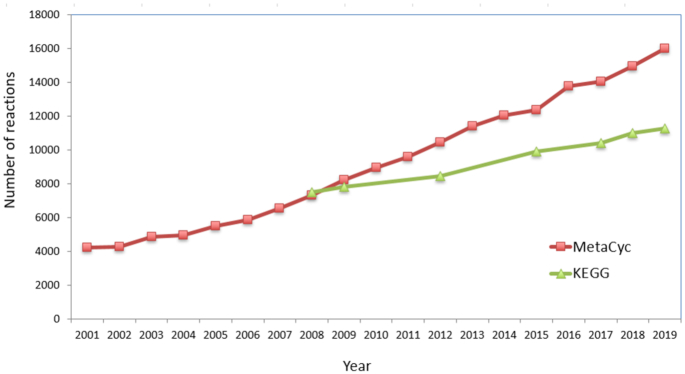
MetaCyc and KEGG (20) are both large metabolic pathway database projects that have been under development for more than two decades. Both projects provide reference pathways that are used to predict the metabolic pathways present in an organism from the annotated genome of that organism. The KEGG project consists of both a reference pathway database and the resulting projection of the reference pathways onto organisms with sequenced genomes. It is useful to compare occasionally the data content of MetaCyc and the KEGG reference pathway database, and a thorough comparison was last published in 2013 (21). Comparing the pathway content of the two databases is non-trivial because, unlike in MetaCyc, just a small fraction of the KEGG metabolic data is contained in objects that correspond to specific pathways. These objects are called ‘modules’, and as of 20 August 2019 KEGG contains 391 metabolic modules (other modules describe other processes such as transport). Most of the metabolic information in KEGG is presented in large diagrams called ‘maps’ that integrate many related pathways from multiple organisms into a single diagram. As of 20 August 2019 KEGG contains 536 such maps. Arguably a better way to compare the metabolic data content of the two databases is a comparison of the reaction count, since this circumvents the problem of arranging reactions within pathways. As of 20 August 2019, KEGG contains 11 289 biochemical reactions, as opposed to 16 031 reactions in MetaCyc (not including transport reactions, binding reactions and redox half-reactions). Figure 3 compares the reaction count in the two databases over time.
Figure 3.
A comparison of the reactions content of MetaCyc and the KEGG database. Data for KEGG are available only since 2008. Since that time the number of reactions in MetaCyc has been growing at about twice the rate of the comparable rate in KEGG. As of 20 August 2019 KEGG contains 11,289 biochemical reactions, as opposed to 16,031 reactions in MetaCyc.
SUBSCRIPTION MODEL FOR BioCyc ACCESS AND DATABASE CURATION
In past papers in the database issue of Nucleic Acids Research we described the MetaCyc database together with the BioCyc PGDB collection (6). As of 2017, SRI International has adopted a subscription-based model for BioCyc access, preventing its description in the database issue. MetaCyc and EcoCyc [the PGDB for Escherichia coli K-12, (22)] remain freely available to all and do not require a subscription.
Because of ongoing difficulties in securing government funds for database curation, we moved to a subscription-based model in the hope of generating funds that would permit us to curate high-quality databases for more organisms, such as important pathogens, biotechnology workhorses, model organisms and promising hosts for biofuels development. In the last 2 years, we used subscription revenues to improve the quality of the PGDBs of the following organisms: Bacillus subtilis 168, Corynebacterium glutamicum ATCC 13032, Homo sapiens, Lactobacillus rhamnosus GG, Mycobacterium tuberculosis H37Rv, Pseudomonas putida KT2440, Saccharomyces cerevisiae S288c, Salmonella enterica enterica 14028S, Salmonella enterica enterica LT2 and Staphylococcus aureus NCTC 8325.
More information about the subscription model is available at http://www.phoenixbioinformatics.org/biocyc/index.html.
HOW TO LEARN MORE ABOUT MetaCyc AND BioCyc
The MetaCyc.org website provides several informational resources, including an online guide for MetaCyc (http://www.metacyc.org/MetaCycUserGuide.shtml), a guide to the science behind the Pathway/Genome Databases (http://biocyc.org/PGDBConceptsGuide.shtml), and instructional webinar videos that describe the usage of MetaCyc, BioCyc and Pathway Tools (http://biocyc.org/webinar.shtml). We routinely host workshops and tutorials (on site and at conferences) that provide training and in-depth discussion of our software for both beginning and advanced users. To stay informed about the most recent changes and enhancements to our software, please join the BioCyc mailing list at http://biocyc.org/subscribe.shtml. A list of our publications is available online at http://biocyc.org/publications.shtml.
DATA AVAILABILITY
The MetaCyc database is openly available to all. See http://biocyc.org/download.shtml for download information. New versions of the downloadable data files and the MetaCyc website are released three times per year. Access to the website is free; users are encouraged to register for a free account as having an account makes it possible to customize preferences and save personalized data such as organism lists and SmartTables.
ACKNOWLEDGEMENTS
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
FUNDING
National Institute of General Medical Sciences, National Institutes of Health (NIH) [GM080746]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Karp P.D., Riley M., Saier M., Paulsen I.T., Paley S.M., Pellegrini-Toole A.. The EcoCyc and MetaCyc databases. Nucleic Acids Res. 2000; 28:56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caspi R., Billington R., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Midford P.E., Ong Q., Ong W.K. et al.. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018; 46:D633–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karp P.D., Paley S.M., Krummenacker M., Latendresse M., Dale J.M., Lee T.J., Kaipa P., Gilham F., Spaulding A., Popescu L. et al.. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief. Bioinform. 2010; 11:40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karp P.D., Latendresse M., Paley S.M., Krummenacker M., Ong Q.D., Billington R., Kothari A., Weaver D., Lee T., Subhraveti P. et al.. Pathway Tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief. Bioinform. 2016; 17:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karp P.D., Latendresse M., Caspi R.. The pathway tools pathway prediction algorithm. Stand. Genomic Sci. 2011; 5:424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karp P.D., Billington R., Caspi R., Fulcher C.A., Latendresse M., Kothari A., Keseler I.M., Krummenacker M., Midford P.E., Ong Q. et al.. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2017; doi:10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vallenet D., Calteau A., Cruveiller S., Gachet M., Lajus A., Josso A., Mercier J., Renaux A., Rollin J., Rouy Z. et al.. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017; 45:D517–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mazourek M., Pujar A., Borovsky Y., Paran I., Mueller L., Jahn M.M.. A dynamic interface for capsaicinoid systems biology. Plant Physiol. 2009; 150:1806–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schlapfer P., Zhang P., Wang C., Kim T., Banf M., Chae L., Dreher K., Chavali A.K., Nilo-Poyanco R., Bernard T. et al.. Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017; 173:2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walsh J.R., Schaeffer M.L., Zhang P., Rhee S.Y., Dickerson J.A., Sen T.Z.. The quality of metabolic pathway resources depends on initial enzymatic function assignments: a case for maize. BMC Syst. Biol. 2016; 10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evsikov A.V., Dolan M.E., Genrich M.P., Patek E., Bult C.J.. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009; 10:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Varki A., Cummings R.D., Aebi M., Packer N.H., Seeberger P.H., Esko J.D., Stanley P., Hart G., Darvill A., Kinoshita T. et al.. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology. 2015; 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herget S., Ranzinger R., Maass K., Lieth C.W.. GlycoCT-a unifying sequence format for carbohydrates. Carbohydr. Res. 2008; 343:2162–2171. [DOI] [PubMed] [Google Scholar]
- 14. Jankowski M.D., Henry C.S., Broadbelt L.J., Hatzimanikatis V.. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 2008; 95:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alberty R.A. Thermodynamics of Biochemical Reactions. 2003; Hoboken: Wiley InterScience. [Google Scholar]
- 16. McDonald A.G., Tipton K.F.. Fifty-five years of enzyme classification: advances and difficulties. FEBS J. 2014; 281:583–592. [DOI] [PubMed] [Google Scholar]
- 17. Green M.L., Karp P.D.. Genome annotation errors in pathway databases due to semantic ambiguity in partial EC numbers. Nucleic Acids Res. 2005; 33:4035–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ceroni A., Dell A., Haslam S.M.. The GlycanBuilder: a fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol. Med. 2007; 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Damerell D., Ceroni A., Maass K., Ranzinger R., Dell A., Haslam S.M.. The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biol. Chem. 2012; 393:1357–1362. [DOI] [PubMed] [Google Scholar]
- 20. Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M.. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019; 47:D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altman T., Travers M., Kothari A., Caspi R., Karp P.D.. A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinformatics. 2013; 14:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karp P.D., Ong W.K., Paley S., Billington R., Caspi R., Fulcher C., Kothari A., Krummenacker M., Latendresse M., Midford P.E. et al.. The EcoCyc Database. EcoSal Plus. 2018; 8:doi:10.1128/ecosalplus.ESP-0006-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MetaCyc database is openly available to all. See http://biocyc.org/download.shtml for download information. New versions of the downloadable data files and the MetaCyc website are released three times per year. Access to the website is free; users are encouraged to register for a free account as having an account makes it possible to customize preferences and save personalized data such as organism lists and SmartTables.